Abstract
As the first de novo actin nucleator discovered, the Arp2/3 complex has been a central player in models of protrusive force production via the dynamic actin network. Here, we review recent studies on the functional role of the Arp2/3 complex in the migration of diverse cell types in different migratory environments. These findings have revealed an unexpected level of plasticity, both in how cells rely on the Arp2/3 complex for migration and other physiological functions and in the intricate modulation of the Arp2/3 complex by other actin regulators and upstream signaling cascades.
Introduction
Actin polymerization drives the morphological changes that allow cells to undergo dynamic processes, such as division, phagocytosis, and migration. The formation of new actin filaments from actin monomers is regulated by three classes of nucleating proteins, including the formins, tandem-monomer-binding nucleators, and the Arp2/3 complex [1–3]. Formins, such as mDia1 and 2, generate unbranched filaments by stabilizing actin trimers and promoting elongation through associations with both the growing barbed end and actin-bound profilin. The tandem-monomer-binding nucleators, including Spire, cordon bleu, and leiomodin, form unbranched filaments by bringing together actin monomers, via a series of monomer-binding WH2 domains, in a configuration that mimics the stable actin trimer. The Arp2/3 complex is unique in that it nucleates branched actin filaments.
The first-identified actin nucleator, the Arp2/3 complex has been studied extensively using in vitro reconstitution assays, biochemical and structural analyses, and in vivo genetics-based functional experiments [4,5]. The Arp2/3 complex consists of seven subunits, two of which (Arp2 and Arp3) are actin-related proteins that serve as a nucleus for the new actin filament. Other Arp2/3 complex subunits bind to existing actin filaments to generate a branch at a ~78° angle from the mother filament [6–8]. Nucleation by Arp2/3 is regulated by many proteins, most prominently the Wiskott-Aldrich syndrome family of nucleation promoting factors (NPFs), such as WASP, N-WASP, Scar/WAVE, and WASH. The WASP family NPFs share a consensus WCA domain in their C-termini that binds monomeric actin through the WH2 domain (W) and the Arp2/3 complex through the cofilin homology region and acidic tail (CA) [9,10]. Thus, in a spatiotemporally-regulated manner, these NPFs facilitate the transition of the Arp2/3 complex from its splayed, inactive conformation to its closed, active conformation and also supply an actin monomer to form the growing barbed end. The WISH/DIP/SPIN90 proteins are NPFs that activate the Arp2/3 complex without binding to F- or G-actin, promoting the formation of unbranched filaments that may serve as the seed for subsequent branch nucleation [11].
Cells must be able to rapidly modify their actin network to adapt to their surroundings. It is well-established that cells can switch between multiple modes of migration depending on their gene expression, the dimensionality of their environment (1D vs 2D vs 3D), and the extracellular matrix (ECM) stiffness and composition [12–24]. This is accomplished in part by complex Arp2/3 regulation combined with the multitude of other mechanisms that promote actin nucleation during cell migration, allowing for tight spatiotemporal control of the formation of different actin architectures. Furthermore, applied mechanical resistance alters the actin network density, geometry, power and stiffness, suggesting that variations in force may influence the organization of actin filaments in vivo [25]. Geometric constraints also dictate the structure of the actin network [26–28]. Nonetheless, variability in experimental conditions and the intricacy of the underlying regulatory mechanisms has made it difficult to dissect the role of Arp2/3 in cell migration. This review will focus on recent advances in our understanding of Arp2/3-mediated actin polymerization during cell migration and the mechanisms by which cells fine-tune their actin networks to adapt to internal perturbations or extracellular environments.
Studies of Arp2/3 Subunit-Disruption in Migration
Because of its central role in the actin polymerization required for cell migration, endocytosis, and other vital processes, functional studies of the Arp2/3 complex were initially hindered by the lethality of Arp2 or Arp3 null cells and animals [29,30]. Early studies with Arp2/3 complex subunit knock-down or NPF inhibition supported the notion that the Arp2/3 complex mediates actin polymerization during lamellipodia formation and migration in cells such as fibroblasts, lymphocytes, mammary carcinoma cells, and the amoeba Dictyostelium discoideum [31–42]. Improvements in genetic techniques over the past decade have allowed for more direct tests of Arp2/3 function in migration in a variety of cell types (Table 1).
Table 1.
Functional consequences of Arp2/3 depletion on migration in various cell types and environments
| Cell Type | Method of Arp2/3 Depletion | Migratory Environment Tested | Arp2/3 Function or Phenotype | Reference |
|---|---|---|---|---|
| Fibroblasts | ES cells from Arpc3-null mice were differentiated into fibroblasts | 2D closed microfluidic chambers | Null cells lost lamellipodia but migrated by combining formin-based filopodia formation with myosin II-based contractility to advance the leading edge; null cells were unable to migrate directionally during wound healing or chemotaxis toward EGF | Suraneni et al, 2012; Suraneni et al, 2015 |
| Mouse embryonic fibroblasts with combined knock-down of Arpc2 and Arp2 subunits in the Ink4a/Arf −/− genetic background; inhibition with CK666 | 2D continual-flow microfluidic chambers | Knock-down cells lost lamellipodia, showed focal adhesion and spreading defects, and migrated with reduced speed via filopodial structures; knock-down cells were deficient in haptotaxis but showed normal chemotaxis if media was replaced; knock-down cells, via NF-κB activation, secreted growth factors that disrupted chemotaxis in a closed system | Wu et al, 2012; Wu et al 2013 | |
| Mouse fibroblasts with conditional Arpc2 knock-out in the Ink4a/Arf −/− genetic background | 2D continual-flow microfluidic chambers | Null cells were able to chemotax toward PDGF | Asokan et al, 2014 | |
| Null cells lost lamellipodia, showed an increase in filopodial protrusions, and migrated with reduced speed | Rotty et al, 2015 | |||
| Neurons and glia | Neural stem cells from Arpc2 knock-out mice were differentiated into oligodendrocyte precursor cells (OPCs) | Oligospheres plated on glass chambers with applied electric fields | Null cells had shorter, filopodial extensions and moved with reduced speed compared to wild-type OPCs; Electric fields enhanced the migration and biased the directionality of wild-type OPCs but did not elicit these responses in null cells | Li et al, 2015 |
| Radial glial cells from Arpc2 knock-out mice | Cortical slices were embedded in matrigel for ex vivo imaging | Null cells had disorganized processes that lacked dynamic ruffles and were shorter than control cell protrusions due to frequent retraction; nulls showed altered cell fates due to premature differentiation, increased apoptosis, and decreased proliferation; these defects contributed to abnormal cortical architecture | Wang et al, 2015 | |
| Neuronal precursor cells from Arpc2 knock-out mice | Wild-type or null neurospheres cultured ex vivo with wild-type brain slices; glass or elastic laminin-coated surfaces in vitro | Null cells could not migrate in the ex vivo system or on soft or low-laminin surfaces in vitro ; null cells lacked lamellipodia but could migrate, with reduced speed, on stiff or high-laminin surfaces | Wang et al, 2015 | |
| Hematopoietic cells | Drosophila pupal macrophages with Arp2 or WAVE RNAi knock-down | 2D glass surfaces and in vivo (WAVE knock-down only) | Knock-down cells lost lamellipodia and generate filopodial protrusions; WAVE knock-down cells extend processes in the direction of a chemotactic gradient but fail to migrate | Sander et al, 2013 |
| Drosophila embryonic macrophages with Arp3 or WASH RNAi knock-down | In vivo | Partial failure of knock-down cells to migrate anteriorly during development due to reduced protrusion size; knock-down cells could migrate posteriorly, but were disorganized | Verboon et al, 2015 | |
| OT-I CD8+ T cells with chemical inhibition of Arp2/3 via CK666 | Microchannels coated with pMHC antigenic complexes | Inhibition of Arp2/3 did not affect fast T cell migration (unbound by antigen) or exploratory migration during kinapse formation (bound by low-affinity antigen), but impaired T cell deceleration for synapse formation (bound by high-affinity antigen) | Moreau et al, 2015 | |
| Dendritic cells (immature and mature, via LPS induction) with conditional Arpc2 knock-out, Arpc4 siRNA knock-down, or chemical inhibition via CK666 | 1D confined microchannels; under agarose; collagen gels; ex vivo in ear epidermal sheets | Arp2/3 depletion enhanced migration speed in immature cells and reduced F-actin at the cell front, leading to defective antigen macropinocytosis; Arp2/3 was not required for F-actin enrichment at the cell rear or for chemotaxis in collagen gels or to lymphatic vessels ex vivo | Vargas et al, 2016 | |
| Immature dendritic cells with Arpc4 siRNA knock-down or chemical inhibition with CK666 | Microchannels with constrictions ≥ 1.5 μm | Arp2/3 depletion did not affect normal migration but prevented cells from passing through narrow confinements (3 μm or less); Arp2/3 depletion prevented perinuclear actin accumulation and nuclear deformation required for passage through narrow pores | Thiam et al, 2016 | |
| Epithelial cells | MCF10A mammary epithelial cells with siRNA knock-down of Arp3 or chemical inhibition via CK869 and CK666 | 2D coverslips | Arp2/3 depletion disrupted lamellipodia formation, reduced migration speed and directional persistence, and impaired focal adhesion assembly and attachment to the ECM; Arp2/3 depletion also weakened the coupling between the cytoskeletal cortex and plasma membrane | Beckham et al, 2014 |
| Cancer cell lines | Human glioma cell lines U251, LN229 and SNB19 with chemical inhibition via CK666 | 2D wound healing; transwell invasion through Matrigel-coated Boyden chambers | Arp2/3 inhibition reduced lamellipodia size, disrupted polarity, and reduced migration and invasion in all cell lines; Arp2/3 expression is correlated with malignancy of patient tumors | Liu et al, 2014 |
| Rat C6 glioma cells or human patient-derived glioma propagating cells with chemical inhibition via CK666 | 2D surfaces or thin (3 – 7 μm), linear laminin tracks | Arp2/3 inhibition blocked 2D migration but enhanced linear/saltatory migration by altering cell morphology, increasing cell velocity, and improving cell alignment in antiparallel arrays | Monzo et al, 2016 | |
| A2780 human ovarian carcinomas with siRNA knock-down of Arpc2 or Arpc3 or chemical inhibition via CK666 | 2D surfaces or elastic 3D cell-derived matrix | Arp2/3 depletion did not affect α5β1 integrin recycling-mediated protrusion morphology, migration, or invasion | Paul et al, 2015 |
For simplicity, this table does not include the many studies involving depletion of Arp2/3 regulators, including NPFs
Fibroblasts
Our group and others have shown that loss of Arp2/3 activity in mouse embryonic fibroblasts through the use of inhibitors, complex subunit knock-outs, or RNAi results in the loss of lamellipodia [43–47]. Surprisingly, these Arp2/3-deficient cells are still migratory, though with reduced speed, through the generation of formin-containing filopodial structures of bundled actin [45,46]. These actin structures coordinate with myosin II-mediated contractility in the cortex to drive leading edge extension [44]. Although these studies found similar alterations in F-actin structures at the leading edge, they disagreed on the effect of Arp2/3 complex loss on migration toward chemical cues: our studies showed reduced directional persistence and a cell-autonomous defect in chemotaxis toward growth factors, while Wu et al found that Arp2/3 knock-down cells only exhibit a chemotactic deficiency if there is interference by secreted inflammatory cytokines [44,47]. The same group did find that haptotaxis toward surface-bound ECM molecules is impaired in the knock-down cells and later repeated their findings with an Arp2/3 subunit knock-out [43,46]. These discrepancies could be caused by differences in genetic background, the manner by which the cells were produced, the targeted Arp2/3 complex subunit, or experimental conditions.
Neural and glial progenitors
Recent studies have examined the role of the Arp2/3 complex in the migration of progenitor cells found in the brain. For example, Arp2/3 is required for the directed migration of neural stem cell-derived oligodendrocyte precursor cells (OPCs) in an electric field, which has implications for neural stem cell transplantation for remyelination [48]. Similar to mouse embryonic fibroblasts, Arpc2-null OPCs were slower, with shorter, filopodial processes than Arpc2-expressing OPCs. We recently found that Arp2/3 is required for neural cell migration in vivo: Arpc2-null mice have impaired cortical architecture due to defects in both radial glial cell processes and the migration of neural progenitors along these tracks [49]. In vitro experiments showed that the migration defect of neural progenitor cells is particularly pronounced in soft or low-laminin environments that mimic brain tissue.
Hematopoietic cells
As opposed to fibroblasts and neural progenitor cells, evidence shows that Arp2/3-mediated actin polymerization inhibits the migration of both dendritic and T cells, which will be discussed below [50,51]. However, a recent study of dendritic cells suggests that successful passage through narrow pores, which is determined by the physical constraints of the nucleus, is associated with an increase in actin polymerization around the nucleus as it reaches the point of constriction [52]. This perinuclear actin, as well as migration under confinement, is reduced by Arp2/3 inhibition or knock-down, but is not required for the confined movement of cells with low lamin levels and softer nuclei, suggesting that actin filaments facilitate migration through narrow spaces by promoting nuclear deformation [52]. In hemocytes, the Drosophila macrophage-like immune cells, depletion of Rho1, WASH, or Arp2/3 subunits reduces the formation of cellular protrusions and prevents a subset of migration events during development [53]; similar studies suggest that lamellipodia formation and migration of hemocytes in vivo require Scar/WAVE [54,55].
Epithelial cells
In MCF10a mammary epithelial cells, inhibition or knock-down of Arp2/3 results in the disruption of lamellipodia and the formation of unstable or bleb-like protrusions, corresponding to a decrease in directional persistence and migratory speed [56]. Furthermore, Arp2/3 inhibition impairs nascent focal adhesion assembly and decreases the coupling between the actin cortex and cell membrane [56].
Cancer cells
Because of the links to metastatic migration, the role of Arp2/3 has been examined in a number of cancer cell lines. The results are conflicting, which is unsurprising given the tremendous genetic diversity and heterogeneity of cancer cells. Several studies have shown that the Arp2/3 complex or Arp2/3-stimulating factors, such as cortactin, are upregulated in malignant gliomas, and inhibition of Arp2/3 activity reduces lamellipodia formation and invasion [57,58]. Similar trends have been shown for Arp2/3 and its NPFs in models of mammary carcinoma [59–61] . However, other studies of glioma motility concluded that Arp2/3 activity is unnecessary, or even inhibitory, for migration in confined environments [62–65]. The mechanisms of Arp2/3-independent migration are unclear, as studies alternatively conclude that either motility is dependent on formin-mediated actin polymerization or is independent of actin polymerization entirely based on observed migration in the presence of actin depolymerizing agents. Similarly, endocytosis of α5β1 integrin enhances the invasion of ovarian carcinoma cells into the 3D matrix by promoting the formation of actin spikes via RhoA-mediated formin activity, independent of Arp2/3 [66].
Multifaceted Regulation of the Arp2/3 Complex Adapts the Actin Network to Diverse Modes of Migration
Proteins that regulate the Arp2/3 complex
In addition to NPFs and upstream signaling factors, a number of proteins function in the regulation of Arp2/3 activity to control cell migration [10,67–77]. Recent studies have centered on a newly-discovered negative regulator of Arp2/3, Arpin [78]. This protein is recruited to the lamellipod by activated Rac, where it directly inhibits Arp2/3 activity to destabilize protrusions [78,79]. Arpin helps control directional persistence and migration speed by inducing pauses in motility [79]. Subsequent studies showed an inverse correlation between Arpin expression and breast cancer metastasis; low levels of Arpin are also associated with elevated expression of WAVE complex subunits and poor recurrence-free survival [80,81]. Another negative regulator, Gadkin, sequesters the Arp2/3 complex to endosomal vesicles, thereby inhibiting cell spreading and affecting the migration speed of dendritic cells in vitro [82,83]. In sum, the numerous potential interactions between Arp2/3 and components of the actin machinery leads to a highly regulated and tunable actin network that can be specifically tailored to a variety of dynamic cellular behaviors.
Differential activation of NPFs fine-tunes the structure of the actin network
Evidence suggests that the differential utilization of various Arp2/3-activating proteins, including NPFs, can alter the migratory behavior of cells depending on environmental conditions. For example, recent work has demonstrated that loss of the Scar/WAVE complex in both carcinoma and normal epithelial cells decreases migration in 2D wound healing assays but promotes N-WASP- and Arp2/3-mediated invasion into 3D matrices through the activation of focal adhesion kinase (FAK) at the leading edge [84,85]. Thus, Arp2/3 plays different roles in 2D vs 3D migration depending on whether it is activated by Scar/WAVE or N-WASP. FAK, heavily implicated in cancer cell migration, interacts with Arp2/3, recruiting it to sites of nascent focal adhesions [86, 87]. The interaction between FAK and Arp2/3 also couples cell adhesion to leading edge protrusions and is required for migration of fibroblasts in response to ECM gradients in 2D [87,88]. Similarly, depletion of N-WASP, WAVE1, or cortactin in fibrosarcoma cells undergoing migration in different environments showed that the regulatory machinery is alternatively activated for 2D vs 3D Arp2/3-based migration, resulting in different morphologies and migratory behaviors [89]. During migration in 3D environments, these cells form thin, dendritic extensions that branch to generate new protrusions that are molecularly distinct from the original, suggesting that the formation of different types of actin structures is coordinated [89].
Balance between Arp2/3 and other actin polymerizing factors
Studies of cells lacking the Arp2/3 complex have indicated that these cells can still generate protrusive actin networks that sustain migration through other actin nucleators [12,43–48,90]. These studies also showed that different actin polymerizing factors lead to different modes of migration [12,43–48,70,90–92] (Figure 1). A specific example of how the utilization of multiple nucleators contributes to cellular function comes from studies of dendritic cells (DCs) in confining microchannels. DCs have two competing pools of actin: a Cdc42- and Arp2/3-mediated accumulation of F-actin at the cell front that slows motility but is required for micropinocytosis and antigen uptake in immature DCs, and a RhoA- and mDia-mediated F-actin enrichment at the cell rear that is necessary for the fast migration and chemotaxis used by mature DCs to travel to the lymph nodes [51]. Stimulation with LPS, which triggers DC maturation, induces a switch from Arp2/3- to formin-derived actin networks with corresponding changes in migratory behavior [51]. However, previous studies reported that depletion of Arp2/3 activators, including Cdc42 and WASP, impairs migration of DCs to the lymph nodes [93–95]. These contradictions may be due to experimental differences in migratory environments. Similarly to DCs, Arp2/3 activity inhibits T cell migration to promote synapse formation upon high-affinity antigen binding, but is not required for the fast motility of T cells that are unbound by antigen [50]. Homeostasis between Arp2/3- and formin-based polymerization in fission yeast is controlled by competition for actin monomers, ensuring the proper assembly of the contractile ring and endocytic actin patches [96]. These studies and others suggest that the balance between different actin nucleators is critical for specific cell function and can be regulated by extracellular stimuli and intracellular signaling cascades [12,50,51,96].
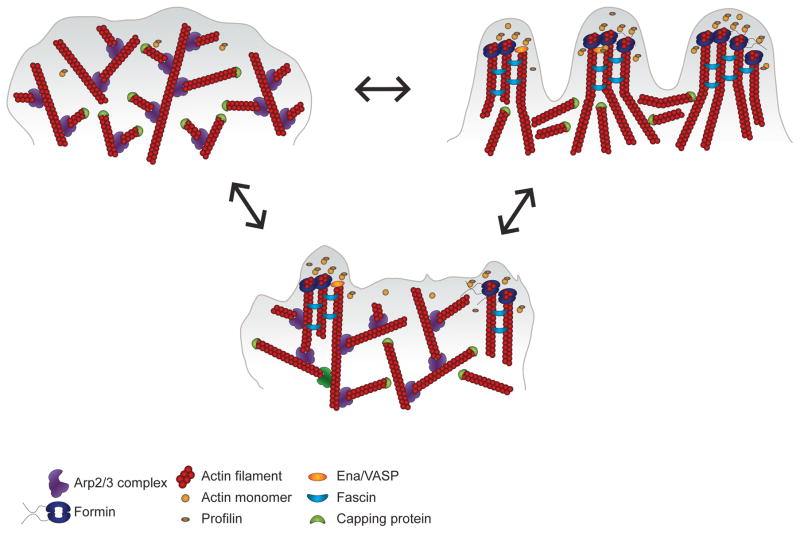
Figure 1. Cells use different actin networks for migration.
The plasticity underlying the regulation of the actin machinery allows cells to adapt to diverse migratory environments. The migration of fibroblasts and other cell types on 2D surfaces is characterized by the formation of broad lamellipodia at the leading edge (upper left). These structures are composed of Arp2/3-mediated branched actin networks. In confined environments or in the absence of the Arp2/3 complex, many cell types generate filopodial structures of bundled linear actin filaments generated by formins (upper right). Properties of the extracellular matrix and inputs from upstream signaling pathways can mediate a switch between these different actin machineries. However, it is likely that cells migrating in diverse 3D environments in vivo use a combination of actin nucleators in order to fine-tune their cytoskeletal networks (lower center); these nucleators have been shown to both cooperate with and antagonize each other, depending on environmental context. Competition for free actin monomers, influenced by profilin, can modulate the balance between Arp2/3- and formin-mediated actin polymerization. Likewise, competition between formins, Ena/VASP, capping protein, profilin, and even NPFs for free barbed ends can influence the length and structure of the actin filaments. Thus, the migration machinery can be thought of as a spectrum, and cells can shift along this continuum from one mode to the next depending on regulatory protein concentrations, genetic factors or environmental cues. It should be noted that this Figure does not include contractility- or pressure-based modes of migration, such as blebbing or lobopodia.
Although switching between distinct types of actin networks is important for regulating migratory behavior under different environmental conditions, evidence suggests that the interplay between various actin polymerizing factors within the same structure also contributes to the fine-tuning of the actin assembly. For example, formins have been observed in lamellipodia, where they modulate the structure of the dendritic actin network by competing with capping protein for free barbed ends and promoting filament elongation [97–99]. Ultrastructural analysis of cells depleted of the formin mDia1, Arp2/3, or both further suggests that these nucleators are spatially intertwined with distinct consequences on cortical actin structure [90]. Other work has shown that fascin-1-containing bundles of actin may serve as a template for lamellipodia formation [88]. This is supported by the finding, generated by a combination of in vitro actin polymerization assays and spatially- and temporally-defined mDia1 inactivation in Hela cells, that mDia1 stimulates Arp2/3 branching activity by providing mother filaments for nucleation [100]. Furthermore, increases in one nucleating factor can compensate for depletion of the other, supporting the conclusion that mDia1 and Arp2/3 activities cooperate to influence the impact of the other on actin meshwork structure [100]. Other studies demonstrating that formins can assemble filopodia using dendritic actin generated by the Arp2/3 complex also highlight the cooperative nature of these nucleating factors [101–104].
Role of profilin as an integrator of multiple actin nucleation factors
Several recent studies have pointed to a role for the actin monomer-binding protein profilin as a negative modulator of Arp2/3-based actin nucleation. In Arpc2-null mouse fibroblasts, only profilin-bound actin monomers can incorporate into barbed ends, and excess profilin can inhibit Arp2/3-mediated actin polymerization both in wild-type cells and in vitro [105]. Profilin is known to enhance formin-mediated barbed end elongation; however, depletion or sequestration of either profilin or Ena/VASP, but not chemical inhibition of formins, which may not be complete, in the Arpc2-null background prevents protrusion formation and spreading [105]. The authors concluded that profilin antagonizes Arp2/3-mediated nucleation and actin branching by facilitating actin polymerization via Ena/VASP, thereby competing away actin monomers from the Arp2/3 complex. In contrast, Ena/VASP has also been proposed to interact with the WAVE regulatory complex to stimulate Arp2/3 activation, enhance migration, and maintain normal lamellipodia formation in Drosophila hemocytes and C. elegans epidermal cells, suggesting that this family of actin assembly proteins may have alternative functions in diverse cell types or under different conditions [106,107]. Work combining analysis of the contractile ring in S. pombe with in vitro actin reconstitution assays also shows that profilin inhibits Arp2/3 function, thereby maintaining the balance between formin- and Arp2/3-mediated actin polymerization that is required for cytokinesis [108]. These studies conclude that profilin tunes the actin network through the competition for actin monomers [105,108]; however, another study suggests that profilin competes with formins, VASP, capping protein, and Arp2/3-mediated end branching by binding to the barbed ends of F-actin, thereby regulating the length of actin filaments [109]. Although the mechanisms may be complex, profilin expression is emerging as an important regulator of actin network dynamics during migration by fine-tuning the activities of various actin polymerization proteins.
Conclusions and outlook
Recent findings on the function of the Arp2/3 complex, an actin nucleator long thought to play a key role in actin-based protrusive force generation, during the migration of diverse cell types in various experimental settings have been both fascinating and confusing. Underneath the conflicting functional consequences of Arp2/3 inhibition, it is becoming apparent that there is an unexpected level of complexity and plasticity in actin network formation, regulation, and force production to drive cell motility in response to specific geometric or mechanical properties of the environment. Particularly important is the intricate interplay, both competitive and collaborative, between different actin nucleators. Through gene expression and distinct subcellular localizations or activating signaling molecules, maintaining a balance between the Arp2/3 complex, its regulators, and other nucleating proteins is crucial for modulating the dynamic actin structures found during cell migration. In cancer, the mechanistic adaptability of the actin network is influenced not only by environments but also by the genetic heterogeneity of tumor cells, highlighting the considerable challenge in understanding and preventing cancer cell migration during dissemination and metastasis. Because of the extraordinary plasticity of cell motility machineries, inconsistencies in experimental observations associated with different cell lines or environmental conditions could shed light on the adaptive principles and regulatory complexity of the actin network in cell migration.
Acknowledgments
The authors would like to thank Dr. Marc Edwards for critical reading and suggestions. We would also like to thank members of the Li lab for helpful discussion. This work is supported by National Institute of Health grants RO1GM057063 and R35GM118172 to RL.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chesarone MA, Goode BL. Actin nucleation and elongation factors: mechanisms and interplay. Curr Opin Cell Biol. 2009;21:28–37. doi: 10.1016/j.ceb.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Skau CT, Waterman CM. Specification of Architecture and Function of Actin Structures by Actin Nucleation Factors. Annu Rev Biophys. 2015;44:285–310. doi: 10.1146/annurev-biophys-060414-034308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Firat-Karalar EN, Welch MD. New mechanisms and functions of actin nucleation. Curr Opin Cell Biol. 2011;23:4–13. doi: 10.1016/j.ceb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rotty JD, Wu C, Bear JE. New insights into the regulation and cellular functions of the ARP2/3 complex. Nat Rev Mol Cell Biol. 2013;14:7–12. doi: 10.1038/nrm3492. [DOI] [PubMed] [Google Scholar]
- 5.Goley ED, Welch MD. The ARP2/3 complex: an actin nucleator comes of age. Nat Rev Mol Cell Biol. 2006;7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- 6.Rouiller I, Xu XP, Amann KJ, Egile C, Nickell S, Nicastro D, Li R, Pollard TD, Volkmann N, Hanein D. The structural basis of actin filament branching by the Arp2/3 complex. J Cell Biol. 2008;180:887–895. doi: 10.1083/jcb.200709092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Volkmann N, Amann KJ, Stoilova-McPhie S, Egile C, Winter DC, Hazelwood L, Heuser JE, Li R, Pollard TD, Hanein D. Structure of Arp2/3 complex in its activated state and in actin filament branch junctions. Science. 2001;293:2456–2459. doi: 10.1126/science.1063025. [DOI] [PubMed] [Google Scholar]
- 8.Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci U S A. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kollmar M, Lbik D, Enge S. Evolution of the eukaryotic ARP2/3 activators of the WASP family: WASP, WAVE, WASH, and WHAMM, and the proposed new family members WAWH and WAML. BMC Res Notes. 2012;5:88. doi: 10.1186/1756-0500-5-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Insall RH, Machesky LM. Actin dynamics at the leading edge: from simple machinery to complex networks. Dev Cell. 2009;17:310–322. doi: 10.1016/j.devcel.2009.08.012. [DOI] [PubMed] [Google Scholar]
- 11.Wagner AR, Luan Q, Liu SL, Nolen BJ. Dip1 defines a class of Arp2/3 complex activators that function without preformed actin filaments. Curr Biol. 2013;23:1990–1998. doi: 10.1016/j.cub.2013.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guetta-Terrier C, Monzo P, Zhu J, Long H, Venkatraman L, Zhou Y, Wang P, Chew SY, Mogilner A, Ladoux B, et al. Protrusive waves guide 3D cell migration along nanofibers. J Cell Biol. 2015;211:683–701. doi: 10.1083/jcb.201501106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doyle AD, Petrie RJ, Kutys ML, Yamada KM. Dimensions in cell migration. Curr Opin Cell Biol. 2013;25:642–649. doi: 10.1016/j.ceb.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedl P, Sahai E, Weiss S, Yamada KM. New dimensions in cell migration. Nat Rev Mol Cell Biol. 2012;13:743–747. doi: 10.1038/nrm3459. [DOI] [PubMed] [Google Scholar]
- 15.Friedl P, Wolf K. Plasticity of cell migration: a multiscale tuning model. J Cell Biol. 2010;188:11–19. doi: 10.1083/jcb.200909003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lammermann T, Sixt M. Mechanical modes of ‘amoeboid’ cell migration. Curr Opin Cell Biol. 2009;21:636–644. doi: 10.1016/j.ceb.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 17.Wolf K, Friedl P. Molecular mechanisms of cancer cell invasion and plasticity. Br J Dermatol. 2006;154(Suppl 1):11–15. doi: 10.1111/j.1365-2133.2006.07231.x. [DOI] [PubMed] [Google Scholar]
- 18.Sahai E, Marshall CJ. Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nat Cell Biol. 2003;5:711–719. doi: 10.1038/ncb1019. [DOI] [PubMed] [Google Scholar]
- 19.Doyle AD, Wang FW, Matsumoto K, Yamada KM. One-dimensional topography underlies three-dimensional fibrillar cell migration. J Cell Biol. 2009;184:481–490. doi: 10.1083/jcb.200810041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bergert M, Chandradoss SD, Desai RA, Paluch E. Cell mechanics control rapid transitions between blebs and lamellipodia during migration. Proc Natl Acad Sci U S A. 2012;109:14434–14439. doi: 10.1073/pnas.1207968109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parekh A, Ruppender NS, Branch KM, Sewell-Loftin MK, Lin J, Boyer PD, Candiello JE, Merryman WD, Guelcher SA, Weaver AM. Sensing and modulation of invadopodia across a wide range of rigidities. Biophys J. 2011;100:573–582. doi: 10.1016/j.bpj.2010.12.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petrie RJ, Gavara N, Chadwick RS, Yamada KM. Nonpolarized signaling reveals two distinct modes of 3D cell migration. J Cell Biol. 2012;197:439–455. doi: 10.1083/jcb.201201124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ulrich TA, de Juan Pardo EM, Kumar S. The mechanical rigidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells. Cancer Res. 2009;69:4167–4174. doi: 10.1158/0008-5472.CAN-08-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24••.Charras G, Sahai E. Physical influences of the extracellular environment on cell migration. Nat Rev Mol Cell Biol. 2014;15:813–824. doi: 10.1038/nrm3897. This review highlights how the properties of the extracellular environment, including dimensionality, stiffness, and confinement, influence the mechanisms of cell migration. [DOI] [PubMed] [Google Scholar]
- 25••.Bieling P, Li TD, Weichsel J, McGorty R, Jreij P, Huang B, Fletcher DA, Mullins RD. Force Feedback Controls Motor Activity and Mechanical Properties of Self-Assembling Branched Actin Networks. Cell. 2016;164:115–127. doi: 10.1016/j.cell.2015.11.057. Using in vitro actin polymerization assays with purified proteins, the authors show that mechanical force alters the structure and function of the actin network. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vignaud T, Galland R, Tseng Q, Blanchoin L, Colombelli J, Thery M. Reprogramming cell shape with laser nano-patterning. J Cell Sci. 2012;125:2134–2140. doi: 10.1242/jcs.104901. [DOI] [PubMed] [Google Scholar]
- 27•.Letort G, Politi AZ, Ennomani H, Thery M, Nedelec F, Blanchoin L. Geometrical and mechanical properties control actin filament organization. PLoS Comput Biol. 2015;11:e1004245. doi: 10.1371/journal.pcbi.1004245. Through a combination of simulations and in vitro validation experiments, this study demonstrates that actin nucleation geometry, as well as the mechanical properties of the resulting filaments, determines the overall actin network structure. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reymann AC, Martiel JL, Cambier T, Blanchoin L, Boujemaa-Paterski R, Thery M. Nucleation geometry governs ordered actin networks structures. Nat Mater. 2010;9:827–832. doi: 10.1038/nmat2855. [DOI] [PubMed] [Google Scholar]
- 29.Zaki M, King J, Futterer K, Insall RH. Replacement of the essential Dictyostelium Arp2 gene by its Entamoeba homologue using parasexual genetics. BMC Genet. 2007;8:28. doi: 10.1186/1471-2156-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yae K, Keng VW, Koike M, Yusa K, Kouno M, Uno Y, Kondoh G, Gotow T, Uchiyama Y, Horie K, et al. Sleeping beauty transposon-based phenotypic analysis of mice: lack of Arpc3 results in defective trophoblast outgrowth. Mol Cell Biol. 2006;26:6185–6196. doi: 10.1128/MCB.00018-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nicholson-Dykstra SM, Higgs HN. Arp2 depletion inhibits sheet-like protrusions but not linear protrusions of fibroblasts and lymphocytes. Cell Motil Cytoskeleton. 2008;65:904–922. doi: 10.1002/cm.20312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rogers SL, Wiedemann U, Stuurman N, Vale RD. Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J Cell Biol. 2003;162:1079–1088. doi: 10.1083/jcb.200303023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steffen A, Faix J, Resch GP, Linkner J, Wehland J, Small JV, Rottner K, Stradal TE. Filopodia formation in the absence of functional WAVE- and Arp2/3-complexes. Mol Biol Cell. 2006;17:2581–2591. doi: 10.1091/mbc.E05-11-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Machesky LM, Insall RH. Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr Biol. 1998;8:1347–1356. doi: 10.1016/s0960-9822(98)00015-3. [DOI] [PubMed] [Google Scholar]
- 35.Bailly M, Ichetovkin I, Grant W, Zebda N, Machesky LM, Segall JE, Condeelis J. The F-actin side binding activity of the Arp2/3 complex is essential for actin nucleation and lamellipod extension. Curr Biol. 2001;11:620–625. doi: 10.1016/s0960-9822(01)00152-x. [DOI] [PubMed] [Google Scholar]
- 36.Yamaguchi H, Lorenz M, Kempiak S, Sarmiento C, Coniglio S, Symons M, Segall J, Eddy R, Miki H, Takenawa T, et al. Molecular mechanisms of invadopodium formation: the role of the N-WASP-Arp2/3 complex pathway and cofilin. J Cell Biol. 2005;168:441–452. doi: 10.1083/jcb.200407076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamazaki D, Suetsugu S, Miki H, Kataoka Y, Nishikawa S, Fujiwara T, Yoshida N, Takenawa T. WAVE2 is required for directed cell migration and cardiovascular development. Nature. 2003;424:452–456. doi: 10.1038/nature01770. [DOI] [PubMed] [Google Scholar]
- 38.Yan C, Martinez-Quiles N, Eden S, Shibata T, Takeshima F, Shinkura R, Fujiwara Y, Bronson R, Snapper SB, Kirschner MW, et al. WAVE2 deficiency reveals distinct roles in embryogenesis and Rac-mediated actin-based motility. EMBO J. 2003;22:3602–3612. doi: 10.1093/emboj/cdg350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi CH, Thomason PA, Zaki M, Insall RH, Barber DL. Phosphorylation of actin-related protein 2 (Arp2) is required for normal development and cAMP chemotaxis in Dictyostelium. J Biol Chem. 2013;288:2464–2474. doi: 10.1074/jbc.M112.435313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blagg SL, Stewart M, Sambles C, Insall RH. PIR121 regulates pseudopod dynamics and SCAR activity in Dictyostelium. Curr Biol. 2003;13:1480–1487. doi: 10.1016/s0960-9822(03)00580-3. [DOI] [PubMed] [Google Scholar]
- 41.Ura S, Pollitt AY, Veltman DM, Morrice NA, Machesky LM, Insall RH. Pseudopod growth and evolution during cell movement is controlled through SCAR/WAVE dephosphorylation. Curr Biol. 2012;22:553–561. doi: 10.1016/j.cub.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bear JE, Rawls JF, Saxe CL., 3rd SCAR, a WASP-related protein, isolated as a suppressor of receptor defects in late Dictyostelium development. J Cell Biol. 1998;142:1325–1335. doi: 10.1083/jcb.142.5.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43••.Asokan SB, Johnson HE, Rahman A, King SJ, Rotty JD, Lebedeva IP, Haugh JM, Bear JE. Mesenchymal chemotaxis requires selective inactivation of myosin II at the leading edge via a noncanonical PLCgamma/PKCalpha pathway. Dev Cell. 2014;31:747–760. doi: 10.1016/j.devcel.2014.10.024. Unlike Suraneni et al, the authors find that fibroblasts lacking Arp2/3 activity can undergo chemotaxis to PDGF by inactivating myosin II at the cell front via PKCα-mediated, non-canonical phosphorylation of the regulatory light chain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44••.Suraneni P, Fogelson B, Rubinstein B, Noguera P, Volkmann N, Hanein D, Mogilner A, Li R. A mechanism of leading-edge protrusion in the absence of Arp2/3 complex. Mol Biol Cell. 2015;26:901–912. doi: 10.1091/mbc.E14-07-1250. This study of fibroblasts derived from Arp2/3 null mice shows that cells can compensate for the lack of Arp2/3 activity through a combination of formin-based filopodial protrusions and myosin II-based contractility. Although these Arp2/3-deficient cells are migratory, they are unable to chemotax on 2D, contrary to the findings of Asokan et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suraneni P, Rubinstein B, Unruh JR, Durnin M, Hanein D, Li R. The Arp2/3 complex is required for lamellipodia extension and directional fibroblast cell migration. J Cell Biol. 2012;197:239–251. doi: 10.1083/jcb.201112113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu C, Asokan SB, Berginski ME, Haynes EM, Sharpless NE, Griffith JD, Gomez SM, Bear JE. Arp2/3 is critical for lamellipodia and response to extracellular matrix cues but is dispensable for chemotaxis. Cell. 2012;148:973–987. doi: 10.1016/j.cell.2011.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu C, Haynes EM, Asokan SB, Simon JM, Sharpless NE, Baldwin AS, Davis IJ, Johnson GL, Bear JE. Loss of Arp2/3 induces an NF-kappaB-dependent, nonautonomous effect on chemotactic signaling. J Cell Biol. 2013;203:907–916. doi: 10.1083/jcb.201306032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Y, Wang PS, Lucas G, Li R, Yao L. ARP2/3 complex is required for directional migration of neural stem cell-derived oligodendrocyte precursors in electric fields. Stem Cell Res Ther. 2015;6:41. doi: 10.1186/s13287-015-0042-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang PSCFS, Guo F, Suraneni P, Xia S, Ramachandran S, Li R. Crucial Roles of the Arp2/3 Complex during Mammalian Corticogenesis. 2015 doi: 10.1242/dev.130542. Submitted. doi: http://dx.doi.org/10.1101/026161. [DOI] [PMC free article] [PubMed]
- 50•.Moreau HD, Lemaitre F, Garrod KR, Garcia Z, Lennon-Dumenil AM, Bousso P. Signal strength regulates antigen-mediated T-cell deceleration by distinct mechanisms to promote local exploration or arrest. Proc Natl Acad Sci U S A. 2015;112:12151–12156. doi: 10.1073/pnas.1506654112. Arp2/3 complex activity inhibits the fast migration of T cells and, in response to high-affinity antigen-binding, triggers a deceleration that facilitates immunological synapse formation. However, Arp2/3 inhibition has little effect on the switch between fast and exploratory modes of migration during kinapse formation, suggesting that different actin nucleators may control migration in response to external stimuli by distinct mechanisms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51••.Vargas P, Maiuri P, Bretou M, Saez PJ, Pierobon P, Maurin M, Chabaud M, Lankar D, Obino D, Terriac E, et al. Innate control of actin nucleation determines two distinct migration behaviours in dendritic cells. Nat Cell Biol. 2016;18:43–53. doi: 10.1038/ncb3284. This work shows that competing pools of actin, generated by different nucleation factors, control distinct migratory behaviors in dendritic cells. Chemical stimuli that elicit dendritic cell maturation alter the balance between Arp2/3 and formin activities, causing a corresponding shift in migratory potential necessary for mature dendritic cell function. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thiam HR, Vargas P, Carpi N, Crespo CL, Raab M, Terriac E, King MC, Jacobelli J, Alberts AS, Stradal T, et al. Perinuclear Arp2/3-driven actin polymerization enables nuclear deformation to facilitate cell migration through complex environments. Nat Commun. 2016;7:10997. doi: 10.1038/ncomms10997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Verboon JM, Rahe TK, Rodriguez-Mesa E, Parkhurst SM. Wash functions downstream of Rho1 GTPase in a subset of Drosophila immune cell developmental migrations. Mol Biol Cell. 2015;26:1665–1674. doi: 10.1091/mbc.E14-08-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sander M, Squarr AJ, Risse B, Jiang X, Bogdan S. Drosophila pupal macrophages--a versatile tool for combined ex vivo and in vivo imaging of actin dynamics at high resolution. Eur J Cell Biol. 2013;92:349–354. doi: 10.1016/j.ejcb.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 55.Evans IR, Ghai PA, Urbancic V, Tan KL, Wood W. SCAR/WAVE-mediated processing of engulfed apoptotic corpses is essential for effective macrophage migration in Drosophila. Cell Death Differ. 2013;20:709–720. doi: 10.1038/cdd.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beckham Y, Vasquez RJ, Stricker J, Sayegh K, Campillo C, Gardel ML. Arp2/3 inhibition induces amoeboid-like protrusions in MCF10A epithelial cells by reduced cytoskeletal-membrane coupling and focal adhesion assembly. PLoS One. 2014;9:e100943. doi: 10.1371/journal.pone.0100943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Z, Yang X, Chen C, Liu B, Ren B, Wang L, Zhao K, Yu S, Ming H. Expression of the Arp2/3 complex in human gliomas and its role in the migration and invasion of glioma cells. Oncol Rep. 2013;30:2127–2136. doi: 10.3892/or.2013.2669. [DOI] [PubMed] [Google Scholar]
- 58.Wang L, Zhao K, Ren B, Zhu M, Zhang C, Zhao P, Zhou H, Chen L, Yu S, Yang X. Expression of cortactin in human gliomas and its effect on migration and invasion of glioma cells. Oncol Rep. 2015;34:1815–1824. doi: 10.3892/or.2015.4156. [DOI] [PubMed] [Google Scholar]
- 59.Oser M, Yamaguchi H, Mader CC, Bravo-Cordero JJ, Arias M, Chen X, Desmarais V, van Rheenen J, Koleske AJ, Condeelis J. Cortactin regulates cofilin and N-WASp activities to control the stages of invadopodium assembly and maturation. J Cell Biol. 2009;186:571–587. doi: 10.1083/jcb.200812176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Monteiro P, Rosse C, Castro-Castro A, Irondelle M, Lagoutte E, Paul-Gilloteaux P, Desnos C, Formstecher E, Darchen F, Perrais D, et al. Endosomal WASH and exocyst complexes control exocytosis of MT1-MMP at invadopodia. J Cell Biol. 2013;203:1063–1079. doi: 10.1083/jcb.201306162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tania N, Condeelis J, Edelstein-Keshet L. Modeling the synergy of cofilin and Arp2/3 in lamellipodial protrusive activity. Biophys J. 2013;105:1946–1955. doi: 10.1016/j.bpj.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Monzo P, Chong YK, Guetta-Terrier C, Krishnasamy A, Sathe SR, Yim EK, Ng WH, Ang BT, Tang C, Ladoux B, et al. Mechanical Confinement Triggers Glioma Linear Migration Dependent On Formin, FHOD3. Mol Biol Cell. 2016 doi: 10.1091/mbc.E15-08-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Panopoulos A, Howell M, Fotedar R, Margolis RL. Glioblastoma motility occurs in the absence of actin polymer. Mol Biol Cell. 2011;22:2212–2220. doi: 10.1091/mbc.E10-10-0849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stroka KM, Jiang H, Chen SH, Tong Z, Wirtz D, Sun SX, Konstantopoulos K. Water permeation drives tumor cell migration in confined microenvironments. Cell. 2014;157:611–623. doi: 10.1016/j.cell.2014.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Beadle C, Assanah MC, Monzo P, Vallee R, Rosenfeld SS, Canoll P. The role of myosin II in glioma invasion of the brain. Mol Biol Cell. 2008;19:3357–3368. doi: 10.1091/mbc.E08-03-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Paul NR, Allen JL, Chapman A, Morlan-Mairal M, Zindy E, Jacquemet G, Fernandez del Ama L, Ferizovic N, Green DM, Howe JD, et al. alpha5beta1 integrin recycling promotes Arp2/3-independent cancer cell invasion via the formin FHOD3. J Cell Biol. 2015;210:1013–1031. doi: 10.1083/jcb.201502040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aerbajinai W, Liu L, Zhu J, Kumkhaek C, Chin K, Rodgers GP. Glia Maturation Factor-gamma Regulates Monocyte Migration through Modulation of beta1-Integrin. J Biol Chem. 2016 doi: 10.1074/jbc.M115.674200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Edwards M, Zwolak A, Schafer DA, Sept D, Dominguez R, Cooper JA. Capping protein regulators fine-tune actin assembly dynamics. Nat Rev Mol Cell Biol. 2014;15:677–689. doi: 10.1038/nrm3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fujiwara I, Remmert K, Piszczek G, Hammer JA. Capping protein regulatory cycle driven by CARMIL and V-1 may promote actin network assembly at protruding edges. Proc Natl Acad Sci U S A. 2014;111:E1970–1979. doi: 10.1073/pnas.1313738111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Le Clainche C, Carlier MF. Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol Rev. 2008;88:489–513. doi: 10.1152/physrev.00021.2007. [DOI] [PubMed] [Google Scholar]
- 71.Loisel TP, Boujemaa R, Pantaloni D, Carlier MF. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature. 1999;401:613–616. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- 72.Boczkowska M, Rebowski G, Dominguez R. Glia maturation factor (GMF) interacts with Arp2/3 complex in a nucleotide state-dependent manner. J Biol Chem. 2013;288:25683–25688. doi: 10.1074/jbc.C113.493338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73•.Abella JV, Galloni C, Pernier J, Barry DJ, Kjaer S, Carlier MF, Way M. Isoform diversity in the Arp2/3 complex determines actin filament dynamics. Nat Cell Biol. 2016;18:76–86. doi: 10.1038/ncb3286. In this study, the authors conclude that isoform variants of the Arpc1 and Arpc5 subunits give rise to Arp2/3 complexes with distinct properties due to differential regulation by cortactin and coronin. This leads to the possibility that variations in isoform expression across cell types confer different migratory behaviors. [DOI] [PubMed] [Google Scholar]
- 74••.Helgeson LA, Prendergast JG, Wagner AR, Rodnick-Smith M, Nolen BJ. Interactions with actin monomers, actin filaments, and Arp2/3 complex define the roles of WASP family proteins and cortactin in coordinately regulating branched actin networks. J Biol Chem. 2014;289:28856–28869. doi: 10.1074/jbc.M114.587527. Using a variety of purified truncated or chimeric nucleation promoting factors, the authors determine the mechanisms controlling the synergistic activation of Arp2/3 by cortactin and N-WASP. Furthermore, they show that the structural differences between these NPFs lead to distinct modes of Arp2/3 complex activation and corresponding changes in the resulting actin architecture. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gandhi M, Goode BL. Coronin: the double-edged sword of actin dynamics. Subcell Biochem. 2008;48:72–87. doi: 10.1007/978-0-387-09595-0_7. [DOI] [PubMed] [Google Scholar]
- 76.Bravo-Cordero JJ, Magalhaes MA, Eddy RJ, Hodgson L, Condeelis J. Functions of cofilin in cell locomotion and invasion. Nat Rev Mol Cell Biol. 2013;14:405–415. doi: 10.1038/nrm3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Svitkina TM, Borisy GG. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J Cell Biol. 1999;145:1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dang I, Gorelik R, Sousa-Blin C, Derivery E, Guerin C, Linkner J, Nemethova M, Dumortier JG, Giger FA, Chipysheva TA, et al. Inhibitory signalling to the Arp2/3 complex steers cell migration. Nature. 2013;503:281–284. doi: 10.1038/nature12611. [DOI] [PubMed] [Google Scholar]
- 79.Gorelik R, Gautreau A. The Arp2/3 inhibitory protein arpin induces cell turning by pausing cell migration. Cytoskeleton (Hoboken) 2015;72:362–371. doi: 10.1002/cm.21233. [DOI] [PubMed] [Google Scholar]
- 80.Lomakina ME, Lallemand F, Vacher S, Molinie N, Dang I, Cacheux W, Chipysheva TA, Ermilova VD, de Koning L, Dubois T, et al. Arpin downregulation in breast cancer is associated with poor prognosis. Br J Cancer. 2016;114:545–553. doi: 10.1038/bjc.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu X, Zhao B, Wang H, Wang Y, Niu M, Sun M, Zhao Y, Yao R, Qu Z. Aberrant expression of Arpin in human breast cancer and its clinical significance. J Cell Mol Med. 2016;20:450–458. doi: 10.1111/jcmm.12740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maritzen T, Zech T, Schmidt MR, Krause E, Machesky LM, Haucke V. Gadkin negatively regulates cell spreading and motility via sequestration of the actin-nucleating ARP2/3 complex. Proc Natl Acad Sci U S A. 2012;109:10382–10387. doi: 10.1073/pnas.1206468109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schachtner H, Weimershaus M, Stache V, Plewa N, Legler DF, Hopken UE, Maritzen T. Loss of Gadkin Affects Dendritic Cell Migration In Vitro. PLoS One. 2015;10:e0143883. doi: 10.1371/journal.pone.0143883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Spence HJ, Timpson P, Tang HR, Insall RH, Machesky LM. Scar/WAVE3 contributes to motility and plasticity of lamellipodial dynamics but not invasion in three dimensions. Biochem J. 2012;448:35–42. doi: 10.1042/BJ20112206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tang H, Li A, Bi J, Veltman DM, Zech T, Spence HJ, Yu X, Timpson P, Insall RH, Frame MC, et al. Loss of Scar/WAVE complex promotes N-WASP- and FAK-dependent invasion. Curr Biol. 2013;23:107–117. doi: 10.1016/j.cub.2012.11.059. [DOI] [PubMed] [Google Scholar]
- 86.Brunton VG, Frame MC. Src and focal adhesion kinase as therapeutic targets in cancer. Curr Opin Pharmacol. 2008;8:427–432. doi: 10.1016/j.coph.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 87.Swaminathan V, Fischer RS, Waterman CM. The FAK-Arp2/3 interaction promotes leading edge advance and haptosensing by coupling nascent adhesions to lamellipodia actin. Mol Biol Cell. 2016 doi: 10.1091/mbc.E15-08-0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Johnson HE, King SJ, Asokan SB, Rotty JD, Bear JE, Haugh JM. F-actin bundles direct the initiation and orientation of lamellipodia through adhesion-based signaling. J Cell Biol. 2015;208:443–455. doi: 10.1083/jcb.201406102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Giri A, Bajpai S, Trenton N, Jayatilaka H, Longmore GD, Wirtz D. The Arp2/3 complex mediates multigeneration dendritic protrusions for efficient 3-dimensional cancer cell migration. FASEB J. 2013;27:4089–4099. doi: 10.1096/fj.12-224352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bovellan M, Romeo Y, Biro M, Boden A, Chugh P, Yonis A, Vaghela M, Fritzsche M, Moulding D, Thorogate R, et al. Cellular control of cortical actin nucleation. Curr Biol. 2014;24:1628–1635. doi: 10.1016/j.cub.2014.05.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chhabra ES, Higgs HN. The many faces of actin: matching assembly factors with cellular structures. Nat Cell Biol. 2007;9:1110–1121. doi: 10.1038/ncb1007-1110. [DOI] [PubMed] [Google Scholar]
- 92.Mejillano MR, Kojima S, Applewhite DA, Gertler FB, Svitkina TM, Borisy GG. Lamellipodial versus filopodial mode of the actin nanomachinery: pivotal role of the filament barbed end. Cell. 2004;118:363–373. doi: 10.1016/j.cell.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 93.Benvenuti F, Hugues S, Walmsley M, Ruf S, Fetler L, Popoff M, Tybulewicz VL, Amigorena S. Requirement of Rac1 and Rac2 expression by mature dendritic cells for T cell priming. Science. 2004;305:1150–1153. doi: 10.1126/science.1099159. [DOI] [PubMed] [Google Scholar]
- 94.Lammermann T, Renkawitz J, Wu X, Hirsch K, Brakebusch C, Sixt M. Cdc42-dependent leading edge coordination is essential for interstitial dendritic cell migration. Blood. 2009;113:5703–5710. doi: 10.1182/blood-2008-11-191882. [DOI] [PubMed] [Google Scholar]
- 95.de Noronha S, Hardy S, Sinclair J, Blundell MP, Strid J, Schulz O, Zwirner J, Jones GE, Katz DR, Kinnon C, et al. Impaired dendritic-cell homing in vivo in the absence of Wiskott-Aldrich syndrome protein. Blood. 2005;105:1590–1597. doi: 10.1182/blood-2004-06-2332. [DOI] [PubMed] [Google Scholar]
- 96•.Burke TA, Christensen JR, Barone E, Suarez C, Sirotkin V, Kovar DR. Homeostatic actin cytoskeleton networks are regulated by assembly factor competition for monomers. Curr Biol. 2014;24:579–585. doi: 10.1016/j.cub.2014.01.072. This study suggests that actin monomer levels control the balance between formin- and Arp2/3-mediated actin polymerization, resulting in the formation of different actin structures in fission yeast. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Block J, Breitsprecher D, Kuhn S, Winterhoff M, Kage F, Geffers R, Duwe P, Rohn JL, Baum B, Brakebusch C, et al. FMNL2 drives actin-based protrusion and migration downstream of Cdc42. Curr Biol. 2012;22:1005–1012. doi: 10.1016/j.cub.2012.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yang C, Czech L, Gerboth S, Kojima S, Scita G, Svitkina T. Novel roles of formin mDia2 in lamellipodia and filopodia formation in motile cells. PLoS Biol. 2007;5:e317. doi: 10.1371/journal.pbio.0050317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Skau CT, Plotnikov SV, Doyle AD, Waterman CM. Inverted formin 2 in focal adhesions promotes dorsal stress fiber and fibrillar adhesion formation to drive extracellular matrix assembly. Proc Natl Acad Sci U S A. 2015;112:E2447–2456. doi: 10.1073/pnas.1505035112. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 100•.Isogai T, van der Kammen R, Leyton-Puig D, Kedziora KM, Jalink K, Innocenti M. Initiation of lamellipodia and ruffles involves cooperation between mDia1 and the Arp2/3 complex. J Cell Sci. 2015;128:3796–3810. doi: 10.1242/jcs.176768. Through the use of temporally-controlled inactivation of mDia1 in this study, the authors suggest a mechanism by which formins cooperate with Arp2/3 for ruffle and lamellipodia formation. Specifically, formins produce the linear actin filaments necessary for Arp2/3-mediated branching. [DOI] [PubMed] [Google Scholar]
- 101.Young LE, Heimsath EG, Higgs HN. Cell type-dependent mechanisms for formin-mediated assembly of filopodia. Mol Biol Cell. 2015;26:4646–4659. doi: 10.1091/mbc.E15-09-0626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Korobova F, Svitkina T. Arp2/3 complex is important for filopodia formation, growth cone motility, and neuritogenesis in neuronal cells. Mol Biol Cell. 2008;19:1561–1574. doi: 10.1091/mbc.E07-09-0964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Biyasheva A, Svitkina T, Kunda P, Baum B, Borisy G. Cascade pathway of filopodia formation downstream of SCAR. J Cell Sci. 2004;117:837–848. doi: 10.1242/jcs.00921. [DOI] [PubMed] [Google Scholar]
- 104.Vignjevic D, Yarar D, Welch MD, Peloquin J, Svitkina T, Borisy GG. Formation of filopodia-like bundles in vitro from a dendritic network. J Cell Biol. 2003;160:951–962. doi: 10.1083/jcb.200208059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105••.Rotty JD, Wu C, Haynes EM, Suarez C, Winkelman JD, Johnson HE, Haugh JM, Kovar DR, Bear JE. Profilin-1 serves as a gatekeeper for actin assembly by Arp2/3-dependent and -independent pathways. Dev Cell. 2015;32:54–67. doi: 10.1016/j.devcel.2014.10.026. This study shows that fibroblasts derived from Arp2/3 complex subunit knock-out mice require profilin and Ena/VASP for migration, and that exogenous profilin inhibits Arp2/3 activity in vitro or in wild-type cells. The authors show that, possibly through competition for actin monomers, profilin balances the activities of Arp2/3 and other actin nucleators. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106•.Havrylenko S, Noguera P, Abou-Ghali M, Manzi J, Faqir F, Lamora A, Guerin C, Blanchoin L, Plastino J. WAVE binds Ena/VASP for enhanced Arp2/3 complex-based actin assembly. Mol Biol Cell. 2015;26:55–65. doi: 10.1091/mbc.E14-07-1200. An interaction between WAVE and Ena/VASP enhances Arp2/3-mediated actin assembly in vitro and regulates lamellipodia formation during C. elegans ventral closure; thus, cooperativity amongst NPFs and other elongation factors can alter actin network dynamics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107•.Chen XJ, Squarr AJ, Stephan R, Chen B, Higgins TE, Barry DJ, Martin MC, Rosen MK, Bogdan S, Way M. Ena/VASP proteins cooperate with the WAVE complex to regulate the actin cytoskeleton. Dev Cell. 2014;30:569–584. doi: 10.1016/j.devcel.2014.08.001. In this study, the authors map an interaction between Ena/VASP and a member of the WAVE regulatory complex that promotes synergistic activation of the Arp2/3 complex and enhances cell migration. This interaction is required for normal lamellipodial dynamics in Drosophila macrophages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108•.Suarez C, Carroll RT, Burke TA, Christensen JR, Bestul AJ, Sees JA, James ML, Sirotkin V, Kovar DR. Profilin regulates F-actin network homeostasis by favoring formin over Arp2/3 complex. Dev Cell. 2015;32:43–53. doi: 10.1016/j.devcel.2014.10.027. In fission yeast, profilin-mediated inhibition of the Arp2/3 complex is required for formins to properly assemble the contractile ring, suggesting that the homeostasis between actin nucleators is important for cell function. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pernier J, Shekhar S, Jegou A, Guichard B, Carlier MF. Profilin Interaction with Actin Filament Barbed End Controls Dynamic Instability, Capping, Branching, and Motility. Dev Cell. 2016;36:201–214. doi: 10.1016/j.devcel.2015.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]