Abstract
In Taiwan, oral cancer is the fourth leading cancer in males and is associated with exposure to environmental carcinogens. WW domain-containing oxidoreductase (WWOX), a tumor suppressor gene, is associated with the development of various cancers. We hypothesized that genetic variants of WWOX influence the susceptibility to oral cancer. Five polymorphisms of WWOX gene from 761 male patients with oral cancer and 1199 male cancer-free individuals were genotyped. We observed that individuals carrying the polymorphic allele of WWOX rs11545028 are more susceptible to oral cancer. Furthermore, patients with advanced-stage oral cancer were associated with a higher frequency of WWOX rs11545028 polymorphisms with the variant genotype TT than did patients with the wild-type gene. An additional integrated in silico analysis confirmed that rs11545028 affects WWOX expression, which significantly correlates with tumor expression and subsequently with tumor development and aggressiveness. In conclusion, genetic variants of WWOX contribute to the occurrence of oral cancer, and the findings regarding these biomarkers provided a prediction model for risk assessment.
Keywords: single nucleotide polymorphism, WWOX, oral cancer
INTRODUCTION
Oral cancer, a common malignant disease affecting the head and neck region, has a poor prognosis. The incidence of oral cancer is high in Asia, particularly in Taiwan, and it is the fourth common cancer in males [1]. Furthermore, oral cancer was ranked the fifth most common malignancy in Taiwan, annually accounting for more than 2600 deaths in both sexes. Oral squamous cell carcinoma (OSCC) is the most common cancer, accounting for approximately 90% of all oral cancers [2]. Characterizing multiple genetic alterations in OSCC is a critical problem in understanding tumor development and its association with environmental factors including tobacco smoking, alcohol consumption, betel-quid chewing, chronic inflammation, and viral infection [3–6].
The WW domain-containing oxidoreductase (WWOX) gene, a tumor suppressor gene, is located on chromosome 16q23 and encompasses the common fragile site FRA16D [7]. WWOX, which encodes a 414-amino acid protein, possesses 2 N-terminal WW domains and a high homology domain of the short-chain dehydrogenase/reductase family [8–10]. WWOX is emerging as a tumor suppressor that is also involved in metabolic and neurological disorders [11], In vivo studies have indicated that the WWOX gene is alternatively knocked out in mice, causing Leydig cell development failure in the testis and affecting normal prostate function [12]. However, several studies have reported a loss or downregulation of the WWOX protein and homozygous deletion within the WWOX locus in multiple malignant neoplasms such as lung cancer, pancreatic adenocarcinoma, oral cancer, ovarian cancer, and renal cell carcinoma [13–21].
Growing evidence emphasizes the importance of genetic variations, which induce cancer by affecting the functions of oncogenes and tumor suppressor genes or enzyme metabolism. The expression of certain genes may be affected by single-nucleotide polymorphisms (SNPs), which are the most common types of DNA sequence variation. Moreover, previous studies have reported the effect of WWOX gene polymorphisms on human cancer susceptibility, and they have indicated that genotyping-related SNPs may efficiently predict the risk of cancers and other diseases [22–24]. Highly variable intronic and exonic polymorphisms were observed within WWOX in tumor cell lines [25]. In addition, studies have identified several SNPs in WWOX as potential risk factors for several cancers such as thyroid carcinomas, esophageal adenocarcinoma, pancreatic and ovarian cancer [22, 26–28]. Genome-wide scan analysis studies conducted on the rs1079635 which is in intron 7 of WWOX have also reported that this region demonstrated a strong association with prostate cancer susceptibility [29]. Nevertheless, although the effects of WWOX on functional analysis and phenotypic studies are adequately documented, the role of WWOX genetic polymorphism in the association between environmental carcinogens and OSCC and the clinicopathological characteristics of OSCC remain poorly investigated. In this study, we used a case-control study with 2 independent cohorts and analyzed 5 SNPs in WWOX in addition to investigating the associations between the SNPs and environmental factors. We further investigated the association between genetic factors and oral cancer clinicopathological characteristics.
RESULTS
Association between WWOX single nucleotide polymorphisms and OSCC
Table 2 shows the results of the statistical analysis of demographic characteristics. Significant differences were observed in the distribution of betel-quid chewing (p < 0.001), cigarette smoking (p < 0.001), and alcohol consumption (p < 0.001) between the controls and patients with OSCC. Table 3 shows genotype distributions and associations between oral cancer and WWOX gene polymorphisms. Alleles with the highest distribution frequency for rs11545028, rs12918952, rs3764340, rs73569323, and rs383362 polymorphisms of WWOX in both the controls and patients with OSCC were heterozygous for C/C, heterozygous for G/G, homozygous for C/C, homozygous for C/C, and homozygous for G/G, respectively. In these controls, the genotypic frequency of WWOX SNP rs11545028, rs12918952, rs3764340, rs73569323, and rs383362 were in the Hardy-Weinberg equilibrium (p=0.065, χ2 value: 3.402; p=0.093, χ2 value: 2.824; p=0.383, χ2 value: 0.759; p=0.489, χ2 value: 0.479 and p=0.066, χ2 value: 4.947, respectively). Furthermore, after adjustments for several variables, we observed no significant differences in the incidence rates of oral cancer in patients with rs12918952, rs3764340, rs73569323, and rs383362 polymorphisms of WWOX compared with those with the wild-type (WT) gene. However, oral cancer patients with the WWOX polymorphic rs11545028 T/T and combination of CT and TT genotypes exhibited a 1.824-fold (95% CI: 1.224-2.716) and 1.227-fold (95% CI: 1.022-1.473; both p < 0.05) higher risk of OSCC than did patients with the corresponding WT homozygous gene. To clarify the influence of the polymorphic WWOX genotypes on the clinicopathological status, such as TNM clinical staging, tumor size, lymph node involvement, and cell differentiation, the distribution frequency of clinical statuses and WWOX genotype frequencies in patients with oral cancer were estimated. Regarding the genotypic frequency of the SNPs, WWOX rs11545028 demonstrated significant associations with clinical pathological variables in patients with OSCC. The results form Table 4 shown that WWOX rs11545028 gene polymorphism is associated with clinical stage (p= 0.030) and lymph node metastasis (p = 0.010), but no difference was observed in tumor size and cell differentiation (Table 4).
Table 2. The distributions of demographical characteristics in 1199 male controls and 761 male patients with oral cancer.
| Variable | Controls (N=1199) | Patients (N=761) | p value |
|---|---|---|---|
| Age (yrs) | Mean ± S.D. | Mean ± S.D. | |
| 53.90 ± 10.02 | 54.63 ± 11.13 | p=0.133 | |
| Betel quid chewing | |||
| No | 1000 (83.4%) | 149(19.6%) | |
| Yes | 199(16.6%) | 612 (80.4%) | p < 0.001* |
| Cigarette smoking | |||
| No | 563 (47.0%) | 89(11.7%) | |
| Yes | 636 (53.0%) | 672 (88.3%) | p < 0.001* |
| Alcohol drinking | |||
| No | 962 (80.2%) | 327 (43.0%) | |
| Yes | 237 (19.8%) | 434 (57.0%) | p < 0.001* |
| Stage | |||
| I+II | 362 (47.6%) | ||
| III+IV | 399 (52.4%) | ||
| Tumor T status | |||
| T1+T2 | 425 (55.8%) | ||
| T3+T4 | 336 (44.2%) | ||
| Lymph node status | |||
| NO | 509 (66.9%) | ||
| N1+N2+N3 | 252 (33.1%) | ||
| Metastasis | |||
| MO | 752 (98.8%) | ||
| Ml | 9(1.2%) | ||
| Cell differentiation | |||
| Well differentiated | 117(15.4%) | ||
| Moderately or poorly differentiated | 644 (84.6%) |
Mann-Whitney U test or Fisher’s exact test was used between healthy controls and patients with oral cancer.
p value < 0.05 as statistically significant.
Table 3. Odds ratio (OR) and 95% confidence interval (CI) of oral cancer associated with WWOX genotypic frequencies.
| Variable | Controls (N=1199) n (%) | Patients (N=761) n (%) | OR (95% Cl) | p value |
|---|---|---|---|---|
| rsll545028 | ||||
| CC | 700 (58.4%) | 406 (53.4%) | 1.00 | |
| CT | 447 (37.3%) | 300 (39.4%) | 1.157 (0.956-1.400) | p=0.134 |
| TT | 52 (4.3%) | 55 (7.2%) | 1.824(1.224-2.716)* | p=0.003* |
| CT+TT | 499 (41.6%) | 355 (46.6%) | 1.227(1.022-1.473)* | p=0.029* |
| rsl2918952 | ||||
| GG | 1088 (90.7%) | 674 (88.6%) | 1.00 | |
| GA | 111 (9.3%) | 82(10.8%) | 1.193 (0.882-1.612) | p=0.252 |
| AA | 0 (0%) | 5 (0.6%) | --- | ---- |
| GA+AA | 111 (9.3%) | 87(11.4%) | 1.265 (0.940-1.702) | p=0.119 |
| rs3764340 | ||||
| CC | 1016 (84.7%) | 630 (82.8%) | 1.00 | |
| CG | 173 (14.4%) | 123 (16.2%) | 1.147 (0.892-1.475) | p=0.287 |
| GG | 10 (0.9%) | 8 (1.0%) | 1.290 (0.507-3.286) | p=0.593 |
| CG+GG | 183 (15.3%) | 131 (17.2%) | 1.154 (0.903-1.475) | p=0.251 |
| rs73569323 | ||||
| CC | 1152 (96.1%) | 740 (97.2%) | 1.00 | |
| CT | 47 (3.9%) | (2.7%) | 0.662 (0.389-1.127) | p=0.129 |
| TT | 0 (0%) | 1 (0.1%) | ---- | ---- |
| CT+TT | 47 (3.9%) | 21 (2.8%) | 0.696 (0.412-1.173) | p=0.171 |
| rs383362 | ||||
| GG | 887 (74.0%) | 558 (73.3%) | 1.00 | p=0.596 |
| GT | 299 (24.9%) | 199 (26.2%) | 1.058 (0.859-1.303) | p=0.213 |
| TT | 13 (1.1%) | 4 (0.5%) | 0.489 (0.159-1.508) | p=0.749 |
| GT+TT | 312 (26.0%) | 203 (26.7%) | 1.034 (0.842-1.271) |
The odds ratio (OR) with their 95% confidence intervals were estimated by logistic regression models.
p value < 0.05 as statistically significant.
Table 4. Odds ratio (OR) and 95% confidence intervals (CI) of clinical statuses associated with genotypic frequencies of WWOX rs11545028 in male oral cancer patients (n=761).
| Variable | OR (95% CI) | p value | ||
|---|---|---|---|---|
| Clinical Stage | ||||
| rsll545028 | Stage I+II | Stage III+IV | ||
| (n=362) (%) | (n=399) (%) | |||
| CC | 196 (54.1%) | 210(52.6%) | 1.00 | |
| CT | 148 (40.9%) | 152 (38.1%) | 0.947 (0.701-1.280) | p=0.781 |
| TT | 18(5.0%) | 37 (9.3%) | 1.919(1.057-3.482) | p=0.030* |
| Tumor size | ||||
| rsll545028 | ≦ T2 | >T2 | ||
| (n=425) (%) | (n=336) (%) | |||
| CC | 226 (53.2%) | 180 (53.6%) | 1.00 | |
| CT | 171 (40.2%) | 129 (38.4%) | 0.947 (0.701-1.280) | p=0.724 |
| TT | 28 (6.6%) | 27 (8.0%) | 1.211 (0.689-2.128) | p=0.506 |
| Lymph node metastasis | ||||
| rsll545028 | No | Yes | ||
| (n=509) (%) | (n=252) (%) | |||
| CC | 278 (54.6%) | 128 (50.8%) | 1.00 | |
| CT | 203 (39.9%) | 97 (38.5%) | 1.038 (0.754-1.429) | p=0.820 |
| TT | 28 (5.5%) | 27 (10.7%) | 2.0974(1.186-3.698) | p=0.010* |
| Cell differentiated grade | ||||
| rsll545028 | ≦ Grade I | >Grade I | ||
| (n=117) (%) | (n=644) (%) | |||
| CC | 59 (50.4%) | 347 (53.9%) | 1.00 | |
| CT | 48 (41.0%) | 252 (39.1%) | 0.893 (0.590-1.351) | p=0.591 |
| TT | 10 (8.5%) | 45 (7.0%) | 0.765 (0.366-1.602) | p=0.476 |
Cell differentiate grade:
grade I: well differentiated; grade II: moderately differentiated; grade III: poorly differentiated.
p value < 0.05 as statistically significant.
Functional analysis of the WWOX rs11545028 locus
We also investigated whether rs11545028 was associated with the differential expression of WWOX as a preliminary assessment of the putative functional role of the SNP. We obtained human WWOX from the NCBI gene database, selected its respective longest transcript, and defined its promoters as 1-kb upstream to 1-kb downstream of the predicted transcription start sites (Figure 1A). Moreover, we identified the putative functional role of rs11545028, as indicated by the functional annotations in the ENCODE data. We determined that rs11545028 was situated at a locus with TF binding, histone modification patterns, DNase hypersensitivity, and CpG islands that were characterized as promoters or enhancers in several cell types (Figure 1B). The effect of rs11545028 may be attributed to the suboptimal Kozak context surrounding the initiation codon of upstream open reading frames of human WWOX (Figure 1C), which enables the modulation of initiation rates in response to the translational status. In addition, the GTEx database revealed a statistically significant downregulation of WWOX mRNA expression in the whole blood, muscle skeletal and esophagus mucosa of rs11545028-variant genotypes (CT or TT) compared with that of the WT homozygous CC genotype (p = 0.011, p = 0.0016 and p = 0.027, respectively) (Figure 1D-1E).
Figure 1. Exon and intron structure of WWOX gene in human and the features of SNPs of WWOX gene (NM_016373.3), which were used to analyze in this study.
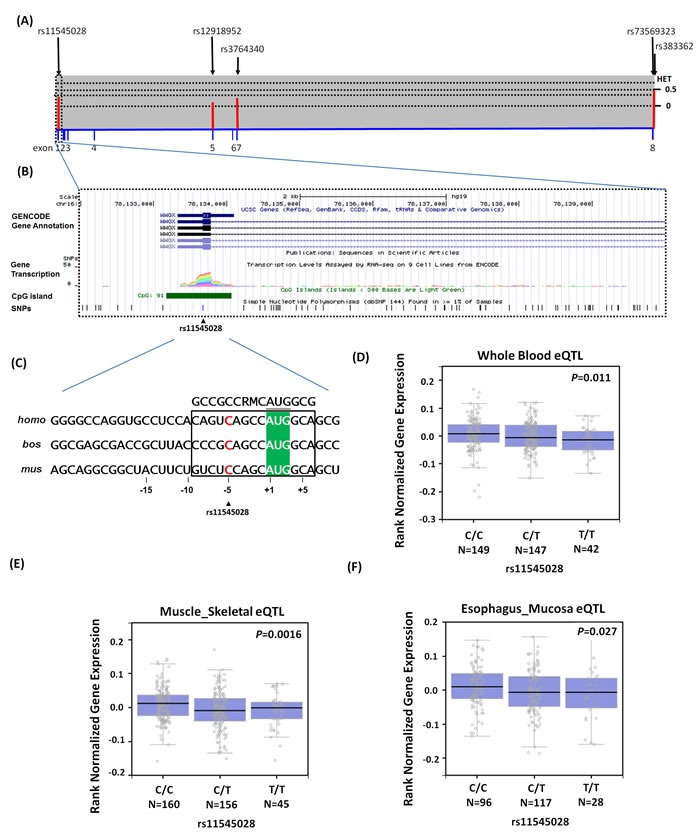
(A) Exons are shown by the filled blue boxes and are number 1 to 8 from the chromosome positions chr16:78,133,310 to 79,246,567 (reference genome GRCh37.p13). The lower panel shows population-specific heterozygosity frequencies of this polymorphism in East Asian population (HAPMAP-CHB); and the SNPs of WWOX gene are indicated by the black arrows and labeled reference SNP ID number. (B) Expanded view of the ENCODE data for the 5′UTR block containing the WWOX rs11545028 using UCSC genome browser on GRCh37/hg19 assembly. Chromatin State Segmentation track displays chromatin state segmentations by integrating ChIP-seq data using a Hidden Markov Model for H1 embryonic stem cells (color orange), HepG2 hepatocellular carcinoma cells (color green), HUVEC umbilical vein endothelial cells (color deep blue), HSMM (color blue), skeletal muscle myoblasts (color yellow), NHEK epidermal keratinocytes (color purple), and NHLF lung fibroblasts (color red). CpG islands are typically common near transcription start sites and may be associated with promoter regions. (C) Upstream open reading frames in WWOX transcripts of human (homo, NM_016373.3), cow (bos, NM_001078092), and mouse (mus, NM_019573.3) sequences shown in this alignment. Initiation codons of WWOX protein are highlighted by green background color. Consensus residues of the core Kozak context (residues at -9 or +3) are above this alignment in box, where M denotes A or C; R denotes A or G. (D-F) Expression quantitative trait locus association between rs11545028 and WWOX expression in (D) whole blood, (E) muscle skeletal and (F) esophagus mucosa (GTEx data set). Numbers in parentheses indicate the number of cases.
Functional analysis of the WWOX rs11545028 locus in clinical sample
To determine the functional effect of the rs11545028 polymorphism on WWOX expression, we generated luciferase reporter vectors with either the rs11545028 C allele or the rs11545028 T allele. We used these vectors for transfection of HSC-3, OECM-1 and SCC-9 oral cancer cells lines. As shown in Figure 2A, the vectors with the rs11545028 T allele had significantly lower luciferase activity compared to the vectors with the rs11545028 C allele among these three cell lines (p<0.05). Furthermore, to realize correlation between the mRNA, protein level of WWOX and rs11545028 polymorphism, quantitative real time-PCR (qPCR) and Immunohistochemical (IHC) staining were used to analyze WWOX mRNA and protein expression in cancer tissue of 34 and 51 OSCC patients, respectively. We found that OSCC patient who carry C/T or T/T of rs11545028 polymorphism have significantly lower mRNA levels of WWOX compare to C/C genotype (Figure 2B). Furthermore, when WWOX expression was classified into a two-tier grading system of weak (−/1+) (Figure 2C) and strong (2+/3+) (Figure 2D) WWOX staining, our analysis shown that specimens with rs11545028 C/C have higher WWOX expression, while specimens with rs11545028 C/T or T/T have lower WWOX expression (p=0.022) (Figure 2E). Overall, rs11545028 C to T substitution might affect the translational initiation and reduce WWOX mRNA and protein expression and the risk of oral cancer.
Figure 2. WWOX expression was correlated with rs11545028 genotypes in oral cancer patients.
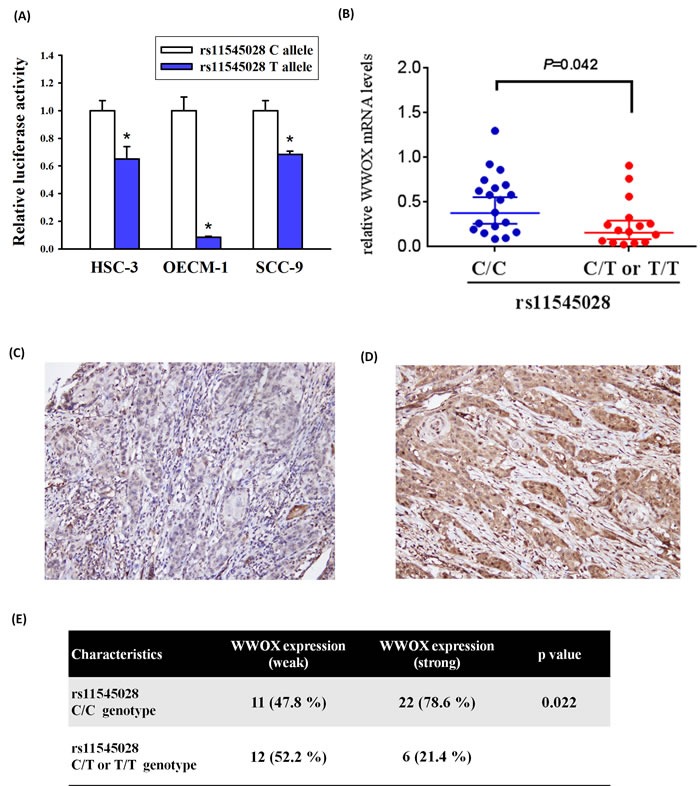
(A) Two luciferase reporter vectors with either the rs11545028 C allele or the rs11545028 T allele were transfected to HSC-3, OECM-1 and SCC-9 oral cancer cells lines. Data are mean values with standard deviation from at least three independent experiments. (B) WWOX mRNA expression in cancer tissue of 34 OSCC patients was analyzed by quantitative real time-PCR assay. Numbers in parentheses indicate the number of cases. (C) Weak cytoplasmic WWOX expression in OSCC. (D) Strong cytoplasmic WWOX expression in OSCC. (E) The OSCC specimens with rs11545028 C/C have higher WWOX expression, while specimens with rs11545028 C/T or T/T have lower WWOX expression (p=0.022).
DISCUSSION
Several studies have suggested that chromosome 16q23 contains a tumor suppressor gene involved in multiple tumor types. WWOX was mapped to this region, and the loss of function of WWOX in cancer cells was associated with mucinous histologies and a poor prognosis, suggesting that WWOX suppresses tumor progression [30]. Recent studies have reported that WWOX polymorphism is associated with the susceptibility to several carcinomas including lung, breast, bladder, colorectal, and pancreatic cancers [31, 32]. Five SNPs were included in a case-control study with 2 independent cohorts design. One of the SNPs (rs11545028) is located in exon 1 of WWOX. Our data reveal an increased risk of OSCC among patients with the WWOX polymorphic rs11545028 T/T compared with those with homozygous C/C. Only few studies have examined the functional role of rs11545028, and its functional importance has yet to be examined. An association of the risk of OSCC with the location of the analyzed variant is proposed.
In lung cancer, exonic polymorphisms within WWOX were revealed to exhibit a high incidence of the deletion of exon 6-8, which may result from amino acid changes and thus the loss of the tumor suppression function of WWOX. Furthermore, missense polymorphisms of WWOX, including Arg−314→His, Lys−182→Glu, Arg−120→Trp, and Thr−111→Ser, were detected in blood specimens from 15 and 34 patients with ovarian and colorectal cancers, respectively, but not in healthy participants [25]. By contrast, in our study, 2 of the missense SNPs located in exon 6-8 did not confer a risk of OSCC. In contrast to other tumors, the effect of missense polymorphisms in exon 6-8 did not alter the frequency of DNA strand breakage in OSCC, and this factor might be associated with the cancer type. Therefore, we suggested the presence of another major regulating mechanism associated with the downregulation of WWOX expression in OSCC.
Notably, we observed that the nonsense polymorphism rs11545028 C > T located in exon 1 conferred an increased risk of OSCC. Previous studies have reported that rs11545028 (C121T) in the data set indicated no significant difference between each tumor cell line and normal cell lines, even when the frequency of T/T in patients was lower. In this study, we determined whether the genetic variant rs11545028 C > T contributed to oral cancer susceptibility. Functional annotations from the ENCODE data indicate that rs11545028 is located in the region of an open chromatin, which probably corresponds to the promoters and CpG islands of WWOX. Several studies have documented the importance of transcriptional regulation between WWOX polymorphisms and cancer risk [23, 33]. Moreover, the common modifications of epigenetic changes in chromatin include DNA methylation, which has been considered a crucial mechanism underlying the inactivation of tumor suppressor genes as well as the loss of heterozygosity and mutation. An abnormal DNA methylation mainly occurs in the promoter region (CpG islands), which is associated with the transcriptional inactivation of tumor suppressor genes during tumor progression. The methylation rate of the WWOX promoter has been reported to be associated with the loss of WWOX expression in breast, lung, bladder, pancreatic, and prostate cancers [14, 32, 34]. The CpG methylation status in the WWOX promoter region was significantly higher in late-stage epithelial ovarian cancer tissues than in early stage epithelial ovarian cancer tissues [35]. In head and neck squamous cell carcinoma, it has also been reported that WWOX expression was decreased by miR-134 and promoter methylation [36, 37]. Liu et al. showed miR-134 expression contributes to head and neck carcinogenesis by targeting the WWOX [36]. Moreover, Ekizoglu et al. reported that decreased WWOX expression in advanced-stage tumor samples or in tumors with OSCC was associated with methylation of the WWOX promoter region [37]. Pimenta et al. also shown that the WWOX gene alteration is an early genetic alteration and may contribute to oral carcinogenesis [38]. Nevertheless, the frequency of the methylation of the WWOX promoter was not explained in our study, and the methylation rate must be examined further. Critical evidence indicates the importance of the methylation of the WWOX promoter. The highest methylation at the CpG site (approximately 60%) was observed in the promoter region (−328 to −41 bp) and exon 1 (−27 to +334 bp) of WWOX [39]. Furthermore, an aberrant methylation of these WWOX regions may occur at the early stage of cancer and, more precisely, at the advanced stage of esophageal squamous cell carcinoma, thus coinciding with the rs11545028 (C121T) region. This observation suggests that WWOX methylation is a critical event in the development of OSCC and that silencing through WWOX methylation is a pivotal mechanism underlying WWOX inactivation.
In a previous study, rs11545028 was predicted to lie within the Kozak translation initiation site, which comprises 6−8 nucleotides surrounding the initiation codon [40]. Studies conducted on the optimal Kozak sequence at positions −3 and +4 have proposed a valuable method for determining gene expression. However, the Kozak sequence has been increasingly demonstrated to be capable of altering translational machinery in response to the regulation of gene expression [41, 42]. A recent study revealed that an SNP located at position −1C/T in the Kozak sequence of CD40 was highly meaningful because the CD40expression levels were significantly higher in −1C/C carriers than in −1C/Tand −1T/T carriers [43]. Consistent with this observation, the less consensus Kozak sequence involving the nucleotide T at position −4 may markedly affect protein expression [44]. Figure 1C shows that the WWOX polymorphism rs11545028 at position −5 that involves the original consensus Kozak sequence contains the nucleotide C. Previous studies have demonstrated an association between the C allele of the Kozak polymorphism and gene expression both in vitro and in vivo. In the current study, the sequence containing the nucleotide C at position −5 more closely approximated the Kozak consensus, suggesting that the mRNA with the nucleotide T at 121 was associated with a markedly diminished efficiency. The GTEx database also revealed a significant drop in the WWOX mRNA expression in carriers of a genotype involving the variant T at rs11545028. In addition, we observed a high frequency of the homozygous 121 TT genotype and its combination with the heterozygous WWOX CT in patients, suggesting that changes in the translation initiation rate generally explains the differences in protein expression among the participants. The WWOX expression also suppresses tumor growth and induces cell apoptosis [45]. We observed that rs11545028 was associated with a higher risk of stage III and IV cancers, lymph node metastasis, and the cell differentiation grade. Overall, our findings suggest that the rs11545028 T allele reduced the translation initiation rate, which subsequently reduced the WWOX expression, thus contributing to a more aggressive phenotype in OSCC.
In conclusion, examining the complete medical information and conducting additional bioinformative analyses of a high number of patients provided comprehensive evidence of WWOX polymorphism in OSCC. Our results suggest that the WWOX polymorphic rs11545028 C/T in the suboptimal Kozak context is associated with clinical statuses and susceptibility to OSCC. The coeffects of WWOX polymorphism and environmental carcinogens markedly facilitate OSCC development. Overall, our analyses provide deeper insights into naturally occurring TIS variants. Comprehensive data on such types of variant are required for developing therapeutic approaches that can eventually ameliorate the clinical phenotype in patients harboring the corresponding lesions.
MATERIALS AND METHODS
Patient specimens
In 2007–2014, for the case group, we recruited 761 male patients at Chung Shan Medical University Hospital in Taichung and Changhua Christian Hospital in Changhua, Taiwan. For the control group, we randomly chose 1199 male non-cancer individuals from Taiwan Biobank and these control groups had neither self-reported history of cancer of any sites. For both groups, we administered a questionnaire to obtain information on their exposure to betel quid chewing, tobacco use, and alcohol consumption. Medical information of the patients, including TNM clinical staging, primary tumor size, lymph node involvement, and histologic grade, was obtained from their medical records. All participants provided written consent, and the Chung-Shan Medical University Hospital ethics committees approved the research protocol and informed consent was obtained from all subjects (CSMUH No: CS13214-1). All the methods applied in the study were carried out in accordance with the approved guidelines.
DNA extraction
DNA was extracted from buffy coats (white blood cells) using a QIAamp DNA blood mini kits (Qiagen, Valencia, California) as described in detail previously [46]. DNA was dissolved in TE buffer and used as the template in polymerase chain reactions
SNP selection and genotyping
In this study, the selection of 5 well-characterized common polymorphisms from WWOX gene is based on their wide associations with the development of cancer (Table 1, Figure 1A) [23–26, 33]. We included rs11545028 in the 5′UTR region. Rs12918952 and rs3764340, which are located in the exon of WWOX, were selected in this study since these 2 SNPs may result from amino acid changes and thus the loss of the tumor suppression function of WWOX [25]. The allelic discrimination of WWOX rs11545028, rs12918952, rs3764340, rs73569323, and rs383362 polymorphisms were assessed using an ABI StepOne TM Real-Time PCR System (Applied Biosystems, Foster City, CA) and analyzed using SDS v3.0 software (Applied Biosystems, Foster City, CA) as previously described [47].
Table 1. Variants, position, function, amino acid and changes of observed WWOX sequence variations.
| Exon (chromosome position ||) | |||||
|---|---|---|---|---|---|
| 1 (78,099,774) |
5 (78,386,878) |
7 (78,432,540) |
8 (79,211,868) |
8 (79,211,923) |
|
| nucleotide change | C>T | G>A | C>G | C>T | G>C |
| mRNA position † | 362 | 901 | 1,210 | 1,683 | 1,738 |
| Protein position ‡ | 179 | 282 | - | - | |
| domain | - | SDR domain | SDR domain | - | |
| dbSNP (rs number) | rsl 1545028 | rsl 2918952 | rs3 764340 | rs73569323 | rs383362 |
| Function | 5′UTR | Nonsynonymous | Nonsynonymous | 3′UTR | 3′UTR |
| dbSNP allele | - | GCA>ACA | CCA>GCA | - | - |
| Protein residue | - | Ala>Thr | Pro>Ala | - | - |
| Codon position | - | 1 | 1 | - | - |
| Heterozygous (%) † | 26.8 | 9.8 | 17.1 | - | 40.8 |
CRCh38.p2
HapMap-CHB
SDR domain: short-chain dehydrogenase/reductase domain
Construction of luciferase reporter plasmids
A luciferase reporter plasmid encompassing the major allele (C) and minor allele (T) of rs11545028 in the promoter region of the WWOX gene was cloned into the pGL3-Enhancer Luciferase Reporter Vectors (Promega Corp., Madison, WI, USA), according to manufacturer instructions. The vectors were sequenced to confirm the orientation and integrity.
Transient transfections and luciferase assay
HSC-3 cells were purchased by the Japanese Collection of Research Bioresources Cell Bank (JCRB, Shinjuku, Japan) [48]. SCC-9 cells were purchased from and validated by the American Type Culture Collection (ATCC, Manassas, VA, USA). Both cell lines maintained in DMEM/F12 supplemented with 10% FBS, 400 ng/ml hydrocortisone and 0.1 mM non-essential amino acids (NEAA; Life Technologies). OECM-1 cells were obtained from Dr Meng's group where the cell line is originally established and authenticated and maintained in RPMI (Gibco) supplemented with 10% FBS [49]. All the cells were cultured and maintained at 37 °C in a 5% CO2 and 95% air atmosphere. Each cell was seeded per well in a 24-well plate, and each well was transfected with 0.75 μg of the vector DNA containing either the rs11545028 C allele or the rs11545028 T allele by using the Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA), according to manufacturer instructions. Cells were collected 48 h after transfection and analyzed for luciferase activity by using the Luciferase Reporter Assay System (Promega, Madison, WI, USA). All transfections were performed in duplicate and repeated three times.
RNA preparation, TaqMan quantitative real-time PCR
Total RNA was isolated from oral cancer tissues using RNeasy Mini Kit (Qiagen, Valencia, CA, USA). Quantitative real-time PCR analysis was performed using TaqMan one-step PCR Master Mix (Applied Biosystems, Foster City, CA, USA). Total cDNA (2 μg) was added per 9 μl reactions with WWOX or GAPDH primers and TaqMan probes. The WWOX (Hs03044790_m1) and GAPDH (Hs99999905_m1) primers and probes were designed using commercial software (ABI PRISM Sequence Detection System; Applied Biosystems, Foster City, CA, USA) as previously described [50].
Immunohistochemistry
OSCC tissue microarray block slides were deparaffinised, as stated in our previous study [51]. The slides were incubated with 1:200 diluted anti-WWOX antibodies (Santa Cruz Biotechnology, Santa Cruz, CA, USA) for 60 min at room temperature. After thoroughly washing with PBS, the conventional streptavidin–biotin peroxidase method (LSAB Kit K675; Dako, Copenhagen, Denmark) using 3,3′-diaminobenzidine (DAB) was employed for assessing signal development. Two pathologists blinded to the clinical outcomes semiquantitatively assessed WWOX expression based on the staining intensity; they independently scored sections through light microscopy.
Statistical analysis
Mann–Whitney U-test and Fisher's exact test were used to compare the age differences and demographic characteristic distributions between the controls and patients with oral cancer. The odds ratio and 95% CIs of the association between the genotype frequencies and oral cancer risk and the clinical pathological characteristics were estimated using multiple logistic regression models. p < 0.05 was considered significant. The data were analyzed on SAS statistical software (Version 9.1, 2005; SAS Institute, Cary, NC).
Bioinformatics analysis
We used several semiautomated bioinformatics tools for assessing whether rs11545028 or its related genetic variants were associated with a putative function that might affect patient outcomes. HaploReg [52] v4 and Genotype-Tissue Expression (GTEx) [53] from the Encyclopedia of DNA Elements (ENCODE) [54] project were used for identifying the regulatory potential of candidate functional variants to examine factors of interest such as transcription factor (TF)–chromatin immunoprecipitation signals, DNase peaks, DNase footprints, and predicted DNA sequence motifs for TFs. The GTEx data were used for identifying the associations between the SNPs and whole blood-specific gene expression levels. Moreover, the publicly available cBioPortal for Cancer Genomics [55] and UCSC Cancer Genomics Browser [56] for hepatocellular adenocarcinomas were used for analyzing WWOX expression, DNA methylation, molecular features, and clinical outcomes.
Acknowledgments
This study was also supported by a research grant from Health and welfare surcharge of tobacco products, Ministry of Health and Welfare (MOHW103-TD-B-111-08; MOHW104-TDU-B-212-124-005 and MOHW105-TDU-B-212-134002).
Footnotes
CONFLICTS OF INTEREST
The authors declared no conflict of interest.
Author contributions
Yu-Fan Liu and Chiao-Wen Lin conceived and designed the experiments; Hsin-Lin Cheng, Shun-Fa Yang and Shih-Chi Su performed the experiments; Yu-Fan Liu, Chun-Wen Su and Shun-Fa Yang analyzed the data; Mu-Kuan Chen contributed samples; Hsin-Lin Cheng and Chiao-Wen Lin wrote the paper.
REFERENCES
- 1.Lin CW, Yang SF, Chuang CY, Lin HP, Hsin CH. Association of matrix metalloproteinase-11 polymorphisms with susceptibility and clinicopathologic characteristics for oral squamous cell carcinoma. Head Neck. 2015;37:1425–1431. doi: 10.1002/hed.23771. [DOI] [PubMed] [Google Scholar]
- 2.Chien MH, Lin CW, Cheng CW, Wen YC, Yang SF. Matrix metalloproteinase-2 as a target for head and neck cancer therapy. Expert Opin Ther Targets. 2013;17:203–216. doi: 10.1517/14728222.2013.740012. [DOI] [PubMed] [Google Scholar]
- 3.Califano J, van der Riet P, Westra W, Nawroz H, Clayman G, Piantadosi S, Corio R, Lee D, Greenberg B, Koch W, Sidransky D. Genetic progression model for head and neck cancer: implications for field cancerization. Cancer Res. 1996;56:2488–2492. [PubMed] [Google Scholar]
- 4.Nagaraj NS, Beckers S, Mensah JK, Waigel S, Vigneswaran N, Zacharias W. Cigarette smoke condensate induces cytochromes P450 and aldo-keto reductases in oral cancer cells. Toxicol Lett. 2006;165:182–194. doi: 10.1016/j.toxlet.2006.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biolchini F, Pollastri G, Figurelli S, Chiarini L. Carcinogen metabolism, DNA damage repair and oral head and neck squamocellular carcinoma (HNSCC). A review. Minerva Stomatol. 2005;54:405–414. [PubMed] [Google Scholar]
- 6.Yen CY, Liu SY, Chen CH, Tseng HF, Chuang LY, Yang CH, Lin YC, Wen CH, Chiang WF, Ho CH, Chen HC, Wang ST, et al. Combinational polymorphisms of four DNA repair genes XRCC1, XRCC2, XRCC3, and XRCC4 and their association with oral cancer in Taiwan. J Oral Pathol Med. 2008;37:271–277. doi: 10.1111/j.1600-0714.2007.00608.x. [DOI] [PubMed] [Google Scholar]
- 7.Aqeilan RI, Abu-Remaileh M, Abu-Odeh M. The common fragile site FRA16D gene product WWOX: roles in tumor suppression and genomic stability. Cell Mol Life Sci. 2014;71:4589–4599. doi: 10.1007/s00018-014-1724-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bednarek AK, Laflin KJ, Daniel RL, Liao Q, Hawkins KA, Aldaz CM. WWOX, a novel WW domain-containing protein mapping to human chromosome 16q23.3-24.1, a region frequently affected in breast cancer. Cancer Res. 2000;60:2140–2145. [PubMed] [Google Scholar]
- 9.Chang NS, Pratt N, Heath J, Schultz L, Sleve D, Carey GB, Zevotek N. Hyaluronidase induction of a WW domain-containing oxidoreductase that enhances tumor necrosis factor cytotoxicity. J Biol Chem. 2001;276:3361–3370. doi: 10.1074/jbc.M007140200. [DOI] [PubMed] [Google Scholar]
- 10.Ried K, Finnis M, Hobson L, Mangelsdorf M, Dayan S, Nancarrow JK, Woollatt E, Kremmidiotis G, Gardner A, Venter D, Baker E, Richards RI. Common chromosomal fragile site FRA16D sequence: identification of the FOR gene spanning FRA16D and homozygous deletions and translocation breakpoints in cancer cells. Hum Mol Genet. 2000;9:1651–1663. doi: 10.1093/hmg/9.11.1651. [DOI] [PubMed] [Google Scholar]
- 11.Abu-Remaileh M, Joy-Dodson E, Schueler-Furman O, Aqeilan RI. Pleiotropic Functions of Tumor Suppressor WWOX in Normal and Cancer Cells. J Biol Chem. 2015;290:30728–30735. doi: 10.1074/jbc.R115.676346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aqeilan RI, Hagan JP, de Bruin A, Rawahneh M, Salah Z, Gaudio E, Siddiqui H, Volinia S, Alder H, Lian JB, Stein GS, Croce CM. Targeted ablation of the WW domain-containing oxidoreductase tumor suppressor leads to impaired steroidogenesis. Endocrinology. 2009;150:1530–1535. doi: 10.1210/en.2008-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finnis M, Dayan S, Hobson L, Chenevix-Trench G, Friend K, Ried K, Venter D, Woollatt E, Baker E, Richards RI. Common chromosomal fragile site FRA16D mutation in cancer cells. Hum Mol Genet. 2005;14:1341–1349. doi: 10.1093/hmg/ddi144. [DOI] [PubMed] [Google Scholar]
- 14.Qin HR, Iliopoulos D, Semba S, Fabbri M, Druck T, Volinia S, Croce CM, Morrison CD, Klein RD, Huebner K. A role for the WWOX gene in prostate cancer. Cancer Res. 2006;66:6477–6481. doi: 10.1158/0008-5472.CAN-06-0956. [DOI] [PubMed] [Google Scholar]
- 15.Gourley C, Paige AJ, Taylor KJ, Scott D, Francis NJ, Rush R, Aldaz CM, Smyth JF, Gabra H. WWOX mRNA expression profile in epithelial ovarian cancer supports the role of WWOX variant 1 as a tumour suppressor, although the role of variant 4 remains unclear. Int J Oncol. 2005;26:1681–1689. doi: 10.3892/ijo.26.6.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donati V, Fontanini G, Dell'Omodarme M, Prati MC, Nuti S, Lucchi M, Mussi A, Fabbri M, Basolo F, Croce CM, Aqeilan RI. WWOX expression in different histologic types and subtypes of non-small cell lung cancer. Clin Cancer Res. 2007;13:884–891. doi: 10.1158/1078-0432.CCR-06-2016. [DOI] [PubMed] [Google Scholar]
- 17.Nunez MI, Rosen DG, Ludes-Meyers JH, Abba MC, Kil H, Page R, Klein-Szanto AJ, Godwin AK, Liu J, Mills GB, Aldaz CM. WWOX protein expression varies among ovarian carcinoma histotypes and correlates with less favorable outcome. BMC Cancer. 2005;5:64. doi: 10.1186/1471-2407-5-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pimenta FJ, Gomes DA, Perdigao PF, Barbosa AA, Romano-Silva MA, Gomez MV, Aldaz CM, De Marco L, Gomez RS. Characterization of the tumor suppressor gene WWOX in primary human oral squamous cell carcinomas. Int J Cancer. 2006;118:1154–1158. doi: 10.1002/ijc.21446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuroki T, Trapasso F, Shiraishi T, Alder H, Mimori K, Mori M, Croce CM. Genetic alterations of the tumor suppressor gene WWOX in esophageal squamous cell carcinoma. Cancer Res. 2002;62:2258–2260. [PubMed] [Google Scholar]
- 20.Lin JT, Tzai TS, Liao CY, Wang JS, Wu TT, Wang HY, Wu CH, Yu CC, Lu PJ. WWOX protein expression varies among RCC histotypes and downregulation of WWOX protein correlates with less-favorable prognosis in clear RCC. Ann Surg Oncol. 2013;20:193–199. doi: 10.1245/s10434-012-2371-x. [DOI] [PubMed] [Google Scholar]
- 21.Iliopoulos D, Guler G, Han SY, Druck T, Ottey M, McCorkell KA, Huebner K. Roles of FHIT and WWOX fragile genes in cancer. Cancer Lett. 2006;232:27–36. doi: 10.1016/j.canlet.2005.06.048. [DOI] [PubMed] [Google Scholar]
- 22.Schirmer MA, Luske CM, Roppel S, Schaudinn A, Zimmer C, Pfluger R, Haubrock M, Rapp J, Gungor C, Bockhorn M, Hackert T, Hank T, Strobel O, Werner J, Izbicki JR, Johnsen SA, Gaedcke J. Brockmoller J and Ghadimi BM. Relevance of Sp Binding Site Polymorphism in WWOX for Treatment Outcome in Pancreatic Cancer. J Natl Cancer Inst. 2016:108. doi: 10.1093/jnci/djv387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo W, Dong Z, Dong Y, Guo Y, Kuang G, Yang Z. Genetic and epigenetic alterations of WWOX in the development of gastric cardia adenocarcinoma. Environ Mol Mutagen. 2013;54:112–123. doi: 10.1002/em.21748. [DOI] [PubMed] [Google Scholar]
- 24.Yang L, Liu B, Huang B, Deng J, Li H, Yu B, Qiu F, Cheng M, Wang H, Yang R, Yang X, Zhou Y, Lu J. A functional copy number variation in the WWOX gene is associated with lung cancer risk in Chinese. Hum Mol Genet. 2013;22:1886–1894. doi: 10.1093/hmg/ddt019. [DOI] [PubMed] [Google Scholar]
- 25.Paige AJ, Taylor KJ, Taylor C, Hillier SG, Farrington S, Scott D, Porteous DJ, Smyth JF, Gabra H, Watson JE. WWOX: a candidate tumor suppressor gene involved in multiple tumor types. Proc Natl Acad Sci U S A. 2001;98:11417–11422. doi: 10.1073/pnas.191175898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cancemi L, Romei C, Bertocchi S, Tarrini G, Spitaleri I, Cipollini M, Landi D, Garritano S, Pellegrini G, Cristaudo A, Pinchera A, Barale R, Elisei R, Landi S, Gemignani F. Evidences that the polymorphism Pro-282-Ala within the tumor suppressor gene WWOX is a new risk factor for differentiated thyroid carcinoma. Int J Cancer. 2011;129:2816–2824. doi: 10.1002/ijc.25937. [DOI] [PubMed] [Google Scholar]
- 27.Gu J, Ajani JA, Hawk ET, Ye Y, Lee JH, Bhutani MS, Hofstetter WL, Swisher SG, Wang KK, Wu X. Genome-wide catalogue of chromosomal aberrations in barrett's esophagus and esophageal adenocarcinoma: a high-density single nucleotide polymorphism array analysis. Cancer Prev Res (Phila) 2010;3:1176–1186. doi: 10.1158/1940-6207.CAPR-09-0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paige AJ, Zucknick M, Janczar S, Paul J, Mein CA, Taylor KJ, Stewart M, Gourley C, Richardson S, Perren T, Ganesan TS, Smyth JF, Brown R, Gabra H. WWOX tumour suppressor gene polymorphisms and ovarian cancer pathology and prognosis. Eur J Cancer. 2010;46:818–825. doi: 10.1016/j.ejca.2009.12.021. [DOI] [PubMed] [Google Scholar]
- 29.Lange EM, Beebe-Dimmer JL, Ray AM, Zuhlke KA, Ellis J, Wang Y, Walters S, Cooney KA. Genome-wide linkage scan for prostate cancer susceptibility from the University of Michigan Prostate Cancer Genetics Project: suggestive evidence for linkage at 16q23. Prostate. 2009;69:385–391. doi: 10.1002/pros.20891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang M, Gu J, Wang Y, Gong B. Loss of WWOX expression in human extrahepatic cholangiocarcinoma. J Cancer Res Clin Oncol. 2009;135:39–44. doi: 10.1007/s00432-008-0449-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zelazowski MJ, Pluciennik E, Pasz-Walczak G, Potemski P, Kordek R, Bednarek AK. WWOX expression in colorectal cancer--a real-time quantitative RT-PCR study. Tumour Biol. 2011;32:551–560. doi: 10.1007/s13277-010-0150-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuroki T, Yendamuri S, Trapasso F, Matsuyama A, Aqeilan RI, Alder H, Rattan S, Cesari R, Nolli ML, Williams NN, Mori M, Kanematsu T, Croce CM. The tumor suppressor gene WWOX at FRA16D is involved in pancreatic carcinogenesis. Clin Cancer Res. 2004;10:2459–2465. doi: 10.1158/1078-0432.ccr-03-0096. [DOI] [PubMed] [Google Scholar]
- 33.Huang D, Qiu F, Yang L, Li Y, Cheng M, Wang H, Ma G, Wang Y, Hu M, Ji W, Zhou Y, Lu J. The polymorphisms and haplotypes of WWOX gene are associated with the risk of lung cancer in southern and eastern Chinese populations. Mol Carcinog. 2013;52(Suppl 1):E19–27. doi: 10.1002/mc.21934. [DOI] [PubMed] [Google Scholar]
- 34.Iliopoulos D, Guler G, Han SY, Johnston D, Druck T, McCorkell KA, Palazzo J, McCue PA, Baffa R, Huebner K. Fragile genes as biomarkers: epigenetic control of WWOX and FHIT in lung, breast and bladder cancer. Oncogene. 2005;24:1625–1633. doi: 10.1038/sj.onc.1208398. [DOI] [PubMed] [Google Scholar]
- 35.Yan H, Sun J. Methylation status of WWOX gene promoter CpG islands in epithelial ovarian cancer and its clinical significance. Biomed Rep. 2013;1:375–378. doi: 10.3892/br.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu CJ, Shen WG, Peng SY, Cheng HW, Kao SY, Lin SC, Chang KW. miR-134 induces oncogenicity and metastasis in head and neck carcinoma through targeting WWOX gene. Int J Cancer. 2014;134:811–821. doi: 10.1002/ijc.28358. [DOI] [PubMed] [Google Scholar]
- 37.Ekizoglu S, Bulut P, Karaman E, Kilic E, Buyru N. Epigenetic and genetic alterations affect the WWOX gene in head and neck squamous cell carcinoma. PLoS One. 2015;10:e0115353. doi: 10.1371/journal.pone.0115353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pimenta FJ, Cordeiro GT, Pimenta LG, Viana MB, Lopes J, Gomez MV, Aldaz CM, De Marco L, Gomez RS. Molecular alterations in the tumor suppressor gene WWOX in oral leukoplakias. Oral Oncol. 2008;44:753–758. doi: 10.1016/j.oraloncology.2007.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo W, Wang G, Dong Y, Guo Y, Kuang G, Dong Z. Decreased expression of WWOX in the development of esophageal squamous cell carcinoma. Mol Carcinog. 2013;52:265–274. doi: 10.1002/mc.21853. [DOI] [PubMed] [Google Scholar]
- 40.Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 41.Frank MB, Reiner AP, Schwartz SM, Kumar PN, Pearce RM, Arbogast PG, Longstreth WT, Jr, Rosendaal FR, Psaty BM, Siscovick DS. The Kozak sequence polymorphism of platelet glycoprotein Ibalpha and risk of nonfatal myocardial infarction and nonfatal stroke in young women. Blood. 2001;97:875–879. doi: 10.1182/blood.v97.4.875. [DOI] [PubMed] [Google Scholar]
- 42.Jacobson EM, Concepcion E, Oashi T, Tomer Y. A Graves’ disease-associated Kozak sequence single-nucleotide polymorphism enhances the efficiency of CD40 gene translation: a case for translational pathophysiology. Endocrinology. 2005;146:2684–2691. doi: 10.1210/en.2004-1617. [DOI] [PubMed] [Google Scholar]
- 43.Tian C, Qin W, Li L, Zheng W, Qiu F. A common polymorphism in CD40 Kozak sequence (-1C/T) is associated with acute coronary syndrome. Biomed Pharmacother. 2010;64:191–194. doi: 10.1016/j.biopha.2009.09.021. [DOI] [PubMed] [Google Scholar]
- 44.Kanaji T, Okamura T, Osaki K, Kuroiwa M, Shimoda K, Hamasaki N, Niho Y. A common genetic polymorphism (46 C to T substitution) in the 5′-untranslated region of the coagulation factor XII gene is associated with low translation efficiency and decrease in plasma factor XII level. Blood. 1998;91:2010–2014. [PubMed] [Google Scholar]
- 45.Wang M, Li Y, Wu M, Wang W, Gong B, Wang Y. WWOX suppresses cell growth and induces cell apoptosis via inhibition of P38 nuclear translocation in cholangiocarcinoma. Cell Physiol Biochem. 2014;34:1711–1722. doi: 10.1159/000366372. [DOI] [PubMed] [Google Scholar]
- 46.Su SC, Hsieh MJ, Liu YF, Chou YE, Lin CW, Yang SF. ADAMTS14 Gene Polymorphism and Environmental Risk in the Development of Oral Cancer. PLoS One. 2016;11:e0159585. doi: 10.1371/journal.pone.0159585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang SF, Yeh CB, Chou YE, Lee HL, Liu YF. Serpin peptidase inhibitor (SERPINB5) haplotypes are associated with susceptibility to hepatocellular carcinoma. Sci Rep. 2016;6:26605. doi: 10.1038/srep26605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yeh CM, Lin CW, Yang JS, Yang WE, Su SC, Yang SF. Melatonin inhibits TPA-induced oral cancer cell migration by suppressing matrix metalloproteinase-9 activation through the histone acetylation. Oncotarget. 2016;7:21952–21967. doi: 10.18632/oncotarget.8009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin CW, Yang WE, Lee WJ, Hua KT, Hsieh FK, Hsiao M, Chen CC, Chow JM, Chen MK, Yang SF, Chien MH. Lipocalin 2 prevents oral cancer metastasis through carbonic anhydrase IX inhibition and is associated with favourable prognosis. Carcinogenesis. 2016;37:712–722. doi: 10.1093/carcin/bgw050. [DOI] [PubMed] [Google Scholar]
- 50.Cheng HL, Lin CW, Yang JS, Hsieh MJ, Yang SF, Lu KH. Zoledronate blocks geranylgeranylation not farnesylation to suppress human osteosarcoma U2OS cells metastasis by EMT via Rho A activation and FAK-inhibited JNK and p38 pathways. Oncotarget. 2016;7:9742–9758. doi: 10.18632/oncotarget.7138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang JS, Lin CW, Chuang CY, Su SC, Lin SH, Yang SF. Carbonic anhydrase IX overexpression regulates the migration and progression in oral squamous cell carcinoma. Tumour Biol. 2015;36:9517–9524. doi: 10.1007/s13277-015-3692-8. [DOI] [PubMed] [Google Scholar]
- 52.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pazin MJ. Using the ENCODE Resource for Functional Annotation of Genetic Variants. Cold Spring Harb Protoc. 2015;2015:522–536. doi: 10.1101/pdb.top084988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goldman M, Craft B, Swatloski T, Cline M, Morozova O, Diekhans M, Haussler D, Zhu J. The UCSC Cancer Genomics Browser: update 2015. Nucleic Acids Res. 2015;43:D812–817. doi: 10.1093/nar/gku1073. [DOI] [PMC free article] [PubMed] [Google Scholar]


