Abstract
Nuclear factor erythroid 2-related factor 2 (Nrf2) provides a cellular defense against oxidative stress by inducing the expression of antioxidant and detoxification enzymes. The calcium antagonist, verapamil, is an FDA-approved drug prescribed for the treatment of hypertension. Here, we show that verapamil acts as a potent Nrf2 activator without causing cytotoxicity, through degradation of Kelch-like ECH-associated protein 1 (Keap1), a Nrf2 repressor. Furthermore, verapamil-induced Keap1 degradation is prominently mediated by a p62-dependent autophagic pathway. Correspondingly, verapamil protects cells from acetaminophen-induced oxidative damage through Nrf2 activation. These results demonstrated the underlying mechanisms for the protective role of verapamil against acetaminophen-induced cytotoxicity.
Keywords: APAP, Keap1, Nrf2, Oxidative stress, Verapamil
INTRODUCTION
The nuclear factor erythroid 2-related factor 2 (Nrf2)-Kelch-like ECH-associated protein 1 (Keap1) pathway protects cells against oxidative stress-mediated damage through the increase of antioxidant and detoxification enzymes (1–3). Keap1, a negative regulator of Nrf2, steadily leads to proteasomal degradation of Nrf2 in unstressed cells (1, 2, 4, 5). Recently, Nrf2 activators have been developed as promising therapeutic drugs for various diseases such as liver disease (6, 7), neuro-degenerative disease (8), and chronic renal disease (9).
Verapamil has been reported to inhibit calcium uptake in vitro and in vivo (10) and is used as a Food and Drug Administration (FDA)-approved drug for the treatment of hypertension (11). Notably, recent studies have reported that verapamil induces autophagy in colon adenocarcinoma (12) and attenuates the accumulation of p62/sqstm1 (hereafter referred to as p62) and ubiquitinated proteins through an increase in the autophagic flux (13, 14).
Acetaminophen (APAP) is widely used as an analgesic and antipyretic drug. However, an overdose of APAP induces hepatotoxicity in humans and rodents (15, 16). Notably, APAP-induced hepatotoxicity is associated with oxidative stress induced by N-Acetyl-p-benzoquinone imine (NAPQI), a reactive metabolite of APAP (17).
In this study, we demonstrate that verapamil acts as a potent Nrf2 activator and protects cells from APAP-induced cell death. In addition, we elucidate the underlying mechanisms for verapamil-mediated Nrf2 activation.
RESULTS
Verapamil activates Nrf2 without causing cytotoxicity
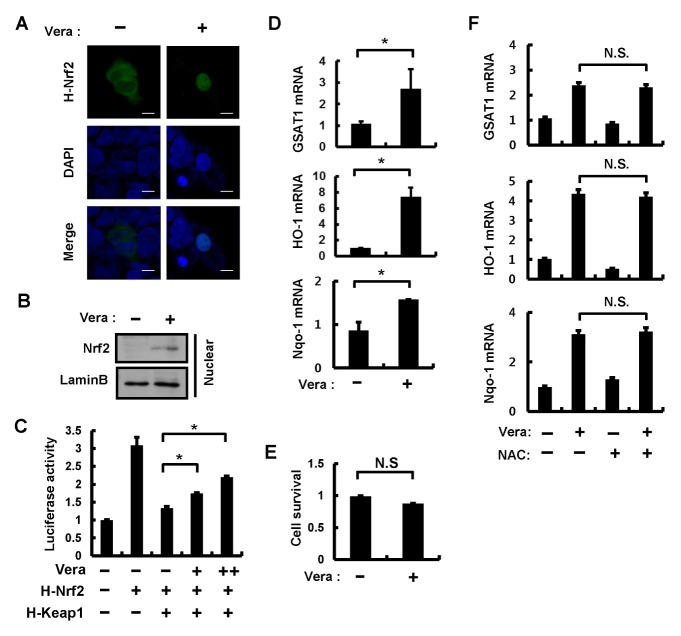
Nrf2 is known to function as an important transcription factor that translocates to the nucleus and induces the transcription of genes for antioxidant or detoxification proteins (3, 18). As our previous research suggested that autophagic Keap1 degradation might represent a possible mechanism of Nrf2 activation, we speculated that the pro-autophagic effects of verapamil might, therefore, influence Nrf2 function. To verify nuclear translocation of Nrf2, HEK293T cells were transfected with HA-tagged Nrf2 (H-Nrf2) and HA-tagged Keap1 (H-Keap1) with concomitant treatment of 100 μM verapamil for 18 h. Immunofluorescence analysis revealed that verapamil increased the amount of Nrf2 in the nucleus (Fig. 1A). To further examine whether verapamil mediated the accumulation of nuclear Nrf2, mouse hepatoma (Hepa1c1c7) cells and MEF cells (data not shown) were treated with 100 μM verapamil for 18 h and the nuclear fractions were isolated. Immunoblot analysis showed that the nuclear amounts of Nrf2 were increased (Fig. 1B). Consistent with this result, we demonstrated verapamil-mediated up-regulation of the transactivation activity of Nrf2 using a luciferase reporter assay (Fig. 1C). This verapamil-induced increase in Nrf2 activity was accompanied by an upregulation of Nrf2 target genes including GSTA1, HO-1, and Nqo-1 in Hepa1c1c7 (Fig. 1D) and MEF (Supplementary Fig. 1) cells.
Fig. 1.
Verapamil activates Nrf2 without causing cytotoxicity. (A) HEK 293T cells treated with DMSO or verapamil (100 μM) for 18 h. HEK 293T cells expressing HA-Nrf2 were subjected to immunofluorescence analysis using an anti-HA antibody. Cell nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Scale bars, 10 μm. (B) Nuclear fractions of Hepa1c1c7 cells treated with verapamil (100 μM) for 18 h were subjected to immunoblot analysis with antibodies against Nrf2 and Lamin B. (C) Hepa1c1c7 cells transfected with vectors for H-Nrf2, H-Keap1, as well as with the luciferase reporter plasmid pNqo1-ARE and pRL-TK (internal control) were treated with verapamil for 18 h. The transfected cells were lysed and we determined the luciferase activity. (D) Total RNA isolated from Hepa1c1c7 cells treated as described in (B) was subjected to quantitative qRT-PCR analysis for the mRNAs of GSTA1, HO-1, and Nqo-1. (E) Cell viability was estimated using a Cell titer-Glo assay kit. The live cell number was expressed as the absorbance at luminescence in Hepa1c1c7 cells. (F) Hepa1c1c7 cells treated with or without NAC (1 mM) prior to verapamil (100 μM) treatment were subjected to qRT-PCR analysis for mRNAs of GSTA-1, HO-1, and Nqo-1. Data are presented as the means ± SD from three independent experiments. *P < 0.05; N.S.: not significant.
To examine whether verapamil treatment resulted in cytotoxicity, verapamil-treated cells were subjected to cell viability analysis. We found that verapamil did not cause cytotoxicity in Hepa1c1c7 (Fig. 1E) or MEF cells (Supplementary Fig. 1). Next, as reactive oxygen species (ROS) can activate Nrf2 by oxidative modification of Keap1, a negative regulator of Nrf2 (19), we examined whether verapamil-induced upregulation of Nrf2 target genes was caused by ROS-mediated Nrf2 activation. Hepa1c1c7 cells were co-treated with verapamil (100 μM) and N-acetyl-L-cysteine (NAC), as a known ROS scavenger. However, the results showed that NAC did not affect the verapamil-mediated upregulation of Nrf2 target genes (Fig. 1F). Taken together, these findings indicate that verapamil acts as a potent Nrf2 activator in an ROS-independent manner.
Verapamil mediates Keap1 degradation via an autophagic pathway
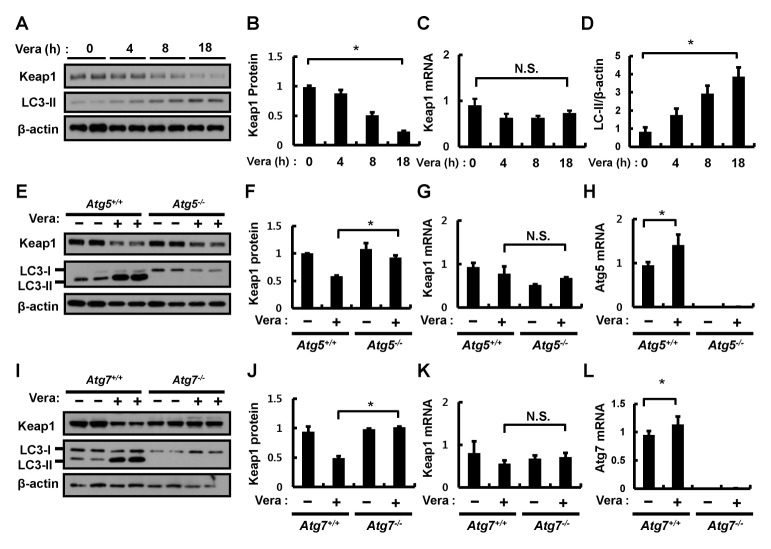
To identify whether verapamil activation of the Nrf2-Keap1 pathway is related to the known autophagic Keap1 degradation mechanism (1, 3, 5, 19), Hepa1c1c7 cells were treated with verapamil (100 μM) for the indicated times. Keap1 protein and mRNA levels were determined by immunoblotting and quantitative qRT-PCR assays, respectively. Verapamil reduced the abundance of Keap1 protein in a time-dependent manner, whereas the amount of Keap1 mRNA remained unaffected (Fig. 2A–C). We also found that verapamil increased the amount of LC3-II, an autophagy marker protein, in Hepa1c1c7 (Fig. 2D) and MEF cells (Supplementary Fig. 2). As shown in Fig. 1, we observed that the verapamil-induced degradation of Keap1 was accompanied by an upregulation of the mRNA levels of Nrf2 target genes, including GSTA1, HO-1, and Nqo-1 in Hepa1c1c7 (Supplementary Fig. 3) and MEF cells (Supplementary Fig. 2).
Fig. 2.
Verapamil mediates Keap1 degradation via an autophagic pathway. (A) Hepa1c1c7 cells were treated with verapamil (100 μM) for the indicated times. Lysates of Hepa1c1c7 cells were subjected to immunoblot analysis with antibodies against Keap1, LC3-II, and β-actin (loading control). (B) Densitometric analysis of Keap1 immunoblots was obtained in (A). Total RNA isolated from cells treated as described was subjected to qRT-PCR analysis for the mRNA of Keap1 (C). (D) Densitometric analysis of LC3-II/β-actin immunoblots was obtained in (A). (E) ATG5+/+ or ATG5−/− MEF cells were incubated in the absence or presence of verapamil (100 μM) for 18 h. Lysates of ATG5+/+ or ATG5−/− MEF cells were subjected to immunoblot analysis with antibodies against Keap1, LC3-II, and β-actin (loading control). (F) Densitometric analysis of Keap1 immunoblots was obtained in (E). Total RNA isolated from cells treated as described in (E) was subjected to qRT-PCR analysis for mRNAs of Keap1 (G) and ATG5 (H). (I) ATG7+/+ or ATG7−/− MEF cells were incubated in the absence or presence of verapamil (100 μM) for 18 h. Lysates of ATG7+/+ or ATG7−/− MEF cells were subjected to immunoblot analysis with antibodies against Keap1, LC3-II, and β-actin (loading control). (J) Densitometric analysis of Keap1 immunoblots was obtained in (H). Total RNA isolated from cells treated as described in (I) was subjected to qRT-PCR analysis for mRNAs of Keap1 (K) and ATG7 (L). Data are the mean ± SD from three independent experiments. *P < 0.05.
Based on these results, we examined whether the verapamil-induced Keap1 degradation was mediated by an autophagic process. To do this, we first examined the autophagic activity of verapamil with the use of an autophagic flux assay (Supplementary Fig. 4). We treated wild-type ATG5 (ATG5+/+) and autophagy-defective ATG5 (ATG5−/−) MEF cells with verapamil (100 μM). The results showed that verapamil-induced Keap1 degradation was markedly inhibited in ATG5−/− MEF cells compared to that in ATG5+/+ MEF cells, whereas the levels of Keap1 mRNA were not altered in either cell type (Fig. 2E–H). Similar experiments with another autophagy-defective ATG7 (ATG7−/−) MEF cell line confirmed that verapamil (100 μM) did not affect Keap1 degradation (Fig. 2I–L). Accordingly, these results indicated that verapamil-mediated Keap1 degradation is mediated by an autophagic process.
p62 is required for verapamil-induced Keap1 degradation and Nrf2 activation
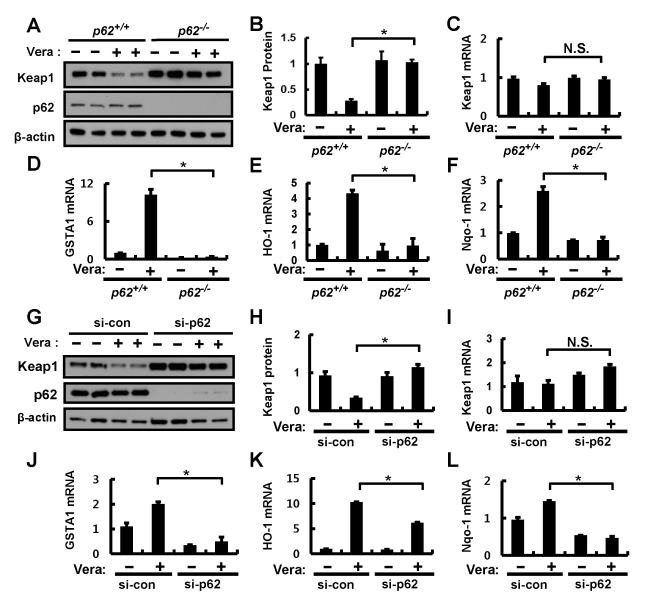
The autophagy adaptor protein, p62, has recently been identified as being essential for Nrf2 activation (2, 5, 20). To examine whether verapamil-induced Nrf2 activation is dependent on p62, wild-type (p62+/+) and p62-deficient (p62−/−) MEF cells were treated with verapamil (100 μM). Immunoblot analysis indicated that verapamil-induced Keap1 degradation was markedly blocked in p62−/− MEF cells compared to p62+/+ MEF cells (Fig. 3A, B). However, the levels of Keap1 mRNAs were not changed in p62 MEF cells (Fig. 3C). Correspondingly, we showed that a p62-deficiency downregulated verapamil-mediated induction of Nrf2 target genes including GSTA1, HO-1, and Nqo-1 (Fig. 3D–F). In line with these observations, small interfering RNA (siRNA)-mediated knockdown of p62 showed effects similar to those of verapamil on Keap1 degradation associated with Nrf2 target gene upregulation in Hepa1c1c7 cells (Fig. 3G–L). It is reported that p62 phosphorylation (p-S351) activates the Nrf2 pathway (21). We also found that verapamil is significantly induced by phosphorylation of p62 at Ser351 (Supplementary Fig. 5). Together, our observations suggest that verapamil-induced Nrf2 activation is mediated by a p62-dependent autophagic pathway.
Fig. 3.
p62 is required for verapamil-induced Keap1 degradation and Nrf2 activation. (A) p62+/+ or p62−/− MEF cells were incubated in the absence or presence of verapamil (100 μM) for 18 h. Lysates of p62+/+ or p62−/− MEF cells were subjected to immunoblot analysis with antibodies against Keap1, p62, and β-actin (loading control). (B) Densitometric analysis of Keap1 immunoblots was obtained in (A). Total RNA isolated from cells treated as described in (A) was subjected to qRT-PCR analysis for mRNAs of Keap1 (C), GSTA1 (D), HO-1 (E), and Nqo-1 (F). (G) Hepa1c1c7 cells transfected with control siRNA (si-control) or p62 siRNA were treated with DMSO or verapamil (100 μM) for 18 h. The cells were lysed and subjected to immunoblot analysis with antibodies against Keap1, p62, and β-actin (loading control). (H) Densitometric analysis of Keap1 immunoblots was obtained in (G). qRT-PCR analysis of mRNAs of Keap1 (I), GSTA1 (J), HO-1 (K), and Nqo-1 (L). Data are presented as the means ± SD from three independent experiments. *P < 0.05.
Verapamil protects cells from APAP-induced cytotoxicity via Nrf2 activation
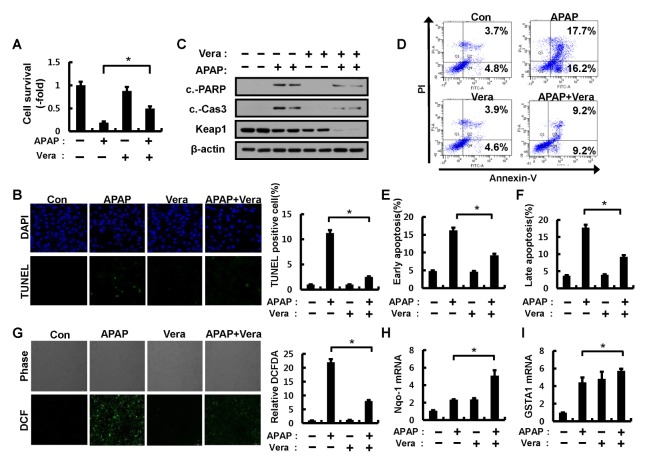
It is well known that an overdose of APAP leads to ROS-mediated cell death (22, 23). Notably, Nrf2 has been reported as an important transcription factor for protecting cells from APAP-mediated cell death (24, 25). To explore the antioxidant function of verapamil as a potent Nrf2 activator in the pathological setting, we examined the effects of verapamil on APAP-mediated cell death. First, we tested whether verapamil protects against APAP-induced apoptotic cell death using a cell viability assay, flow cytometric analysis, and TUNEL analyses. The results showed that verapamil inhibited APAP-induced cell death (Fig. 4A and Fig. 4D–F) and decreased TUNEL-positive cells (Fig. 4B). Consistent with these results, the expression levels of cleaved poly(ADP)ribose polymerase (PARP) and cleaved caspase-3 proteins were partially inhibited in response to verapamil (Fig. 4C). To determine whether this antioxidant role is caused by Keap1 degradation-induced Nrf2 activation in this context, we examined the expression levels of Keap1 protein and Nrf2 target genes. The combination of verapamil and APAP showed additive effects on the induction of Nrf2 target genes (Fig. 4H–I and Supplementary Fig. 6) associated with the degradation of Keap1 (Fig. 4C). These results suggest that verapamil prevents APAP-induced apoptotic cell death by Nrf2 activation through Keap1 degradation. Given that APAP leads to intracellular ROS accumulation and elicits cell death, we next examined the effect of verapamil on APAP-induced ROS accumulation in Hepa1c1c7 cells. We found that verapamil significantly blocked APAP-induced ROS accumulation (Fig. 4G).
Fig. 4.
Verapamil alleviates APAP-induced cytotoxicity via Nrf2 activation. (A) Hepa1c1c7 cells were pretreated with APAP (10 mM) for 1 h and incubated in the absence or presence of verapamil (100 μM) for 18 h. Cell viability was estimated using a Cell titer-Glo assay kit. The live cell number was expressed as the absorbance at luminescence. (B) TUNEL analysis of Hepa1c1c7 cells treated as in (A) and quantitation of TUNEL analysis. (C) Hepa1c1c7 cells treated as described in (A) were lysed and subjected to immunoblot analysis with antibodies against cleaved PARP, cleaved caspase-3, Keap1, and β-actin (loading control). (D) Apoptosis in Hepa1c1c7 cells pretreated with APAP and incubated in the absence or presence of verapamil were detected by FACS analysis for Annexin V and PI staining. (E) Early apoptosis. (F) Late apoptosis. (G) ROS levels were determined using CM-H2DCFH-DA. Representative images are shown. Quantitative analysis of cells treated as described in (A). The relative dichlorofluorescein fluorescence was calculated by averaging the levels of fluorescence from 80–100 cells after subtracting the background fluorescence obtained using a fluorescence microscope. Total RNA isolated from cells treated as described in (A) was subjected to qRT-PCR analysis for mRNAs of GSTA1 (H) and Nqo-1 (I). Data are presented as the means ± SD from three independent experiments. *P < 0.05.
DISCUSSION
A Nrf2-Keap1 pathway is an essential tool for protecting cells against oxidative stress. The underlying molecular mechanism of Nrf2 activation can be explained by a “hinge and latch model” in which the Kelch-repeat domain of the Keap1 homodimer interacts with one entity of Nrf2 via a low-affinity DLG motif (latch) or a high-affinity ETGE motif (hinge) in the Neh2 domain of Nrf2 (2, 26). In response to oxidative stress, Keap1 is modified at one or more of its cysteine residues, which induces a conformational change that interrupts its binding to the low-affinity DLG motif of Nrf2. The unbound Nrf2 is then translocated to the nucleus and induces the transcription of its target genes, including GSTA1, HO-1, and Nqo-1 (2).
Numerous natural or synthetic compounds have been shown to activate Nrf2 and protect cells from oxidative damage in human and rodent tissuse. Most of these electrophilic compounds modify cysteine residues of Keap1, leading to Nrf2 activation (7). One such compound is oltipraz, a synthetic Nrf2 activator that is currently undergoing clinical trials and appears to represent a potential therapeutic drug for diverse diseases, including nonalcoholic steatohepatitis, multiple sclerosis, muscular dystrophy, and skin cancer (27). However, this electrophilic Nrf2 activator also modifies cysteine residues of other proteins and may therefore exhibit potentially unwanted effects some Nrf2 activator (7).
To address this concern, it is necessary to develop non-electrophilic drugs for specific Nrf2 activators. Recently, the autophagy adaptor protein, p62, has been identified as a crucial protein in the activation of the Nrf2-Keap1 pathway (5, 20, 28). The p62 protein specifically interacts with Keap1 and competitively inhibits the interaction between Nrf2 and Keap1 (20, 29). This disruption leads to the stabilization of Nrf2 with concomitant increased in the expression of Nrf2 antioxidant target genes. Furthermore, phosphorylation of Ser351 on p62 mediated by the mammalian target of rapamycin complex 1 (mTORC1) or other kinases strongly facilitates the interaction with Keap1 and activates Nrf2 (21).
Recently, in a search for non-electrophilic Nrf2 activators we identified that ezetimibe, an FDA-approved drug for the treatment of hyperlipidemia, also acts as a potent Nrf2 activator without causing cytotoxicity. The underlying mechanism of ezetimibe on Nrf2 activation was found to be mediated through AMP-activated protein kinase (AMPK)-dependent phosphorylation of the Ser351 site on p62 (30). We also previously identified that Sestrin2, a stress-responsive protein, activates Nrf2 by promoting p62-dependent autophagic Keap1 degradation through its interaction with the p62-Keap1-Nrf2 complex (5). Together, these findings indicate that p62-dependent autophagic Keap1 degradation in addition to Nrf2 stabilization represent likely mechanisms of Nrf2 activation.
Accordingly, in the current study, we found that verapamil, an FDA-approved drug for the treatment of hypertension, also activates Nrf2 by promoting p62-dependent autophagic Keap1 degradation. Verapamil is known to be a L-type calcium channel blocker and inhibits cytosolic Ca2+ levels (31). Verapamil is widely used in the treatment of cardiac vascular diseases such as cardiac arrhythmias, hypertension, and angina pectoris (32). Verapamil has also been reported to induce the autophagic pathway; for example, in colon cancer, verapamil treatment induces massive autophagy-like processes and leads to necrotic cell death (12). Furthermore, verapamil was shown to suppress saturated fatty acid-induced protein inclusion (p62 and ubiquitinated protein) by restoring the autophagic flux (13). Consistent with these reports, we found that verapamil-induced Keap1 degradation is mediated by an autophagic pathway.
We also found that verapamil has a protective role against APAP overdose-induced cell death, an effect also previously demonstrated to be associated with Nrf2 activation. APAP is commonly used as an analgesic and antipyretic drug (33); however, it induces hepatotoxicity when taken in overdose (15). APAP is metabolized by cytochrome P450 (CYP2E1) to generate NAPQI, which in turn may interrupt the repression of Nrf2 by modifying the cysteine residues of Keap1. In addition, NAPQI induces the depletion of glutathione, resulting in ROS accumulation. Furthermore, APAP significantly induces the accumulation of mitochondrial ROS by inhibiting the mitochondrial respiratory chain (16, 34). Notably, several groups have reported that Nrf2 has a protective role against hepatotoxicity produced by APAP in vivo and in vitro (24, 25, 35).
Accordingly, our current finding that verapamil mediated Nrf2 activation by promoting p62-dependent autophagic degradation of Keap1 and attenuated ROS accumulation, provides a suitable explanation for its protective role in APAP-induced cell death. Therefore, we propose that verapamil represents a promising FDA-approved drug for the treatment of oxidative stress-mediated damage through its action as a potent Nrf2 activator. Further studies are needed to investigate verapamil’s effect on the Nrf2 activation using in vivo hepatotoxicity model.
MATERIALS AND METHODS
See supplementary information for Materials and Methods.
Supplementary Information
ACKNOWLEDGEMENTS
This work was supported by the National Research Foundation of Korea (NRF-2013R1A1A2059087; S. H. Bae) and a Faculty Research Grant of the Yonsei University College of Medicine (6-2014-0068; S. H. Bae). It was also supported by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (HI16C0257; S. H. Bae and HI14C2476; Y. H. Lee).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting financial interests.
REFERENCES
- 1.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 2.Itoh K, Mimura J, Yamamoto M. Discovery of the negative regulator of Nrf2, Keap1: a historical overview. Antioxid Redox Signal. 2010;13:1665–1678. doi: 10.1089/ars.2010.3222. [DOI] [PubMed] [Google Scholar]
- 3.Taguchi K, Motohashi H, Yamamoto M. Molecular mechanisms of the Keap1-Nrf2 pathway in stress response and cancer evolution. Genes Cells. 2011;16:123–140. doi: 10.1111/j.1365-2443.2010.01473.x. [DOI] [PubMed] [Google Scholar]
- 4.Itoh K, Wakabayashi N, Katoh Y, et al. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bae SH, Sung SH, Oh SY, et al. Sestrins activate Nrf2 by promoting p62-dependent autophagic degradation of Keap1 and prevent oxidative liver damage. Cell Metab. 2013;17:73–84. doi: 10.1016/j.cmet.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 6.Bataille AM, Manautou JE. Nrf2: a potential target for new therapeutics in liver disease. Clin Pharmacol Ther. 2012;92:340–348. doi: 10.1038/clpt.2012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Musso G, Cassader M, Gambino R. Nonalcoholic steatohepatitis: emerging molecular targets and therapeutic strategies. Nat Rev Drug Discov. 2016;15:249–274. doi: 10.1038/nrd.2015.3. [DOI] [PubMed] [Google Scholar]
- 8.Johnson JA, Johnson DA, Kraft AD, et al. The Nrf2-ARE pathway: an indicator and modulator of oxidative stress in neurodegeneration. Ann N Y Acad Sci. 2008;1147:61–69. doi: 10.1196/annals.1427.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zoja C, Benigni A, Remuzzi G. The Nrf2 pathway in the progression of renal disease. Nephrol Dial Transplant. 2014;29(Suppl 1):i19–i24. doi: 10.1093/ndt/gft224. [DOI] [PubMed] [Google Scholar]
- 10.Goligorsky MS, Chaimovitz C, Rapoport J, Goldstein J, Kol R. Calcium metabolism in uremic nephrocalcinosis: preventive effect of verapamil. Kidney Int. 1985;27:774–779. doi: 10.1038/ki.1985.79. [DOI] [PubMed] [Google Scholar]
- 11.Rosei EA, Dal Palu C, Leonetti G, Magnani B, Pessina A, Zanchetti A. Clinical results of the Verapamil inHypertension and Atherosclerosis Study. VHAS Investigators. J Hypertens. 1997;15:1337–1344. doi: 10.1097/00004872-199715110-00019. [DOI] [PubMed] [Google Scholar]
- 12.Pajak B, Kania E, Gajkowska B, Orzechowski A. Verapamil-induced autophagy-like process in colon adenocarcinoma COLO 205 cells; the ultrastructural studies. Pharmacol Rep. 2012;64:991–996. doi: 10.1016/S1734-1140(12)70896-4. [DOI] [PubMed] [Google Scholar]
- 13.Park HW, Park H, Semple IA, et al. Pharmacological correction of obesity-induced autophagy arrest using calcium channel blockers. Nat Commun. 2014;5:4834. doi: 10.1038/ncomms5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park HW, Lee JH. Calcium channel blockers as potential therapeutics for obesity-associated autophagy defects and fatty liver pathologies. Autophagy. 2014;10:2385–2386. doi: 10.4161/15548627.2014.984268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.James LP, Mayeux PR, Hinson JA. Acetaminophen-induced hepatotoxicity. Drug Metab Dispos. 2003;31:1499–1506. doi: 10.1124/dmd.31.12.1499. [DOI] [PubMed] [Google Scholar]
- 16.Ni HM, Williams JA, Yang H, Shi YH, Fan J, Ding WX. Targeting autophagy for the treatment of liver diseases. Pharmacol Res. 2012;66:463–474. doi: 10.1016/j.phrs.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Copple IM, Goldring CE, Jenkins RE, et al. The hepatotoxic metabolite of acetaminophen directly activates the Keap1-Nrf2 cell defense system. Hepatology. 2008;48:1292–1301. doi: 10.1002/hep.22472. [DOI] [PubMed] [Google Scholar]
- 18.Li N, Alam J, Venkatesan MI, et al. Nrf2 is a key transcription factor that regulates antioxidant defense in macrophages and epithelial cells: protecting against the proinflammatory and oxidizing effects of diesel exhaust chemicals. J Immunol. 2004;173:3467–3481. doi: 10.4049/jimmunol.173.5.3467. [DOI] [PubMed] [Google Scholar]
- 19.Taguchi K, Fujikawa N, Komatsu M, et al. Keap1 degradation by autophagy for the maintenance of redox homeostasis. Proc Natl Acad Sci U S A. 2012;109:13561–13566. doi: 10.1073/pnas.1121572109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komatsu M, Kurokawa H, Waguri S, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 21.Ichimura Y, Waguri S, Sou YS, et al. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol Cell. 2013;51:618–631. doi: 10.1016/j.molcel.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 22.Hinson JA, Roberts DW, James LP. Mechanisms of acetaminophen-induced liver necrosis. Handb Exp Pharmacol. 2010:369–405. doi: 10.1007/978-3-642-00663-0_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGill MR, Sharpe MR, Williams CD, Taha M, Curry SC, Jaeschke H. The mechanism underlying acetaminophen-induced hepatotoxicity in humans and mice involves mitochondrial damage and nuclear DNA fragmentation. J Clin Invest. 2012;122:1574–1583. doi: 10.1172/JCI59755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldring CE, Kitteringham NR, Elsby R, et al. Activation of hepatic Nrf2 in vivo by acetaminophen in CD-1 mice. Hepatology. 2004;39:1267–1276. doi: 10.1002/hep.20183. [DOI] [PubMed] [Google Scholar]
- 25.Chan K, Han XD, Kan YW. An important function of Nrf2 in combating oxidative stress: detoxification of acetaminophen. Proc Natl Acad Sci U S A. 2001;98:4611–4616. doi: 10.1073/pnas.081082098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McMahon M, Thomas N, Itoh K, Yamamoto M, Hayes JD. Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a “tethering” mechanism: a two-site interaction model for the Nrf2-Keap1 complex. J Biol Chem. 2006;281:24756–24768. doi: 10.1074/jbc.M601119200. [DOI] [PubMed] [Google Scholar]
- 27.Yu Z, Shao W, Chiang Y, et al. Oltipraz upregulates the nuclear factor (erythroid-derived 2)-like 2 [corrected] (NRF2) antioxidant system and prevents insulin resistance and obesity induced by a high-fat diet in C57BL/6J mice. Diabetologia. 2011;54:922–934. doi: 10.1007/s00125-010-2001-8. [DOI] [PubMed] [Google Scholar]
- 28.Katsuragi Y, Ichimura Y, Komatsu M. p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J. 2015;282:4672–4678. doi: 10.1111/febs.13540. [DOI] [PubMed] [Google Scholar]
- 29.Lau A, Wang XJ, Zhao F, et al. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol. 2010;30:3275–3285. doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee DH, Han DH, Nam KT, et al. Ezetimibe, an NPC1L1 inhibitor, is a potent Nrf2 activator that protects mice from diet-induced nonalcoholic steatohepatitis. Free Radic Biol Med. 2016;99:520–532. doi: 10.1016/j.freeradbiomed.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 31.Sarkar S. Regulation of autophagy by mTOR-dependent and mTOR-independent pathways: autophagy dysfunction in neurodegenerative diseases and therapeutic application of autophagy enhancers. Biochem Soc Trans. 2013;41:1103–1130. doi: 10.1042/BST20130134. [DOI] [PubMed] [Google Scholar]
- 32.Schmieder RE, Messerli FH, Garavaglia GE, Nunez BD. Cardiovascular effects of verapamil in patients with essential hypertension. Circulation. 1987;75:1030–1036. doi: 10.1161/01.CIR.75.5.1030. [DOI] [PubMed] [Google Scholar]
- 33.Prescott LF, Critchley JA. The treatment of acetaminophen poisoning. Annu Rev Pharmacol Toxicol. 1983;23:87–101. doi: 10.1146/annurev.pa.23.040183.000511. [DOI] [PubMed] [Google Scholar]
- 34.Jiang J, Briede JJ, Jennen DG, et al. Increased mitochondrial ROS formation by acetaminophen in human hepatic cells is associated with gene expression changes suggesting disruption of the mitochondrial electron transport chain. Toxicol Lett. 2015;234:139–150. doi: 10.1016/j.toxlet.2015.02.012. [DOI] [PubMed] [Google Scholar]
- 35.Liu J, Wu KC, Lu YF, Ekuase E, Klaassen CD. Nrf2 protection against liver injury produced by various hepatotoxicants. Oxid Med Cell Longev. 20132013:305861. doi: 10.1155/2013/305861. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.