ABSTRACT
The Abl tyrosine kinase signaling network controls cell migration, epithelial organization, axon patterning and other aspects of development. Although individual components are known, the relationships among them remain unresolved. We now use FRET measurements of pathway activity, analysis of protein localization and genetic epistasis to dissect the structure of this network in Drosophila. We find that the adaptor protein Disabled stimulates Abl kinase activity. Abl suppresses the actin-regulatory factor Enabled, and we find that Abl also acts through the GEF Trio to stimulate the signaling activity of Rac GTPase: Abl gates the activity of the spectrin repeats of Trio, allowing them to relieve intramolecular repression of Trio GEF activity by the Trio N-terminal domain. Finally, we show that a key target of Abl signaling in axons is the WAVE complex that promotes the formation of branched actin networks. Thus, we show that Abl constitutes a bifurcating network, suppressing Ena activity in parallel with stimulation of WAVE. We suggest that the balancing of linear and branched actin networks by Abl is likely to be central to its regulation of axon patterning.
KEY WORDS: Abl, Disabled, Rac, Trio, FRET, Axon guidance
Summary: Genetic epistasis, protein localization and FRET assays reveal the functional organization of the Abl signaling network - a highly conserved and ubiquitous system that controls cell morphology and motility.
INTRODUCTION
During development of the nervous system, cell surface receptors regulate cytoskeletal dynamics to direct axon growth (Dickson, 2002; Song and Poo, 2001, 1999; Tessier-Lavigne and Goodman, 1996). The transformation of extracellular cues into cytoskeletal dynamics is executed by cytoplasmic signaling networks. Among these, the Abl tyrosine kinase signaling pathway plays a leading role in regulation of the neuronal cytoskeleton (Bradley and Koleske, 2009; Krause et al., 2003; Lanier and Gertler, 2000; Moresco and Koleske, 2003). Components of the Abl pathway link to growth cone guidance receptors and actin-regulatory proteins (Le Gall et al., 2008; Liebl et al., 2003; Rhee et al., 2002; Wills et al., 1999a,b). Indeed, nearly all of the common, phylogenetically conserved families of axon guidance receptors signal through Abl. This includes receptors for Netrin, Slit, Semaphorin, Ephrin and others (Bashaw et al., 2000; Deinhardt et al., 2011; Forsthoefel et al., 2005; Garbe et al., 2007; Gupton et al., 2012; Hsouna et al., 2003; Yu et al., 2001). The mechanism of Abl signaling, therefore, is fundamental for understanding growth cone guidance by each of these receptors, as well as the integration of signals from multiple receptors (Crowner et al., 2003; Kuzina et al., 2011; Wills et al., 1999a; Winberg et al., 1998).
The Abl tyrosine kinase pathway was among the first of the conserved signal transduction pathways to be identified, and many of its components have long been known (Dai and Pendergast, 1995; Gertler et al., 1995, 1996; Henkemeyer et al., 1987; Howell et al., 1997; Liebl et al., 2000; Luo, 2000; Wang and Baltimore, 1983). A core set of pathway components cooperate with Abl in neurons, cultured fibroblasts and transformed cell lines (Bradley and Koleske, 2009; Hoffmann, 1991). Disabled (Dab) is an adaptor protein that associates directly with receptors that regulate motility and guidance (Howell et al., 1999a,b; Le Gall et al., 2008). Trio is a guanine nucleotide exchange factor (GEF) that activates both Rac and Rho GTPase and is essential for neuronal morphogenesis (Awasaki et al., 2000; Newsome et al., 2000; Schmidt and Debant, 2014; Steven et al., 1998). Abi is a component of the WAVE complex that activates Arp2/3 to extend branched actin networks (Dai and Pendergast, 1995; Gautreau et al., 2004). Enabled (Ena) extends and bundles linear actin filaments (Gertler et al., 1995, 1996). Dab, Trio and Abi work synergistically with Abl in many contexts (Gertler et al., 1993; Luo, 2000), whereas Ena activity is suppressed by Abl (Gates et al., 2007; Gertler et al., 1995; Grevengoed et al., 2003).
Despite the long history of studies of Abl, however, the functional organization of the pathway has remained obscure. Nearly every component has been linked, genetically, biochemically or both, to nearly every other component (Forsthoefel et al., 2005; Gertler et al., 1993; Hill et al., 1995; Liebl et al., 2000; Sonoshita et al., 2015). Many experiments have yielded contradictory pictures of the relationships among Abl signaling components and of their roles in motility (Bashaw et al., 2000; Bear et al., 2000; Forsthoefel et al., 2005; Hsouna et al., 2003; Juang and Hoffmann, 1999; Krause et al., 2002; Liebl et al., 2003; Lin et al., 2009; Song et al., 2010; Trichet et al., 2008; Wills et al., 1999a,b). Thus, as yet there is no clear and testable model of which components lie upstream or downstream of which other components, an essential requisite for us to interpret the extensive data linking Abl to growth cone motility and neuronal migration in culture and in vivo (Bradley and Koleske, 2009).
We therefore set out to establish the functional relationships among the core proteins of the Abl pathway in vivo in neurons, using direct measures of protein activity. We deployed fluorescence resonance energy transfer (FRET) biosensor probes that allowed us to assay directly, in living cells, the activity of the two key outputs of the Abl pathway in axons: Rac GTPase signaling (Itoh et al., 2002; Wang et al., 2010) and Abl kinase activity (Sterne et al., 2015; Ting et al., 2001). We further validated the resulting model for the Abl pathway both by investigating how each component of the pathway regulates the subcellular localization of other components, and by using genetic epistasis to investigate the dependent relationships of mutations in genes encoding these proteins.
We now reveal the functional relationships among the core components of the Abl signaling network in Drosophila neurons. We show that the adaptor protein Dab stimulates the kinase activity of Abl as well as regulating its subcellular localization. Consistent with this, Dab is found in a biochemical complex with Abl in Drosophila embryos. The kinase activity of Abl stimulates the signaling activity of Rac GTPase, primarily by acting through the GEF Trio. Abl does not act directly on the Rac GEF domain of Trio; rather, Abl gates the action of the spectrin repeats of Trio, allowing them to counteract a repressive intramolecular interaction between the Trio N-terminal domain (NTD) and GEF1 domain, thereby allowing Trio to stimulate Rac. In parallel with the inhibition by Abl of the actin polymerization factor Ena, the Abl-Trio-Rac pathway activates the Abi/WAVE complex that promotes the extension of branched actin networks. We suggest that this combination of effects executes the Abl-dependent regulation of axon growth and guidance.
RESULTS
Dab regulates Abl kinase activity
Previous experiments showed that Dab is a core component of the Abl pathway and suggested that Dab is functionally upstream of Abl: UAS-Abl suppresses the phenotype of a Dab mutant but UAS-Dab cannot compensate for lack of Abl (Song et al., 2010). Moreover, Dab controls the subcellular localization of Abl, but not vice versa (Song et al., 2010). We wondered, however, whether Dab also controls Abl kinase activity. We therefore expressed a FRET probe that measures Abl kinase activity in vivo.
We expressed in Drosophila a unimolecular CFP-YFP FRET biosensor for Abl kinase activity that was originally constructed for use in mammalian cells, employing sequences from the mammalian CrkII protein (Sterne et al., 2015; Ting et al., 2001). Multiple lines of evidence demonstrate that FRET activity of this bioprobe faithfully reports Abl kinase activity in Drosophila. (1) By ratiometric imaging, the reporter gives a FRET signal in cultured photoreceptor (PR) neurons that is decreased in cells that are mutant for Abl and increased upon overexpression of wild-type (WT) Abl or expression of Bcr-Abl transgenes (Fig. 1A,B). (2) The FRET signal is inhibited acutely by treatment of PRs with the Abl-specific drug STI-571 (Gleevec), and this inhibition is reversible upon washout of the drug (Fig. 1C,D). The residual FRET signal in the presence of the drug is slightly less than that in Abl mutant PRs, which is likely to be because of perdurance of maternal Abl. We cannot rule out the possibility that the effect of STI-571 is incomplete since the FRET signal in treated Abl mutant cells appears to be slightly less than that in treated WT cells, but the difference is not statistically significant. (3) Photobleaching of the YFP acceptor reveals a FRET signal that is consistent with the FRET efficiency calculated by ratiometric imaging (Fig. 1E). (4) A mutant of the reporter that lacks its phosphorylatable tyrosine (YF mutant) does not give a FRET signal (Fig. 1E).
Fig. 1.
Validation of the Abl FRET biosensor. A CFP-YFP unimolecular FRET biosensor for Abl kinase activity was expressed in Drosophila photoreceptor neurons under the control of GMR-GAL4, and FRET was imaged and quantified in cultured primary photoreceptors. (A) Single optical section of one or two photoreceptors showing biosensor distribution (YFP, top) and FRET (bottom) in cells from larval eye discs of the indicated genotypes. Quantified cell is outlined in red, and the pseudocolor scale for FRET signal is shown with limiting values indicated, in this and all other FRET images. Note that the WT photoreceptor in A is the 30 min (pre-drug) image from C. (B) Mean FRET ratio (MFR) for each genotype. (C) Single photoreceptor showing Abl FRET probe distribution and timecourse of FRET channel upon treatment for the indicated time with the Abl kinase inhibitor STI-571, and drug washout. (D) Timecourse of Abl inhibition by STI-571 and washout (n=32). MFR has been normalized to 1 at time 0. (E) Quantification of Abl reporter FRET by YFP photobleaching for the indicated genotypes. Error bars indicate s.e.m. and the number of cells imaged (n) is shown. ns, not statistically significant; ***P<0.001; in all cases two-tailed t-test, with Bonferroni correction for multiple testing. Scale bar: 10 μm in A,C.
We next used the Abl FRET probe to test whether Dab modulates Abl kinase activity (Fig. 2A,B). We assayed PRs from WT or Dab mutant flies that express the Abl FRET probe and found that Abl kinase activity was suppressed in Dab mutant neurons [mean FRET ratio (MFR)=1.55±0.1 in WT versus 0.93±0.2 in Dabmz; mean±s.e.m. in this and all subsequent FRET values; P<0.01]. We also overexpressed Dab in photoreceptors bearing the probe and found that Abl kinase activity was stimulated (MFR=2.1±0.1 in UAS-Dab; P<0.01). Control western analysis verified that the Dab genotype does not alter the level of Abl protein (not shown). Together, these data show that Dab enhances Abl kinase activity.
Fig. 2.
Dab stimulates Abl kinase activity. (A) Cultured photoreceptors of the indicated genotypes expressing the Abl FRET bioprobe. The left panel of each pair shows the probe distribution (YFP) and the right panel shows FRET. Area used for quantification is outlined and a pseudocolored activity key is shown. Scale bar: 10 μm. (B) Mean Abl FRET ratio for each genotype. Error bars indicate s.e.m. **P<0.01. (C) Anti-Dab western of WT embryo lysates. Dab runs as a family of two or three products of ∼250-280 kDa (arrows). Molecular weight markers are indicated. Similar results were obtained in six independent experiments, and in extracts of S2 cells (not shown).
It has been suggested that the Abl and Dab orthologs associate in mammalian cells (Sonoshita et al., 2015). We therefore tested Abl/Dab association in Drosophila by co-immunoprecipitation (co-IP) from embryo extracts and found that immunoprecipitation of Abl co-precipitated Dab (Fig. 2C). Co-precipitation was also observed from extracts of cultured Drosophila S2 cells (not shown). A control documenting immunoprecipitation of Abl by anti-Abl antibody is provided in Fig. S1. These data demonstrate that Abl and Dab are associated in common complexes in vivo in Drosophila.
Abl kinase regulates the signaling activity of Rac GTPase
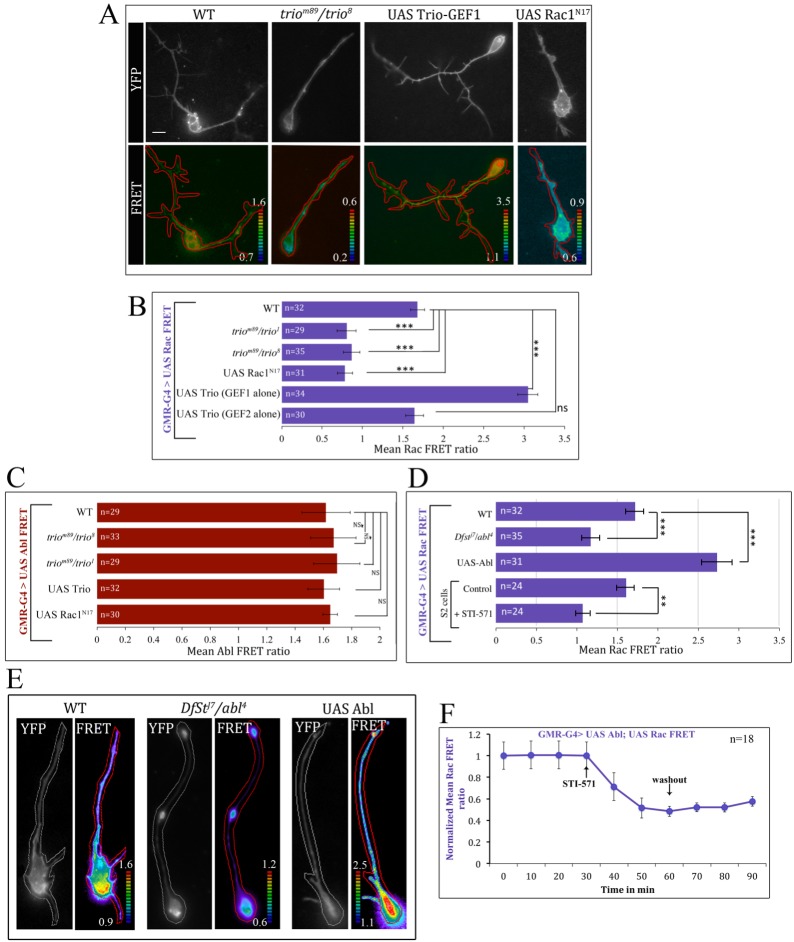
In neurons, Abl cooperates with Rho family GTPases, particularly Rac. However, it is not clear whether Rac and Abl are both downstream of a regulator that acts on both or whether one of these proteins controls the other. We deployed a second FRET probe to assay Rac activity in living cells. Raichu-Rac was developed as a FRET probe of Rac signaling in mammalian cells, and it was shown also to function in the Drosophila ovary (Itoh et al., 2002; Wang et al., 2010), so we validated the probe in Drosophila PRs and S2 cells. In photoreceptors, Rac biosensor activity decreased substantially in the background of mutations that inactivate the GEF Trio (MFR=0.8±0.12 in triom89/trio1; 0.9±0.1 in triom89/trio8) or upon co-expression of a dominant-negative form of Rac (0.79±0.1 in UAS-Rac1N17) compared with WT controls (1.68±0.08; P<0.001 in each comparison; Fig. 3A,B). We further validated these results in a gain-of-function background by co-expressing the biosensor with either the GEF1 domain or the GEF2 domain of Trio in WT PRs. The GEF1 domain of Trio activates Rac signaling, whereas the GEF2 domain does not (Newsome et al., 2000). Consistent with this, co-expression of GEF1 gave a substantial increase in the FRET signal of the Rac bioprobe (MFR=3.04±0.12) compared with the control (1.68±0.08), whereas co-expression of GEF2 had no effect (1.64±0.11; Fig. 3B). These results verify the reliability of the Rac biosensor for reporting endogenous Rac activity in Drosophila PRs.
Fig. 3.
Validation of Rac FRET probe and Abl/Rac epistasis. Abl-responsive or Rac-responsive CFP-YFP unimolecular FRET probes were expressed in Drosophila under the control of GMR-GAL4 and ratiometric FRET was imaged. (A) Single cultured photoreceptors of the indicated genotypes expressing the Raichu-Rac FRET probe. Top panel of each pair shows probe distribution (YFP) and bottom panel shows pseudocolored FRET channel with color scale. Scale bar: 10 μm. (B) Mean Rac FRET ratio for each genetic background. (C) Mean Abl FRET ratio in genotypes that alter trio or Rac1 activity. (D) Mean Rac FRET ratio in genotypes that alter Abl activity, or in WT with and without 30 min treatment with STI-571. (E) Single cultured photoreceptors expressing the Raichu-Rac FRET bioprobe. (F) Timecourse of Raichu-Rac FRET signal in photoreceptors expressing UAS-Abl upon treatment with the Abl inhibitor STI-571 and washout. Signals are normalized to FRET ratio at start of experiment. Error bars indicate s.e.m. *P<0.05, **P<0.01, ***P<0.001.
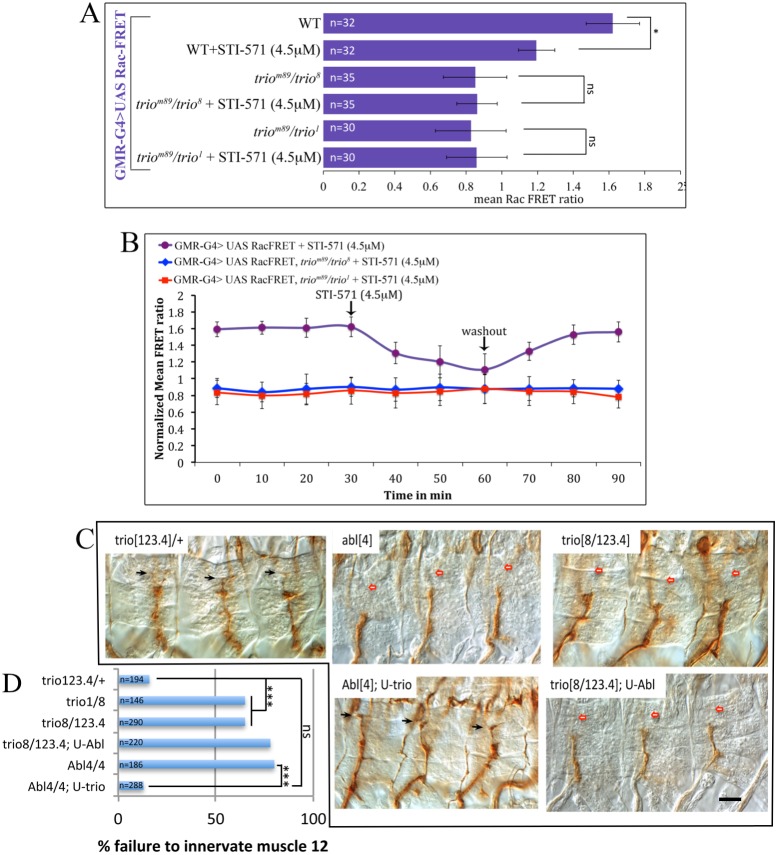
We next used the Abl and Rac FRET biosensors to perform parallel epistasis experiments, which revealed that Abl kinase regulates Rac signaling activity but that Rac does not regulate Abl kinase. We first assayed Abl kinase activity while we manipulated Rac activity in PRs (Fig. 3C). In WT PRs, Abl FRET activity is 1.6±0.2, and this remained unaltered in the background of different trio mutant alleles (1.7±0.14 in triom89/trio8; 1.6±0.11 in triom89/trio1) or upon co-expression of dominant-negative UAS-Rac1N17 (1.64±0.1) or UAS-trio (1.6±0.2). By contrast, altering endogenous Abl activity exerted a strong influence on endogenous Rac, both in PRs and S2 cells (Fig. 3D,E). First, Rac biosensor activity diminished in Abl mutant photoreceptors (1.17±0.11; P<0.01) and was hyperactivated by UAS-Abl (2.74±0.2; P<0.001) compared with basal expression in controls (1.7±0.11). Acute inhibition of Abl kinase by STI-571 also suppressed Rac activity within minutes, both in cultured S2 cells (Rac MFR=1.1±0.08 after 30 min of treatment with STI-571 versus 1.6±0.1 before treatment; P<0.05; Fig. 3D) and in PRs (Rac MFR=1.1±0.16 after treatment versus 1.6±0.08 before treatment; P<0.05; Fig. 4A, Fig. 3F). Note that absolute FRET ratio values measured in S2 cells should not be compared directly with those in PRs. These data demonstrate that Abl kinase activity is required to stimulate Rac. Moreover, the acute suppression of Rac by an Abl inhibitor argues against the hypothesis that suppression of Rac in an Abl mutant arises from indirect compensation for a chronic genetic manipulation, and rather suggests that the effect is relatively direct. For reasons that we do not understand, in a small fraction of experiments suppression of Rac signal by Abl inhibition was not reversed by washout of the drug (Fig. 3F, but compare with Fig. 4B). This was observed occasionally in both the WT background and in the context of Abl overexpression.
Fig. 4.
Abl regulation of Rac is mediated through Trio. (A) Mean Rac FRET ratio for the indicated genotypes, with or without 30 min treatment with the Abl inhibitor STI-571. *P<0.05. (B) Timecourse of Raichu-Rac FRET signal of the indicated genotypes upon treatment with STI-571 and washout. FRET signals for the timecourses are not normalized to the starting values in order to allow comparison of trio mutant to control. (C) Nomarski images showing three hemisegments of stage 17 embryos of the indicated genotypes, immunostained with anti-Fas2 and filet-mounted to reveal the ISNb motonerve. Black arrows indicate muscle 12 neuromuscular synapses; red arrows indicate positions of missing synapses (‘stall' phenotype). Scale bar: 10 µm. (D) Quantification of ISNb stall phenotypes (failure to innervate muscle 12). ***P<0.001; χ2, with Bonferroni correction. Error bars indicate s.e.m.
Abl acts through Trio to stimulate Rac signaling in neurons
There are a number of Rac GEFs in Drosophila, but one in particular, Trio, behaves like a core component of the Abl signaling pathway. trio mutants produce axonal phenotypes that mimic Abl mutations, and trio interacts genetically with Abl: heterozygous mutations of trio enhance the phenotype of Abl and vice versa, and ena heterozygotes suppress the trio phenotype (Awasaki et al., 2000; Bateman et al., 2000; Liebl et al., 2000; Newsome et al., 2000). Our FRET experiments now show that Abl-dependent regulation of Rac in photoreceptors is mediated by Trio. Rac FRET activity is inhibited significantly either by inhibition of Abl or mutation of trio, as shown above. Combining both, however, does not further depress Rac activity (MFR=0.89±0.1 in triom89/trio8 versus 0.86±0.2 with STI-571; and MFR=0.82±0.11 in triom89/trio1 versus 0.85±0.2 with STI-571; Fig. 4A). This is clearest in a timecourse, where addition of STI-571 had no effect on the Rac FRET ratio in a trio mutant genetic background (Fig. 4B). This demonstrates that Abl and Trio act serially, in a common pathway, and not in parallel. The failure to observe an additive effect of trio and STI-571 is not due to a limitation of the dynamic range of the assay, since other experimental paradigms do produce significantly lower Rac FRET ratios (Fig. 5D, see below).
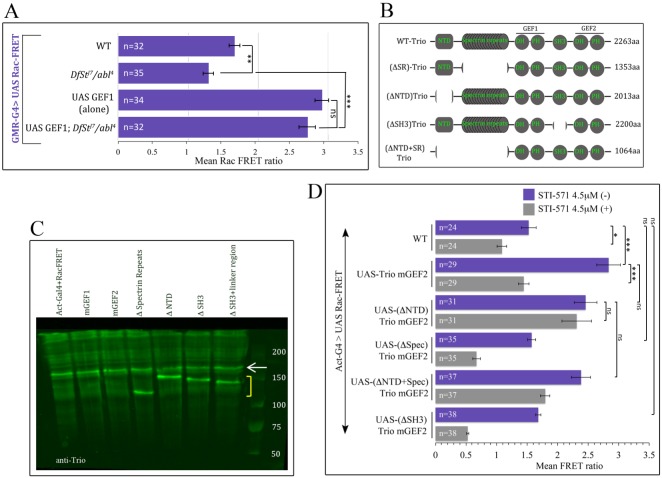
Fig. 5.
Structure/function analysis of Trio and its regulation by Abl. (A) Mean Rac FRET ratio in cultured photoreceptors for the indicated genotypes. (B) Schematic of Drosophila Trio protein domain structure and the deletions tested here. Deleted codons are indicated. SR, spectrin repeats. (C) Anti-Trio western of Drosophila S2 cells expressing the indicated Trio derivatives. Endogenous WT Trio is seen in all lanes (arrow). Deleted derivatives are visible in lanes 4-7 (bracket); derivatives bearing point mutations (lanes 2 and 3) cannot be distinguished from the endogenous protein. (D) Mean Rac FRET ratio assayed after co-transfection of Raichu-Rac reporter into S2 cells with the indicated transgenes. Gray bars show FRET activity after 30 min treatment with STI-571; purple bars show FRET activity after 30 min mock treatment. WT refers to results of transfection with FRET reporter transgene plus carrier DNA, but no trio transgene. Note that the data for control with and without STI-571 (top two bars) were also presented in Fig. 3D. Error bars indicate s.e.m. *P<0.05, **P<0.01, ***P<0.001.
We verified by genetic epistasis that Abl acts linearly upstream of trio. Abl and trio mutants each cause stalling of the ISNb motor nerve at the junction of muscles 6 and 13, with failure to innervate muscle 12, as reported previously (Bateman et al., 2000; Song and Giniger, 2011). We found, however, that overexpression of trio suppressed the ISNb phenotype of Abl mutants, but overexpression of Abl did not suppress trio (Fig. 4C,D). Thus, in trio123.4/8 there was a failure to innervate muscle 12 in 65% of hemisegments (n=290), and this frequency was nearly the same upon overexpression of Abl (78% stall, n=220). By contrast, whereas Abl4/4 mutants showed the ISNb stall phenotype in 80% of hemisegments (n=186), overexpression of trio reduced that to just 13% (n=288; P<0.001, χ2), which is not significantly different from the percentage innervation of control embryos at this stage (trio123.4/+, muscle 12 not yet innervated in 16% of hemisegments; n=194; P=0.37). These data are consistent with a simple, linear dependence pathway with Abl upstream of trio, as implied by the FRET results.
Abl controls derepression of the Trio GEF1 domain via the Trio spectrin repeats
We wondered whether Abl stimulates Trio GEF activity directly through an effect on the Rac-specific GEF1 domain or by some other mechanism. Expression of Trio GEF1 by itself in WT photoreceptors increased the Rac FRET ratio from 1.7±0.08 to 2.9±0.19 (Fig. 5A). Expression of Trio GEF1 in Abl mutant PRs, however, decreased this ratio only slightly (to 2.7±0.23), consistent with Abl suppressing endogenous Trio but not the expressed Trio; if the expressed GEF1 also required Abl, then the reduction of the FRET ratio would have been far greater (Fig. 5D). We therefore inferred that Abl does not regulate Trio via a direct effect on the GEF1 domain.
To map the region of Trio that mediates the effect of Abl we analyzed Trio derivatives by co-expressing them with the Rac bioprobe in S2 cells. To provide an ideal control, each trio mutation was introduced in parallel into two backbones, one bearing a point mutation that inactivates the Rac-specific GEF1 domain (trio mGEF1) and the other with the same amino acid change but in the Rho-specific GEF2 domain (trio mGEF2). trio mGEF1 provides a negative control backbone that should be unable to activate Rac FRET activity regardless of other mutations; trio mGEF2 provides a backbone that is fully active for Rac FRET activity and is therefore appropriate for assaying the effects of modifications to other domains, but bears the identical amino acid change in the GEF2 domain in order to control for nonspecific effects of the GEF mutation through unrelated mechanisms (Shivalkar and Giniger, 2012; Song and Giniger, 2011); it also rules out any background contributions of GEF2 to the Rac FRET signal.
Rac FRET analysis revealed (1) that the SH3 domain is required for all activity of full-length Trio, (2) that the NTD of Trio represses the Rac GEF activity of Trio in the absence of Abl kinase, and (3) that in the presence of Abl kinase, the Trio spectrin repeats relieve that NTD-dependent repression (Fig. 5D). As described above, the Rac bioprobe gave a baseline FRET ratio of 1.6±0.1 in S2 cells, and addition of STI-571 suppressed this to 1.1±0.08 (Fig. 3D). Expression of trio mGEF2 in cells bearing the Rac FRET biosensor enhanced the Rac FRET ratio to 2.8±0.2 in the absence of drug, and addition of STI-571 suppressed that FRET signal to 1.5±0.11 (P<0.001 for the comparison with and without drug). This establishes the dynamic range of the assay.
Next, we tested the functions of various Trio domains by deletion (Fig. 5B,D, Table S1). First, expression of trioΔSH3 (in the mGEF2 backbone) did not enhance the Rac FRET ratio (MFR=1.7±0.07; P>0.4 versus control with no trio transgene). This shows that the SH3 domain is essential for Trio activity. Second, expression of trio mGEF2ΔNTD (MFR=2.4±0.19) increased Rac activity nearly as much as trio mGEF2 itself (MFR=2.8±0.2; P>0.1), showing that the NTD domain is not needed for activation of Rac. Strikingly, however, addition of STI-571 did not affect the Rac signal of trio mGEF2ΔNTD (MFR=2.3±0.23; P>0.8 for the comparison with and without drug), showing that this derivative no longer requires Abl kinase for activity. A plausible explanation for this observation came from analyzing the effects of deleting the spectrin repeats. Expression of trio mGEF2Δspec, which deletes only the spectrin repeats, abolished GEF activity (MFR=1.6±0.06; P>0.8 versus no transgene control). By contrast, expressing a derivative that lacked both the spectrin repeats and the NTD, termed trio mGEF2Δ(NTD+spec), yielded as much activity (MFR=2.4±0.17) as trio mGEF2Δ(NTD) itself (P>0.8). The simplest hypothesis, therefore, is that in the native protein the NTD suppresses GEF activity in the absence of Abl kinase, and in the presence of Abl this suppression is counteracted by a mechanism that employs sequences within the spectrin repeat region. Consistent with this hypothesis, trio mGEF2Δ(NTD+spec) was also largely insensitive to Abl (MFR=1.8±0.12 in the presence of STI-571, reflecting inhibition of endogenous Trio but little effect on the expressed protein). Potential molecular mechanisms for these effects are considered in the Discussion. In these experiments, all trio derivatives were expressed at similar levels (Fig. 5C).
Trio/Rac acts in parallel to Ena, downstream of Abl
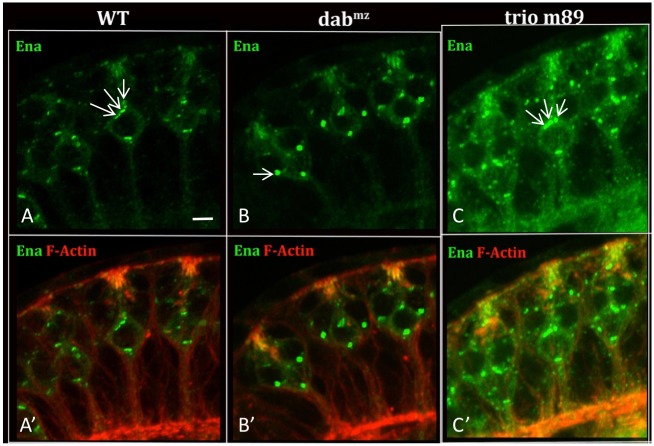
Ena is a direct regulator of actin organization that functions downstream of Abl (Gates et al., 2007; Gertler et al., 1995; Grevengoed et al., 2003; Kannan et al., 2014), but it was not known whether Trio acts between Abl and Ena in a single linear pathway or in parallel to Ena. We have shown previously that loss of Abl or Dab causes Ena protein to coalesce in large aggregates at the most basal part of the soma of PRs (Kannan et al., 2014). If this is mediated via Trio then a trio mutant should show the same phenotype. We therefore examined Ena localization in trio mutant eye discs and found that Ena protein remains distributed throughout the cytoplasm of the cell soma, and does not settle at the axon exit site of the PR soma (Fig. 6). This excludes the hypothesis that Trio is upstream of Ena in a simple, linear dependence pathway, and instead argues that the signaling network bifurcates downstream of Abl, with suppression of Ena and activation of Trio/Rac forming parallel branches.
Fig. 6.
Ena and Trio act in parallel downstream of Abl. Eye imaginal discs were isolated from third instar larvae of the indicated genotypes, stained with anti-Ena (green) and phalloidin (red) and imaged by confocal microscopy. (A-C) Ena channel; (A′-C′) Ena and phalloidin. Arrows highlight the disc-like appearance of Ena immunoreactivity associated with cis-Golgi, distributed throughout the photoreceptor soma in (A,A′) WT and (C,C′) trio−. (B,B′) Ena immunoreactivity in the Dab mutant concentrates in a single, large blob at the most basal point in the soma of each photoreceptor. Scale bar: 5 µm.
Rac interacts with WAVE to controls ISNb axon patterning
Rac has many biochemical targets, but one of its key effectors is the WAVE complex. In the absence of Rac activity, the VCA motif of WAVE protein (SCAR in Drosophila) is sequestered (Chen et al., 2010; Gautreau et al., 2004). Binding of SCAR to GTP-Rac releases that interaction, allowing recruitment of Arp2/3 and promotion of branched actin networks (Blanchoin et al., 2000; Machesky and Insall, 1998; Marchand et al., 2001). We therefore performed genetic interaction tests and discovered that Trio/Rac interacts functionally with WAVE components to control Drosophila axon patterning. We knocked down scar expression by RNAi (Zallen et al., 2002) in neurons and found that this produced the ISNb stalling phenotype characteristic of Abl pathway mutants: ISNb stalls in 24% of RNAi-expressing hemisegments (n=377) versus 7% in control (n=177; P<0.001, χ2; Fig. 7B,C,F). This is supported by analysis of embryos doubly heterozygous for both a scar mutation and a mutation in Abl-interacting protein (Abi), which encodes another core component of the WAVE complex; the doubly heterozygous embryos displayed the same axon stalling phenotype in 79% of hemisegments (n=214) versus 16% stalling for scarΔ37/+ (n=210) and 31% stalling for AbiKO/+ (n=256; P<0.001 compared with either heterozygous mutation alone, χ2; Fig. 7A,D,F). The high stall frequency in Abi heterozygotes was unexpected, but it was observed with two independent Abi alleles and in multiple genetic backgrounds. Apparently, Abi is partly haploinsufficient for ISNb extension. In ISNb extension, SCAR evidently interacts intimately with Trio and Rac, as embryos that are doubly heterozygous for mutations in trio and scar also show a strongly synergistic ISNb stalling phenotype [67% stall (n=86) for trio123.4/+; Df(scar)/+ and 51% stall (n=187) for trio123.4/+; scarΔ37/+ versus 16% stall (n=254) for trio123.4/+; in each case P<0.001, χ2] (Fig. 7E,F). Together, these data show that the functional interaction of Trio/Rac with the WAVE complex is essential for a characteristic Abl-dependent axon guidance decision.
Fig. 7.
Genetic interaction of Trio/Rac with WAVE components and model for the Abl pathway. (A-E) Nomarski images showing three hemisegments of stage 17 embryos of the indicated genotypes, immunostained with anti-Fas2 and filet-mounted to reveal the ISNb motonerve. Black arrows indicate muscle 12 neuromuscular synapses; red arrows indicate positions of missing synapses (‘stall' phenotype). Scale bar: 10 µm. (F) Quantification of ISNb stall phenotypes (failure to innervate muscle 12). ***P<0.001, χ2 with Bonferroni correction. (G) Model for the functional organization of the Abl signaling pathway. Arrow indicates a stimulatory interaction, T-bar indicates an inhibitory interaction, dashed line indicates a physical association (co-IP) without specifying epistatic directionality.
DISCUSSION
We show here that Abl tyrosine kinase and its interacting co-factors form a bifurcating protein network that controls axon patterning in Drosophila (Fig. 7G). Dab, which associates with guidance and motility receptors, is an upstream regulator of Abl localization and activity. Abl, in turn, regulates the localization and suppresses the activity of the actin regulator Ena. Abl also stimulates Rac GTPase signaling through regulation of the Rac GEF Trio. Trio GEF activity is repressed by its own NTD in the absence of Abl, but in the presence of Abl the spectrin repeats of Trio relieve that NTD-dependent repression. Finally, a key target of Abl-dependent Rac activity in axon patterning is a second actin-regulatory complex, WAVE: Trio/Rac interacts genetically with WAVE components, including WAVE/SCAR itself and the Abl-interacting protein Abi, to promote Abl-dependent axon patterning. Together, these interactions allow the Abl signaling network to control a wide variety of axon patterning decisions throughout animal phylogeny, perhaps by coordinating the Ena-dependent dynamics of linear actin bundles with the WAVE-dependent dynamics of branched actin networks.
The relationship of Dab to Abl has long been controversial (Gertler et al., 1993; Howell et al., 1997; Liebl et al., 2003; Song et al., 2010; Sonoshita et al., 2015). For some time, it was unclear whether Dab is even a core component of the Abl pathway, although more recently we established the central role of Dab in Abl pathway function (Liebl et al., 2003; Song et al., 2010). Moreover, a recent report suggested that Dab can co-IP with Abl from mammalian cells (Sonoshita et al., 2015), whereas previous investigators only detected binding by in vitro pulldown assays (Gertler et al., 1993; Howell et al., 1997). We reinvestigated this question and found that, indeed, Dab and Abl co-IP from both cultured Drosophila S2 cells and WT embryo lysates. We also found that Dab enhances the kinase activity of Abl in vivo, as assayed by FRET, in addition to controlling Abl subcellular localization. These data provide a potential biochemical basis for the functional interaction of these proteins.
Here we demonstrate that Dab physically associates with Abl, and we have shown previously that Dab associates with Trio (Le Gall et al., 2008). The most parsimonious interpretation, therefore, is that Abl, Dab and Trio form a single, trimeric complex. We have as yet been unable, however, to show direct interaction of Abl with Trio or co-IP of Abl and Trio from tissue or cell lysates. Although we cannot exclude the possibility that Dab forms separate complexes with Abl and with Trio, we think it more likely that this reflects technical limitations of the experiment. It might be that the trimeric complex is labile in vitro, preventing us from detecting co-IP of Abl with Trio.
It is unclear how Abl kinase induces the Trio spectrin repeats to relieve repression of Trio GEF1 activity by the Trio NTD. A previous study reported direct binding between specific Abl and Trio domains in vitro, as assayed using pulldowns with protein fragments (Forsthoefel et al., 2005). We were unable to confirm this by co-IP of the full-length proteins from cell or embryo lysates, but that could reflect insufficient sensitivity of the assay, particularly if association with specific activated receptors is necessary for complex formation. Previous studies have also failed to provide rigorous evidence of direct phosphorylation of Trio by Abl (Forsthoefel et al., 2005). Our FRET results, however, are consistent with the idea that an additional protein may be involved in the stimulation of Trio by Abl – one that can be titrated into inactive complexes by expression of non-inducible Trio derivatives (Fig. 5D). It might be, for example, that this hypothetical third component is the direct target of Abl kinase, and then associates with Trio or signals to it. Testing this hypothesis awaits the isolation of such a putative co-factor. In addition, although our data show a strong requirement for Abl kinase activity in Trio activation, we cannot rule out the possibility that Abl also has a scaffolding role that is separate from its kinase activity (Hoffmann, 1991; Rogers et al., 2016). We could not use transgenic overexpression of Abl derivatives to query possible non-kinase functions of Abl, or to challenge the trio dependence of activation of Rac FRET by Abl, since overexpression of Abl caused non-physiological activation of other Rac GEFs, including SOS (R.K. and E.G., unpublished observations).
It is striking that conceptually analogous, but molecularly very different, interactions have now been observed between Abl and Trio in different contexts. In experiments here, we show that Abl interacts functionally with Trio to stimulate Rac signaling, and this requires Abl kinase activity. Analogously, in mammalian tumor cells, Abl phosphorylates the C-terminal portion of Trio to stimulate its Rho GEF activity, and this promotes metastasis (Sonoshita et al., 2015). The domain of Trio that becomes phosphorylated by Abl in the mouse tumor model, however, and by Fyn in mouse neurons (DeGeer et al., 2013) does not exist in Drosophila Trio. Moreover, previous experiments have shown that the Rho GEF domain of Trio is dispensable for axon and dendrite development in Drosophila and C. elegans, and that it is the Rac GEF activity of Trio that controls neuronal morphogenesis (Iyer et al., 2012; Shivalkar and Giniger, 2012; Song and Giniger, 2011; Steven et al., 1998). Nonetheless, it is remarkable that in both contexts Abl acts through stimulation of Trio to control motility events that depend on Rho family GTPases, albeit by different molecular mechanisms.
Ena and Rac/WAVE are two key outputs of the Abl pathway for regulation of the neuronal cytoskeleton. Ena regulates actin via three activities (Krause et al., 2003; Trichet et al., 2008): it promotes actin polymerization, bundles actin filaments through its multimerization domain, and prevents capping of the actin barbed end. Mutant analysis suggests that all three activities contribute to Ena-dependent axon patterning. For example, mutation of the dimerization domain generates a null ena allele in Drosophila, and mutation of capping protein β can suppress ena phenotypes (Gates et al., 2009). Together, the different activities of Ena conspire to extend and bundle parallel actin filaments, as are found in filopodia. WAVE/SCAR is the core component of a large protein complex also comprising Sra1, Nap1, Abi and HSPC300 (Chen et al., 2010; Gautreau et al., 2004). This complex promotes branched actin networks by recruiting the Arp2/3 complex to the side of an existing actin filament and stimulating its ability to nucleate a daughter filament at a 70° angle to the mother filament (Blanchoin et al., 2000; Machesky and Insall, 1998). The WAVE complex is initially inactive, and must be activated by association with GTP-Rac (Eden et al., 2002). Once activated, WAVE complexes extend branched actin networks, for example in lamellipodia. The contrasting activities of Ena and Rac/WAVE can be seen in their effects on cell morphology, where overexpression of Ena promotes a spiky, filopodial morphology, whereas activation of Rac produces a broad, lamellar morphology (Insall and Machesky, 2009; Lacayo et al., 2007; Rogers et al., 2003).
Our data demonstrating that Abl simultaneously suppresses the activity of Ena, but stimulates WAVE/Rac, therefore reveal that the structure of the Abl network intrinsically introduces an antagonism between the two major classes of actin structures in the cell (Burke et al., 2014; Chen et al., 2014). A receptor that evokes the activity of the Abl pathway will automatically induce a default balance between the two major classes of actin structures in the growth cone, linear actin bundles and branched actin networks. Receptors that modulate the two legs of the pathway separately, by contrast, will modify that balance. It is attractive to speculate that the ability of Abl to automatically balance, and rebalance, the different kinds of actin structures in the cell might be why so many motility and guidance receptors have evolved to signal through the Abl network (Bradley and Koleske, 2009; Lanier and Gertler, 2000).
For more than two decades, the Abl signaling cassette has been one of the key systems used to interrogate axon growth and guidance and neuronal migration (Goodman and Shatz, 1993; Hoffmann, 1991; Lanier and Gertler, 2000; Wills et al., 1999a). Its explanatory value, however, has been severely limited by the lack of a molecular schema for interpreting the interactions among its components. The results that we report here provide a detailed molecular model for the Abl signaling network in neurons that can now be used to design and interpret cellular and molecular studies of the mechanisms of neuronal morphogenesis and motility.
MATERIALS AND METHODS
Drosophila stocks
Drosophila bearing P[UAS Rac FRET] were obtained from D. Montell (UC Santa Barbara, CA, USA); P[UAS BCR-p210] and P[UAS BCR-p185] were from M. Peifer (UNC-Chapel Hill, NC, USA); Abi mutants were from J.-L. Juang (NHRI, Taipei, Taiwan); and P[UAS-SCAR RNAi] was from J. Zallen (MSKCC, New York, NY, USA). All other fly stocks were described previously (Kannan et al., 2014; Song and Giniger, 2011; Song et al., 2010) or obtained from the Bloomington Drosophila Stock Center.
Construction of Abl FRET biosensor and generation of Drosophila transgenics
Plasmid for mammalian CFP-YFP Abl biosensor (GenBank accession number AF440203; 2334 bp) was obtained from Dr Roger Tsien (UCSD, San Diego, CA, USA). Site-directed mutagenesis was performed to construct a Y221F mutation as a negative control. cDNA of both the WT and Y221F biosensors was subcloned into pUAS-T with BamHI/XhoI, and transgenic fly lines were generated by BestGene.
Drosophila S2 cell growth and transfection
Drosophila S2 cells (E. Serpe, NIH, Bethesda, MD, USA) were cultured in Schneider's medium (Gibco, Thermo Fisher) with 10% heat-inactivated fetal bovine serum (JRH Biosciences) and 1% penicillin-streptomycin (1:100; Gibco). Medium was filter sterilized and cells grown at 23-25°C. S2 cell transfection was by standard methods using DDAB reagent (Sigma) plus 0.7-4 µg plasmid DNA. Transgene expression was driven with actin-GAL4 (pRK241). For FRET experiments, measurements were performed 48 h after transfection.
Drosophila PR culture
Culture of larval photoreceptors was performed by published methods (Newsome et al., 2000). Briefly, 100-150 eye-antennal imaginal discs were dissected from third instar larvae in S2 cell culture media, and the eye portion of each was isolated with a tungsten needle and transferred to a microcentrifuge tube using a silanized pipet. Discs were incubated for 30 min at room temperature in 400 μl collagenase plus 100 μl liberase I, then triturated to homogeneity. Cells were pelleted for 5 min at 5000 g, washed three times with culture medium, suspended in 100 μl culture medium in a MatTek dish coated with poly L-lysine and conconavalin-A, and incubated at 25°C in the dark in a humid chamber.
Ratio FRET imaging in S2 cells and PRs
Ratiometric FRET imaging was carried out by the method of Koga et al. (2006). Images were collected using a DeltaVision microscope with 60×, 1.42 NA lens, 2× zoom and 2×2 binning (for PRs) or a Zeiss LSM 510 confocal microscope using 63×, 1.4 NA, 2× zoom (for S2 cells). On the DeltaVision, stacks of ∼50-75 z-sections were acquired with 0.15 µm step size sequentially in YFP, CFP, FRET, and DIC. On the LSM 510, a 405 nm laser was used to excite and a stack of ∼14 z-sections was collected simultaneously in CFP and FRET. Measurements were performed from z-projections of the image stack. Regions of interest (ROIs) were drawn manually in MetaMorph (Molecular Devices) and average intensity values for the same ROI in CFP (Icfp) and FRET (Ifret) were exported into Excel (Microsoft). Background average intensity values (Ibg-cfp and Ibg-fret) were measured from the same images. The FRET ratio (Rf) was calculated as Rf=(Icfp−Ibg-cfp)/(Ifret−Ibg-fret). CFP/FRET ratio images were presented for display purposes in the intensity modified display mode (Wang et al., 2005). 23-38 cells were imaged for each value reported, except for STI-571 timecourse experiments (n=17-20).
Acceptor photobleaching for Abl FRET
FRET measurement by acceptor photobleaching was performed as described (Karpova et al., 2003). Single plane sequential images in CFP, then YFP, were collected using a Zeiss LSM 780 with 63×, 1.46 NA objective, 4× zoom. GaASP detector was adjusted to eliminate cross-talk and optimize dynamic range. Photobleaching of the whole cell was performed with a 514 nm laser. We collected three reference images before photobleach and five images after. Circular ROIs encompassing the whole cell were drawn manually in background-subtracted CFP channel images to measure average intensity values (In) before and after photobleach. FRET efficiency (Ef) was calculated as Ef=(I6−I5)×100/I6, where In represents average intensity at the nth time point. As a control, we performed mock photobleach with the laser switched off without changing other FRET imaging parameters.
STI-571 preparation and treatment in PR and S2 cell cultures
A 100 mg pill of STI-571 (Novartis, obtained from NIH Clinical Center Pharmacy) was dissolved in 20 ml PBS to provide a 10 mM stock, and aliquots were stored at −20°C. STI-571 was added to media at time of imaging. Washout was initiated with de-oxygenated culture medium using an automatic pump.
trio mutant constructs
Equivalent domain deletions were introduced into UAS-Trio mGEF1 and UAS-Trio mGEF2 backbones by PCR using Expand high-fidelity PCR Taq DNA polymerase (Roche) and verified by sequencing.
Immunostaining and antibodies
Larval eye discs
Third instar larval eye discs with brain lobes attached were dissected, fixed, mounted in Vectashield (Vector Labs) and imaged by standard methods (Kannan et al., 2014). z-sections were acquired using a Zeiss LSM 510 confocal at 63× magnification and deconvoluted using AutoQuant (Media Cybernetics).
Embryo staining for ISNb motor axon defects
Embryo fixation, staining, imaging and quantification of ISNb phenotypes were as described previously (Song and Giniger, 2011; Song et al., 2010). Abdominal hemisegments 2-7 of early stage 17 embryos were scored by Nomarski microscopy for presence or absence of a neuromuscular junction on muscle 12. Data from independent experiments were pooled for each genotype to derive the number of hemisegments in which ISNb did, or did not, form the muscle 12 neuromuscular junction. Significance was assessed by χ2 test.
Antibodies and phalloidin
Mouse anti-Ena (5G2; 1:50), rat anti-Elav (7E8A10; 1:20), anti-Fasciclin 2 (1D4 concentrate; 1:100), anti-Dab (P4D11 for immunoprecipitation; P6E11 for western analysis) and anti-Trio (9A; 1:50) were from the Developmental Studies Hybridoma Bank (Iowa City, IA, USA). Rabbit anti-Dab and rabbit anti-Abl were described previously (Song et al., 2010). Mouse anti-Drosophila Abl polyclonal serum was prepared by the Antibody Development Laboratory of the Fred Hutchinson Cancer Research Center, using the same immunogen as for rabbit anti-Abl. Specificity was verified by staining of Abl null mutant imaginal discs. Rabbit anti-β-galactosidase (55976; 1:10,000) was from Cappel. Secondary antibodies (1:250) were from Jackson ImmunoResearch. Alexa 488-conjugated phalloidin (Life Technologies) was used at 1:300.
Biochemical methods
Co-IP experiments were performed as described previously (Le Gall et al., 2008).
Western blotting
Western blotting was performed by standard methods. Signals were visualized using secondary antibodies coupled with IR-Dye 700 or 800 and scanned with the Odyssey infrared imaging system (Li-Cor Biosciences), or using peroxidase-coupled secondary antibodies and ECL reagents (Lumigen).
Image preparation
Image adjustments, including brightness, contrast, gamma and color balance, were applied to entire figure panels, as necessary. DIC embryo images were montaged from multiple focal planes and in some cases an unsharp mask was applied to the final image to clarify anatomical features.
Statistical methods
Numerical data in all experiments were corrected for multiple testing by the Bonferroni method. For embryo axonal phenotypes, statistical significance of planned comparisons between genotypes was assessed by a two-class (innervated versus non-innervated) χ2 test with one degree of freedom, with P-value calculated in Excel (CHITEST), followed by Bonferroni correction.
Acknowledgements
We are grateful to everyone who gave us advice, assistance and reagents for these experiments. We particularly thank Roger Tsien and Alice Ting for the mammalian Abl FRET probe, and Denise Montell and members of her lab for DNA and flies bearing the Rac FRET probe. In addition to the members of our lab, especially Ginger Hunter and Josh Spurrier for comments on the manuscript, we are particularly grateful to Chi-Hon Lee, Ela Serpe and members of their labs for many helpful discussions and for invaluable assistance, and to Steven Vogel and Clare Waterman for advice on FRET. We also thank Stephen Wincovitch of the National Human Genome Research Institute (NHGRI) Microscopy Core for much help with imaging, and Kory Johnson of the NINDS Biostatistics Core for assistance with statistical analysis.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: R.K., J.-K.S., M.S., L.K. and E.G.; Special Methodology: T.K.; Investigation: R.K., J.-K.S., T.K., A.C., M.S., B.W., L.K., I.K., Q.G. and E.G.; Writing: R.K. and E.G.; Visualization: R.K., J.-K.S. and E.G.; Supervision: E.G.
Funding
These experiments were supported by the Basic Neuroscience Program of the National Institute of Neurological Disorders and Stroke (NINDS) Intramural Research Program (grant Z01-NS003013) to E.G. R.K. was supported in part by a Department of Biotechnology Ramalingaswami re-entry fellowship from the Government of India. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.143776.supplemental
References
- Awasaki T., Saito M., Sone M., Suzuki E., Sakai R., Ito K. and Hama C. (2000). The Drosophila trio plays an essential role in patterning of axons by regulating their directional extension. Neuron 26, 119-131. 10.1016/S0896-6273(00)81143-5 [DOI] [PubMed] [Google Scholar]
- Bashaw G. J., Kidd T., Murray D., Pawson T. and Goodman C. S. (2000). Repulsive axon guidance: Abelson and Enabled play opposing roles downstream of the roundabout receptor. Cell 101, 703-715. 10.1016/S0092-8674(00)80883-1 [DOI] [PubMed] [Google Scholar]
- Bateman J., Shu H. and Van Vactor D. (2000). The guanine nucleotide exchange factor trio mediates axonal development in the Drosophila embryo. Neuron 26, 93-106. 10.1016/S0896-6273(00)81141-1 [DOI] [PubMed] [Google Scholar]
- Bear J. E., Loureiro J. J., Libova I., Fässler R., Wehland J. and Gertler F. B. (2000). Negative regulation of fibroblast motility by Ena/VASP proteins. Cell 101, 717-728. 10.1016/S0092-8674(00)80884-3 [DOI] [PubMed] [Google Scholar]
- Blanchoin L., Amann K. J., Higgs H. N., Marcahnd J.-B., Kaiser D. A. and Pollard T. D. (2000). Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature 404, 1007-1011. 10.1038/35010008 [DOI] [PubMed] [Google Scholar]
- Bradley W. D. and Koleske A. J. (2009). Regulation of cell migration and morphogenesis by Abl-family kinases: emerging mechanisms and physiological contexts. J. Cell Sci. 122, 3441-3454. 10.1242/jcs.039859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke T. A., Christensen J. R., Barone E., Suarez C., Sirotkin V. and Kovar D. R. (2014). Homeostatic actin cytoskeleton networks are regulated by assembly factor competition for monomers. Curr. Biol. 24, 579-585. 10.1016/j.cub.2014.01.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Borek D., Padrick S. B., Gomez T. S., Metlagel Z., Ismail A. M., Umetani J., Billadeau D. D., Otwinowski Z. and Rosen M. K. (2010). Structure and control of the actin regulatory WAVE complex. Nature 468, 533-538. 10.1038/nature09623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. J., Squarr A. J., Stephan R., Chen B., Higgins T. E., Barry D. J., Martin M. C., Rosen M. K., Bogdan S. and Way M. (2014). Ena/VASP proteins cooperate with the WAVE complex to regulate the actin cytoskeleton. Dev. Cell 30, 569-584. 10.1016/j.devcel.2014.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowner D., Le Gall M., Gates M. A. and Giniger E. (2003). Notch steers Drosophila ISNb motor axons by regulating the Abl signaling pathway. Curr. Biol. 13, 967-972. 10.1016/S0960-9822(03)00325-7 [DOI] [PubMed] [Google Scholar]
- Dai Z. and Pendergast A. M. (1995). Abi-2, a novel SH3-containing protein interacts with the c-Abl tyrosine kinase and modulates c-Abl transforming activity. Genes Dev. 9, 2569-2582. 10.1101/gad.9.21.2569 [DOI] [PubMed] [Google Scholar]
- DeGeer J., Boudeau J., Schmidt S., Bedford F., Lamarche-Vane N. and Debant A. (2013). Tyrosine phosphorylation of the Rho guanine nucleotide exchange factor Trio regulates netrin-1/DCC-mediated cortical axon outgrowth. Mol. Cell. Biol. 33, 739-751. 10.1128/MCB.01264-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deinhardt K., Kim T., Spellman D. S., Mains R. E., Eipper B. A., Neubert T. A., Chao M. V. and Hempstead B. L. (2011). Neuronal growth cone retraction relies on proneurotrophin receptor signaling through Rac. Sci. Signal. 4, ra82 10.1126/scisignal.2002060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson B. J. (2002). Molecular mechanisms of axon guidance. Science 298, 1959-1964. 10.1126/science.1072165 [DOI] [PubMed] [Google Scholar]
- Eden S., Rohatgi R., Podtelejnikov A. V., Mann M. and Kirschner M. W. (2002). Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature 418, 790-793. 10.1038/nature00859 [DOI] [PubMed] [Google Scholar]
- Forsthoefel D. J., Liebl E. C., Kolodziej P. A. and Seeger M. A. (2005). The Abelson tyrosine kinase, the Trio GEF and Enabled interact with the Netrin receptor Frazzled in Drosophila. Development 132, 1983-1994. 10.1242/dev.01736 [DOI] [PubMed] [Google Scholar]
- Garbe D. S., O'Donnell M. and Bashaw G. J. (2007). Cytoplasmic domain requirements for Frazzled-mediated attractive axon turning at the Drosophila midline. Development 134, 4325-4334. 10.1242/dev.012872 [DOI] [PubMed] [Google Scholar]
- Gates J., Mahaffey J. P., Rogers S. L., Emerson M., Rogers E. M., Sottile S. L., Van Vactor D., Gertler F. B. and Peifer M. (2007). Enabled plays key roles in embryonic epithelial morphogenesis in Drosophila. Development 134, 2027-2039. 10.1242/dev.02849 [DOI] [PubMed] [Google Scholar]
- Gates J., Nowotarski S. H., Yin H., Mahaffey J. P., Bridges T., Herrera C., Homem C. C. F., Janody F., Montell D. J. and Peifer M. (2009). Enabled and Capping protein play important roles in shaping cell behavior during Drosophila oogenesis. Dev. Biol. 333, 90-107. 10.1016/j.ydbio.2009.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautreau A., Ho H. -H., Li J., Steen H., Gygi S. P. and Kirschner M. W. (2004). Purification and architecture of the ubiquitous Wave complex. Proc. Natl. Acad. Sci. USA 101, 4379-4383. 10.1073/pnas.0400628101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertler F. B., Hill K. K., Clark M. J. and Hoffmann F. M. (1993). Dosage-sensitive modifiers of Drosophila abl tyrosine kinase function: prospero, a regulator of axonal outgrowth, and disabled, a novel tyrosine kinase substrate. Genes Dev. 7, 441-453. 10.1101/gad.7.3.441 [DOI] [PubMed] [Google Scholar]
- Gertler F. B., Comer A. R., Juang J. L., Ahern S. M., Clark M. J., Liebl E. C. and Hoffmann F. M. (1995). enabled, a dosage-sensitive suppressor of mutations in the Drosophila Abl tyrosine kinase, encodes an Abl substrate with SH3 domain-binding properties. Genes Dev. 9, 521-533. 10.1101/gad.9.5.521 [DOI] [PubMed] [Google Scholar]
- Gertler F. B., Niebuhr K., Reinhard M., Wehland J. and Soriano P. (1996). Mena, a relative of VASP and Drosophila Enabled, is implicated in the control of microfilament dynamics. Cell 87, 227-239. 10.1016/S0092-8674(00)81341-0 [DOI] [PubMed] [Google Scholar]
- Goodman C. S. and Shatz C. J. (1993). Developmental mechanisms that generate precise patterns of neuronal connectivity. Cell 72 Suppl., 77-98. 10.1016/s0092-8674(05)80030-3 [DOI] [PubMed] [Google Scholar]
- Grevengoed E. E., Fox D. T., Gates J. and Peifer M. (2003). Balancing different types of actin polymerization at distinct sites: roles for Abelson kinase and Enabled. J. Cell Biol. 163, 1267-1279. 10.1083/jcb.200307026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupton S. L., Riquelme D., Hughes-Alford S. K., Tadros J., Rudina S. S., Hynes R. O., Lauffenburger D. and Gertler F. B. (2012). Mena binds alpha5 integrin directly and modulates alpha5beta1 function. J. Cell Biol. 198, 657-676. 10.1083/jcb.201202079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkemeyer M. J., Gertler F. B., Goodman W. and Hoffmann F. M. (1987). The Drosophila Abelson proto-oncogene homolog: identification of mutant alleles that have pleiotropic effects late in development. Cell 51, 821-828. 10.1016/0092-8674(87)90105-X [DOI] [PubMed] [Google Scholar]
- Hill K. K., Bedian V., Juang J. L. and Hoffmann F. M. (1995). Genetic interactions between the Drosophila Abelson (Abl) tyrosine kinase and failed axon connections (fax), a novel protein in axon bundles. Genetics 141, 595-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann F. M. (1991). Drosophila abl and genetic redundancy in signal transduction. Trends Genet. 7, 351-355. 10.1016/0168-9525(91)90254-F [DOI] [PubMed] [Google Scholar]
- Howell B. W., Gertler F. B. and Cooper J. A. (1997). Mouse disabled (mDab1): a Src binding protein implicated in neuronal development. EMBO J. 16, 121-132. 10.1093/emboj/16.1.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell B. W., Herrick T. M. and Cooper J. A. (1999a). Reelin-induced tyrosine phosphorylation of disabled 1 during neuronal positioning. Genes Dev. 13, 643-648. 10.1101/gad.13.6.643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell B. W., Lanier L. M., Frank R., Gertler F. B. and Cooper J. A. (1999b). The disabled 1 phosphotyrosine-binding domain binds to the internalization signals of transmembrane glycoproteins and to phospholipids. Mol. Cell. Biol. 19, 5179-5188. 10.1128/MCB.19.7.5179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsouna A., Kim Y.-S. and VanBerkum M. F. A. (2003). Abelson tyrosine kinase is required to transduce midline repulsive cues. J. Neurobiol. 57, 15-30. 10.1002/neu.10232 [DOI] [PubMed] [Google Scholar]
- Insall R. H. and Machesky L. M. (2009). Actin dynamics at the leading edge: from simple machinery to complex networks. Dev. Cell 17, 310-322. 10.1016/j.devcel.2009.08.012 [DOI] [PubMed] [Google Scholar]
- Itoh R. E., Kurokawa K., Ohba Y., Yoshizaki H., Mochizuki N. and Matsuda M. (2002). Activation of rac and cdc42 video imaged by fluorescent resonance energy transfer-based single-molecule probes in the membrane of living cells. Mol. Cell. Biol. 22, 6582-6591. 10.1128/MCB.22.18.6582-6591.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer S. C., Wang D., Iyer E. P. R., Trunnell S. A., Meduri R., Shinwari R., Sulkowski M. J. and Cox D. N. (2012). The RhoGEF trio functions in sculpting class specific dendrite morphogenesis in Drosophila sensory neurons. PLoS ONE 7, e33634 10.1371/journal.pone.0033634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juang J.-L. and Hoffmann F. M. (1999). Drosophila abelson interacting protein (dAbi) is a positive regulator of abelson tyrosine kinase activity. Oncogene 18, 5138-5147. 10.1038/sj.onc.1202911 [DOI] [PubMed] [Google Scholar]
- Kannan R., Kuzina I., Wincovitch S., Nowotarski S. H. and Giniger E. (2014). The Abl/enabled signaling pathway regulates Golgi architecture in Drosophila photoreceptor neurons. Mol. Biol. Cell 25, 2993-3005. 10.1091/mbc.E14-02-0729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpova T. S., Baumann C. T., He L., Wu X., Grammer A., Lipsky P., Hager G. L. and McNally J. G. (2003). Fluorescence resonance energy transfer from cyan to yellow fluorescent protein detected by acceptor photobleaching using confocal microscopy and a single laser. J. Microsc. 209, 56-70. 10.1046/j.1365-2818.2003.01100.x [DOI] [PubMed] [Google Scholar]
- Koga F., Xu W., Karpova T. S., McNally J. G., Baron R. and Neckers L. (2006). Hsp90 inhibition transiently activates Src kinase and promotes Src-dependent Akt and Erk activation. Proc. Natl. Acad. Sci. USA 103, 11318-11322. 10.1073/pnas.0604705103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M., Bear J. E., Loureiro J. J. and Gertler F. B. (2002). The Ena/VASP enigma. J. Cell Sci. 115, 4721-4726. 10.1242/jcs.00218 [DOI] [PubMed] [Google Scholar]
- Krause M., Dent E. W., Bear J. E., Loureiro J. J. and Gertler F. B. (2003). Ena/VASP proteins: regulators of the actin cytoskeleton and cell migration. Annu. Rev. Cell Dev. Biol. 19, 541-564. 10.1146/annurev.cellbio.19.050103.103356 [DOI] [PubMed] [Google Scholar]
- Kuzina I., Song J. K. and Giniger E. (2011). How Notch establishes longitudinal axon connections between successive segments of the Drosophila CNS. Development 138, 1839-1849. 10.1242/dev.062471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacayo C. I., Pincus Z., VanDuijn M. M., Wilson C. A., Fletcher D. A., Gertler F. B., Mogilner A. and Theriot J. A. (2007). Emergence of large-scale cell morphology and movement from local actin filament growth dynamics. PLoS Biol. 5, e233 10.1371/journal.pbio.0050233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier L. M. and Gertler F. B. (2000). From Abl to actin: Abl tyrosine kinase and associated proteins in growth cone motility. Curr. Opin. Neurobiol. 10, 80-87. 10.1016/S0959-4388(99)00058-6 [DOI] [PubMed] [Google Scholar]
- Le Gall M., De Mattei C. and Giniger E. (2008). Molecular separation of two signaling pathways for the receptor, Notch. Dev. Biol. 313, 556-567. 10.1016/j.ydbio.2007.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebl E. C., Forsthoefel D. J., Franco L. S., Sample S. H., Hess J. E., Cowger J. A., Chandler M. P., Shupert A. M. and Seeger M. A. (2000). Dosage-sensitive, reciprocal genetic interactions between the Abl tyrosine kinase and the putative GEF trio reveal trio's role in axon pathfinding. Neuron 26, 107-118. 10.1016/S0896-6273(00)81142-3 [DOI] [PubMed] [Google Scholar]
- Liebl E. C., Rowe R. G., Forsthoefel D. J., Stammler A. L., Schmidt E. R., Turski M. and Seeger M. A. (2003). Interactions between the secreted protein Amalgam, its transmembrane receptor Neurotactin and the Abelson tyrosine kinase affect axon pathfinding. Development 130, 3217-3226. 10.1242/dev.00545 [DOI] [PubMed] [Google Scholar]
- Lin T.-Y., Huang C.-H., Kao H.-H., Liou G.-G., Yeh S.-R., Cheng C.-M., Chen M.-H., Pan R.-L. and Juang J.-L. (2009). Abi plays an opposing role to Abl in Drosophila axonogenesis and synaptogenesis. Development 136, 3099-3107. 10.1242/dev.033324 [DOI] [PubMed] [Google Scholar]
- Luo L. (2000). Trio quartet in D. (melanogaster). Neuron 26, 1-2. 10.1016/S0896-6273(00)81129-0 [DOI] [PubMed] [Google Scholar]
- Machesky L. M. and Insall R. H. (1998). Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr. Biol. 8, 1347-1356. 10.1016/S0960-9822(98)00015-3 [DOI] [PubMed] [Google Scholar]
- Marchand J.-B., Kaiser D. A., Pollard T. D. and Higgs H. N. (2001). Interaction of WASP/Scar proteins with actin and vertebrate Arp2/3 complex. Nat. Cell Biol. 3, 76-82. 10.1038/35050590 [DOI] [PubMed] [Google Scholar]
- Moresco E. M. and Koleske A. J. (2003). Regulation of neuronal morphogenesis and synaptic function by Abl family kinases. Curr. Opin. Neurobiol. 13, 535-544. 10.1016/j.conb.2003.08.002 [DOI] [PubMed] [Google Scholar]
- Newsome T. P., Schmidt S., Dietzl G., Keleman K., Åsling B., Debant A. and Dickson B. J. (2000). Trio combines with dock to regulate Pak activity during photoreceptor axon pathfinding in Drosophila. Cell 101, 283-294. 10.1016/S0092-8674(00)80838-7 [DOI] [PubMed] [Google Scholar]
- Rhee J., Mahfooz N. S., Arregui C., Lilien J., Balsamo J. and VanBerkum M. F. A. (2002). Activation of the repulsive receptor Roundabout inhibits N-cadherin-mediated cell adhesion. Nat. Cell Biol. 4, 798-805. 10.1038/ncb858 [DOI] [PubMed] [Google Scholar]
- Rogers S. L., Wiedemann U., Stuurman N. and Vale R. D. (2003). Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J. Cell Biol. 162, 1079-1088. 10.1083/jcb.200303023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers E. M., Spracklen A. J., Bilancia C. G., Sumigray K. D., Allred S. C., Nowotarski S. H., Schaefer K. N., Ritchie B. J. and Peifer M. (2016). Abelson kinase acts as a robust, multifunctional scaffold in regulating embryonic morphogenesis. Mol. Biol. Cell 27, 2613-2631. 10.1091/mbc.e16-05-0292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S. and Debant A. (2014). Function and regulation of the Rho guanine nucleotide exchange factor Trio. Small GTPases 5, e29769 10.4161/sgtp.29769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivalkar M. and Giniger E. (2012). Control of dendritic morphogenesis by Trio in Drosophila melanogaster. PLoS ONE 7, e33737 10.1371/journal.pone.0033737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J. K. and Giniger E. (2011). Noncanonical Notch function in motor axon guidance is mediated by Rac GTPase and the GEF1 domain of Trio. Dev. Dyn. 240, 324-332. 10.1002/dvdy.22525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H.-J. and Poo M.-M. (1999). Signal transduction underlying growth cone guidance by diffusible factors. Curr. Opin. Neurobiol. 9, 355-363. 10.1016/S0959-4388(99)80052-X [DOI] [PubMed] [Google Scholar]
- Song H. and Poo M. (2001). The cell biology of neuronal navigation. Nat. Cell Biol. 3, E81-E88. 10.1038/35060164 [DOI] [PubMed] [Google Scholar]
- Song J. K., Kannan R., Merdes G., Singh J., Mlodzik M. and Giniger E. (2010). Disabled is a bona fide component of the Abl signaling network. Development 137, 3719-3727. 10.1242/dev.050948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoshita M., Itatani Y., Kakizaki F., Sakimura K., Terashima T., Katsuyama Y., Sakai Y. and Taketo M. M. (2015). Promotion of colorectal cancer invasion and metastasis through activation of NOTCH-DAB1-ABL-RHOGEF protein TRIO. Cancer Discov. 5, 198-211. 10.1158/2159-8290.CD-14-0595 [DOI] [PubMed] [Google Scholar]
- Sterne G. R., Kim J. H. and Ye B. (2015). Dysregulated Dscam levels act through Abelson tyrosine kinase to enlarge presynaptic arbors. Elife 4, e05196 10.7554/elife.05196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steven R., Kubiseski T. J., Zheng H., Kulkarni S., Mancillas J., Morales A. R., Hogue C. W. V., Pawson T. and Culotti J. (1998). UNC-73 activates the Rac GTPase and is required for cell and growth cone migrations in C. elegans. Cell 92, 785-795. 10.1016/S0092-8674(00)81406-3 [DOI] [PubMed] [Google Scholar]
- Tessier-Lavigne M. and Goodman C. S. (1996). The molecular biology of axon guidance. Science 274, 1123-1133. 10.1126/science.274.5290.1123 [DOI] [PubMed] [Google Scholar]
- Ting A. Y., Kain K. H., Klemke R. L. and Tsien R. Y. (2001). Genetically encoded fluorescent reporters of protein tyrosine kinase activities in living cells. Proc. Natl. Acad. Sci. USA 98, 15003-15008. 10.1073/pnas.211564598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trichet L., Sykes C. and Plastino J. (2008). Relaxing the actin cytoskeleton for adhesion and movement with Ena/VASP. J. Cell Biol. 181, 19-25. 10.1083/jcb.200710168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. Y. and Baltimore D. (1983). Cellular RNA homologous to the Abelson murine leukemia virus transforming gene: expression and relationship to the viral sequence. Mol. Cell. Biol. 3, 773-779. 10.1128/MCB.3.5.773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Botvinick E. L., Zhao Y., Berns M. W., Usami S., Tsien R. Y. and Chien S. (2005). Visualizing the mechanical activation of Src. Nature 434, 1040-1045. 10.1038/nature03469 [DOI] [PubMed] [Google Scholar]
- Wang X., He L., Wu Y. I., Hahn K. M. and Montell D. J. (2010). Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nat. Cell Biol. 12, 591-597. 10.1038/ncb2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills Z., Bateman J., Korey C. A., Comer A. and Van Vactor D. (1999a). The tyrosine kinase Abl and its substrate enabled collaborate with the receptor phosphatase Dlar to control motor axon guidance. Neuron 22, 301-312. 10.1016/S0896-6273(00)81091-0 [DOI] [PubMed] [Google Scholar]
- Wills Z., Marr L., Zinn K., Goodman C. S. and Van Vactor D. (1999b). Profilin and the Abl tyrosine kinase are required for motor axon outgrowth in the Drosophila embryo. Neuron 22, 291-299. 10.1016/S0896-6273(00)81090-9 [DOI] [PubMed] [Google Scholar]
- Winberg M. L., Mitchell K. J. and Goodman C. S. (1998). Genetic analysis of the mechanisms controlling target selection: complementary and combinatorial functions of netrins, semaphorins, and IgCAMs. Cell 93, 581-591. 10.1016/S0092-8674(00)81187-3 [DOI] [PubMed] [Google Scholar]
- Yu H.-H., Zisch A. H., Dodelet V. C. and Pasquale E. B. (2001). Multiple signaling interactions of Abl and Arg kinases with the EphB2 receptor. Oncogene 20, 3995-4006. 10.1038/sj.onc.1204524 [DOI] [PubMed] [Google Scholar]
- Zallen J. A., Cohen Y., Hudson A. M., Cooley L., Wieschaus E. and Schejter E. D. (2002). SCAR is a primary regulator of Arp2/3-dependent morphological events in Drosophila. J. Cell Biol. 156, 689-701. 10.1083/jcb.200109057 [DOI] [PMC free article] [PubMed] [Google Scholar]