Abstract
Mouse double minute 4 (MDM4) is a p53-interacting oncoprotein that plays an important role in the p53 tumor suppressor pathway. The common rs4245739 A > C polymorphism creates a miR-191 binding site in the MDM4 gene transcript. Numerous studies have investigated the association between this MDM4 polymorphism and cancer risk, but have failed to reach a definitive conclusion. To address this issue, we conducted a meta-analysis by selecting eligible studies from MEDLINE, EMBASE, and Chinese Biomedical databases. Odds ratios (ORs) and 95% confidence intervals (CIs) were used to assess the strength of the associations. We also performed genotype-based mRNA expression analysis using data from 270 individuals retrieved from public datasets. A total of 15 studies with 19796 cases and 49681 controls were included in the final meta-analysis. The pooled results revealed that the MDM4 rs4245739C allele is associated with a decreased cancer risk in the heterozygous (AC vs. AA: OR = 0.82, 95% CI = 0.73−0.93), dominant (AC/CC vs. AA: OR = 0.82, 95% CI = 0.72−0.93), and allele contrast models (C vs. A: OR = 0.84, 95% CI = 0.76−0.94). The association was more prominent in Asians and population-based studies. We also found that the rs4245739C allele was associated with decreased MDM4 mRNA expression, especially for Caucasians. Thus the MDM4 rs4245739 A > C polymorphism appears to be associated with decreased cancer risk. These findings would be strengthened by new studies with larger sample sizes and encompassing additional ethnicities.
Keywords: MDM4, polymorphism, cancer susceptibility, meta-analysis
INTRODUCTION
Based on the latest GLOBOCAN estimates, there were approximately 14.1 million new cancer cases and 8.2 million cancer-related deaths worldwide in 2012 [1]. Developing countries accounted for almost 57% of new cancer cases and 65% of cancer-related deaths [1]. According to the trend in cancer incidence, the expected number of new cancer cases will reach 22.2 million worldwide in 2030 [2]. Leading risk factors for cancer development include tobacco use, overweight/obesity, physical inactivity, and infection [1]. Moreover, molecular epidemiological studies have demonstrated that genetic factors including single nucleotide polymorphisms (SNPs), may also play an important role in carcinogenesis [3–9].
As the gatekeeper for cellular growth and division, the tumor suppressor protein p53 maintains genomic stability and regulating cell growth, division, and apoptosis. Dysfunctional p53 protein can lead to the initiation and progression of tumors [10]. Mouse double minute 4 (MDM4) protein is a structural homolog of MDM2, which contains a p53 binding domain at the N-terminus and a RING finger domain at the C-terminus. MDM4 has been shown to inhibit p53 transcriptional activity directly by binding to its transcriptional activation domain. Overactive MDM4 reduces p53 tumor suppression function and contributes to tumor formation and progression [11]. The MDM4 protein can also inhibit the degradation of MDM2 by interacting with its RING finger domain [11]. Overexpression of MDM4 is associated with tumor progression and poor prognosis [12–14]. Previous molecular epidemiology studies suggest that genetic variations in MDM4 gene are associated with risk of various types of cancer [15–20].
Among the many MDM4 polymorphisms, a common genetic variant rs4245739 A > C has been widely investigated for its association with cancer susceptibility [21–28]. This polymorphism is located in the 3′ untranslated region (UTR) of the MDM4 gene, and creates a miR-191 target site that can lead to decreased expression of MDM4. However, the studies have generated controversial results regarding the association between this polymorphism and cancer risk. The possible reasons for the inconsistencies include differences in ethnicity and geographic location, as well as the limited sample size. To date, no meta-analysis has been conducted to comprehensively investigate the association of MDM4 rs4245739 A > C with overall cancer risk. To address the controversy regarding this association, we performed the current meta-analysis to precisely define the effect of MDM4 rs4245739 A > C polymorphism on overall cancer risk.
RESULTS
Characteristics of eligible studies
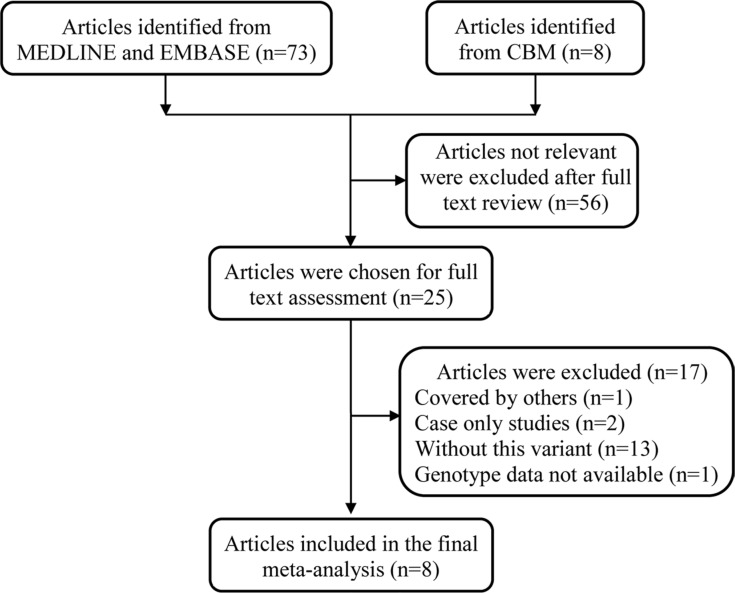
A total of 81 articles were retrieved after an initial literature search in MEDLINE, EMBASE, and Chinese Biomedical (CBM) databases (Figure 1). After full text review, 73 articles were excluded for the following reasons: review articles, duplicate studies, non-case-control study design, genotype distributions were not available, or no evaluation of the association between MDM4 rs4245739 A > C polymorphism and cancer risk. Ultimately, we found that only eight articles [21–28] met the inclusion criteria (Table 1). Among the eight articles, several investigations involving subjects from different areas were divided by area [22, 23, 27] and investigations were also separated by cancer type [26, 28]. As a result, a total of 15 case-control studies with 19796 cases and 49681 controls were included in the final meta-analysis. Of these, sample sizes ranged from 200 to 6512 for cases, and from 400 to 41451 for controls. The genotype distributions of the MDM4 rs4245739 A > C polymorphism were in accordance with Hardy-Weinberg equilibrium (HWE) in the controls in all 15 studies. Studies were performed on various types of cancer. Four studies focused on breast cancer [21, 22, 26], three on lung cancer [26, 27], two on esophageal squamous cell carcinoma [23], and one each on non-Hodgkin lymphoma [24], gastric cancer [25], colon cancer [26], prostate cancer [26], ovarian cancer [28] and endometrial cancer [28]. Seven studies were conducted among Caucasians [21, 26, 28], and eight among Asians [22–25, 27]. All 15 studies were considered high quality; one was scored as 10, eleven as 12 and three as 13.
Figure 1. Flow diagram of included studies for the association between MDM4 rs4245739 A > C polymorphism and overall cancer risk.
Table 1. Characteristics of studies included in the current meta-analysis.
| Surname | Year | Cancer type | Country | Ethnicity | Control source | Genotype method | Case | Control | MAF | HWE | Score | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AC | CC | All | AA | AC | CC | All | ||||||||||
| Garcia-Closas | 2013 | Breast | Multi-center | Caucasian | Mixed | Illumina array | 3318 | 2637 | 557 | 6512 | 22825 | 15798 | 2828 | 41451 | 0.26 | 0.183 | 12 |
| Liu | 2013 | Breast | China | Asian | PB | PCR-RFLP | 733 | 67 | 0 | 800 | 686 | 111 | 3 | 800 | 0.07 | 0.505 | 13 |
| Liu | 2013 | Breast | China | Asian | PB | PCR-RFLP | 278 | 22 | 0 | 300 | 501 | 96 | 3 | 600 | 0.09 | 0.483 | 12 |
| Zhou | 2013 | ESCC | China | Asian | PB | PCR-RFLP | 501 | 37 | 2 | 540 | 478 | 70 | 2 | 550 | 0.07 | 0.740 | 13 |
| Zhou | 2013 | ESCC | China | Asian | PB | PCR-RFLP | 529 | 56 | 3 | 588 | 510 | 88 | 2 | 600 | 0.08 | 0.379 | 13 |
| Fan | 2014 | NHL | China | Asian | PB | PCR-RFLP | 187 | 13 | 0 | 200 | 346 | 53 | 1 | 400 | 0.07 | 0.487 | 12 |
| Feng | 2014 | Gastric | China | Asian | HB | PCR-RFLP | 208 | 209 | 51 | 468 | 210 | 219 | 64 | 493 | 0.35 | 0.561 | 10 |
| Gansmo | 2015 | Breast | Norway | Caucasian | PB | LightSNiP assay | 966 | 643 | 108 | 1717 | 1021 | 703 | 146 | 1870 | 0.27 | 0.106 | 12 |
| Gansmo | 2015 | Colon | Norway | Caucasian | PB | LightSNiP assay | 823 | 600 | 108 | 1531 | 2042 | 1439 | 266 | 3747 | 0.26 | 0.566 | 12 |
| Gansmo | 2015 | Lung | Norway | Caucasian | PB | LightSNiP assay | 715 | 515 | 101 | 1331 | 2042 | 1439 | 266 | 3747 | 0.26 | 0.566 | 12 |
| Gansmo | 2015 | Prostate | Norway | Caucasian | PB | LightSNiP assay | 1412 | 927 | 161 | 2500 | 1021 | 736 | 120 | 1877 | 0.26 | 0.410 | 12 |
| Gao | 2015 | Lung | China | Asian | PB | PCR-RFLP | 297 | 22 | 1 | 320 | 548 | 90 | 2 | 640 | 0.07 | 0.399 | 12 |
| Gao | 2015 | Lung | China | Asian | PB | PCR-RFLP | 183 | 17 | 0 | 200 | 321 | 77 | 2 | 400 | 0.10 | 0.248 | 12 |
| Gansmo | 2016 | Ovarian | Norway | Caucasian | HB | LightSNiP assay | 716 | 564 | 105 | 1385 | 1021 | 703 | 146 | 1870 | 0.27 | 0.106 | 12 |
| Gansmo | 2016 | Endometrial | Norway | Caucasian | HB | LightSNiP assay | 757 | 541 | 106 | 1404 | 1021 | 703 | 146 | 1870 | 0.27 | 0.106 | 12 |
MAF, minor allele frequency; HWE, Hardy-Weinberg equilibrium, ESCC, esophageal squamous cell carcinoma; NHL, non-Hodgkin lymphoma; PB, population based; HB, hospital based; PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism.
Meta-analysis results
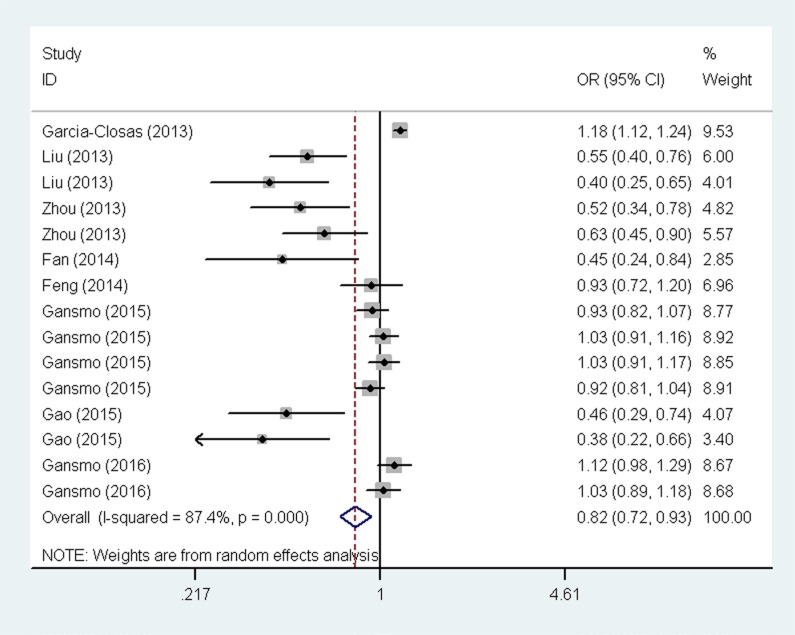
The overall analysis results are shown in Figure 2 and Table 2. We found the presence of significant heterogeneity under all genetic models (P het< 0.10); thus, we chose the random-effects model because it can generate wider confidence intervals (CIs). We found that the MDM4 rs4245739C carriers had a significantly decreased overall cancer risk under the heterozygous [AC vs. AA: odds ratio (OR) = 0.82, 95% CI = 0.73–0.93], dominant (AC + CC vs. AA: OR = 0.82, 95% CI = 0.72–0.93), and allele contrast models (C vs. A: OR = 0.84, 95% CI = 0.76–0.94). In the subgroup analysis by ethnicity, similar results were found among Asians (AC vs. AA: OR = 0.55, 95% CI = 0.43–0.70; AC + CC vs. AA: OR = 0.54, 95% CI = 0.43–0.69; and C vs. A: OR = 0.56, 95% CI = 0.44–0.72), but not among Caucasians. When analyses were stratified by the source of controls, significant association with decreased cancer risk was found among population-based studies (AC vs. AA: OR = 0.70, 95% CI = 0.58–0.83; AC/CC vs. AA: OR = 0.70, 95% CI = 0.59–0.83; and C vs. A: OR = 0.73, 95% CI = 0.63–0.85). In the stratified analysis by quality score, significant associations were found among studies with scores ≥12 (AC vs. AA: OR = 0.81, 95% CI = 0.71–0.93; AC + CC vs. AA: OR = 0.81, 95% CI = 0.71–0.92; and C vs. A: OR = 0.84, 95% CI = 0.75–0.93).
Figure 2. Forest plot for the association between the MDM4 rs4245739 A > C polymorphism and cancer risk under the dominant model.
The horizontal lines represent the ORs and 95% CIs of single studies. The diamond represents the pooled OR and 95% CI.
Table 2. Meta-analysis of the association between MDM4 rs4245739 A > C polymorphism and cancer risk.
| Variables | No. of studies | Sample size | Homozygous | Heterozygous | Recessive | Dominant | Allele | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC vs. AA | AC vs. AA | CC vs. AC+AA | AC+CC vs. AA | C vs. A | |||||||||||||
| OR (95% CI) | P het | I2 (%) | OR (95% CI) | P het | I2 (%) | OR (95% CI) | P het | I2 (%) | OR (95% CI) | P het | I2 (%) | OR (95% CI) | P het | I2 (%) | |||
| All a | 15 | 19796/49681 | 1.01 (0.87–1.17) | 0.005 | 54.9 | 0.82 (0.73–0.93) | < 0.001 | 85.7 | 1.00 (0.89–1.14) | 0.041 | 42.6 | 0.82 (0.72–0.93) | < 0.001 | 87.4 | 0.84 (0.76–0.94) | < 0.001 | 87.9 |
| Cancer type | |||||||||||||||||
| Breast | 4 | 9329/44721 | 0.94 (0.55–1.61) | < 0.001 | 83.3 | 0.78 (0.57–1.06) | < 0.001 | 92.1 | 0.95 (0.59–1.51) | 0.002 | 79.3 | 0.76 (0.55–1.05) | < 0.001 | 93.7 | 0.77 (0.57–1.03) | < 0.001 | 94.3 |
| Lung | 3 | 1851/4787 | 1.08 (0.84–1.37) | 0.762 | 0.0 | 0.58 (0.29–1.19) | < 0.001 | 90.0 | 1.07 (0.84–1.35) | 0.814 | 0.0 | 0.59 (0.29–1.20) | < 0.001 | 90.5 | 0.61 (0.31–1.19) | < 0.001 | 90.0 |
| ESCC | 2 | 1128/1150 | 1.20 (0.32–4.50) | 0.759 | 0.0 | 0.57 (0.43–0.74) | 0.484 | 0.0 | 1.27 (0.34–4.79) | 0.763 | 0.0 | 0.58 (0.45–0.76) | 0.464 | 0.0 | 0.62 (0.48–0.79) | 0.438 | 0.0 |
| Others | 6 | 7488/4640 | 0.98 (0.87–1.10) | 0.953 | 0.0 | 1.00 (0.89–1.11) | 0.036 | 58.1 | 0.97 (0.86–1.09) | 0.970 | 0.0 | 0.99 (0.89–1.10) | 0.041 | 56.9 | 0.99 (0.92–1.06) | 0.080 | 49.2 |
| Ethnicity | |||||||||||||||||
| Caucasians | 7 | 16380/45198 | 1.03 (0.88–1.22) | < 0.001 | 75.3 | 1.04 (0.97–1.12) | 0.012 | 63.1 | 1.02 (0.89–1.18) | 0.005 | 67.8 | 1.04 (0.95–1.13) | < 0.001 | 75.1 | 1.03 (0.95–1.11) | < 0.001 | 80.4 |
| Asians | 8 | 3416/4483 | 0.78 (0.54–1.14) | 0.908 | 0.0 | 0.55 (0.43–0.70) | 0.007 | 64.0 | 0.81 (0.56–1.16) | 0.918 | 0.0 | 0.54 (0.43–0.69) | 0.007 | 64.1 | 0.56 (0.44–0.72) | 0.001 | 71.2 |
| Source of control | |||||||||||||||||
| PB | 11 | 10027/7737 | 0.96 (0.85–1.08) | 0.763 | 0.763 | 0.70 (0.58–0.83) | < 0.001 | 83.4 | 0.96 (0.85–1.09) | 0.807 | 0.0 | 0.70 (0.59–0.83) | < 0.001 | 84.0 | 0.73 (0.63–0.85) | < 0.001 | 84.3 |
| HB | 3 | 3257/2363 | 0.97 (0.81–1.15) | 0.623 | 0.623 | 1.07 (0.97–1.18) | 0.461 | 0.0 | 0.94 (0.79–1.11) | 0.757 | 0.0 | 1.05 (0.96–1.16) | 0.387 | 0.0 | 1.02 (0.95–1.10) | 0.371 | 0.0 |
| Mixed | 1 | 6512/41451 | 1.36 (1.23–1.49) | / | / | 1.15 (1.09–1.21) | / | / | 1.28 (1.16–1.41) | / | / | 1.18 (1.12–1.24) | / | / | 1.16 (1.11–1.21) | / | / |
| Quality score | |||||||||||||||||
| ≥ 12 | 14 | 19596/49281 | 1.03 (0.88–1.19) | 0.009 | 53.9 | 0.81 (0.71–0.93) | < 0.001 | 86.6 | 1.02 (0.90–1.16) | 0.055 | 41.0 | 0.81 (0.71–0.92) | < 0.001 | 88.2 | 0.84 (0.75–0.93) | < 0.001 | 88.5 |
| < 12 | 1 | 200/400 | 0.81 (0.53–1.22) | / | / | 0.96 (0.74–1.26) | / | / | 0.82 (0.55–1.21) | / | / | 0.93 (0.72–1.20) | / | / | 0.92 (0.76–1.11) | / | / |
Het, heterogeneity; ESCC, esophageal squamous cell carcinoma; HB, hospital based; PB, population based.
The MDM4 mRNA expression by genotypes
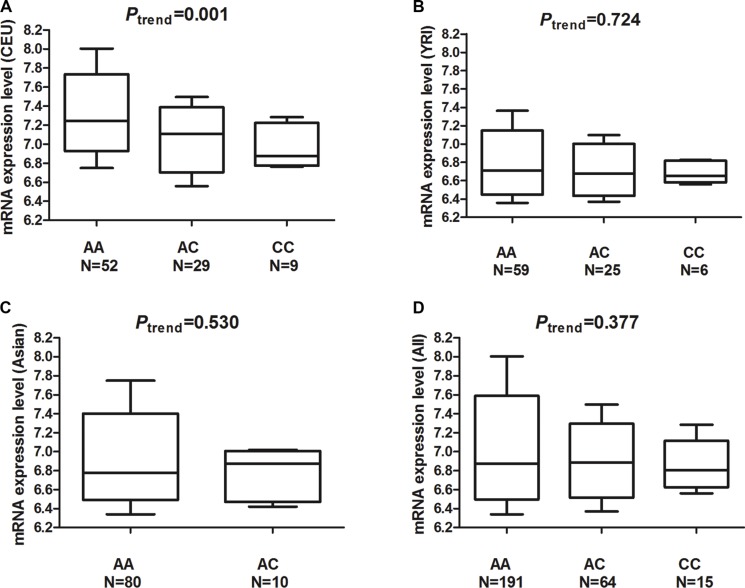
In the genotype-based mRNA expression analysis (Table 3 and Figure 3) using public datasets, we found the rs4245739C allele carriers had trends toward decreased mRNA expression level among Caucasians, Asians, Africans, and all subjects. The decrease in the MDM4 mRNA expression reached a statistical significance among the Caucasians (AC vs. AA: P = 0.002; CC vs. AA: P = 0.004, and AC + CC vs. AA: P = 0.0002), but not among other populations.
Table 3. MDM4 mRNA expression by the genotypes of rs4245739 A > C, using data from the HapMapa.
| Population | genotypes | No. | Mean ± SD | Pb | Ptrendc |
|---|---|---|---|---|---|
| CEU | AA | 52 | 7.28 ± 0.31 | 0.001 | |
| AC | 29 | 7.06 ± 0.27 | 0.002 | ||
| CC | 9 | 6.96 ± 0.20 | 0.004 | ||
| Dominant | 38 | 7.03 ± 0.26 | 0.0002 | ||
| YRI | AA | 59 | 6.75 ± 0.23 | 0.724 | |
| AC | 25 | 6.72 ± 0.22 | 0.578 | ||
| CC | 6 | 6.68 ± 0.11 | 0.271 | ||
| Dominant | 31 | 6.71 ± 0.20 | 0.459 | ||
| Asian | AA | 80 | 6.86 ± 0.31 | 0.530 | |
| AC | 10 | 6.79 ± 0.23 | 0.530 | ||
| CC | 0 | / | / | ||
| Dominant | 10 | 6.79 ± 0.23 | 0.530 | ||
| All | AA | 191 | 6.94 ± 0.36 | 0.377 | |
| AC | 64 | 6.88 ± 0.29 | 0.271 | ||
| CC | 15 | 6.85 ± 0.22 | 0.165 | ||
| Dominant | 79 | 6.88 ± 0.28 | 0.135 |
CEU, 90 Utah residents with ancestry from northern and western Europe; YRI, 90 Yoruba in Ibadan, Nigeria.
Genotype data and mRNA expression levels for MDM4 by genotypes were obtained from the HapMap phase II release 23 data from EBV-transformed lymphoblastoid cell lines from 270 individuals.
Two-sided Student's t test within the stratum.
P values for the trend test of MDM4 mRNA expression among 3 genotypes for each SNP from a general linear model.
Figure 3. mRNA expression level of the MDM4 gene in Epstein-Barr virus (EBV)-transformed lymphoblastoid cell lines.
(A) mRNA expression in 90 cell lines from unrelated CEU (Utah residents with ancestry from northern and western Europe) individuals. (B) mRNA expression in 90 cell lines of unrelated YRI (Yoruba in Ibadan, Nigeria) individuals. (C) mRNA expression in 90 cell lines of unrelated Asian individuals. (D) mRNA expression in 270 cell lines of all individuals.
Sensitivity analysis and publication bias
We conducted sensitivity analysis to assess the influence of each individual study on the pooled ORs and 95% CIs by omitting one study each time. No individual study could alter the pooled ORs significantly, which demonstrated that the studies were relatively statistically robust. Additionally, we found that no single study could alter the publication bias in an obvious manner (data not shown).
DISCUSSION
In the present meta-analysis comprising 69477 subjects from 15 studies, we completed the first comprehensive evaluation of the association between MDM4 gene rs4245739 A > C polymorphism and overall cancer risk. The pooled results indicate that the MDM4 rs4245739 A > C polymorphism was significantly associated with decreased overall cancer risk, which was consistent with the results of our genotype-based mRNA expression analysis.
SNPs are the most common type of genetic variations. The majority of SNPs are silent or have limited influence on the function and expression of genes. Only a small fraction of SNPs have been reported to be potentially functional and associated with cancer susceptibility [29–32], in accordance with the theory of the driver and passenger somatic mutations in human cancer genome [33]. The influence of genetic variations, particularly SNPs, on an individual's cancer susceptibility under similar environmental exposures, has been widely investigated and has become a hot research topic worldwide [34]. The association between SNPs and cancer risk may be strongly cancer-specific [35]. Numerous previous studies have investigated the association between MDM4 gene polymorphisms and cancer susceptibility [21–28].
The MDM4 gene (also known as HDMX or MDMX) is located at chromosome 1q32, a region that is frequently found to be amplified in cancer [36]. This gene contains 11 exons and encodes a protein of 490 amino acids [37], in which at least 2709 SNPs have been identified (http://www.ncbi.nlm.nih.gov/projects/SNP). Among these SNPs, a potentially functional polymorphism (rs4245739 A > C) has received a great deal of recent attention. The rs4245739 A > C polymorphism was first identified in 40 German patients with familial breast cancer through sequencing the whole coding and flanking untranslated regions of MDM4 gene [38]. This polymorphism, located in the 3′ UTR of MDM4 gene, generates a miR-191 target binding site. As a result, miR-191 selectively binds to mRNA harboring the MDM4 rs4245739C allele to decrease the expression of MDM4 gene at both mRNA and protein levels, but not mRNA with MDM4 rs4245739A allele (wild-type). The decreased MDM4 expression caused by miR-191 binding might increase the activity of p53 and consequentially modify an MDM4 rs4245739A allele carriers' susceptibility to ovarian cancer and retinoblastoma [39, 40]. Additionally, the MDM4 rs4245739 AC genotype may be associated with increased overall survival in non-small cell lung cancer, when compared to the AA genotype [41]. Despite the biological plausibility, studies investigating the association between this polymorphism and cancer risk have yielded inconclusive results [21–28]. For instance, some studies found that the rs4245739 A > C polymorphism was significantly associated with decreased cancer risk [22–24, 27]; in contrast, Garcia-Closas et al. [21]. reported that this polymorphism was associated with increased breast cancer risk. Moreover, others found that this polymorphism may have weak or no effect on cancer risk [25, 26, 28]. It is widely recognized that different cancer types have unique characteristics and involve differing signal pathways. Even among the same cancer type, cancers from different patients display significant heterogeneity. The possible reasons for discrepancies regarding cancer susceptibility may be ascribed to tumor specificity, differences in ethnicity, and variations in sample sizes included in each investigation. When we combined all available investigations, we found that the rs4245739C allele carriers had decreased cancer risk, especially among Asians. Moreover, we also found that the rs4245739C allele was associated with decreased mRNA expression of MDM4 by genotype-based mRNA expression analysis, which could provide further biological evidence of the possible mechanism of this polymorphism.
Although this is the first meta-analysis investigating the association between MDM4 gene rs4245739 A > C polymorphism and overall cancer risk, some limitations should be discussed. First, although all of the eligible studies were pooled together, the total number of studies and the sample sizes for most types of cancer were still relatively small. As a result, statistical power might be limited while evaluating the association of interest, especially in the subgroup analysis. For instance, there was only one study available for several types of cancer including non-Hodgkin lymphoma, gastric cancer, colon cancer, and prostate cancer. No pooled study could be performed for these cancers. Second, there was heterogeneity among the included studies, which might stem from the inconsistent results derived from different cancers and ethnicities. Third, nearly all of the studies included in this meta-analysis were conducted among Asians and Caucasians. In light of genetic and geographical differences, more investigations from different areas and ethnicities are required to verify our findings. Fourth, the lack of original data limited the further evaluation of potential gene-gene and gene-environment interactions that may modulate cancer risk. Fifth, we may have missed some publications, especially studies without genotype data and those with negative results that were not published. For example, the genotype data for the control subjects were not available in the investigation carried out by Wynendaele et al. [39]. As a result, this study was not included in the current meta-analysis. Finally, publication bias may exist since only published studies were included in our meta-analysis. So, the conclusions drawn from the current study should be interpreted with caution.
In conclusion, our meta-analysis revealed that MDM4 gene rs4245739 A > C polymorphism was associated with a reduction in overall cancer susceptibility. Due to the limitations of the current meta-analysis, future studies with larger sample size and different ethnicities and cancer types are needed to confirm these findings.
MATERIALS AND METHODS
Publication search
We conducted a comprehensive literature search for all relevant publications concerning the association between MDM4 rs4245739 A > C polymorphism and cancer risk from MEDLINE, EMBASE, and CBM database (prior to 5 September, 2016). The following search terms were used: “MDM4 or HDMX or MDMX or MRP1 or rs4245739”, “cancer or carcinoma or tumor or neoplasm”, and “polymorphism or variant or variation”. We also searched for additional relevant studies from the references of retrieved publications.
Inclusion and exclusion criteria
Studies meeting the following inclusion criteria were included: (1) evaluated the association between MDM4 rs4245739 A > C polymorphism and cancer risk; (2) case-control study or cohort study; (3) genotype distributions were available for both cases and controls; (4) published in English or Chinese.
Exclusion criteria included: (1) not case-control study design; (2) did not evaluate the association between MDM4 rs4245739 A > C polymorphism and cancer risk; (3) studies with overlapping participants; (4) conference abstracts, review articles, comments, meta-analyses, or editorial articles. In the case of duplicate or overlapping studies, only the most complete one was included.
Data extraction
Two authors (Chaoyi Xu and Jinhong Zhu) independently extracted the following information from each investigation: the first author's surname, publication year, country, ethnicity, control source, genotyping method, as well as number of case and control with AA, AC and CC genotypes. All disagreements were resolved through discussion between these two investigators until a consensus was reached.
Genotype-based mRNA expression analysis
We performed genotype-based mRNA expression analysis as we described previously [3, 42–45]. Genotypes data of MDM4 rs4245739 A > C polymorphism for 270 individuals with three ethnicities were obtained from HapMap (http://www.hapmap.org). The MDM4 gene mRNA expression data for the same 270 individuals were downloaded from SNPexp (http://app3.titan.uio.no/biotools/tool.php?app=snpexp).
Statistical analysis
The strength of the association between MDM4 rs4245739 A > C polymorphism and overall cancer risk were assessed using crude OR and 95% CI under the homozygous (CC vs. AA), heterozygous (AC vs. AA), recessive (CC vs. AC + AA), dominant (AC + CC vs. AA), and allele contrast models (C vs. A). Goodness-of-fit χ2 test was adopted to test deviation from HWE for the genotypes of control subjects. Heterogeneity was assessed using χ2-based Q test, and was considered as significant when P < 0.10. We also qualified the heterogeneity using I2 statistics, a value with a range from 0% to 100%. A higher I2 value indicates a greater degree of heterogeneity [46]. When significant heterogeneity was found, random-effects model [47] would be adopted; otherwise, fixed-effects model (the Mantel-Haenszel method) [48] would be used. The quality of each investigation was evaluated by quality assessment criteria (Supplementary Table S1) as we described previously [43]. Subgroup analysis was conducted by cancer type (investigations with only one study would be merged into the “others” group), ethnicity, source of control and quality score of investigations. Sensitivity analysis was conducted to evaluate the stability of the results. The pooled ORs and 95% CIs were estimated by excluding one study at a time to evaluate the influence of single investigation. The potential publication bias was estimated using Begg's funnel plot [49] and Egger's linear regression test [50]. In terms of genotype-based mRNA expression, two-sided Student's t test was used for the comparison of two groups, and one-way ANOVA was adopted for comparison among three different genotypes. The statistical analysis was performed with STATA software (version 11.0; Stata Corporation, College Station, TX). All the statistics were two-sided, with P < 0.05 indicating statistical significance.
SUPPLEMENTARY MATERIALS TABLES
Footnotes
CONFLICTS OF INTEREST
The authors declare no competing financial interests.
FUNDING
This study was supported by grants from the National Natural Science Foundation of China (Grant No. 81502046), Special Financial Grant from the China Postdoctoral Science Foundation (Grant No. 2014T70836), Scientific Research Foundation of Wenzhou (Y20100175), Zhejiang Provincial Medical and Health Science and Technology plan (2013RCA035), Project of Science and Technology Department of Zhejiang Province (2014C37003), and Zhejiang Provincial Natural Science Foundation of China (LY16H160054).
REFERENCES
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Jemal A, Grey N, Ferlay J, Forman D. Global cancer transitions according to the Human Development Index (2008–2030): a population-based study. Lancet Oncol. 2012;13:790–801. doi: 10.1016/S1470-2045(12)70211-5. [DOI] [PubMed] [Google Scholar]
- 3.He J, Shi TY, Zhu ML, Wang MY, Li QX, Wei QY. Associations of Lys939Gln and Ala499Val polymorphisms of the XPC gene with cancer susceptibility: a meta-analysis. Int J Cancer. 2013;133:1765–1775. doi: 10.1002/ijc.28089. [DOI] [PubMed] [Google Scholar]
- 4.Hua RX, Li HP, Liang YB, Zhu JH, Zhang B, Ye S, Dai QS, Xiong SQ, Gu Y, Sun XZ. Association between the PARP1 Val762Ala polymorphism and cancer risk: evidence from 43 studies. PLoS One. 2014;9:e87057. doi: 10.1371/journal.pone.0087057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xie S, Shan XF, Shang K, Xu H, He J, Cai ZG. Relevance of LIG4 gene polymorphisms with cancer susceptibility: evidence from a meta-analysis. Sci Rep. 2014;4:6630. doi: 10.1038/srep06630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xue WQ, He YQ, Zhu JH, Ma JQ, He J, Jia WH. Association of BRCA2 N372H polymorphism with cancer susceptibility: a comprehensive review and meta-analysis. Sci Rep. 2014;4:6791. doi: 10.1038/srep06791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Wu XS, He J, Ma T, Lei W, Shen ZY. A novel TP53 variant (rs78378222 A > C) in the polyadenylation signal is associated with increased cancer susceptibility: Evidence from a meta-analysis. Oncotarget. 2016;7:32854–65. doi: 10.18632/oncotarget.9056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu B, Tian J, Zhong R, Tian Y, Chen W, Qian J, Zou L, Xiao M, Shen N, Yang H, Lou J, Qiu Q, Ke J, et al. Genetic variants in the SWI/SNF complex and smoking collaborate to modify the risk of pancreatic cancer in a Chinese population. Mol Carcinog. 2015;54:761–768. doi: 10.1002/mc.22140. [DOI] [PubMed] [Google Scholar]
- 9.Ke J, Lou J, Chen X, Li J, Liu C, Gong Y, Yang Y, Zhu Y, Zhang Y, Tian J, Chang J, Zhong R, Gong J, et al. Identification of a functional variant for colorectal cancer risk mapping to chromosome 5q31.1. Oncotarget. 2016;7:35199–207. doi: 10.18632/oncotarget.9298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 11.Shvarts A, Steegenga WT, Riteco N, van Laar T, Dekker P, Bazuine M, van Ham RC, van der Houven van Oordt W, Hateboer G, van der Eb AJ, Jochemsen AG. MDMX: a novel p53-binding protein with some functional properties of MDM2. EMBO J. 1996;15:5349–5357. [PMC free article] [PubMed] [Google Scholar]
- 12.Bartel F, Schulz J, Blumke K, Kappler M, Bache M, Schmidt H, Taubert H. HDMX amplification and high levels of HDMX-S splice variant are correlated with a poor prognosis in soft tissue sarcomas. [Article in German] Verh Dtsch Ges Pathol. 2004;88:199–206. [PubMed] [Google Scholar]
- 13.Bartel F, Schulz J, Bohnke A, Blumke K, Kappler M, Bache M, Schmidt H, Wurl P, Taubert H, Hauptmann S. Significance of HDMX-S (or MDM4) mRNA splice variant overexpression and HDMX gene amplification on primary soft tissue sarcoma prognosis. Int J Cancer. 2005;117:469–475. doi: 10.1002/ijc.21206. [DOI] [PubMed] [Google Scholar]
- 14.Liu L, Fan L, Fang C, Zou ZJ, Yang S, Zhang LN, Li JY, Xu W. S-MDM4 mRNA overexpression indicates a poor prognosis and marks a potential therapeutic target in chronic lymphocytic leukemia. Cancer Sci. 2012;103:2056–2063. doi: 10.1111/cas.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atwal GS, Kirchhoff T, Bond EE, Montagna M, Menin C, Bertorelle R, Scaini MC, Bartel F, Bohnke A, Pempe C, Gradhand E, Hauptmann S, Offit K, et al. Altered tumor formation and evolutionary selection of genetic variants in the human MDM4 oncogene. Proc Natl Acad Sci USA. 2009;106:10236–10241. doi: 10.1073/pnas.0901298106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kulkarni DA, Vazquez A, Haffty BG, Bandera EV, Hu W, Sun YY, Toppmeyer DL, Levine AJ, Hirshfield KM. A polymorphic variant in human MDM4 associates with accelerated age of onset of estrogen receptor negative breast cancer. Carcinogenesis. 2009;30:1910–1915. doi: 10.1093/carcin/bgp224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun T, Lee GS, Oh WK, Pomerantz M, Yang M, Xie W, Freedman ML, Kantoff PW. Single-nucleotide polymorphisms in p53 pathway and aggressiveness of prostate cancer in a Caucasian population. Clin Cancer Res. 2010;16:5244–5251. doi: 10.1158/1078-0432.CCR-10-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu H, Wang LE, Liu Z, Wei S, Li G, Sturgis EM, Wei Q. Polymorphisms of MDM4 and risk of squamous cell carcinoma of the head and neck. Pharmacogenet Genomics. 2011;21:388–396. doi: 10.1097/FPC.0b013e32834632e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z, Sturgis EM, Zhang Y, Huang Z, Zhou Q, Wei Q, Li G. Combined p53-related genetic variants together with HPV infection increase oral cancer risk. Int J Cancer. 2012;131:E251–258. doi: 10.1002/ijc.27335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu H, Sturgis EM, Liu Z, Wang LE, Wei Q, Li G. Modifying effect of MDM4 variants on risk of HPV16-associated squamous cell carcinoma of oropharynx. Cancer. 2012;118:1684–1692. doi: 10.1002/cncr.26423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia-Closas M, Couch FJ, Lindstrom S, Michailidou K, Schmidt MK, Brook MN, Orr N, Rhie SK, Riboli E, Feigelson HS, Le Marchand L, Buring JE, Eccles D, et al. Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nat Genet. 2013;45:392–398. doi: 10.1038/ng.2561. 398e391–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J, Tang X, Li M, Lu C, Shi J, Zhou L, Yuan Q, Yang M. Functional MDM4 rs4245739 genetic variant, alone and in combination with P53 Arg72Pro polymorphism, contributes to breast cancer susceptibility. Breast Cancer Res Treat. 2013;140:151–157. doi: 10.1007/s10549-013-2615-x. [DOI] [PubMed] [Google Scholar]
- 23.Zhou L, Zhang X, Li Z, Zhou C, Li M, Tang X, Lu C, Li H, Yuan Q, Yang M. Association of a genetic variation in a miR-191 binding site in MDM4 with risk of esophageal squamous cell carcinoma. PLoS One. 2013;8:e64331. doi: 10.1371/journal.pone.0064331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan C, Wei J, Yuan C, Wang X, Jiang C, Zhou C, Yang M. The functional TP53 rs1042522 and MDM4 rs4245739 genetic variants contribute to Non-Hodgkin lymphoma risk. PLoS One. 2014;9:e107047. doi: 10.1371/journal.pone.0107047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng JB, Nie YQ, Lin Y, Li YF, Du YL. Association between single nucleotide polymorphism at miR-191 target site and susceptibility to gastric cancer. [Article in Chinese] The Journal of Practical Medicine. 2014;30:1217–1220. [Google Scholar]
- 26.Gansmo LB, Romundstad P, Birkeland E, Hveem K, Vatten L, Knappskog S, Lonning PE. MDM4 SNP34091 (rs4245739) and its effect on breast-, colon-, lung-, and prostate cancer risk. Cancer Med. 2015;4:1901–1907. doi: 10.1002/cam4.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao F, Xiong X, Pan W, Yang X, Zhou C, Yuan Q, Zhou L, Yang M. A Regulatory MDM4 Genetic Variant Locating in the Binding Sequence of Multiple MicroRNAs Contributes to Susceptibility of Small Cell Lung Cancer. PLoS One. 2015;10:e0135647. doi: 10.1371/journal.pone.0135647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gansmo LB, Bjornslett M, Halle MK, Salvesen HB, Dorum A, Birkeland E, Hveem K, Romundstad P, Vatten L, Lonning PE, Knappskog S. The MDM4 SNP34091 (rs4245739) C-allele is associated with increased risk of ovarian-but not endometrial cancer. Tumour Biol. 2016 doi: 10.1007/s13277-016-4940-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi TY, Cheng X, Yu KD, Sun MH, Shao ZM, Wang MY, Zhu ML, He J, Li QX, Chen XJ, Zhou XY, Wu X, Wei Q. Functional variants in TNFAIP8 associated with cervical cancer susceptibility and clinical outcomes. Carcinogenesis. 2013;34:770–778. doi: 10.1093/carcin/bgt001. [DOI] [PubMed] [Google Scholar]
- 30.Zhu ML, He J, Wang M, Sun MH, Jin L, Wang X, Yang YJ, Wang JC, Zheng L, Xiang JQ, Wei QY. Potentially functional polymorphisms in the ERCC2 gene and risk of esophageal squamous cell carcinoma in Chinese populations. Sci Rep. 2014;4:6281. doi: 10.1038/srep06281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang MY, He J, Zhu ML, Teng XY, Li QX, Sun MH, Wang XF, Yang YJ, Wang JC, Jin L, Wang YN, Wei QY. A Functional Polymorphism (rs2494752) in the AKT1 Promoter Region and Gastric Adenocarcinoma Risk in an Eastern Chinese Population. Sci Rep. 2016;6:20008. doi: 10.1038/srep20008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gong J, Tian J, Lou J, Ke J, Li L, Li J, Yang Y, Gong Y, Zhu Y, Zhang Y, Zhong R, Chang J, Miao X. A functional polymorphism in lnc-LAMC2–1:1 confers risk of colorectal cancer by affecting miRNA binding. Carcinogenesis. 2016;37:443–451. doi: 10.1093/carcin/bgw024. [DOI] [PubMed] [Google Scholar]
- 33.Tan H, Bao J, Zhou X. A novel missense-mutation-related feature extraction scheme for ‘driver’ mutation identification. Bioinformatics. 2012;28:2948–2955. doi: 10.1093/bioinformatics/bts558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pharoah PD, Dunning AM, Ponder BA, Easton DF. Association studies for finding cancer-susceptibility genetic variants. Nat Rev Cancer. 2004;4:850–860. doi: 10.1038/nrc1476. [DOI] [PubMed] [Google Scholar]
- 35.Tan H, Bao J, Zhou X. Genome-wide mutational spectra analysis reveals significant cancer-specific heterogeneity. Sci Rep. 2015;5:12566. doi: 10.1038/srep12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ried T, Just KE, Holtgreve-Grez H, du Manoir S, Speicher MR, Schrock E, Latham C, Blegen H, Zetterberg A, Cremer T, Auer G. Comparative genomic hybridization of formalin-fixed, paraffin-embedded breast tumors reveals different patterns of chromosomal gains and losses in fibroadenomas and diploid and aneuploid carcinomas. Cancer Res. 1995;55:5415–5423. [PubMed] [Google Scholar]
- 37.Shvarts A, Bazuine M, Dekker P, Ramos YF, Steegenga WT, Merckx G, van Ham RC, van der Houven van Oordt W, van der Eb AJ, Jochemsen AG. Isolation and identification of the human homolog of a new p53-binding protein, Mdmx. Genomics. 1997;43:34–42. doi: 10.1006/geno.1997.4775. [DOI] [PubMed] [Google Scholar]
- 38.Reincke S, Govbakh L, Wilhelm B, Jin H, Bogdanova N, Bremer M, Karstens JH, Dork T. Mutation analysis of the MDM4 gene in German breast cancer patients. BMC Cancer. 2008;8:52. doi: 10.1186/1471-2407-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wynendaele J, Bohnke A, Leucci E, Nielsen SJ, Lambertz I, Hammer S, Sbrzesny N, Kubitza D, Wolf A, Gradhand E, Balschun K, Braicu I, Sehouli J, et al. An illegitimate microRNA target site within the 3′ UTR of MDM4 affects ovarian cancer progression and chemosensitivity. Cancer Res. 2010;70:9641–9649. doi: 10.1158/0008-5472.CAN-10-0527. [DOI] [PubMed] [Google Scholar]
- 40.McEvoy J, Ulyanov A, Brennan R, Wu G, Pounds S, Zhang J, Dyer MA. Analysis of MDM2 and MDM4 single nucleotide polymorphisms, mRNA splicing and protein expression in retinoblastoma. PLoS One. 2012;7:e42739. doi: 10.1371/journal.pone.0042739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Y, Gao W, Ding X, Xu W, Liu D, Su B, Sun Y. Variations within 3′-UTR of MDM4 gene contribute to clinical outcomes of advanced non-small cell lung cancer patients following platinum-based chemotherapy. Oncotarget. 2016 doi: 10.18632/oncotarget.10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He J, Xi B, Ruiter R, Shi TY, Zhu ML, Wang MY, Li QX, Zhou XY, Qiu LX, Wei QY. Association of LEP G2548A and LEPR Q223R polymorphisms with cancer susceptibility: evidence from a meta-analysis. PLoS One. 2013;8:e75135. doi: 10.1371/journal.pone.0075135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He J, Liao XY, Zhu JH, Xue WQ, Shen GP, Huang SY, Chen W, Jia WH. Association of MTHFR C677T and A1298C polymorphisms with non-Hodgkin lymphoma susceptibility: evidence from a meta-analysis. Sci Rep. 2014;4:6159. doi: 10.1038/srep06159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi TY, He J, Wang MY, Zhu ML, Yu KD, Shao ZM, Sun MH, Wu X, Cheng X, Wei Q. CASP7 variants modify susceptibility to cervical cancer in Chinese women. Sci Rep. 2015;5:9225. doi: 10.1038/srep09225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He J, Wang F, Zhu J, Zhang R, Yang T, Zou Y, Xia H. Association of potentially functional variants in the XPG gene with neuroblastoma risk in a Chinese population. J Cell Mol Med. 2016;20:1481–1490. doi: 10.1111/jcmm.12836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 47.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 48.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 49.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 50.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.