Abstract
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is a long non-coding RNA (lncRNA) that contributes to the initiation and development of many solid tumors, including osteosarcoma (OS). Here, we showed that MALAT1 was increased in human OS cell lines and tissues and promoted OS cell growth, while MALAT1 knockdown suppressed OS cell growth. We also detected downregulation of MIR376A, a suppressor of OS growth, and upregulation of TGFA, a promoter of OS growth, in OS tissues. TGFA expression was positively correlated with MALAT1 expression, and both were negatively correlated with MIR376A expression. There was a direct interaction between MIR376A and MALAT1 via a putative MIR376A binding site within the MALAT1 3′-untranslated region (3′-UTR). There was also a direct interaction between MIR376A and the TGFA 3′-UTR. Thus, MALAT1 may promote OS cell growth through inhibition of MIR376A, leading to increased expression of TGFA. Our results suggest a MALAT1/MIR376A/TGFA axis mediates OS cell proliferation and tumor progression.
Keywords: IncRNA, MALAT1, MIR376A, osteosarcoma, TGFA
INTRODUCTION
More than 90% of the human DNA sequence is actively transcribed but only 2% of it encodes protein. The majority of transcripts are referred to as non-coding RNAs (ncRNAs) [1, 2]. Small non-coding RNAs, especially, microRNAs, have been studied extensively and their roles in gene regulation and cellular function have been elucidated in numerous cancers [2]. Recently, long non-coding RNAs (lncRNAs) have been reported to play important roles during development and in diseases, including cancer [3, 4]. LncRNAs can be oncogenic or function as tumor suppressors [5, 6]. Several lncRNAs play an oncogenic role in breast, gastric, colorectal, and cervical cancers [7], while others function as tumor suppressors in hepatocellular carcinoma and gastric cancer [8, 9]. In osteosarcoma (OS), expression of several lncRNAs may be upregulated [10, 11].
Recently, the lncRNA, MALAT1 was reported to be upregulated in lung, breast, pancreas, liver, colon, gastric, uterus, cervix and prostate cancers [12, 13]. MALAT1 may also serve as an independent prognostic biomarker for survival of these cancers. MALAT1 expression is associated with OS cell fate, as MALAT1 knockdown delays tumor growth in an OS xenograft model, suggesting its oncogenic role and potential as a therapeutic target [14]. MALAT1 also promotes OS cell growth and metastasis, possibly via activation of the PI3K/AKT signaling pathway [15]. While these findings demonstrate a clear correlation between MALAT1 and OS, the specific effect of MALAT1 on OS tumorigenesis and the mechanisms involved remain to be determined.
In this study, we measured expression of MALAT1, MIR376A and TGFA in OS cell lines and tissues. We found a negative correlation between MIR376A and MALAT1 or TGFA expression. To understand the mechanisms of MALAT1 in the OS tumorigenesis, we detected a direct interaction between MIR376A and both MALAT1 and TGFA, suggesting a novel mechanism of MALAT1, MIR376A, and TGFA in the regulation of OS cell growth.
RESULTS
Upregulation of MALAT1 expression in human osteosarcoma tissues and cells
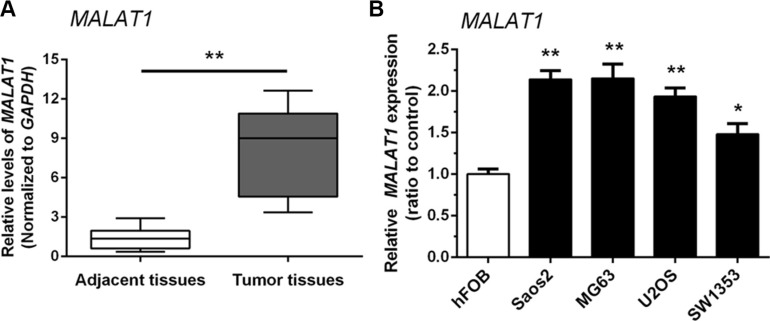
The expression of MALAT1 in 38 paired samples (OS specimens and corresponding adjacent non-tumor tissues) was examined by real-time qPCR. MALAT1 expression was higher in tumor tissues compared with adjacent non-tumor tissues (Figure 1A). We also examined MALAT1 expression in four human OS cell lines: Saos2, MG63, SW1353, U2OS, and compared them with normal human osteoblast (hFOB) cells. As shown in Figure 1B, the expression of MALAT1 was higher in all four human OS cell lines than in hFOB cells.
Figure 1. Upregulation of MALAT1 expression in human osteosarcoma tissues and cells.
(A) MALAT1 expression was higher in tumor tissues compared with adjacent non-tumor tissues. (B) MALAT1 expression in four human OS cell lines: Saos2, MG63, SW1353 and U2OS, was upregulated compared to normal human osteoblast (hFOB) cells. Data are presented as mean ± SD of three independent experiments.
MALAT1 promotion of osteosarcoma cell growth in vitro
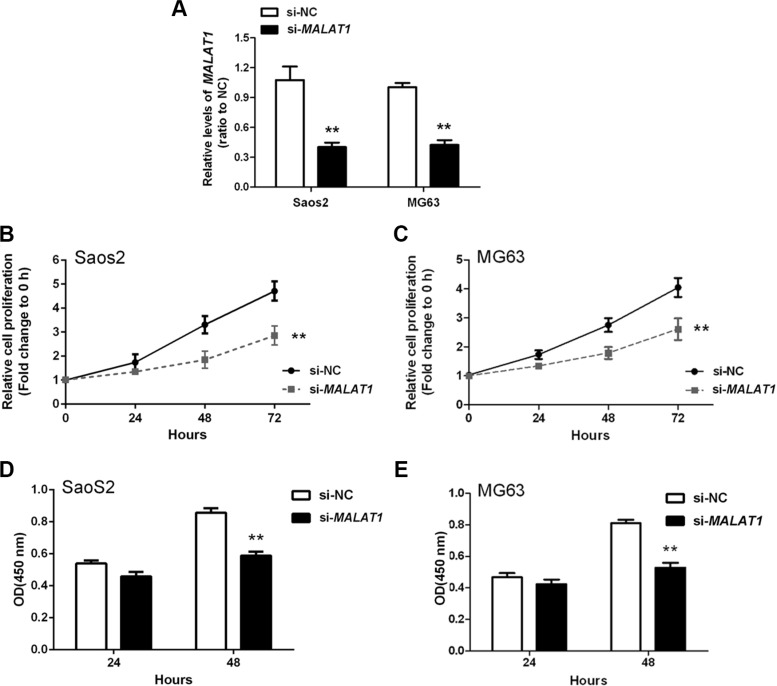
To determine the association of MALAT1 expression with OS cell proliferation and DNA synthesis, MALAT1 siRNA (si-MALAT1), as well as negative control (si-NC), were transfected into two human OS cell lines: Saos2 and MG63. Compared with the si-NC group, MALAT1 expression was decreased in cells transfected with si-MALAT1 as measured by real-time qPCR (Figure 2A). Cell proliferation and DNA synthesis were determined by MTT and BrdU assays, respectively. When compared with the si-NC group, knockdown of MALAT1 attenuated the growth of both Saos2 and MG63 cells up to three days (Figure 2B and 2C). Knockdown of MALAT1 also reduced DNA synthesis in both Saos2 and MG63 cells (Figure 2D and 2E). Together, these data indicated that si-MALAT1 successfully knocked-down MALAT1 expression, and that MALAT1 promotes OS cell growth and DNA synthesis.
Figure 2. MALAT1 promotion of osteosarcoma cell growth in vitro.
(A) MALAT1 knockdown was achieved by si-MALAT1 and the inhibitory efficiency was verified by real-time PCR. (B and C) MTT assays revealed that knockdown of MALAT1 attenuated the growth of both Saos2 and MG63 cell lines up to three days, compared with si-NC group. (D and E) BrdU assays revealed that knockdown of MALAT1 markedly reduced DNA synthesis in both Saos2 and MG63 cell lines. Data are presented as mean ± SD of three independent experiments.
MIR376A suppression of osteosarcoma cell growth in vitro
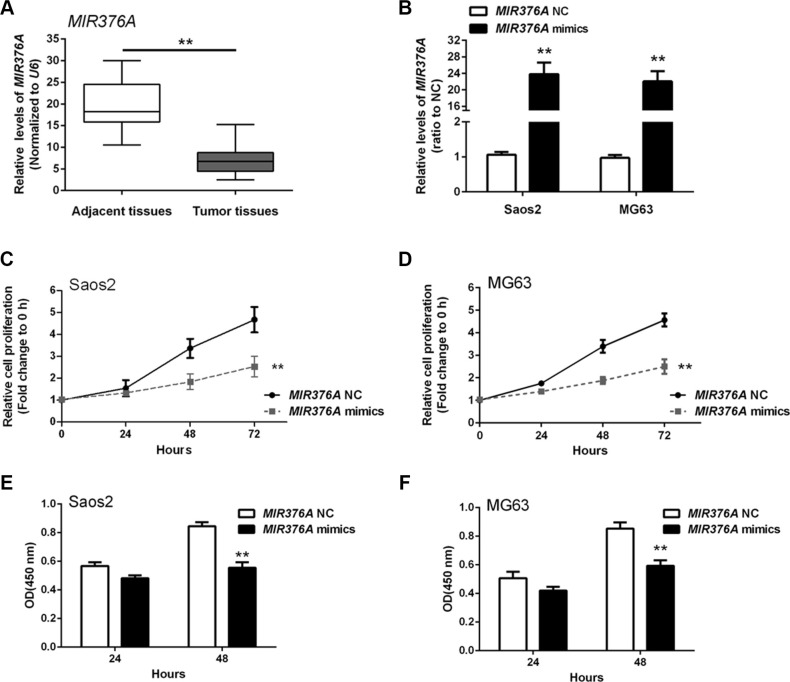
MIR376A was recently shown to function as a tumor suppressor in several cancers [16, 17]. To investigate the role of MIR376A in OS, we first examined MIR376A expression in OS tissues. MIR376A was downregulated in OS tissues compared with adjacent normal tissues (Figure 3A). MIR376A mimics were then transfected into Saos2 and MG63 cells to achieve MIR376A overexpression as confirmed by real-time qPCR (Figure 3B). MTT assays on the MIR376A overexpressing Saos2 and MG63 cells revealed that overexpression of MIR376A reduced cell growth when compared with the MIR376A-NC group (Figure 3C and 3D). In addition, MIR 376A overexpression reduced DNA synthesis in both Saos2 and MG63 cells compared with the MIR376A-NC group, as indicated by BrdU incorporation (Figure 3E and 3F). These data demonstrated that MIR376A inhibited OS cell growth and proliferation.
Figure 3. MIR376A suppression of osteosarcoma cell growth in vitro.
(A) Expression of MIR376A was downregulated in OS tissues compared with adjacent normal tissues. (B) MIR376A mimics were used to achieve MIR376A overexpression as confirmed by real-time qPCR in Saos2 and MG63 cell lines. (C and D) Cell growth of both Saos2 and MG63 cell lines were reduced in response to MIR376A overexpression compared with MIR376A NC group. (E and F) Results from BrdU incorporation assays indicated that MIR376A overexpression markedly reduced DNA synthesis in both Saos2 and MG63 cell lines compared with the MIR376A-NC group. Data are presented as mean ± SD of three independent experiments.
Inverse correlation of MIR376A and MALAT1 expression in osteosarcoma tissues and direct interaction between MIR376A and the 3′UTR of MALAT1 in vitro
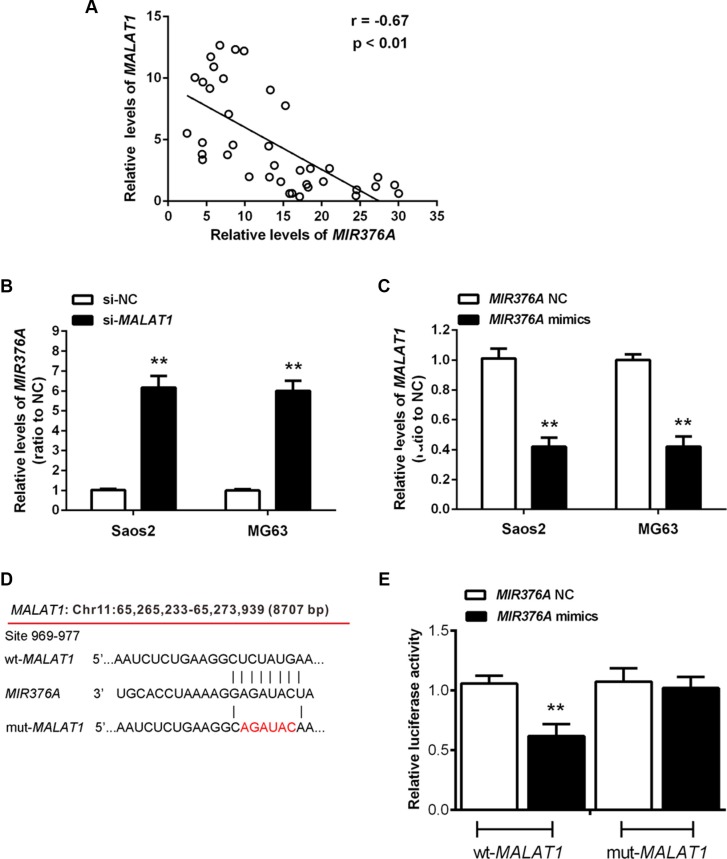
According to previous studies, MIR376 family members play a suppressive role in OS [18, 19]. To investigate whether MIR376A correlates with MALAT1 in regulation of OS cell growth, we performed expression analysis and found an inverse correlation between MALAT1 and MIR376A expression in OS tissues (Figure 4A). Knockdown of MALAT1 caused upregulation of MIR376A (Figure 4B), whereas MIR376A overexpression resulted in decreased MALAT1 expression compared with the MIR376A-NC group (Figure 4C). Together, these data suggested that MIR376A expression is negatively correlated with MALAT1 expression in OS.
Figure 4. Inverse association of MIR376A with MALAT1 expression in osteosarcoma tissues and direct interaction between MIR376A and the 3′UTR of MALAT1 in vitro.
(A) An inverse correlation between MALAT1 and MIR376A expression was observed. (B) Real-time PCR assay showed that knockdown of MALAT1 caused upregulation of MIR376A. (C) MALAT1 expression was decreased in response to MIR376A overexpression, compared with the MIR376A-NC group. Data are presented as mean ± SD of three independent experiments. (D) Generation of wt-MALAT1 and mut-MALAT1 containing luciferase reporter vectors by sequentially mutating the predicted MIR376A binding site in the MALAT1 3′ untranslated region. (E) The wt-MALAT1/mut-MALAT1 vectors and MIR376A-NC/MIR376A mimics were co-transfected into Saos2 cells, respectively. Luciferase activity of the wt-MALAT1 vector was reduced in cells co-transfected with MIR376A mimics. Repression of luciferase activity by MIR376A was not shown in cells transfected with mut-MALAT1. Data are presented as mean ± SD of three independent experiments.
To investigate whether MIR376A directly binds to MALAT1, we generated two luciferase reporter constructs: a wt-MALAT1 and a mut-MALAT1. The mut-MALAT1 contained a 6 bp mutation in the putative MIR376A binding site within the MALAT1 3′-UTR (Figure 4D). These wt-MALAT1 and mut-MALAT1 vectors and MIR376A-NC or MIR376A mimics were co-transfected into Saos2 cells. When compared with the control groups, luciferase activity of the wt-MALAT1 vector was reduced in cells transfected with MIR376A mimics (Figure 4E). The repression of luciferase activity by MIR376A was not seen in cells transfected with mut-MALAT1 (Figure 4E). These results suggested a direct interaction between MIR376A and MALAT1 via the 6-bp putative MIR376A binding site within the 3′UTR of MALAT1.
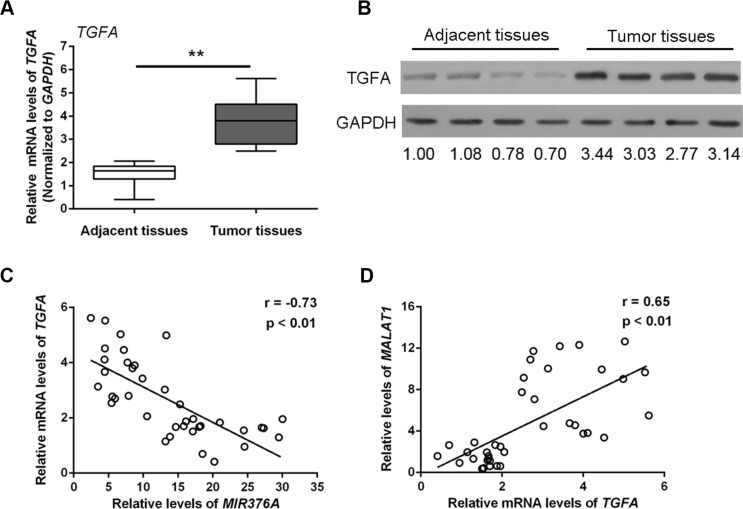
Upregulation of TGFA expression in osteosarcoma tissues and cells and its correlation with MIR376A and MALAT1
TGFA is a direct target of MIR376C, one of the MIR376 family members that regulate OS cell growth [18]. To investigate whether TGFA associates with MIR376A and MALAT1 and plays a role in OS, we examined TGFA mRNA and protein expression in OS tissues. Both TGFA mRNA expression (Figure 5A) and TGFA protein expression (Figure 5B) were upregulated in tumor tissues compared with adjacent normal tissues. Thus, there was an inverse correlation between TGFA and MIR376A (Figure 5C), whereas TGFA expression was positively correlated with MALAT1 expression (Figure 5D).
Figure 5. Upregulation of TGFA expression in osteosarcoma tissues and cells and its correlation with MIR376A and MALAT1.
(A) Expression of TGFA mRNA was upregulated in tumor tissues compared with adjacent normal tissues. (B) Expression of TGFA protein was upregulated in tumor tissues compared with adjacent normal tissues. (C) An inverse correlation between TGFA and MIR376A expression was observed. (D) A positive correlation between MALAT1 and TGFA expression was observed. Data are presented as mean ± SD of three independent experiments.
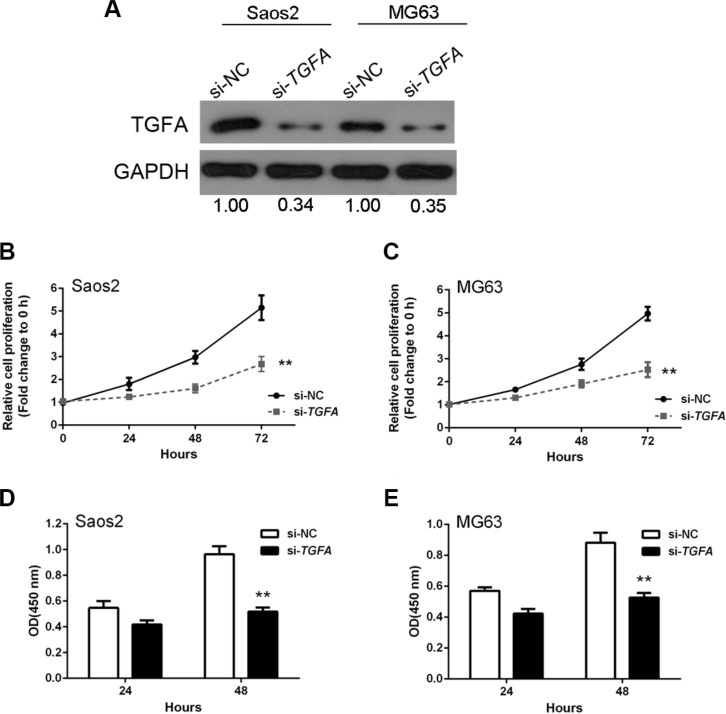
TGFA promotion of osteosarcoma cell growth in vitro
We next investigated the effect of TGFA on OS cell growth by knockdown of TGFA in OS cells. TGFA was successfully knocked-down by si-TGFA, as demonstrated by western blots showing less TGFA protein expression (Figure 6A). MTT assays revealed that OS cell growth was attenuated in response to TGFA inhibition by si-TGFA (Figure 6B and 6C). DNA synthesis in both Saos2 and MG63 cells were also reduced after TGFA inhibition, as indicated by BrdU incorporation (Figure 6D and 6E). Together, these results suggested that TGFA promotes OS cell growth and proliferation.
Figure 6. TGFA promotion of osteosarcoma cell growth in vitro.
(A) TGFA knockdown was achieved by si-TGFA as demonstrated by Western blot assay which showed much less protein expression of TGFA. (B and C) MTT assay results showed that OS cell growth was attenuated in response to TGFA inhibition by si-TGFA. (D and E) BrdU results showed that DNA synthesis capacities were reduced after TGFA inhibition. Data are presented as mean ± SD of three independent experiments.
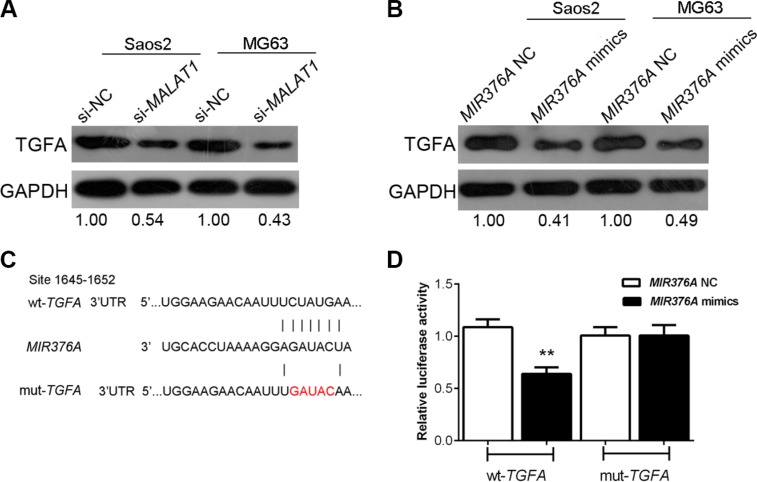
Regulation of TGFA by MALAT1 and MIR376A in human OS cells and TGFA as a direct target of MIR376A
TGFA expression was negatively correlated with MIR376A but positively correlated with MALAT1 expression (Figure 5B and 5C). To further determine their relationship, we measured the expression of TGFA in response to MIR376A overexpression and MALAT1 knockdown in human OS cells. TGFA was downregulated by overexpression of MIR376A as demonstrated by western blot (Figure 7A). Knockdown of MALAT1 also downregulated TGFA in Saos2 and MG63 cells (Figure 7B).
Figure 7. Regulation of TGFA by manipulation of MALAT1 and MIR376A in human OS cells and TGFA as a direct target of MIR376A.
(A) Western blot assay showed that the expression of TGFA was downregulated by MIR376A overexpression in both Saos2 and MG63 cell lines. (B) Western blot results showed that knockdown of MALAT1 also downregulated TGFA in both Saos2 and in MG63 cells. (C) Generation of wt-TGFA and a corresponding mut-TGFA containing a 5 bp mutation in a putative MIR376A binding site within its 3′-UTR. (D) The wt-TGFA/mut-TGFA vectors and MIR376A-NC/MIR376A mimics were co-transfected into Saos2 cells, respectively. The luciferase activity of the wt-TGFA reporter was reduced by co-transfection with MIR376A mimics, as compared with the control groups. No reduction of reporter activity was shown in cells co-transfected with MIR376A and the mut-TGFA reporter. Data are presented as mean ± SD of three independent experiments.
To investigate whether MIR376A repression of OS cell growth is through inhibition of TGFA, we generated two TGFA-containing luciferase reporter constructs: wt-TGFA and a mut-TGFA which contained a 5 bp mutation in a putative MIR376A binding site within its 3′-UTR (Figure 7C). These luciferase reporter constructs were co-transfected into Saos2 cells with MIR376A-NC or MIR376A mimics. The luciferase activity of the wt-TGFA reporter was reduced by transfection of MIR376A mimics when compared with the control groups (Figure 7D). There was no reduction of reporter activity in cells co-transfected with MIR376A and the mut-TGFA reporter, suggesting that MIR376A directly inhibits TGFA through interaction with its 3′-UTR (Figure 7D). These results support a direct correlation of MALAT1, MIR376A, and TGFA expression in the regulation of OS cell growth.
DISCUSSION
The lncRNA MALAT1 is upregulated in many cancers, including OS [12, 13, 20]. MALAT1 has been shown to promote cancer cell proliferation in a variety of malignancies and acts as an oncogene in renal cancer [21]. MALAT1 promotes tumor-driven angiogenesis by upregulating pro-angiogenic gene expression in neuroblastoma [22]. In addition, MALAT1 may also promote colorectal cancer development by directly targeting AKAP9 [23]. In our study, we showed that MALAT1 expression was higher in all four OS cell lines and tissues compared with normal cell lines and adjacent normal tissues. We also found that knockdown of MALAT1 by siRNA resulted in decreased cell proliferation and DNA synthesis in OS cells. These results suggested that MALAT1 functions as an oncogene in OS by promoting OS cell growth, and that MALAT1 might be related with tumor progression.
MIR376A is a tumor-suppressive microRNA associated with multiple cancers, including hepatocellular carcinoma, melanoma, and glioblastoma [16, 17, 24, 25]. MIR376A maps to the 14q32 locus, which harbors a cluster of miRNAs that are known to regulate proliferation, apoptosis, migration, and invasion of several cancers [24, 26]. We found that MIR376A expression was downregulated in OS tissues, and MIR376A overexpression inhibited OS cell growth and proliferation. We also observed an inverse correlation between MALAT1 and MIR376A expression in OS tissues. Knock-down of MALAT1 upregulated MIR376A, while overexpression of MIR376A inhibited MALAT1 expression in vitro, suggesting a direct association between MALAT1 and MIR376A. In support of this, we found that MIR376A may regulate MALAT1 via a putative binding site within its 3′UTR. These results provide direct evidence of a MALAT1-MIR376A interaction in the regulation of OS tumorigenesis. Interestingly, it has been shown that MALAT1 promotes proliferation and metastasis in kidney renal cell carcinoma, possibly through inhibition of MIR200S, as forced overexpression of MIR200S partially attenuated the effects of MALAT1 on growth and metastasis [27]. Moreover, a negative correlation between MIR101 or MIR217 and MALAT1 was previously observed in esophageal squamous cell carcinoma, while knockdown of MALAT1 inhibited cell growth, migration, and invasion [28]. These studies, along with our data, suggest that a lncRNA-miRNA interaction might be important in the process of tumorigenesis.
It has been reported that TGFA regulates OS cell growth as a direct target of MIR376C, one of the MIR376 family members [18]. Our data indicated that TGFA expression was increased in OS tissues compared with adjacent non-tumor tissues and that TGFA promoted OS cell growth in vitro. We also showed that TGFA expression is positively correlated with MALAT1 but negatively correlated with MIR376A. Moreover, overexpression of MIR376A or knockdown of MALAT1 both resulted in markedly reduced TGFA expression. There was a direct interaction between MIR376A and TGFA, with MIR376A regulating TGFA via a 5-bp putative binding site within its 3′-UTR. Previous studies have shown that TGFA promotes OS cell growth, invasion and migration, and support our findings [18, 29]. Recently, other studies have indicated that the interaction between TGFA and miRNAs play an essential role in OS tumorigenesis [18, 29]. Similar to our findings, few studies have also shown that lncRNA-miRNA interactions, for example, the H19-MIR675 interaction and the MALAT1-MIR9 interaction, are important singling pathways in the process of OS tumorigenesis [30, 31]. Thus, our data indicate that MALAT1 may promote OS cell growth through inhibition of MIR376A and by targeting TGFA. This is the first time that lncRNAs, miRNAs, and TGFA have been linked in OS in vitro, which warrants further studies to verify this finding in animal models.
In conclusion, we found differential expression of MALAT1, MIR376A and TGFA in OS cell lines and tissues. All three genes have been associated with OS tumor progression. We showed that TGFA expression correlated with MALAT1 and MIR376A expression in OS. More importantly, there was a direct interaction between MIR376A and MALAT1 or TGFA. Our results support a MALAT1/MIR376A/TGFA axis in OS tumor progression whereby MALAT1 promotes OS cell growth through inhibition of MIR376A and targeting of TGFA.
MATERIALS AND METHODS
Cell lines
Human OS cell lines, Saos2, MG63, U2OS, SW1353, and normal cells, hFOB, were purchased from American Type Culture Collection.
Tissue specimens
Thirty-eight paired OS specimens and corresponding adjacent non-tumor tissues were collected from tumor surgical resection in Xiangya Hospital of Central South University (Changsha, China). All the human tissues were obtained with informed consent and this study was approved by the Clinical Research Ethics Committee of Xiangya Hospital of Central South University.
Cell transfection
Cells were seeded in 6-well plates at a concentration of 2 × 105 cells/well. When cells reached 40–60% confluence, 150-nM MIR376A mimics or negative control (NC) was transfected using Lipofectamine™ 2000 transfection reagent (Invitrogen, USA) following the protocol recommended by the manufacturer. The miRNA mimic and NC were synthesized by Shanghai GenePharma Co. (Shanghai, China). Their sequences were as follows: 5′-UUCUCCGAAC GUGUCACGUT T-3′ (sense) and 5′-ACGUGACACG UUCGGAGAAT T-3′ (antisense) for NC and 5′-AUCAUAGAGG AAAAUCCACG U-3′ (sense) and 5′-GUGGAUUUUC CUCUAUGAUU U-3′ (antisense) for MIR376A mimics. After 48 h transfection, the cells were collected and used for further analyses.
MTT assay
Cell proliferation assay using the MTT kit (Promega Corporation, Madison, WI, USA) was performed according to the manufacturer's instructions. Briefly, cells were seeded into 96-well plates at a density of 5000 cells per well and grown for 24 hours. The cells were then transfected with 100 nM MIR376A mimics, MIR376A-NC, si-NC/si-MALAT1 or si-NC/si-TGFA. After 24 h transfection, 20 μL of 5 mg/mL MTT was added and further incubated for 4 h in a humidified incubator. 200 μL of DMSO was added to dissolve the formazan after supernatant removed. Optical density (OD) was measured at 490 nm.
BrdU incorporation assay
BrdU assays were performed to determine DNA synthesis at 24 h and 48 h after transfection of Saos2 and MG63 cells with designated constructs, miRNAs, or siRNAs. After transfection, cells were incubated with a final concentration of 10 μM BrdU (BD Pharmingen, San Diego, CA, USA) for 2 to 24 h, followed by fixation for 30 min after removing the medium. Cells were then incubated with peroxidase-coupled anti-BrdU-antibody (Sigma-Aldrich) for 60 min, washed with PBS and further incubated with peroxidase substrate (tetramethylbenzidine) for 30 min. Absorbance values were measured at 450 nm.
Western blot
RIPA buffer (Sigma-Aldrich, USA) was used to lyse cells with Complete Protease Inhibitor Cocktail (Roche, USA). Cell lysates were transferred to 1.5 mL tube and kept at −20°C before use. SDS-PAGE was conducted to separate the cellular proteins. Proteins were separated by 5% stacking gel and 10% running gel. The molecular weight of candidate proteins was referred to the Pre-stained SeeBlue rainbow marker (Invitrogen, USA) loaded in parallel. The following antibodies were used: MALAT1 (Santa Cruz, USA), TGFA (Abcam, MA, USA), and β-actin (Sigma, USA). Blots were detected using a Kodak film developer (Fujifilm, Japan).
RNA extraction and real-time PCR
Total RNA was extracted using TRIZOL reagent (Invitrogen, USA) following the manufacturer's instructions. A High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, USA) was used to reversely transcribe RNA samples. Quantitative RT-PCR was performed using the Fast Start Universal SYBR Green Mastermix (Roche, USA). Primers are shown in Table 1. The relative fold changes of candidate genes were analyzed using the 2−ΔΔCT method.
Table 1. The primers used for real-time PCR.
| Name | Sequences | |
|---|---|---|
| miR-376a | Forward | 5′- GTGCAGGGTCCGAGGT-3′ |
| Reverse | 5′- ATCATAGAGGAAAATCCACG -3′ | |
| MALAT1 | Forward | 5′-AAAGCAAGGTCTCCCCACAAG-3′ |
| Reverse | 5′-GGTCTGTGCTAGATCAAAAGGCA-3′ | |
| TGF-α | Forward | 5′-AGCTGCTAGCGCCTAGCGAT-3′ |
| Reverse | 5′-CCCGTCTGATAGCGCATTCGTGT-3′ | |
| GAPDH | Forward | 5′-AGAAGGCTGG GGCTCATTTG-3′ |
| Reverse | 5′-AGGGGCCATC CACAGTCTTC-3′ |
Luciferase reporter assays
The 3′-UTR or mutant 3′-UTR of MALAT1 containing the putative target site for MIR376A was chemically synthesized and inserted downstream of the luciferase gene in the internal control pRSV-β-Galactosidase vector. Saos2 cells cultured in 24-well plates were co-transfected with luciferase reporter plasmids and miRNA mimics as well as the internal control pRSV-β-Galactosidase vector. After transfection for 48 h, Saos2 cells were lysed with lysis buffer (25 mM Tris-phosphate, 1% Triton X-100, 1 mM DTT, 2 mM EDTA, 10% Glycerol, pH = 27.8). Cells were then collected and centrifuged at 14,000 rpm for 3 min, and the supernatant transferred to a new 1.5 μL tube. Luciferase reporter activity was monitored by mixing 50μL supernatant with 50 μL luciferase assay buffer using the Gloxmax 20/20 Luminometer (Promega). O-nitrophenyl-β-galactoside (ONPG) colorimetric assays were performed to measure the β-Galactosidase activity from the pRSV-β-Galactosidase vector, which was used for normalization of the luminescence levels. 50 μL supernatant from aforementioned cell extract was mixed with 100 μL of ONPG solution (0.666 mg/ml ONPG, 40 mM NaH2PO4, 60 mM Na2HPO4, 10 mM KCl, 1 mM MgSO4, 2% β-mercaptoethanol) and β-Galactosidase activity was measured using the ELISA plate reader (Bio-Rad, USA) at the wavelength of 490nm.
Statistical analysis
Experimental results are presented as mean ± SD. Comparisons between two groups were conducted using two-tailed Student's T-test and differences were considered to be statistically significant when the P value was less than 0.05.
ACKNOWLEDGMENTS AND FUNDING
This work was supported by the National Natural Science Foundation of China (No. 81401838 and No. 81402224), the Young Teacher's Boosting Project of the Fundamental Research Funds for the Central Universities in Central South University, China (Project number: 2012QNZT095), the Provincial Science Foundation of Hunan (No. 2015JJ3139), the Development and Reform Commission of Hunan Province ([2014]658–8), the NSFC grants, China (Nos. 31271399 and 81472047), and the innovation program of Jiangsu province, China (2013–480).
Footnotes
CONFLICTS OF INTEREST
None to declare.
REFERENCES
- 1.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martens-Uzunova ES, Böttcher R, Croce CM, Jenster G, Visakorpi T, Calin GA. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur Urol. 2014;65:1140–1151. doi: 10.1016/j.eururo.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 4.Di Gesualdo F, Capaccioli S, Lulli M. A pathophysiological view of the long non-coding RNA world. Oncotarget. 2014;5:10976–10996. doi: 10.18632/oncotarget.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malek E, Jagannathan S, Driscoll JJ. Correlation of long non-coding RNA expression with metastasis, drug resistance and clinical outcome in cancer. Oncotarget. 2014;5:8027–8038. doi: 10.18632/oncotarget.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhan A, Mandal SS. LncRNA HOTAIR: A master regulator of chromatin dynamics and cancer. Biochim Biophys Acta. 2015;1856:151–164. doi: 10.1016/j.bbcan.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Liu Z, Yao B, Dou C, Xu M, Xue Y, Ding L, Jia Y, Zhang H, Li Q, Tu K, Jiao Y, Liu Q, et al. Long non-coding RNA TUSC7 acts a molecular sponge for miR-10a and suppresses EMT in hepatocellular carcinoma. Tumour Biol. 2016;37:11429–41. doi: 10.1007/s13277-016-4892-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qi P, Xu MD, Shen XH, Ni SJ, Huang D, Tan C, Weng WW, Sheng WQ, Zhou XY, Du X. Reciprocal repression between TUSC7 and miR-23b in gastric cancer. Int J Cancer. 2015;137:1269–1278. doi: 10.1002/ijc.29516. [DOI] [PubMed] [Google Scholar]
- 10.Wang B, Su Y, Yang Q, Lv D, Zhang W, Tang K, Wang H, Zhang R, Liu Y. Overexpression of Long Non-Coding RNA HOTAIR Promotes Tumor Growth and Metastasis in Human Osteosarcoma. Mol Cells. 2015;38:432–440. doi: 10.14348/molcells.2015.2327. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Cong M, Li J, Jing R, Li Z. Long non-coding RNA tumor suppressor candidate 7 functions as a tumor suppressor and inhibits proliferation in osteosarcoma. Tumour Biol. 2016;37:9441–50. doi: 10.1007/s13277-015-4414-y. [DOI] [PubMed] [Google Scholar]
- 12.Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339:159–166. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 13.Li T, Mo X, Fu L, Xiao B, Guo J. Molecular mechanisms of long noncoding RNAs on gastric cancer. Oncotarget. 2016;7:8601–12. doi: 10.18632/oncotarget.6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai X, Liu Y, Yang W, Xia Y, Yang C, Yang S, Liu X. Long noncoding RNA MALAT1 as a potential therapeutic target in osteosarcoma. J Orthop Res. 2016;34:932–941. doi: 10.1002/jor.23105. [DOI] [PubMed] [Google Scholar]
- 15.Dong Y, Liang G, Yuan B, Yang C, Gao R, Zhou X. MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the PI3K/Akt pathway. Tumour Biol. 2015;36:1477–1486. doi: 10.1007/s13277-014-2631-4. [DOI] [PubMed] [Google Scholar]
- 16.Zheng Y, Yin L, Chen H, Yang S, Pan C, Lu S, Miao M, Jiao B. MiR-376a suppresses proliferation and induces apoptosis in hepatocellular carcinoma. FEBS Lett. 2012;586:2396–2403. doi: 10.1016/j.febslet.2012.05.054. [DOI] [PubMed] [Google Scholar]
- 17.Fellenberg J, Sähr H, Kunz P, Zhao Z, Liu L, Tichy D, Herr I. Restoration of miR-127–3p and miR376a-3p counteracts the neoplastic phenotype of giant cell tumor of bone derived stromal cells by targeting COA1, GLE1 and PDIA6. Cancer Lett. 2016;371:134–141. doi: 10.1016/j.canlet.2015.10.039. [DOI] [PubMed] [Google Scholar]
- 18.Jin Y, Peng D, Shen Y, Xu M, Liang Y, Xiao B, Lu J. MicroRNA-376c inhibits cell proliferation and invasion in osteosarcoma by targeting to transforming growth factor-alpha. DNA Cell Biol. 2013;32:302–309. doi: 10.1089/dna.2013.1977. [DOI] [PubMed] [Google Scholar]
- 19.Duan Z, Choy E, Harmon D, Liu X, Susa M, Mankin H, Hornicek F. MicroRNA-199a-3p is downregulated in human osteosarcoma and regulates cell proliferation and migration. Mol Cancer Ther. 2011;10:1337–1345. doi: 10.1158/1535-7163.MCT-11-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gutschner T, Hämmerle M, Diederichs S. MALAT1 — a paradigm for long noncoding RNA function in cancer. J Mol Med (Berl) 2013;91:791–801. doi: 10.1007/s00109-013-1028-y. [DOI] [PubMed] [Google Scholar]
- 21.Hirata H, Hinoda Y, Shahryari V, Deng G, Nakajima K, Tabatabai ZL, Ishii N, Dahiya R. Long Noncoding RNA MALAT1 Promotes Aggressive Renal Cell Carcinoma through Ezh2 and Interacts with miR-205. Cancer Res. 2015;75:1322–1331. doi: 10.1158/0008-5472.CAN-14-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tee AE, Liu B, Song R, Li J, Pasquier E, Cheung BB, Jiang C, Marshall GM, Haber M, Norris MD, Fletcher JI, Dinger ME, Liu T. The long noncoding RNA MALAT1 promotes tumor-driven angiogenesis by upregulating pro-angiogenic gene expression. Oncotarget. 2016;7:8663–75. doi: 10.18632/oncotarget.6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang MH, Hu ZY, Xu C, Xie LY, Wang XY, Chen SY, Li ZG. MALAT1 promotes colorectal cancer cell proliferation/migration/invasion via PRKA kinase anchor protein 9. Biochim Biophys Acta. 2015;1852:166–174. doi: 10.1016/j.bbadis.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zehavi L, Avraham R, Barzilai A, Bar-Ilan D, Navon R, Sidi Y, Avni D, Leibowitz-Amit R. Silencing of a large microRNA cluster on human chromosome 14q32 in melanoma: biological effects of mir-376a and mir-376c on insulin growth factor 1 receptor. Mol Cancer. 2012;11:44. doi: 10.1186/1476-4598-11-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choudhury Y, Tay FC, Lam DH, Sandanaraj E, Tang C, Ang BT, Wang S. Attenuated adenosine-to-inosine editing of microRNA-376a* promotes invasiveness of glioblastoma cells. J Clin Invest. 2012;122:4059–4076. doi: 10.1172/JCI62925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Formosa A, Markert EK, Lena AM, Italiano D, Finazzi-Agro’ E, Levine AJ, Bernardini S, Garabadgiu AV, Melino G, Candi E. MicroRNAs, miR-154, miR-299–5p, miR-376a, miR-376c, miR-377, miR-381, miR-487b, miR-485–3p, miR-495 and miR-654–3p, mapped to the 14q32. 31 locus, regulate proliferation, apoptosis, migration and invasion in metastatic prostate cancer cells. Oncogene. 2014;33:5173–5182. doi: 10.1038/onc.2013.451. [DOI] [PubMed] [Google Scholar]
- 27.Xiao H, Tang K, Liu P, Chen K, Hu J, Zeng J, Xiao W, Yu G, Yao W, Zhou H, Li H, Pan Y, Li A, et al. LncRNA MALAT1 functions as a competing endogenous RNA to regulate ZEB2 expression by sponging miR-200s in clear cell kidney carcinoma. Oncotarget. 2015;6:38005–38015. doi: 10.18632/oncotarget.5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, Li M, Wang Z, Han S, Tang X, Ge Y, Zhou L, Zhou C, Yuan Q, Yang M. Silencing of long noncoding RNA MALAT1 by miR-101 and miR-217 inhibits proliferation, migration, and invasion of esophageal squamous cell carcinoma cells. J Biol Chem. 2015;290:3925–3935. doi: 10.1074/jbc.M114.596866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang G, Zhang P, Lv A, Liu Y, Wang G. MiR-205 functions as a tumor suppressor via targeting TGF-a in osteosarcoma. Exp Mol Pathol. 2016;100:160–166. doi: 10.1016/j.yexmp.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 30.He P, Zhang Z, Huang G, Wang H, Xu D, Liao W, Kang Y. miR-141 modulates osteoblastic cell proliferation by regulating the target gene of lncRNA H19 and lncRNA H19-derived miR-675. Am J Transl Res. 2016;8:1780–1788. eCollection 2016. [PMC free article] [PubMed] [Google Scholar]
- 31.Fang D, Yang H, Lin J, Teng Y, Jiang Y, Chen J, Li Y. 17β-estradiol regulates cell proliferation, colony formation, migration, invasion and promotes apoptosis by upregulating miR-9 and thus degrades MALAT-1 in osteosarcoma cell MG-63 in an estrogen receptor-independent manner. Biochem Biophys Res Commun. 2015;457:500–506. doi: 10.1016/j.bbrc.2014.12.114. [DOI] [PubMed] [Google Scholar]