Abstract
The highly variable pharmacokinetics and narrow therapeutic window of tacrolimus (TAC) has hampered its clinical use. Genetic polymorphisms may contribute to the variable response, but the evidence is not compelling, and the explanation is unclear. In this study we attempted to find previously unknown genetic factors that may influence the TAC dose requirements. The association of 105 pathway-related single nucleotide polymorphisms (SNPs) with TAC dose-adjusted concentrations (C0/D) was examined at 7, 30 and 90 d post-operation in 382 Chinese kidney transplant recipients. In CYP3A5 non-expressers, the patients carrying the IL-3 rs181781 AA genotype showed a significantly higher TAC logC0/D than those with the AG genotype at 30 and 90 d post-operation (AA vs AG, 2.21±0.06 vs 2.01±0.03, P=0.004; and 2.17±0.06 vs 2.03±0.03, P=0.033, respectively), and than those with the GG genotype at 30 d (AA vs GG, 2.21±0.06 vs 2.04±0.03, P =0.011). At 30 d, the TAC logC0/D in the grouped AG+GG genotypes of CTLA4 rs4553808 was significantly lower than that in the AA genotype (P =0.041) in CYP3A5 expressers, but it was higher (P=0.008) in the non-expressers. We further validated the influence of CYP3A5 rs776746, CYP3A4 rs2242480 and rs4646437 on the TAC C0/D; other candidate SNPs were not associated with the differences in TAC C0/D. In conclusion, genetic polymorphisms in the immune genes IL-3 rs181781 and CTLA4 rs4553808 may influence the TAC C0/D. They may, together with CYP3A5 rs776746, CYP3A4 rs2242480 and rs4646437, contribute to the variation in TAC dose requirements. When conducting individualized therapy with tacrolimus, these genetic factors should be taken into account.
Keywords: tacrolimus, Chinese kidney transplant recipients, individualized therapy, dose-adjusted concentration, gene polymorphism, CYP, IL-3, CTLA4
Introduction
Tacrolimus (FK506, TAC) is a calcineurin inhibitor that has been widely used in modern immunosuppressive therapy for solid organ transplantation since it was approved in 19941,2. Its efficacy has been confirmed through clinical trials and clinical practice. However, TAC is characterized by an oral bioavailability varying from 4% to 89% (mean approximately 25%) and a narrow therapeutic index3. In addition, TAC doses are poorly correlated with the concentrations in whole blood, and large inter-individual differences exist in the TAC dose required to achieve target blood concentrations1,2,4,5. It was reported in a study of Chinese renal transplant recipients that the maximum area under the curve (AUCmax) was almost 4 times higher than the minimum (AUCmin) after the first oral dose6,7. Therefore, it is very likely that a patient can receive a dose that is either too high or too low, resulting in an increased risk of graft rejection, graft loss, and reduced survival as well as an increased risk of infections and malignancies8. Although therapeutic drug monitoring (TDM) can reduce this risk, it is actually a trial-and-error approach and has no predictive value for the initial TAC dose8. Pharmacogenomics may be helpful in predicting the appropriate TAC dose to use after transplantation, one that will shorten the time to reach a stable drug dose.
Some clinical factors and genetic factors play critical roles in determining TAC pharmacokinetics (PK)9,10,11. Tacrolimus is known to be transported by P-glycoprotein (MDR1 or ABCB1) and metabolized by cytochrome P450 (CYP) 3A4 and CYP3A5. TAC forms a complex with the immunophilin FK-binding protein 12 (FKBP12) and strongly inhibits calcineurin phosphatase activity, IL-2 expression, and subsequently, T-cell activation7. Genetic polymorphisms in TAC pharmacokinetic pathways have been widely investigated (www.pharmgkb.org), and it has been confirmed that a single-nucleotide polymorphism, 6986A>G, within intron 3 of the CYP3A5 gene can affect the pharmacokinetics of tacrolimus5,12. Recent studies have also shown that TAC pharmacokinetics can be affected by polymorphisms in CYP3A4 (*22, *B, *1G), ABCB1 (2677G>A/T, 1236C>T, 3435C>T), P450 (cytochrome) oxidoreductase (POR) *28, nuclear receptor subfamily 1, group I, member 2 (NR1I2), catechol-O-methyltransferase (COMT), Toll-like receptor 4 (TLR4), and IL-1813,14,15,16,17,18,19,20. However, these conclusions are controversial. There are also reports that polymorphisms in genes that participate in the pharmacodynamic (PD) pathway, including IL-2, IL-6, and IL-18, and genes involved in the immune system, for instance TLR4, can influence TAC pharmacokinetics20,21,22. The existing algorithms, including genetic and clinical factors, for TAC dose prediction only account for 40% to 60% of TAC dose variations9,23. The rest of the variations may be caused by unknown genetic factors that are involved in TAC pharmacodynamics, the immune system, or other related pathways. Our study includes common SNPs involved in these pathways, including rs776746 (ie, *3 or 6986A>G) in CYP3A5, rs2242480 (ie, *1G or 20230 C>T) and rs4646437 in CYP3A4, rs181781 in IL-3, and rs4553808 in the cytotoxic T lymphocyte antigen-4 (CTLA4) gene. Because the C/D ratio (the ratio of the lowest concentration of the drug in the blood to the corresponding daily TAC dose) is a predictor of the TAC metabolism rate24, we used the C/D ratio as an index of tacrolimus pharmacokinetics, with the aim of finding previously unknown genetic factors that influence the TAC dose requirement and that may be useful for individualized therapy with tacrolimus.
Materials and methods
Ethics statement
This study was approved by the ethics committee of the Institute of Clinical Pharmacology, Central South University (CTXY-120030-2), and it has been registered at http://www.chictr.org (ChiCTR-RNC-12002894). Written informed consent was obtained from all participants. All allogeneic kidney grafts were from voluntary living donors or from donors after cardiac death (DCD) obtained from 2010 to 2014.
Study population
All stable tacrolimus-treated kidney transplant recipients in the third Xiangya Hospital and the Peking University Third Hospital from 2010 to 2014 were recruited for this retrospective study. We collected routine TDM (whole-blood C0), demographic and clinical data from the Chinese Scientific Registry of Kidney Transplantation (http://www.csrkt.org/). We selected eligible patients using the following inclusion criteria: (1) patients experiencing kidney transplantation for the first time, (2) patients taking TAC as the main immunosuppressive drug for a period of 3 months or longer post operation, and (3) patients volunteering to participate in this study. The exclusion criteria were (1) patients who changed TAC for cyclosporine or other immunosuppressive agent within 3 months post operation, (2) patients for whom important clinical data, eg, weight, TAC doses or blood concentrations, were missing, (3) patients who experienced graft failure or death after the operation, (4) patients for whom the quality of extracted DNA samples did not meet the MassARRAY sequence requirements. In total, 382 Chinese kidney transplant patients were included (for patient selection see Supplementary Figure S1), and all of them were followed for more than 3 months. The characteristics of the patients are given in Table 1. According to the therapeutic regimen, all patients were treated with tacrolimus combined with mycophenolic acid administered as its prodrug, mycophenolate mofetil, and a low dose of methylprednisolone. All patients received induction therapy by either antithymocyte globulin or daclizumab based on the patient risk for rejection. A tacrolimus loading dose of 0.10 mg/kg every 12 h was begun on the day of transplantation, and the tacrolimus dose was subsequently adjusted to achieve low predose blood concentrations of 6–12 ng/mL for the first 3 months. Tacrolimus doses were adjusted according to TDM.
Table 1. Characteristics of patients.
| Variable | Value |
|---|---|
| Age (year), range (mean±SD) | 10–70 (36.4±10.3) |
| Gender, n (%) | |
| Male | 247 (64.8%) |
| Female | 134 (35.2) |
| Weight (kg), range (mean±SD) | 33–92 (58.4±10.0) |
| BMI, (kg/m2), range (mean±SD) | 14.8–38.3 (21.2±3.6) |
| Hypertension, n (%) | |
| Yes | 269 (70.8%) |
| No | 111 (29.2) |
| TAC initial dose (mg/d), range (mean±SD) | 1.8–16.4 (5.2±1.8) |
| TAC initial C (ng/mL), range (mean±SD) | 0.9–21.1 (7.2±13.7) |
| GC dose (mg/d), range (mean±SD) | 2.0–300.0 (21.8±24.3) |
| MMF dose (mg/d), range (mean±SD) | 250–2500 (1272.9±332.6) |
| GLU (mmol/L), range (mean±SD) | 3.5–12.6 (5.6±1.1) |
| BUN (mmol/L), range (mean±SD) | 2.5–33.7 (7.2±4.3) |
| ALT (IU/L), range (mean±SD) | 1.0–98 (22.5±14.9) |
| AST (IU/L), range (mean±SD) | 5.0–54 (18.6±8.5) |
| Hb (g/L), range (mean±SD) | 38.2–196 (127.1±22.0) |
| TP (g/L), range (mean±SD) | 50.7–107.0 (71.2±8.5) |
Abbreviations: ALT, alanine aminotransferase; AST, Aspartate transaminase; BMI, body mass index; BUN, urea nitrogen; C, concentration; GC, Glucocorticoid; GLU, glucose; MMF, mycophenolate mofetil; Hb, hemoglobin; SD, standard deviation; TAC, Tacrolimus; TP, total protein
Study design
We conducted a retrospective study to investigate the impact of pathway related SNPs on tacrolimus dose requirements during a 3 month post-transplantation period. The C/D ratio (ie, dose-adjusted drug C0) as an index of tacrolimus pharmacokinetics was used as an outcome variable. This ratio, defined as the drug C0 concentration per daily weight-adjusted drug dose [(ng/mL)/(mg·kg−1·d−1)], has commonly been used in other studies25. From 64 candidate genes, 105 SNPs (Supplementary SNP distributions) selected by a review of the published literature were genotyped in a cohort of 382 Chinese kidney transplant recipients who received similar therapy regimes. Doses and C0 values at 7, 30, and 90 d were collected and analyzed. Other clinical data, including gender, age, and body mass index, were also analyzed.
Genotyping analysis
Qualified 2 mL blood samples were collected from each patient. Genomic DNA was isolated from whole blood samples using Wizard Genomic DNA Purification Kits (Promega) and stored at -80 °C for analysis. The DNA concentration, purity, and degree of degradation were detected by agarose gel electrophoresis, and the OD260/OD280 values were verified by nucleic acid and protein assays. Participants were genotyped for rs776746 (CYP3A5 *3, 6986A>G), rs2242480 (CYP3A4 *1G, 20230C>T), CYP3A4 rs4646437, IL-3 rs181781, CTLA4 rs4553808 and other candidate SNPs by using the MassARRAY Compact System.
Statistical analysis
The data were analyzed using the IBM SPSS 20.0 statistical software. The data are expressed as the mean±SE (standard error) except when stated otherwise. The outcome variable C0/D at 7, 30, and 90 d post-transplantation were normalized by a logarithmic transformation and were confirmed to be not significantly different from a normal distribution by the Kolmogorov-Smirnov test. The general linear model was used to assess the statistical significance of differences in the logC0/D among different genotypes. Multiple comparison correction by age and gender was appropriately applied. The Hardy-Weinberg equilibrium test was performed using the appropriate 2 test. Pairwise r2 and D' values for linkage disequilibrium were calculated using the online web tool SHEsis software (http://analysis2.biox.cn/myAnalysis.php). Groups were compared using the general linear model ANOVA or the Wilcoxon-Mann-Whitney test for continuous variables and χ2 or Fisher's exact test for categorical variables. The null hypothesis was rejected with a P value <0.05.
Results
Clinical characteristics of recipients
A total of 382 kidney transplant recipients (135 from Peking and 247 from Xiangya) were included in our retrospective study (Supplementary Figure S1). The collected clinical characteristics of all the recipients are listed in Table 1; 64.8% of the recipients were males. The average age was 36.4±10.3 years, and the average weight was 58.4±10.0 kg. The mean TAC initial dose among patients was 5.2±1.8 mg/d (range 3–16 mg/d).
Genotypes
Among the 105 candidate SNPs, we found only 6 SNPs (CYP3A5 rs776746, CYP3A4 rs2242480 and rs4646437, IL-3 rs181781, CTLA4 rs4553808 and ATP1B2 rs1641536) that were significantly associated with TAC logC0/D after the data were adjusted by age and gender. All the 6 SNPs were in Hardy-Weinberg equilibrium (Table 2). There were no significant clinical data variations among the different genotypes of CYP3A4 rs2242480, rs4646437, IL-3 rs181781 and ATP1B2 rs1641536 (Supplementary Table S1, S2, S3, S4). Although the daily doses of mycophenolate mofetil (MMF) at 1 week and 1 month showed differences, they were not significant (Supplementary Table S5). The glucose concentrations at 1 week in the different genotypes of CYP3A5 rs776746 and the urea nitrogen (BUN) in the different genotypes of CTLA4 rs4553808 were significantly different (Supplementary Tables S5, S6). To exclude the influence of the clinical variations, we adjusted them as the covariates in the following analysis. No significant differences were observed in the clinical characteristics of CYP3A5 expressers (rs776746 AA+AG) and non-expressers (rs776746 GG) (Supplementary Table S7).
Table 2. Allelic frequency of recipients according to genotypes.
| Gene | SNP | Genotype | Allelic frequency (%) | P value* | ||
|---|---|---|---|---|---|---|
| CYP3A5 | Rs776746 | AA (n=24) | AG (n=155) | GG (n=192) | A/G (27.4/72.6) | 0.325 |
| ATP1B2 | Rs1641536 | AA (n=42) | AG (n=173) | GG (n=165) | A/G (33.8/66.2) | 0.739 |
| CYP3A4 | Rs2242480 | TT (n=23) | CT (n=160) | CC (n=196) | T/C (27.2/72.8) | 0.730 |
| CYP3A4 | Rs4646437 | TT (n=8) | CT (n=102) | CC (n=270) | T/C (15.5/84.5) | 0.650 |
| IL-3 | Rs181781 | GG (n=155) | AG (n=177) | AA (n=47) | G/A (64.2/35.8) | 0.747 |
| CTLA4 | Rs4553808 | AA (n=236) | AG (n=106) | GG (n=9) | A/G (82.3/17.7) | 0.473 |
*Hardy-Weinberg equilibrium (HWE) Chi-squared test.
The association of IL-3 rs181781, CTLA4 rs4553808 and ATP1B2 rs1641536 with the TAC logC0/D
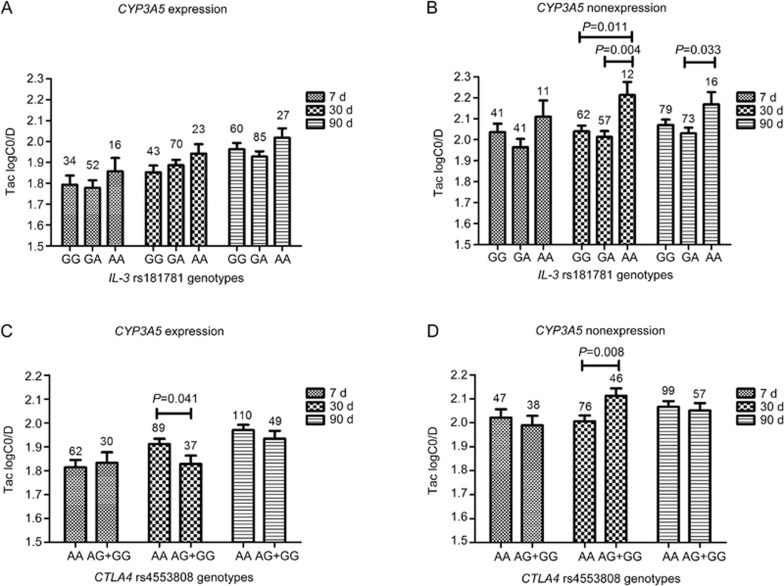
The IL-3 and CTLA4 genes are two major components of the immune system. Our results showed that both IL-3 rs181781 and CTLA4 rs4553808 had significant influences on the TAC logC0/D at 3 months post-transplantation with adjusted P values of 0.033 and 0.012, respectively (Supplementary Figure S2). Considering that CYP3A5 rs776746 is one of the most influential genetic factors and that it showed a strong influence (P value <0.001) at different time points (Figure 1) in our study, we then conducted an analysis stratified by CYP3A5 rs776746 mutations of the influence of IL-3 rs181781 and CTLA4 rs4553808. We classified the patients into CYP3A5 expressers (rs776746 AA+AG genotypes) and CYP3A5 non-expressers (rs776746 GG genotype). No significant influence of IL-3 rs181781 was observed on the TAC logC0/D in the CYP3A5 expression group, whereas among the CYP3A5 non-expressers, the IL-3 rs181781 AA genotype showed a higher TAC logC0/D than the AG genotype at 30 and 90 d (AA vs AG, 2.21±0.06 vs 2.01±0.03, P=0.004; and 2.17±0.06 vs 2.03±0.03, P=0.033, respectively), and the GG genotype at 30 d (AA vs GG, 2.21±0.06 vs 2.04±0.03, P=0.011) post-transplantation (Figure 2). For CTLA4 rs4553808 (-1661 A>G), however, the frequency of the G allele was lower, and it has been reported that the functional effect of CTLA4 can be attributed to the -1661G allele26. In our study, considering the restriction of the small number (n=9) of GG genotypes among the patients, we grouped the G allele carriers (GG+AG genotypes) when conducting the stratified analysis with the CYP3A5 rs776746 mutations. The results of our study showed that the TAC logC0/D among the AG+GG genotypes was lower in CYP3A5 expressers (AG+GG vs AA, 1.83±0.04 vs 1.91±0.02, P=0.041) at 30 d, whereas in non-expressers, it was higher (AG+GG vs AA, 2.11±0.03 vs 2.01±0.02, P=0.008). However, the TAC logC0/D showed no difference at 7 or 90 d post-operation (Figure 2). When we analyzed the influence of ATP1B2 rs1641536, however, although it showed an influence on the TAC logC0/D at 7 d post-operation (Supplementary Figure S2), after validation by the CYP3A5 rs776746 stratified analysis, the effect did not exist in the CYP3A5 expressers or in the non-expressers.
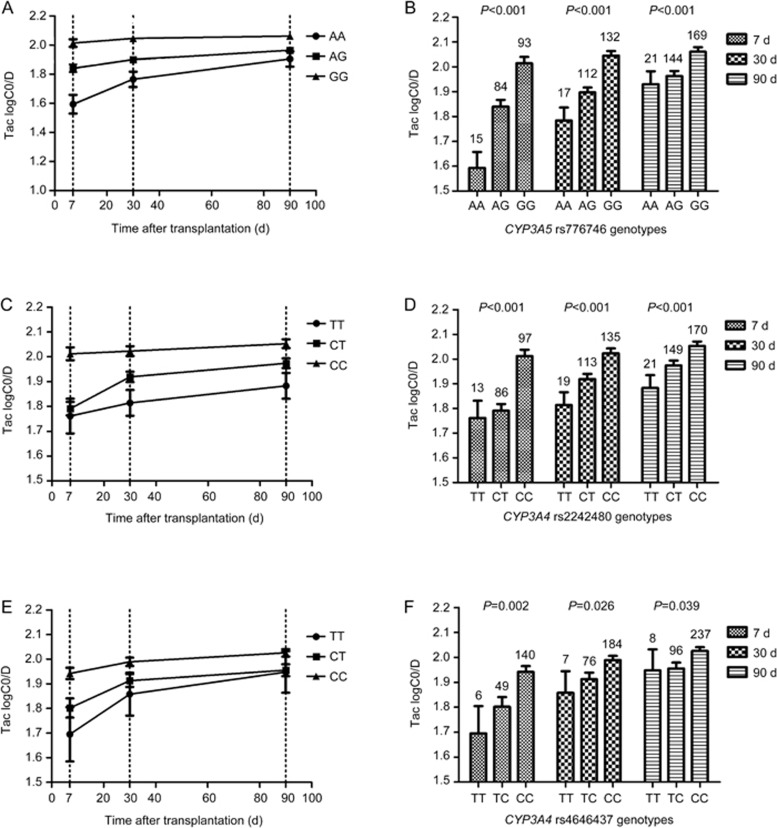
Figure 1.
Effects of CYP3A5 rs776746, CYP3A4 rs2242480 and rs4646437 genetic polymorphisms on the TAC C0/D. Different genotypes of CYP3A5 rs776746 (A, B), CYP3A4 rs2242480 (C, D), and CYP3A4 rs4646437 (E, F) were found to be associated with TAC logC0/D variations at 7, 30, 90 d post transplantation after the data were adjusted for age, gender, glucose concentrations, and BUN.
Figure 2.
The influence of IL-3 rs181781 and CTLA4 rs4553808 on TAC logC0/D in CYP3A5 expressers and non-expressers. (A and B) represent the association of IL3 rs181781 polymorphisms with TAC logC0/D in CYP3A5 expressers and non-expressers, respectively; (C and D) represent the association of CTLA4 rs4553808 polymorphisms with TAC logC0/D in CYP3A5 expressers and non-expressers, respectively.
The association of CYP3A5 rs776746, CYP3A4 rs2242480 and rs4646437 with the TAC logC0/D
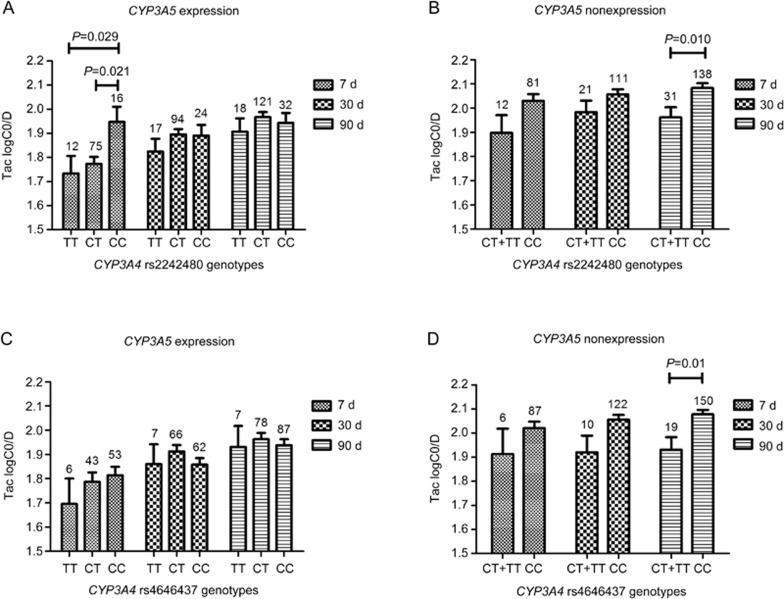
CYP3A5 and CYP3A4 are the two main metabolic enzymes of tacrolimus. We found that CYP3A5 rs776746 and CYP3A4 rs2242480 and rs4646437 were associated with the TAC logC0/D after being adjusted by clinical data (including age and gender) at 7, 30, and 90 d post-transplantation. CYP3A5 rs776746 showed the strongest influence (P value <0.001) at different time points (Figure 1). In the stratified analysis for the influence of CYP3A4 rs2242480 and rs4646437, we found that in the CYP3A5 expression group, a significant difference was observed in the TAC logC0/D of different genotypes of rs2242480 at 7 d post-transplantation. The TAC logC0/D of patients carrying the CC genotype was significantly higher than that of patients with the CT genotype (CC vs CT, 1.95±0.06 vs 1.77±0.03; P=0.021) or the TT genotype (CC vs TT, 1.95±0.06 vs 1.73±0.07; P=0.029), whereas it was not significantly different in patients with different rs4646437 genotypes at different time points (Figure 3). For the CYP3A5 non-expression group, however, because of the limited number of cases of rs2242480 TT (n=3) and rs4646437 TT (n=1), we grouped them together with the rs2242480 CT and rs4646437 CT genotype patients when performing the analysis. The TAC logC0/D was significantly influenced by both rs2242480 (CC vs CT+TT, 2.08±0.02 vs 1.96±0.04; P=0.01) and rs4646437 (CC vs CT+TT, 2.08±0.02 vs 1.93±0.05; P=0.01) mutations at 90 d post-transplantation (Figure 3). The rs2242480 and rs4646437 mutations showed a similar trend. Considering that these 2 SNPs and CYP3A5 rs776746 are closely spaced on chromosome 7, we conducted a linkage disequilibrium (LD) analysis. Our results showed that an LD block exists between rs2242480 and rs4646437 (D'= 0.96, r2= 0.45), whereas the LD of these with CYP3A5 rs776746 is not significant, with a D'= 0.68 for both (Supplementary Figure S3).
Figure 3.
The influence of CYP3A4 rs2242480 and rs4646437 on TAC logC0/D in CYP3A5 expressers and non-expressers. (A and B) represent the association of CYP3A4 rs2242480 polymorphisms with TAC logC0/D in CYP3A5 expressers and non-expressers, respectively; (C and D) represent the association of CYP3A4 rs4646437 polymorphisms with TAC logC0/D in CYP3A5 expressers and non-expressers, respectively.
Discussion
IL-3 is a secreted glycoprotein that participates in the immune response. Previous studies have demonstrated that a promoter SNP of IL-3, rs181781, is associated with acute rejection (AR) after kidney transplantation27. Our study showed that in CYP3A5 non-expressers, the IL-3 rs181781 AA genotype showed higher TAC logC0/Ds than the AG genotypes at 30 and 90 d (P=0.004 and P=0.033, respectively), and the GG genotypes at 30 d (P=0.011) post-transplantation (Figure 2). As reported by Lee et al, in the promoter SNP rs181781 site, 3 transcription factors (Sp1, SRF, and HNF2C) bind to the A-containing sequence, whereas only 1 (SP1) binds to the G-containing sequence; therefore, it is likely that rs181781 may influence the expression of IL-3 by influencing transcription factor binding27. Considering the role of IL-3 in the immune system, differences in the expression of IL-3 may influence the tacrolimus dose requirements.
The human CTLA-4 gene, which is approximately 6.2 kb in size, is located on human chromosome 228. The gene behaves as a critical negative regulator of the T cell-mediated immune response that can inhibit T Helper (TH) 1 and TH2 cytokine production and cell cycle progression in antigen presenting cells (APC)29. Numerous studies have shown the association of CTLA-4 polymorphisms with allograft rejection, end-stage renal disease, delayed graft function and infection after renal transplantation29,30,31,32, and the expression pattern of the protein encoded by CTLA4 was found to be altered by the polymorphisms rs4553808 (-1661A/G) in the CTLA4 gene promoter33. It is reported that a carry of G allele variation will increase CTLA4 mRNA level and expression, thereby promoting consensus sequence in the regulatory region of the CTLA4 promoter in humans26,34. The result of our study revealed that polymorphism of rs4553808 (-1661A/G) in CTLA4 gene promoter may influence TAC dose requirement. Compared with AA genotypes, the TAC logC0/D at 30 d post-transplantation in the grouped AG+GG genotypes is significant lower in CYP3A5 expression group patients (P=0.041; Figure 2); however, when compared with the different genotypes in the CYP3A5 non-expression patients, the result was reversed. The TAC logC0/D at 30 d post-transplantation in the grouped AG+GG genotypes was significantly higher than that of the AA genotypes (P=0.008, Figure 2). The results observed in the CYP3A5 expression group may be related to the expression state of CYP3A5, which is the polymorphism that has the greatest influence on TAC pharmacokinetics5. In the CYP3A5 non-expression patients, the results may be more convincing because they exclude the effect of CYP3A5 expression. However, our results showed no difference at 7 or 90 d post-operation. This result is credible considering the fact that it takes 2 or more weeks to reach a stable maintenance dose of TAC23.
To our knowledge, our study is the first to report that the IL-3 rs181781 and CTLA4 rs4553808 genetic polymorphisms can affect the TAC dose requirement. As key elements in the immune system, changes in the expression of IL-3 and CTLA4 can lead to alterations of the immune response, which in turn can influence the TAC dose required to cause immune suppression. It is difficult to understand the result of a study showing that after TAC treatment for 24 h, the expression level of IL-3 was significantly down-regulated35. In fact, other cytokines, including IL-2, IL-4, IL-6 and IL-18, can be selectively inhibited by tacrolimus36. Previous studies have reported genetic associations of IL-2, IL-4 and IL-6 with blood concentrations of calcineurin inhibitors21. Genetic polymorphisms in IL-18 (eg, rs5744247) and IL-10 (eg, -819 C/T and -592 C/A) can also affect TAC dose requirements37,38. However, the exact mechanism by which these polymorphisms affect TAC dose requirements is unknown. There is a hypothesis that these immune related genes may influence the expression of genes that play important roles in TAC pharmacokinetics, and a study reported that increased serum IL-1beta, IL-6, and IL-10 could lead to a significant decrease in cytochrome P450 (CYP)-mediated drug metabolism39.
Our results also validate with a large sample size (334 cases) that an rs776746 mutation in intron 3 of the CYP3A5 gene is significantly (P<0.001) associated with tacrolimus pharmacokinetics at different times post-transplantation (Figure 1). The CYP3A5 gene codes for the main metabolic enzyme for tacrolimus, and in the main functional variant, CYP3A5 6986A>G (rs776746), a guanine is substituted for adenine. Carriers of the A allele (AA or AG, CYP3A5 expressers) can produce very high levels of functional CYP3A5 compared with homozygous GG carriers (CYP3A5 non-expressers)12. Thus, GG genotype patients need less TAC (high C0/D ratio) to reach the target blood concentration than the A allele carriers. In accordance with this explanation, our data showed a significantly higher level of the logarithmic transformed TAC C0/D in GG than in AG or AA genotype patients at 7, 30 and 90 d post-transplantation. Interestingly, the differences in the logC0/D decreased with time but were still significant at 3 months post transplantation (Figure 1). Our study was restricted by the follow-up time (3 months in all cases); therefore, the effect of the CYP3A5 rs776746 polymorphism on the TAC logC0/D at longer times requires validation.
Tacrolimus can also be metabolized by CYP3A4, which is second to CYP3A5 in importance. Our results show that CYP3A4 rs2242480 (*1G) and rs4646437 are significantly associated with the TAC logC0/D at different times after adjusting for age and gender. The results are consistent with the research results of Li et al19. A previous study suggested that the rs4646437-T allele was in strong LD (r2= 0.82) with the CYP3A5*1 allele in Caucasian renal transplant recipients40. A study of Chinese groups showed a moderate degree (r2= 0.502) of LD between rs2242480 and rs776746, and a low degree (r2= 0.244) of LD between rs4646437 and rs77674619. In our study (r2= 0.46 and r2= 0.22, respectively), an LD block was observed between rs2242480 and rs4646437 (Supplementary Figure S3). These two SNPs should be analyzed together. However, the results of our stratified analysis showed that both rs2242480 and rs4646437 affected the TAC logC0/D at 3 months in CYP3A5 non-expressers (GG). In CYP3A5 expressers (AA+AG), rs2242480 also showed an influence on the TAC logC0/D at 7 d (Figure 3). The discrepancies in the relationships between the rs2242480 and rs4646437 polymorphisms and the logC0/D at d 7 and d 90 may be due to the small sample size analyzed at d 7. No influence on the TAC logC0/D was observed for rs4646437 at different time points in CYP3A5 expressers. This is consistent with another study of CYP3A4 rs224248041. Actually, CYP3A4 rs2242480 is a functional SNP that could increase the activity of CYP3A4, and it resembles rs4646437 that can affect the hepatic CYP3A4 protein expression19,42. We conclude that CYP3A4 rs2242480 and rs4646437 are independent factors that can influence the TAC logC0/D.
We have also found that for the ATPase beta 2 (ATP1B2) SNP rs1641536, the homozygous GG and AA patients showed a trend toward higher logC0/D than the heterozygous patients. The difference was significant at d 7 (P=0.021, Supplementary Figure S2). However, in an analysis stratified for CYP3A5 expression, there was no difference at different times. It is likely that the significance may be caused by the effect of the CYP3A5 rs776746 variation because of its unbalanced distribution in different genotypes of rs1641536. The effect of ATP1B2 rs1641536 should be validated in further studies.
Our study has several limitations. This is a retrospective study, and some personal donor information was not available, although the genotypes of donors have been reported to be important factors influencing TAC dose requirements. In addition, some important clinical data, including the TAC C0 and drug dose, were not always completely recorded at 7, 30 and 90 d after renal transplantation. Consequently, it was difficult to integrate all the samples and the clinical data. Furthermore, we did not include drug combinations, eg, calcium channel blockers, proton pump inhibitors, and anti-fungal drugs, because in some cases, this information was missing; however, some studies have indicated that drug combinations can affect TAC blood concentrations.
Conclusions
The findings of our retrospective study suggest that among the candidate 105 SNPs that function in different pathways, the CYP3A5 rs776746, CYP3A4 rs2242480 and rs4646437, IL-3 rs181781 and CTLA4 rs4553808 genetic polymorphisms probably influence the TAC dose requirements in Chinese kidney transplant recipients.
Our findings may contribute to understanding the genetic basis for individual variations in tacrolimus pharmacokinetics, and may provide information for further studies of the clinical implications of tacrolimus for individualized medicine.
Author contribution
Mou-ze LIU contributed to the study design, research performance, data analysis and manuscript writing; Hua-wen XIN, Ying-zi MING, and Wei ZHANG as the co-corresponding authors were involved in study design, data collection and final approval of the version to be published; Hai-yan HE, Yue-li ZHANG, Fa-zhong HE, Jian-quan LUO, Zhi-ying LUO, and Ming-jie SHAO participated in sample collection, acquisition of data, experiments and revision of the draft; Yong-fang HU, Xiao-ping CHEN, Zhao-qian LIU, and Hong-hao ZHOU contributed to the study design and data analysis.
Acknowledgments
This work was supported by the National Key Research and Development Program (No 2016YFC0905000), the National High Technology Research and Development Program of China, the “863” Project (No 2012AA02A518), the National Natural Science Foundation of China (No 81522048, 81573511, and 81273595), and the Innovation Driven Project of Central South University (No 2016CX024).
Footnotes
Supplementary information is available at website of Acta Pharmacologica Sinica.
Supplementary Information
Consort diagram of patients selection
Clinical characteristics of recipients based on rs2242480 genotypes.
Clinical characteristics of recipients based on rs4646437 genotypes.
Clinical characteristics of recipients based on rs181781 genotypes.
Clinical characteristics of recipients based on rs1641536 genotypes.
Clinical characteristics of recipients based on rs776746 genotypes.
Clinical characteristics of recipients based on rs4553808 genotypes.
Clinical characteristics: CYP 3A5 rs776746 (AA&AG vs GG).
References
- Rath T. Tacrolimus in transplant rejection. Expert Opin Pharmacother 2013; 14: 115–22. [DOI] [PubMed] [Google Scholar]
- Barbarino JM, Staatz CE, Venkataramanan R, Klein TE, Altman RB. PharmGKB summary: cyclosporine and tacrolimus pathways. Pharmacogenet Genomics 2013; 23: 563–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataramanan R, Swaminathan A, Prasad T, Jain A, Zuckerman S, Warty V, et al. Clinical pharmacokinetics of tacrolimus. Clin Pharmacokinet 1995; 29: 404–30. [DOI] [PubMed] [Google Scholar]
- Werk AN, Lefeldt S, Bruckmueller H, Hemmrich-Stanisak G, Franke A, Roos M, et al. Identification and characterization of a defective CYP3A4 genotype in a kidney transplant patient with severely diminished tacrolimus clearance. Clin Pharmacol Ther 2014; 95: 416–22. [DOI] [PubMed] [Google Scholar]
- Birdwell KA, Decker B, Barbarino JM, Peterson JF, Stein CM, Sadee W, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for CYP3A5 genotype and tacrolimus dosing. Clin Pharmacol Ther 2015; 98: 19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Zheng KL, Chen LZ, Dai YP, Fei JG, Qiu J, et al. Clinical pharmacokinetics of tacrolimus after the first oral administration in combination with mycophenolate mofetil and prednisone in Chinese renal transplant recipients. Transplant Proc 2005; 37: 4246–50. [DOI] [PubMed] [Google Scholar]
- Li CJ, Li L. Tacrolimus in preventing transplant rejection in Chinese patients--optimizing use. Drug Des Devel Ther 2015; 9: 473–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elens L, Bouamar R, Shuker N, Hesselink DA, van Gelder T, van Schaik RH. Clinical implementation of pharmacogenetics in kidney transplantation: calcineurin inhibitors in the starting blocks. Br J Clin Pharmacol 2014; 77: 715–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Mao Y, Razo J, Zhou X, Wong ST, Patel S, et al. Using genetic and clinical factors to predict tacrolimus dose in renal transplant recipients. Pharmacogenomics 2010; 11: 1389–402. [DOI] [PubMed] [Google Scholar]
- Kurzawski M, Drozdzik M. Pharmacogenetics in solid organ transplantation: genes involved in mechanism of action and pharmacokinetics of immunosuppressive drugs. Pharmacogenomics 2013; 14: 1099–118. [DOI] [PubMed] [Google Scholar]
- Hesselink DA, Bouamar R, Elens L, van Schaik RH, van Gelder T. The role of pharmacogenetics in the disposition of and response to tacrolimus in solid organ transplantation. Clin Pharmacokinet 2014; 53: 123–39. [DOI] [PubMed] [Google Scholar]
- Rojas L, Neumann I, Herrero MJ, Boso V, Reig J, Poveda JL, et al. Effect of CYP3A5*3 on kidney transplant recipients treated with tacrolimus: a systematic review and meta-analysis of observational studies. Pharmacogenomics J 2015; 15: 38–48. [DOI] [PubMed] [Google Scholar]
- Elens L, Capron A, van Schaik RH, De Meyer M, De Pauw L, Eddour DC, et al. Impact of CYP3A4*22 allele on tacrolimus pharmacokinetics in early period after renal transplantation: toward updated genotype-based dosage guidelines. Ther Drug Monit 2013; 35: 608–16. [DOI] [PubMed] [Google Scholar]
- Moes DJ, Swen JJ, den Hartigh J, van der Straaten T, van der Heide JJ, Sanders JS, et al. Effect of CYP3A4*22, CYP3A5*3, and CYP3A combined genotypes on cyclosporine, everolimus, and tacrolimus pharmacokinetics in renal transplantation. CPT Pharmacometrics Syst Pharmacol 2014; 3: e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde I, Bremer S, Midtvedt K, Mohebi B, Dahl M, Bergan S, et al. The influence of CYP3A, PPARA, and POR genetic variants on the pharmacokinetics of tacrolimus and cyclosporine in renal transplant recipients. Eur J Clin Pharmacol 2014; 70: 685–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge H, Elens L, de Loor H, van Schaik RH, Kuypers DR. The CYP3A4*22 C>T single nucleotide polymorphism is associated with reduced midazolam and tacrolimus clearance in stable renal allograft recipients. Pharmacogenomics J 2015; 15: 144–52. [DOI] [PubMed] [Google Scholar]
- Shi WL, Tang HL, Zhai SD. Effects of the CYP3A4*1B genetic polymorphism on the pharmacokinetics of tacrolimus in adult renal transplant recipients: a meta-analysis. PLoS One 2015; 10: e0127995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusinato DA, Lacchini R, Romao EA, Moyses-Neto M, Coelho EB. Relationship of CYP3A5 genotype and ABCB1 diplotype to tacrolimus disposition in Brazilian kidney transplant patients. Br J Clin Pharmacol 2014; 78: 364–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CJ, Li L, Lin L, Jiang HX, Zhong ZY, Li WM, et al. Impact of the CYP3A5, CYP3A4, COMT, IL-10 and POR genetic polymorphisms on tacrolimus metabolism in Chinese renal transplant recipients. PLoS One 2014; 9: e86206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wu S, Chen D, Guo F, Zhong L, Fan J, et al. Influence of TLR4 rs1927907 locus polymorphisms on tacrolimus pharmacokinetics in the early stage after liver transplantation. Eur J Clin Pharmacol 2014; 70: 925–31. [DOI] [PubMed] [Google Scholar]
- Seyhun Y, Ciftci HS, Kekik C, Karadeniz MS, Tefik T, Nane I, et al. Genetic association of interleukin-2, interleukin-4, interleukin-6, transforming growth factor-beta, tumour necrosis factor-alpha and blood concentrations of calcineurin inhibitors in Turkish renal transplant patients. Int J Immunogenet 2015; 42: 147–60. [DOI] [PubMed] [Google Scholar]
- Xing J, Zhang X, Fan J, Shen B, Men T, Wang J. Association between interleukin-18 promoter variants and tacrolimus pharmacokinetics in Chinese renal transplant patients. Eur J Clin Pharmacol 2015; 71: 191–8. [DOI] [PubMed] [Google Scholar]
- Li L, Li CJ, Zheng L, Zhang YJ, Jiang HX, Si-Tu B, et al. Tacrolimus dosing in Chinese renal transplant recipients: a population-based pharmacogenetics study. Eur J Clin Pharmacol 2011; 67: 787–95. [DOI] [PubMed] [Google Scholar]
- Tholking G, Fortmann C, Koch R, Gerth HU, Pabst D, Pavenstadt H, et al. The tacrolimus metabolism rate influences renal function after kidney transplantation. PLoS One 2014; 9: e111128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesche D, Sigurdardottir V, Setoud R, Oberhansli M, Carrel T, Fiedler GM, et al. CYP3A5*3 and POR*28 genetic variants influence the required dose of tacrolimus in heart transplant recipients. Ther Drug Monit 2014; 36: 710–5. [DOI] [PubMed] [Google Scholar]
- Bouqbis L, Izaabel H, Akhayat O, Perez-Lezaun A, Calafell F, Bertranpetit J, et al. Association of the CTLA4 promoter region (-1661G allele) with type 1 diabetes in the South Moroccan population. Genes Immun 2003; 4: 132–7. [DOI] [PubMed] [Google Scholar]
- Lee DY, Song SB, Moon JY, Jeong KH, Park SJ, Kim HJ, et al. Association between interleukin-3 gene polymorphism and acute rejection after kidney transplantation. Transplant Proc 2010; 42: 4501–4. [DOI] [PubMed] [Google Scholar]
- Teft WA, Kirchhof MG, Madrenas J. A molecular perspective of CTLA-4 function. Annu Rev Immunol 2006; 24: 65–97. [DOI] [PubMed] [Google Scholar]
- Canossi A, Aureli A, Delreno F, Iesari S, Cervelli C, Clemente K, et al. Influence of cytotoxic T-lymphocyte antigen-4 polymorphisms on acute rejection onset of cadaveric renal transplants. Transplant Proc 2013; 45: 2645–9. [DOI] [PubMed] [Google Scholar]
- Misra MK, Kapoor R, Pandey SK, Sharma RK, Agrawal S. Association of CTLA-4 gene polymorphism with end-stage renal disease and renal allograft outcome. J Interferon Cytokine Res 2014; 34: 148–61. [DOI] [PubMed] [Google Scholar]
- Gao JW, Guo YF, Fan Y, Qiu JX, Bao ED, Liu Y, et al. Polymorphisms in cytotoxic T lymphocyte associated antigen-4 influence the rate of acute rejection after renal transplantation in 167 Chinese recipients. Transpl Immunol 2012; 26: 207–11. [DOI] [PubMed] [Google Scholar]
- Ruhi C, Sallakci N, Yegin O, Suleymanlar G, Ersoy FF. The influence of CTLA-4 single nucleotide polymorphisms on acute kidney allograft rejection in Turkish patients. Clin Transplant 2015; 29: 612–8. [DOI] [PubMed] [Google Scholar]
- Guo Y, Guo F, Wei C, Qiu J, Liu Y, Fang Y, et al. CTLA4 gene polymorphisms influence the incidence of infection after renal transplantation in Chinese recipients. PLoS One 2013; 8: e70824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng R, Song F, Yang X, Sun P, Hu J, Zhu C, et al. Association between cytotoxic T lymphocyte antigen-4 +49A/G, -1722T/C, and -1661A/G polymorphisms and cancer risk: a meta-analysis. Tumour Biol 2014; 35: 3627–39. [DOI] [PubMed] [Google Scholar]
- Wen J, Ji ZG, Niu JR. Regulatory effects of cyclosporin A and tacrolimus on the immunological gene expressions in renal transplant recipients. Zhongguo Yi Xue Ke Xue Yuan Xue Bao 2012; 34: 563–6. [DOI] [PubMed] [Google Scholar]
- James DG. A new immunosuppressant: tacrolimus. Postgrad Med J 19963; 72: 586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Zhang X, Ren L, Chen D, Wu S, Guo F, et al. Donor IL-18 rs5744247 polymorphism as a new biomarker of tacrolimus elimination in Chinese liver transplant patients during the early post-transplantation period: results from two cohort studies. Pharmacogenomics 2015; 16: 239–50. [DOI] [PubMed] [Google Scholar]
- Zhang X, Wang Z, Fan J, Liu G, Peng Z. Impact of interleukin-10 gene polymorphisms on tacrolimus dosing requirements in Chinese liver transplant patients during the early posttransplantation period. Eur J Clin Pharmacol 2011; 67: 803–13. [DOI] [PubMed] [Google Scholar]
- Gorski JC, Hall SD, Becker P, Affrime MB, Cutler DL, Haehner-Daniels B. In vivo effects of interleukin-10 on human cytochrome P450 activity. Clin Pharmacol Ther 2000; 67: 32–43. [DOI] [PubMed] [Google Scholar]
- Crettol S, Venetz JP, Fontana M, Aubert JD, Pascual M, Eap CB. CYP3A7, CYP3A5, CYP3A4, and ABCB1 genetic polymorphisms, cyclosporine concentration, and dose requirement in transplant recipients. Ther Drug Monit 2008; 30: 689–99. [DOI] [PubMed] [Google Scholar]
- Zuo XC, Ng CM, Barrett JS, Luo AJ, Zhang BK, Deng CH, et al. Effects of CYP3A4 and CYP3A5 polymorphisms on tacrolimus pharmacokinetics in Chinese adult renal transplant recipients: a population pharmacokinetic analysis. Pharmacogenet Genomics 2013; 23: 251–61. [DOI] [PubMed] [Google Scholar]
- He BX, Shi L, Qiu J, Tao L, Li R, Yang L, et al. A functional polymorphism in the CYP3A4 gene is associated with increased risk of coronary heart disease in the Chinese Han population. Basic Clin Pharmacol Toxicol 2011; 108: 208–13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Consort diagram of patients selection
Clinical characteristics of recipients based on rs2242480 genotypes.
Clinical characteristics of recipients based on rs4646437 genotypes.
Clinical characteristics of recipients based on rs181781 genotypes.
Clinical characteristics of recipients based on rs1641536 genotypes.
Clinical characteristics of recipients based on rs776746 genotypes.
Clinical characteristics of recipients based on rs4553808 genotypes.
Clinical characteristics: CYP 3A5 rs776746 (AA&AG vs GG).