Abstract
Protein glycosylation is a post-translational modification (PTM) responsible for many aspects of proteomic diversity and biological regulation. Assignment of intact glycan structures to specific protein attachment sites is a critical step towards elucidating the function encoded in the glycome. Previously, we developed isotope targeted glycoproteomics (IsoTaG) as a mass-independent mass spectrometry method to characterize azide-labeled intact glycopeptides from complex proteomes. Here we extend the IsoTaG approach with the use of alkynyl sugars as metabolic labels, and employ new probes in analysis of the sialylated glycoproteome from PC-3 cells. Using an Orbitrap Fusion Tribrid mass spectrometer, we identified 699 intact glycopeptides from 192 glycoproteins. These intact glycopeptides represent a total of eight sialylated glycan structures across 126 N- and 576 O-glycopeptides. IsoTaG is therefore an effective platform for identification of intact glycopeptides labeled by alkynyl or azido-sugars and will facilitate further studies of the glycoproteome.
Keywords: Glycoproteomics, chemical proteomics, LC-MS/MS, metabolic labeling, sialic acid
Introduction
Glycosylation is a heterogeneous post-translational modification (PTM) that decorates proteins from the extracellular matrix to the intracellular nucleus in mammalian cells. The glycoproteome influences diverse biological processes, such as immunological regulation [1] or cancer progression[2,3], via modulation of innate biophysical properties[4]. Recent attention has been focused on sialylated glycans due to their prevalence on many cancers and their proposed roles in metastasis [5,6] and immune evasion [7,8]. Likewise, fucosylated glycans have been associated with cancer and may play similar roles [9,10]. Thus, these glycan class are important to understand in molecular details as they may hold keys to new therapeutic and diagnostic strategies.
Until recently, cancer glycosylation was largely characterized at the level of global cell surface abundance using lectins, antibodies, or related detection reagents [11,12]. A more detailed structural analysis of cancer glycoproteomes has been challenging due to the complex and heterogeneous nature of glycosylation [13,14]. Nonetheless, progress has been accelerating. Structural profiling of glycoproteins from complex mixtures has been tackled using various enrichment strategies and mass spectrometry (MS) to obtain information about glycan attachment site and glycan heterogeneity. Glycoprotein enrichment by chemical oxidation followed by hydrazide capture[15], lectin affinity[16], chemical labeling[17], and metabolic labeling [18] approaches have been reported, among other techniques [14,19,20]. In most studies, glycans were separated from their peptide scaffolds for direct observation of the glycosite. This can be accomplished by treatment with PNGase F or Endo H for N-glycosite characterization [21,22], oxidative proteolysis [23], beta elimination for O-glycans [24], or acid hydrolysis of terminal sialic acid .[25]. As well, a genetic engineering approach to identify O-glycosites has been developed [26,27].
Several MS methods show promise for intact glycopeptide analysis [28,29], including correlation of the released glycans with deglycosylated peptides to assign the intact glycopeptide [30]. However, these methods require that glycans and their peptide substrates are characterized in separate analyses [31–33]. Consequently, when these approaches are applied to complex protein mixtures, glycan–protein associations can only be made indirectly, and are therefore less accurate than what might achieved by direct observation. But, current methods for direct observation, while increasing confidence of glycan–protein associations, are burdened with challenging assignments due to potential insufficient tandem MS coverage [34,35]. A directed method to characterize intact glycopeptides is necessary as the assignment of heterogeneous glycoproteins within a complex mixture is computationally challenging.
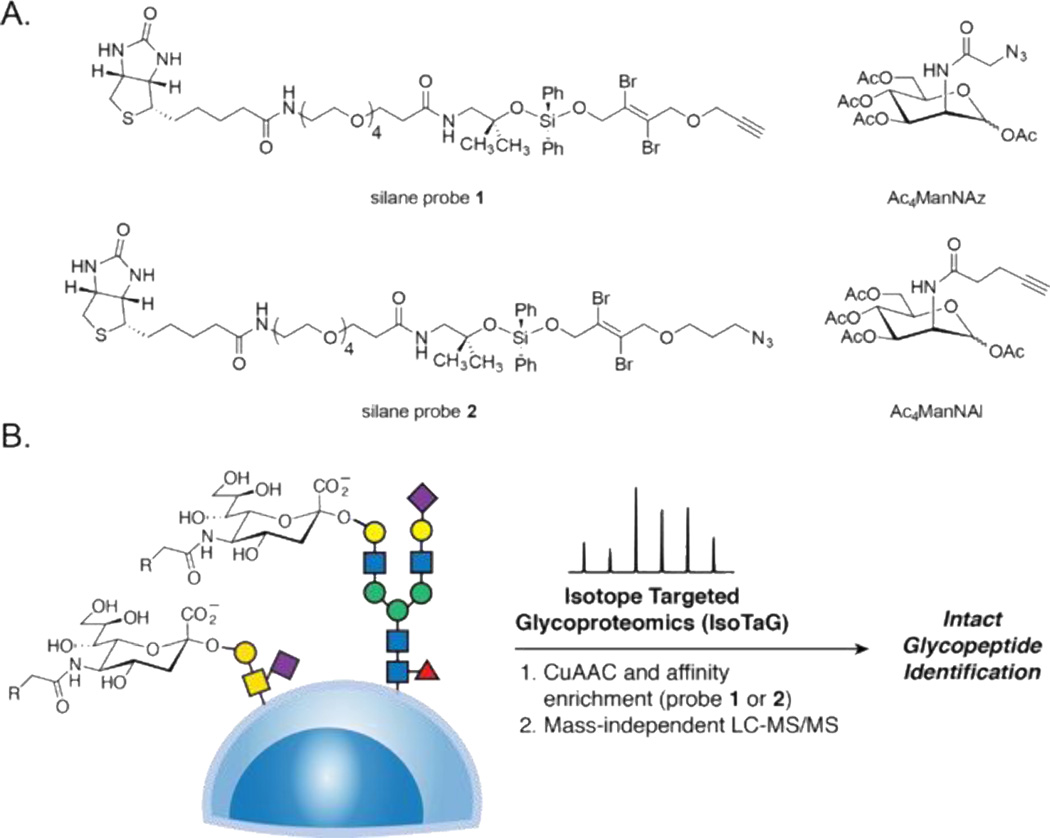
An orthogonal approach to enrich and confidently assign intact glycopeptides involves use of enrichment with cleavable probes together with mass-independent MS. Recent work from our lab has applied this approach to a platform termed isotope targeted glycoproteomics (IsoTaG) to tag and characterize a range of glycopeptides, including those bearing sialylated glycans [36]. The process begins with metabolic labeling of cultured cells with azide-functionalized monosaccharide substrates. For example, peracetylated N-azidoacetylmannosamine (Ac4ManNAz) is metabolized to the corresponding azido sialic acid (SiaNAz) and then labels sialylated glycans. Likewise, peracetylated N-azidoacetylgalactosamine (Ac4GalNAz) metabolically labels glycans bearing GalNAc or GlcNAc, which includes numerous mammalian glycan structures. After metabolic labeling, the glycoproteins are affinity enriched with cleavable alkynyl biotin probe 1, which we attach to the target substrates by copper-catalyzed azide–alkyne cycloaddition (CuAAC, also called “click chemistry”, Figure 1A). Simultaneously, glycopeptides are isotopically recoded by two bromine atoms embedded within the probe for targeted assignment of the glycopeptide by MS. Isotopic recoding enables mass-independent MS where glycopeptides are immediately recognized by full scan MS to guide subsequent targeted assignment.
Figure 1.
Chemical probe design and application in IsoTaG. A. Complementary probe and glycan pairs. Silane probe 1 reacts selectively with azido sugars (e.g., Ac4ManNAz) glycoproteins. Silane probe 2 reacts selectively with alkynyl sugars (e.g., Ac4ManNAl) glycoproteins. B. Application of IsoTaG to sialylated glycans expressed on the cell surface and secreted.
The IsoTaG approach relies on the efficiency of metabolic labeling by azido sugars to install a handle for tagging with probe 1. Metabolic labeling enables selection of glycoproteins as they are biosynthesized and has been performed in cell culture [18], live animals [37,38], and human tissues [39]. Labeling efficiency, however, is highly dependent on the activity of the biosynthetic enzymes. In some cases, metabolic labeling with alkyne-bearing sugar analogs is advantageous. For example, metabolic labeling with peracetylated N-(4-pentynoyl)mannosamine (Ac4ManNAl) was found to proceed at higher efficiency than with Ac4ManNAz in mice [38]. Alkynyl analogs of fucose [e.g., peracetylated 6-alkynyl fucose (Ac4FucAl)] are incorporated into mammalian glycans with less toxicity as compared to azido analogs [40]. Furthermore, alkynyl GlcNAc analogs for specific labeling of O-GlcNAc vs other GlcNAc-containing glycans have been developed [41]. To exploit these alkynyl sugars in the context of IsoTaG will require the development of new probes and methods.
Here we describe the design of an IsoTaG-compatible azide probe (compound 2) that enables analysis of intact alkyne-labeled glycopeptides. Glycoproteins labeled with alkynyl sugars in the secretome of PC-3 cells were structurally characterized by IsoTaG on an Orbitrap Fusion Tribrid, alongside azide-labeled glycoproteins for comparison. Thirty-seven alkyne-labeled and 695 azide-labeled sialoglycopeptides, respectively, were found from the secretome of PC-3 cells. In sum, 699 intact glycopeptides whose structures included eight discrete sialylated glycans were identified from 192 glycoproteins across 126 N- and 576 O-glycopeptides. These results establish IsoTaG as an effective methodology to characterize intact metabolically-labeled glycopeptides across N- and O-glycans.
Materials and Methods
Chemical Materials
Commercial solvents and reagents were used as received with the following exceptions. Dichloromethane was purified according to the method of Pangborn and co-workers [42]. Triethylamine was distilled from calcium hydride under an atmosphere of nitrogen immediately before use. RapiGest was prepared according to the method of Lee and co-workers [43]. 3-[4-({Bis[(1-tert-butyl-1H-1,2,3-triazol-4-yl)methyl]amino}methyl)-1H-1,2,3-triazol-1-yl]propanol (BTTP) was prepared according to the method of Wu and co-workers [44]. Tetraacetylated N-(4-pentynoyl)mannosamine (Ac4ManNAl) was prepared according to the method of Wong and co-workers [40]. Peracetylated 6-alkynyl fucose (Ac4FucAl) was obtained from Thermo Fisher. Tetraacetylated N-azidoacetyl mannosamine (Ac4ManNAz) was prepared according to the method of Bertozzi and co-workers [45]. EDTA-free protease inhibitor cocktail was obtained from Roche Diagnostics (Version 11). Streptavidin–agarose beads were obtained from Thermo Scientific and washed with PBS prior to use. The silane probe 1 was prepared according to the procedure of Bertozzi and co-workers [36]. Bovine serum albumin (BSA) was obtained from Sigma. Biotin-PEG3-azide was obtained from Sigma.
Synthetic Procedures
The silane probe 2 was prepared as shown in Electronic Supplementary Material (ESM) Figure S1. Please see ESM for full synthetic procedures.
Cell Culture and Metabolic Labeling
PC-3 cells were obtained from the American Type Culture Collection (ATCC) and maintained at 37 °C and 5% CO2 in a water-saturated incubator. PC-3 cells were metabolically labeled between passages 17–22. Cell densities were counted using a hemacytometer and seeded at 2 × 105 cells/mL at the start of metabolic labeling experiments. PC-3 cells were maintained in RPMI-1640 supplemented with 10% FBS and 1% penicillin/streptomycin.
Azido and alkynyl sugars (Ac4ManNAz, Ac4FucAl, or Ac4ManNAl) were prepared as 500 mM stock solutions in dimethylsulfoxide (DMSO). Tissue culture dishes (150 mm) were seeded with 100 µM of Ac4ManNAl, Ac4ManNAz, Ac4FucAl, or vehicle control containing DMSO (3.0 µL). Six dishes per condition were prepared. A suspension of cells at a density of 2 × 105 cells/mL in complete media (RPMI supplemented with 10% FBS and 1% penicillin/streptomycin) was added to the dish (15 mL per dish) and the dishes were incubated for 48 h at 37 °C. The media was aspirated and the adherent cells were washed with PBS (1 × 10 mL). Washed dishes were resuspended in RPMI containing 100 µM glycan metabolite without FBS additive (15 mL), and the cells were incubated an additional 48 h at 37 °C in a humidified 5% CO2 incubator.
The conditioned media (100 mL) was harvested and cleared by centrifugation (150 × g, 3 min). Clarified media was spin concentrated (Amicon, 15 mL 10 kDa spin filter) to 1 mL. The concentrated residue was washed with 1% Triton x100/PBS (3 × 15 mL), and transferred to an Eppendorf microcentrifuge tube as the conditioned media fraction. The conditioned media fraction was adjusted to a final concentration of 1% RapiGest/PBS with a 10% RapiGest/PBS stock solution. Protein concentrations from the conditioned media fractions were measured by bicinchonic acid assay (Pierce).
Chemical Enrichment
Conditioned media from PC-3 cells treated with Ac4ManNAl, Ac4ManNAz, Ac4FucAl, or DMSO vehicle were aliquoted to 2.0 mg fractions (667 µL). To Ac4ManNAz-labeled conditioned media, click chemistry reagents (40.0 µL, 200 µM 1, 300 µM CuSO4, 100 mM BTTP, 2.50 mM sodium ascorbate, mixed immediately before addition to lysates) were added and the reaction was incubated for 3 h at 24 °C. To Ac4ManNAl-labeled, Ac4FucAl-labeled, or DMSO vehicle conditioned media, click chemistry reagents (40.0 µL, 200 µM 2, 300 µM CuSO4, 100 mM BTTP, 2.50 mM sodium ascorbate, mixed immediately before addition to lysates) were added and the reaction was incubated for 3 h at 24 °C. Methanol (1 mL) was added to quench the reaction, and proteins were precipitated for 1 h at –80 °C. Precipitated proteins were pelleted by centrifugation (16,100 × g, 10 min, 4 °C) and the supernatant was discarded. Pelleted proteins were air-dried for 10 min at 24 °C. Dried protein pellets were resuspended in 400 µL of 1% RapiGest/PBS and solubilized by probe sonication (Misonix, 1.0 min, 4 °C). Streptavidin–agarose resin [200 µL, washed with PBS (3 × 1 mL)] was added, and the resulting mixture was incubated for 12 h at 24 °C with rotation. The beads were pelleted by centrifugation (3000 × g, 3 min) and the supernatant containing uncaptured proteins was separated. The beads were washed with 1% RapiGest/PBS (1 mL), 6 M urea (2 × 1 mL), and PBS (5 × 1 mL), and the beads were pelleted by centrifugation (3000 × g, 3 min) between washes.
Washed beads were resuspended in 5 mM DTT/PBS (200 µL) and incubated for 30 min at 24 °C with rotation. Ten mM iodoacetamide (4.0 µL, 500 mM stock solution in PBS) was added to the reduced proteins, and the resultant solution allowed to react for 30 min at 24 °C with rotation, in the dark. Beads were pelleted by centrifugation (3000 × g, 3 min) and resuspended in 0.5 M urea/PBS (200 µL). Trypsin (1.5 µg) was added to the resuspended beads, and digestion proceeded for 12 h at 37 °C. Beads were pelleted by centrifugation (3000 × g, 3 min), and the supernatant digest was collected. The beads were washed with PBS (1 × 200 µL) and H2O (2 × 200 µL). Washes were combined with the supernatant digest to form the trypsin digest of nonconjugated peptides. Probe 1 or 2 was cleaved with two treatments of 2% formic acid/H2O (200 µL) for 30 min at 24 °C with rotation and the eluent was collected. The beads were washed with 50% acetonitrile–water + 1% formic acid (2 × 400 µL), and the washes were combined with the eluent to form the cleavage fraction. The trypsin digest and cleavage fraction were concentrated using a vacuum centrifuge (i.e., a speedvac, 40 °C) to 50 µL. Samples were desalted with a ZipTip P10 and stored at –20 °C until analysis.
Western Blotting
Aliquots collected during the enrichment procedure (10.0 µL) were reduced and separated by standard SDS-PAGE (Bio-Rad, Criterion system), electroblotted onto nitrocellulose, blocked in 5% BSA in Tris-buffered saline with Tween (10 mM Tris pH 8.0, 150 mM NaCl, 0.1% Tween-20), and analyzed by standard enhanced chemiluminescence immunoblotting methods (Pierce). The staining agent was streptavidin–HRP (Pierce, 1:100,000).
Liquid Chromatography–Tandem Mass Spectrometry (LC-MS/MS)
Peptides were analyzed by online capillary nanoLC-MS/MS. Samples were separated on an in-house made 20-cm reversed phase column [100 µm inner diameter, packed with ReproSil-Pur C18-AQ 3.0 μm resin (Dr. Maisch, GmbH)] equipped with a laser-pulled nanoelectrospray emitter tip. Peptides were eluted at a flow rate of 400 nL/min using a two-step linear gradient including 3–25% buffer B in 70 min and 25–40% B in 20 min (buffer A: 0.2% formic acid in water; buffer B: 0.2% formic acid in acetonitrile) in a Dionex UltiMate 3000 HPLC system (Thermo Fisher Scientific). Peptides were then analyzed using an Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher Scientific). Data acquisition was executed in data dependent mode (top speed with duty cycle of 3 s) with full MS scans acquired in the Orbitrap mass analyzer with a resolution of 120000 and m/z scan range of 400–1500. Precursor ions with charge state 2–7 and intensity threshold above 5000 counts were selected for fragmentation using higher-energy collisional dissociation (HCD) with quadrupole isolation, isolation window of 4 m/z and collision energy of 30%. The HCD fragments were analyzed in the Orbitrap mass analyzer with a resolution of 15000. Dynamic exclusion was enabled with a repeat count of 1 and exclusion duration of 30 s. The automatic gain control (AGC) target was set to 400000 and 50000 for full FTMS scans and FTMS2 scans, respectively. The maximum injection time was set to 50 s and 250 s for full FTMS scans and FTMS2 scans. Data from both scans were acquired in profile mode. For additional HCD-triggered MS2 scans, targeted loss trigger and targeted product trigger were performed in separate runs. For targeted loss trigger, the masses of both neutral loss species and their corresponding M+H forms were added to the inclusion mass list. For product ion trigger, the m/z values of the M+H products were added to the inclusion mass list. The inclusion mass list was scanned for throughout data collection. Electron transfer dissociation (ETD) reactions were performed for triggered MS2 following previous HCD scans. Triggered precursor ions with charge state 3–7 were subject to ETD with quadrupole isolation, isolation window of 4 m/z, reaction time of 40 ms, reagent target of 200000, and maximum reagent injection time of 200 ms. ETD supplemental activation (EThcD) was enabled with collision energy of 25%. The detection of ETD fragment ions was performed with the same parameters as HCD scans.
Data analysis
The raw data were processed using Proteome Discoverer 1.4 software (Thermo Fisher Scientific) and searched against the human-specific SwissProt database downloaded on April 29, 2016. Indexed databases for tryptic digests were created allowing for up to three missed cleavages, one fixed modification (carboxyamidomethylcysteine, +57.021 Da), and variable modifications (methionine oxidation, +15.995 Da; and others as described below). Precursor ion mass tolerance for spectra acquired using the Orbitrap were set to 10 ppm. The fragment ion mass tolerance was set to 20 ppm. The SEQUEST HT search engine was used to assign nonconjugated peptides from on-bead tryptic digests. Normalized protein assignments at a 1% false discovery rate (FDR) were considered enriched if the fold change was greater than two and the associated p-value was ≤ 0.05 (t-test) in labeled samples than in the DMSO control (ESM Figure S2).
Samples with intact glycopeptides were searched with the Byonic search algorithm v2.8.2 as a node in Proteome Discoverer 1.4. Byonic is a software package that allows definition of the number of occurences for each modification, which prevents a combinatorial explosion when searching for multiple glycan structures simultaneously [46]. The Byonic score is defined by the absolute quality of the peptide– spectrum match over a range of 0 to 1000, where 300 is a good score and 400 a very good score. Example glycan structure input files are provided in the ESM for Ac4ManNAl, Ac4ManNAz, and Ac4FucAl glycans, respectively. Following a search against the SwissProt human proteome, a search against the pool of enriched glycoproteins was performed. Glycopeptide assignments were aggregated and a minimum of two separate assignments at 5% FDR to the same underlying peptide was used as a cutoff for further analysis. The DMSO control sample was searched separately against each of the glycan structure input files and no intact glycopeptide assignments were reported. Assignments of all spectra in Ac4FucAl-labeled samples were validated by manual inspection for the precursor isotope pattern and expected glycan fragments.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [47] partner repository with the dataset identifier PXD004302. Username: reviewer96861@ebi.ac.uk Password: kVzaRFPo
Results and Discussion
The azide biotin probe 2 was designed to closely mimic the IsoTaG compatible alkyne probe 1 (Figure 1A). Preparation of probe 2 was achieved in three steps (ESM Figure S1). Probe 2 preserves the dibromide motif for isotope recoding with a triplet signature (1:2:1 over M, M+2, M+4) and the cleavable silane linker,[48] which is cleaved in acidic conditions (2% formic acid) tolerated by glycoconjugates. The dibromide motif is detectable computationally using a pattern-recognition algorithm developed in house, termed IsoSTAMP [49].
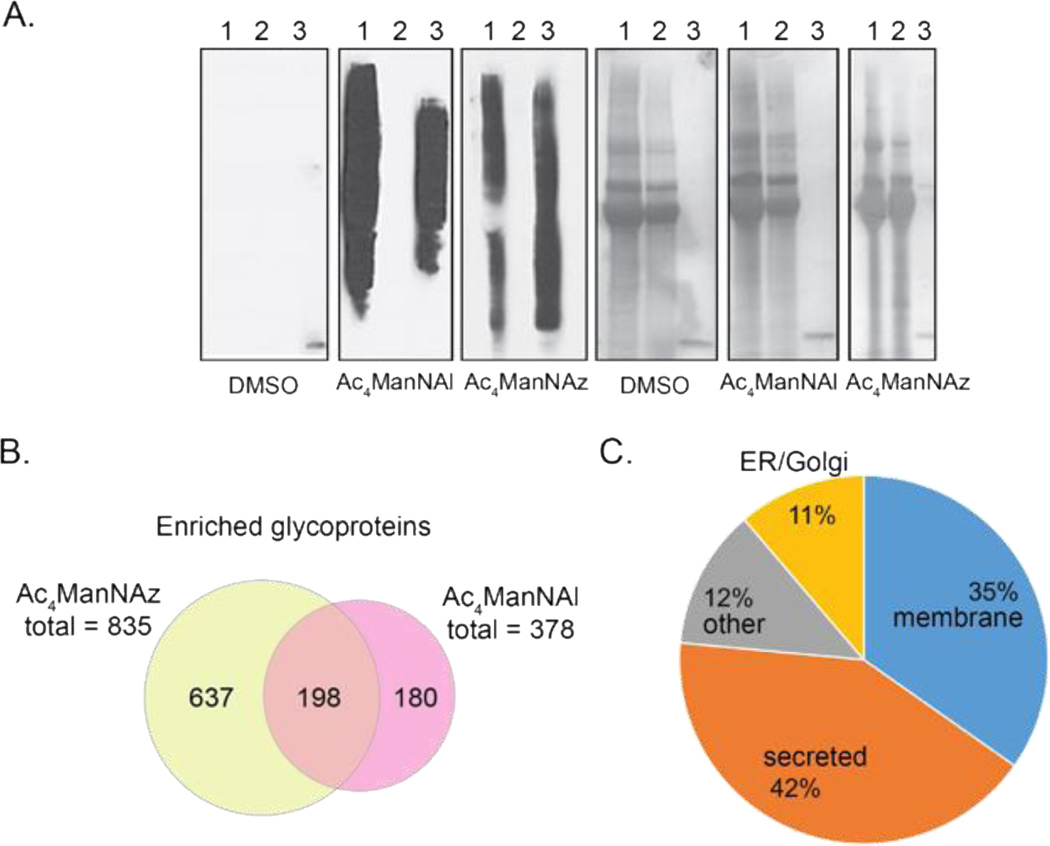
To evaluate the performance of the azide biotin probe 2, PC-3 cells were metabolically labeled with 100 μM Ac4ManNAl for two days in FBS-free media. Ac4ManNAl is intracellularly deacetylated, converted to the corresponding alkynyl sialic acid (SiaNAl), and installed on sialylated glycoproteins [40,38]. The conditioned media were collected and reacted with azide probe 2 by CuAAC. For comparison, PC-3 cells were labeled with 100 µM Ac4ManNAz to produce the corresponding azido sialic acid (SiaNAz), and the conditioned media was reacted with alkyne biotin probe 1 [45,50]. Western blot analysis showed similar tagging and enrichment of conditioned media from cells treated with Ac4ManNAl or Ac4ManNAz, with little off-target tagging from cells treated with the DMSO vehicle (Figure 2A and ESM Figure S3).
Figure 2.
Global analysis of glycoproteins and glycopeptides identified by IsoTaG analysis of conditioned media from metabolically labeled PC-3 cells. A. Anti-biotin Western blot analysis of tagging and enrichment efficiency with DMSO control, Ac4ManNAl, and Ac4ManNAz samples. Total protein levels shown alongside (ponceau). Lanes: (1) protein input following CuAAC reaction with probe 1 or 2, (2) protein supernatant following overnight incubation with streptavidin–agarose beads, (3) aliquot of streptavidin–agarose beads (10 µL) after enrichment. B. Venn diagram of glycoproteins identified from nonconjugated on-bead tryptic peptides from Ac4ManNAz and Ac4ManNAl samples. C. Subcellular localization of proteins identified by direct intact glycopeptide assignments.
Tagged glycoproteins were affinity enriched with streptavidin–agarose beads and glycoproteins were proteolyzed on-bead with trypsin. Nonconjugated peptides from trypsin proteolysis were assigned by SEQUEST HT and analyzed for enrichment (Figure 2B). High-confidence peptide assignments (1% FDR) were aggregated into protein groups. Proteins were considered enriched if a fold change of greater than two and associated p-value ≤ 0.05 was found in metabolically labeled samples as compared to the DMSO control. A pool of 1015 proteins were enriched from a total 1539 identified proteins (66%, ESM Figure S2 and Table S1). Of these glycoproteins, 198 were found in both metabolically labeled samples representing a proteomic overlap of greater than 50% between Ac4ManNAz and Ac4ManNAl samples, with respect to the Ac4ManNAl sample. The greatest number of enriched proteins was identified from Ac4ManNAz conditioned media (835 proteins). Differences in proteomic overlap may reflect the ability of azido and alkynyl sugars to label distinct glycoprotein subsets, as well as the relative permissiveness of PC-3 cells for Ac4ManNAz.
Following release of nonconjugated peptides for glycoprotein identification, beads were treated with 2% formic acid to cleave the silane linker and recover intact glycopeptides for mass-independent MS analysis. The released glycopeptides were analyzed by reversed-phase nanoflow liquid chromatography coupled to an ETD-enabled Orbitrap Fusion Tribrid mass spectrometer. The Fusion Tribrid enables acquisition of HCD and ETD with high mass accuracy to improve fragment assignment by database searching. Acquisition strategies such as HCD product-dependent ETD (HCD-pd-ETD), where ETD is triggered in real-time from observed glycan oxonium ions in HCD spectra, have been shown to optimize MS time for glycopeptide analysis [29]. Thus, an HCD-pd-ETD method with supplemental activation was employed for glycopeptide analysis. Two technical replicates were obtained with ETD triggered from the neutral/charged loss ion or the product ion, respectively.
Dibromide incorporation into the probes 1 and 2 creates an isotopically recoded mass envelope (Figure 1B). We use this isotopic signature as an orthogonal, mass-independent handle for confident glycopeptide identification. The isotopic signature thus introduces a metric to evaluate the efficiency of the product-dependent ETD trigger, and validate glycopeptide false positives and false negatives during database assignment. The isotopic pattern additionally allows rapid assessment of the existence of intact glycopeptides. We applied the pattern searching algorithm IsoStamp to the raw data, and found that without any further analysis, Ac4ManNAz conditioned media possessed approximately 40 times more species carrying the isotopic pattern as compared to the full scan MS from Ac4ManNAl conditioned media. Spectra were searched against the Swiss-Prot human proteome and the database of enriched glycoproteins described above using Byonic v2.8 [46]. Without any further manipulation, up to 928 spectra were assigned as intact glycopeptides in a single MS analysis. A product ion or neutral/charged loss trigger for ETD were equally advantageous for glycopeptide assignment. Glycopeptide assignments with two or more spectral counts at 5% FDR were considered for further analysis. Manual validation for the dibromide precursor of 100 of these glycopeptide assignments in the Ac4ManNAz sample found a 98% true positive rate for the isotopic pattern. Spectra that were assigned as a glycopeptide by database searching, but did not possess an isotopically recoded precursor were thus excluded. As an indication of the confidence that site level analysis enables in glycoproteome studies, no intact glycopeptides were assigned to the DMSO control.
A total of 699 intact glycopeptides were assigned from 192 glycoproteins in PC-3 conditioned media. Intact glycopeptides represented 126 N-glycans and 576 O-glycans. Subcellular localization of the proteins from which they derived was predominantly at the plasma membrane (35%) and secretome (42%), as expected for analysis of conditioned media (Figure 2C). Proteins were additionally annotated to endoplasmic reticulum/Golgi apparatus (11%) and other (12%, e.g., lysosome, vesicles, cytoplasmic) locations within the cell. The molecular function annotation of these glycoproteins includes binding to ions (e.g., calcium, metal), protein binding interactions, and glycan binding (Table 1). The identified glycosites derive from approximately 20% of the glycoproteins observed by nonconjugated peptide analysis (ESM Table S2).
Table 1.
Top 15 molecular function annotations for identified glycoproteins.
| Molecular Function | Protein Count |
|---|---|
| calcium ion binding | 36 |
| heparin binding | 19 |
| metal ion binding | 18 |
| identical protein binding | 15 |
| structural molecule activity | 13 |
| protein homodimerization activity | 12 |
| integrin binding | 11 |
| poly(A) RNA binding | 11 |
| zinc ion binding | 10 |
| cytokine activity | 8 |
| enzyme binding | 7 |
| protease binding | 7 |
| serine-type endopeptidase inhibitor activity | 7 |
| ATP binding | 6 |
| DNA binding | 6 |
Many unassigned spectra were found to derive from isotopically recoded precursors, an indicator that additional assignments may be made on closer analysis. Spectra from the the highest yielding Ac4ManNAz analysis revealed that of 2692 MS2 spectra derived from patterned species, 1542 spectral assignments were made. Spectral unassignment may be caused by low spectrum quality, modification types that were not included in the database search (e.g., acetylation, formylation, alternate glycan structures, amino acid variants) [36], and remaining challenges in automated assignment of glycan, peptide, and particularly glycopeptide fragments. Further analysis using the isotopically recoded precursor may guide assignment of these spectra.
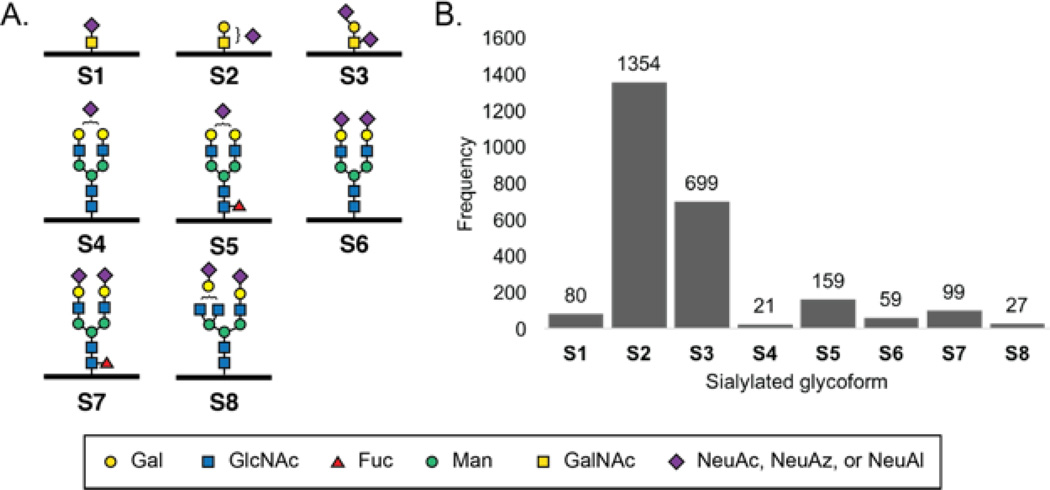
The observed sialylated glycopeptides contained eight discrete sialylated glycan structures (Figure 3A). During database searching, N- and O-glycan structures were searched simultaneously. Sialylated glycans included three O-glycans (S1–S3), moderately complex N-glycans (S4, S6, S8), and their fucosylated counterparts (S5, S7). In particular, the sialyl Tn O-glycan epitope (S1) and Core 1 sialylated glycans (S2, S3) were assigned at a greater frequency than any of the N-glycans. The higher assignment rate of O-glycans may be related to their relative abundance or the relative depth of peptide fragmentation observed with O- vs N-glycopeptides. Glycopeptide assignments were two orders of magnitude higher for conditioned media from azidosugar-labeled PC-3 compared to alkynyl-sugar labeled PC-3 cells (Table S2). This discrepancy is supported by the greater number of glycoproteins enriched from azidosugar-labeled media (ESM Table S1). Due to incorporation differences across metabolites, any quantitative application of IsoTaG would require comparison only when the same metabolite and cell line was used.
Figure 3.
Intact sialylated glycan structures and frequency of occurrence. A. Intact sialylated glycans identified (S1–S8). S1–S3 are O-glycans. S4–S8 are N-glycans. B. Frequency of sialyl glycans observed by spectral counting.
Fucosylated glycopeptide assignments were additionally sought by treatment of PC-3 cells with 100 µM Ac4FucAl [40]. Enrichment and MS analysis of Ac4FucAl-labeled conditioned media led to the identification of 145 intact glycopeptides with a minimum of two spectral counts, and no corresponding assignments found in the DMSO control. However, the assignments corresponding to FucAl-labeled glycopeptides carried low scores (max score = 82, Byonic), and the absence of isotopic recoding due to the dibromide in the full scan MS precursor confirmed these assignments as false positives. Independent analysis by our computational algorithm of the MS1 dataset revealed no isotopically recoded precursors from FucAl-labeled glycopeptides. The incorrect assignment of fucosylated glycopeptides may derive from the fact that fucose, unlike sialic acid, does not generate oxonium ions during tandem MS, and therefore must be inferred indirectly through neutral or charged losses. The low abundance of FucAl-labeled glycopeptides may reflect the low permissivity of the fucose salvage pathway enzymes in PC-3 cells, low rates of fucosylation due to enzymatic suppression [10], or steric inaccessibility of FucAl-labeled glycans to the chemical probe. We investigated the potential for steric inhibition by comparison of tagging efficiency by probe 2 and a biotin-PEG3-azide. By western blot analysis, we found no anti-biotin signal from conditioned media of Ac4FucAl-labeled PC-3 cells using either probe (ESM Figure S4).
Over 80 glycopeptides were identified that exist as multiple glycoforms, a selection of which are highlighted in Table 2. Up to four unique glycoforms were assigned to a single glycopeptide (Entry 9, 15). Several glycoproteins displayed multiple glycosites with high heterogeneity (Entry 9, 12, 15). In some cases, both N- and O-glycosylation was found on the same glycopeptide in separate instances (Entry 2, 9). The full GO molecular function annotation and subcellular localization are presented in ESM Tables S3 and S4. Thus IsoTaG can identify both N- and O-glycans in an unbiased manner by mass-independent MS in a single analysis.
Table 2.
Selected examples of intact glycopeptides with more than three glycoforms identified. Glycosite(s) indicated by lower case letter. IsoTaG enriched glycopeptides carrying SiaNAz were analyzed on an Orbitrap Fusion Tribrid and assigned by database searching (Byonic).
| Entry | Row Labels | Glycan | Protein Name (Accession) |
|---|---|---|---|
| 1 | ALGTHVIHSTHTLPLtVTSQQGVK | S1, S2, S3 | Latent-transforming growth factor beta- binding protein 1 (Q14766) |
| 2 | DHHQAsnSSR | S2, S5, S7 | Dickkopf-related protein 1 (O94907) |
| 3 | DWENQLEASmHSVLSDLHEAVPtVVGIPDG TAVVGR |
S1, S2, S3 | Dystroglycan (Q14118) |
| 4 | EQInITLDHR | S4, S5, S6, S7 |
Procollagen-lysine,2-oxoglutarate 5- dioxygenase 1 (Q02809) |
| 5 | GAPNKEEtPATESPDTGLYYHR | S1, S2, S3 | Nucleobindin-1 (Q02818) |
| 6 | GGGPDPEWGSANtPVPGAPAPHSS | S1, S2, S3 | Prostate-associated microseminoprotein (Q1L6U9) |
| 7 | HnSTGcLR | S2, S5, S7 | Clusterin (P10909) |
| 8 | RtTLSSK | S1, S2, S3 | Dickkopf-related protein 1 (O94907) |
| 9 | sHnR | S3, S4, S5, S7 |
Metalloproteinase inhibitor 1 (P01033) |
| 10 | SHnRSEEFLIAGK | S5, S6, S7 | Metalloproteinase inhibitor 1 (P01033) |
| 11 | StHPPPLPAK | S1, S2, S3 | Latent-transforming growth factor beta- binding protein 1 (Q14766) |
| 12 | STHPPPLPAKEEPVEALtFsR | S1, S2, S3 | Latent-transforming growth factor beta- binding protein 1 (Q14766) |
| 13 | TQTIHSTYsHQQVIPHVYPVAAK | S1, S2, S3 | Latent-transforming growth factor beta- binding protein 1 (Q14766) |
| 14 | VALLQFGGPGEQQVAFPLSHnLTAIHEALET TQYLNSFSHVGAGVVHAINAIVR |
S6, S7, S8 | Collagen alpha-2(VI) chain (P12110) |
| 15 | YIHQnYTK | S4, S5, S6, S8 |
Procollagen-lysine,2-oxoglutarate 5- dioxygenase 1 (Q02809) |
| 16 | YSQAVPAVtEGPIPEVLK | S1, S2, S3 | Cathepsin D (P07339) |
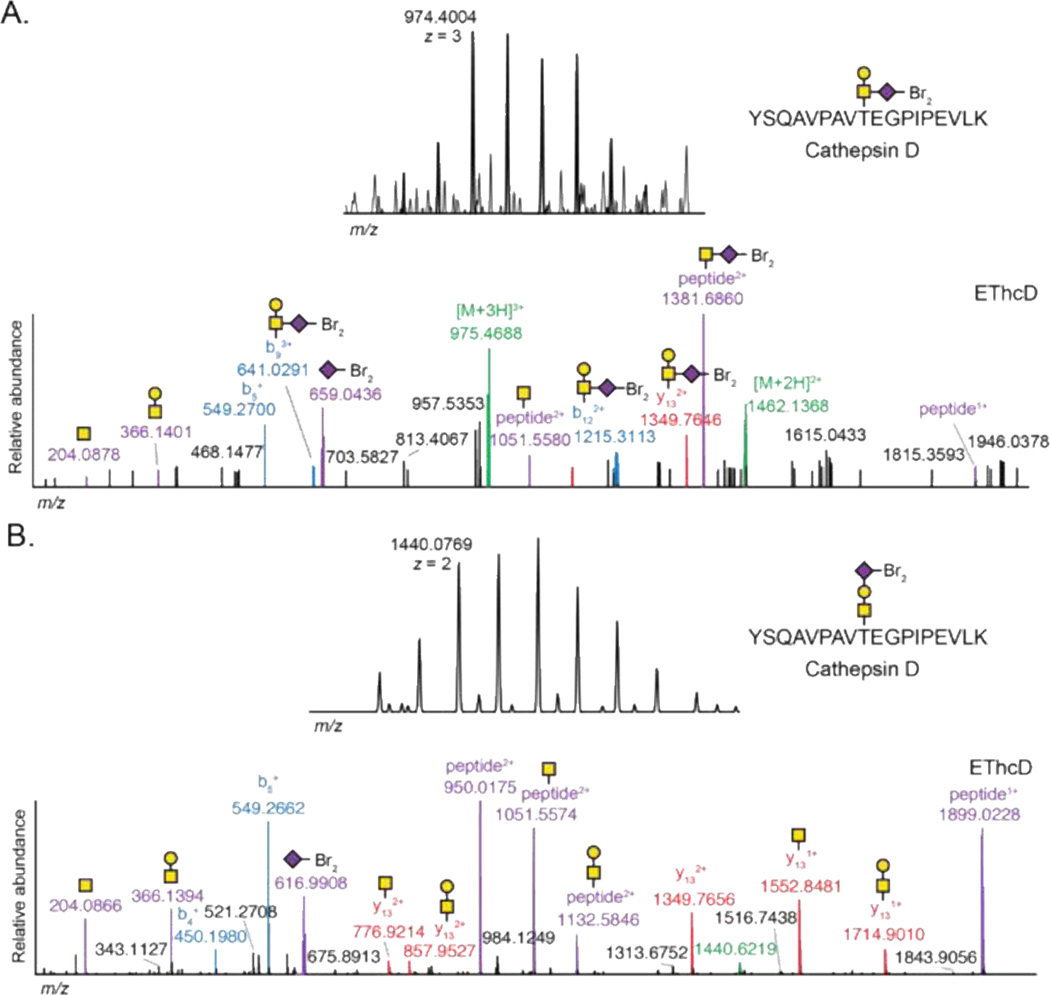
An intact glycoproteomics method enables correlation of the glycan structure to the glycosite. We compared the spectral assignment of YSQAVPAVTEGPIPEVLK carrying glycan S2 from cathepsin D derived from SiaNAl or SiaNAz-labeled PC-3 cells (Figure 4A, 4B, respectively). Both spectra carry isotopically recoded precursors and product ions from the glycan. Closer assessment of the spectra revealed differential localization of the sialic acid residue between the terminal or reducing position of the glycan. Measurement of the relative abundance of oxonium ions revealed differential ratios. The 2,6-sialic acid linkage produced oxonium ions in the ratio of 1:0.3:1 for HexNAc, HexNAcHex, and dibromide-tagged sialic acid (Figure 4A, ESM Table S5), while the corresponding 2,3-sialic acid linkage produced oxonium ions in the ratio of 1:1.2:1.4, respectively (Figure 4B, ESM Table S6). These ratios may reflect specific glycan structures more broadly [28]. IsoTaG delivers the potential to evaluate these intact glycopeptide fragmentation ratios, which may eventually reveal the heterogeneity inherent to glycosylation both within the glycan structure and the peptide modification site.
Figure 4.
Example spectral assignment of the glycopeptide YSQAVPAVTEGPIPEVLK from cathepsin D carrying SiaNAl (A) or SiaNAz (B) incorporation to glycan S2. Assignment of glycan and peptide fragments enables localization of sialic acid in the glycan structure. The relative abundance of highlighted peaks is reported in ESM Tables S1 and S2.
Conclusion
IsoTaG enables the study of intact glycopeptides from azido and alkynyl sugar-labeled samples with high confidence. To evaluate samples labeled by alkynyl sugars (e.g., Ac4ManNAl) by IsoTaG, we developed azide biotin probe 2 for chemical enrichment and isotopic recording. Isotopic recoding imparts an immediate and orthogonal validation of glycopeptide assignments from complex mixtures by full scan MS. Integration of IsoTaG with an Orbitrap Fusion Tribrid led to the assignment of 699 intact glycopeptides from 192 glycoproteins from the PC-3 cell line. These glycopeptides represent both N- and O-glycans (126, 578, respectively), that are modified with eight sialylated glycan structures. These results expand applications of IsoTaG to alkyne-functionalized metabolic labels and will speed the characterization of intact glycoproteins for biomarker discovery and functional studies.
Supplementary Material
Acknowledgments
Financial support from the US National Institutes of Health (CA200423, C.R.B.), Jane Coffin Childs Memorial Fund (C.M.W.), Burroughs Wellcome Fund Career Awards at the Scientific Interface (C.M.W.), Stanford Undergraduate Advising and Research Student Grant (A.F.), the W.M. Keck Foundation Medical Research Program (J.E.E.) and the Bill and Melinda Gates Foundation (J.E.E.) are gratefully acknowledged.
Footnotes
Conflict of Interest: The authors declare no conflict of interest.
References
- 1.Amon R, Reuven EM, Leviatan Ben-Arye S, Padler-Karavani V. Glycans in immune recognition and response. Carbohydr Res. 2014;389:115. doi: 10.1016/j.carres.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 2.Adamczyk B, Tharmalingam T, Rudd PM. Glycans as cancer biomarkers. Biochim Biophys Acta. 2012;1820(9):1347. doi: 10.1016/j.bbagen.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Song E, Hu Y, Hussein A, Yu C-Y, Tang H, Mechref Y. Characterization of the Glycosylation Site of Human PSA Prompted by Missense Mutation using LC–MS/MS. J Proteome Res. 2015;14(7):2872. doi: 10.1021/acs.jproteome.5b00362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paszek MJ, DuFort CC, Rossier O, Bainer R, Mouw JK, Godula K, Hudak JE, Lakins JN, Wijekoon AC, Cassereau L, Rubashkin MG, Magbanua MJ, Thorn KS, Davidson MW, Rugo HS, Park JW, Hammer DA, Giannone G, Bertozzi CR, Weaver VM. The cancer glycocalyx mechanically primes integrin-mediated growth and survival. Nature. 2014;511:319. doi: 10.1038/nature13535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown JR, Fuster MM, Li R, Varki N, Glass CA, Esko JD. A disaccharide-based inhibitor of glycosylation attenuates metastatic tumor cell dissemination. Clin Cancer Res. 2006;12(9):2894. doi: 10.1158/1078-0432.CCR-05-2745. [DOI] [PubMed] [Google Scholar]
- 6.Samraj AN, Pearce OMT, Läubli H, Crittenden AN, Bergfeld AK, Banda K, Gregg CJ, Bingman AE, Secrest P, Diaz SL, Varki NM, Varki A. A red meat-derived glycan promotes inflammation and cancer progression. Proc Natl Acad Sci. 2015;112(2):542. doi: 10.1073/pnas.1417508112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hudak JE, Canham SM, Bertozzi CR. Glycocalyx engineering reveals a Siglec-based mechanism for NK cell immunoevasion. Nat Chem Biol. 2014;10(1):69. doi: 10.1038/nchembio.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao H, Woods EC, Vukojicic P, Bertozzi CR. Precision glycocalyx editing as a strategy for cancer immunotherapy. Proc Natl Acad Sci. 2016 doi: 10.1073/pnas.1608069113. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyoshi E, Moriwaki K, Nakagawa T. Biological function of fucosylation in cancer biology. J Biochem. 2008;143(6):725. doi: 10.1093/jb/mvn011. [DOI] [PubMed] [Google Scholar]
- 10.Okeley NM, Alley SC, Anderson ME, Boursalian TE, Burke PJ, Emmerton KM, Jeffrey SC, Klussman K, Law C-L, Sussman D, Toki BE, Westendorf L, Zeng W, Zhang X, Benjamin DR, Senter PD. Development of orally active inhibitors of protein and cellular fucosylation. Proc Natl Acad Sci. 2013;110(14):5404. doi: 10.1073/pnas.1222263110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liener I. The lectins: properties, functions, and applications in biology and medicine. Elsevier. 1986 [Google Scholar]
- 12.Belardi B, Bertozzi CR. Chemical Lectinology: Tools for Probing the Ligands and Dynamics of Mammalian Lectins In Vivo. Chem Biol. 2015;22(8):983. doi: 10.1016/j.chembiol.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brooks SA. Strategies for Analysis of the Glycosylation of Proteins: Current Status and Future Perspectives. Molecular Biotechnology. 2009;43(1):76. doi: 10.1007/s12033-009-9184-6. [DOI] [PubMed] [Google Scholar]
- 14.Chen C-C, Su W-C, Huang B-Y, Chen Y-J, Tai H-C, Obena RP. Interaction modes and approaches to glycopeptide and glycoprotein enrichment. Analyst. 2014;139(4):688. doi: 10.1039/c3an01813j. [DOI] [PubMed] [Google Scholar]
- 15.Zhang H, Li XJ, Martin DB, Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat Biotechnol. 2003;21(6):660. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 16.Vosseller K, Trinidad JC, Chalkley RJ, Specht CG, Thalhammer A, Lynn AJ, Snedecor JO, Guan S, Medzihradszky KF, Maltby DA, Schoepfer R, Burlingame AL. O-linked N-acetylglucosamine proteomics of postsynaptic density preparations using lectin weak affinity chromatography and mass spectrometry. Mol Cell Proteomics. 2006;5(5):923. doi: 10.1074/mcp.T500040-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Clark PM, Dweck JF, Mason DE, Hart CR, Buck SB, Peters EC, Agnew BJ, Hsieh-Wilson LC. Direct In-Gel Fluorescence Detection and Cellular Imaging of O-GlcNAc-Modified Proteins. J Am Chem Soc. 2008;130(35):11576. doi: 10.1021/ja8030467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saxon E, Bertozzi CR. Cell surface engineering by a modified Staudinger reaction. Science. 2000;287(5460):2007. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 19.Thaysen-Andersen M, Packer NH, Schulz BL. Maturing Glycoproteomics Technologies Provide Unique Structural Insights into the N-glycoproteome and Its Regulation in Health and Disease. Mol Cell Proteomics. 2016;15(6):1773. doi: 10.1074/mcp.O115.057638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chandler KB, Costello CE. Glycomics and glycoproteomics of membrane proteins and cell-surface receptors: Present trends and future opportunities. Electrophoresis. 2016;37(11):1407. doi: 10.1002/elps.201500552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tarentino AL, Gomez CM, Plummer TH. Deglycosylation of asparagine-linked glycans by peptide: N-glycosidase F. Biochemistry. 1985;24(17):4665. doi: 10.1021/bi00338a028. [DOI] [PubMed] [Google Scholar]
- 22.Zielinska DF, Gnad F, Wiśniewski JR, Mann M. Precision Mapping of an In Vivo N-Glycoproteome Reveals Rigid Topological and Sequence Constraints. Cell. 2010;141(5):897. doi: 10.1016/j.cell.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 23.Song X, Ju H, Lasanajak Y, Kudelka MR, Smith DF, Cummings RD. Oxidative release of natural glycans for functional glycomics. Nat Meth. 2016;13(6):528. doi: 10.1038/nmeth.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alfaro JF, Gong C-X, Monroe ME, Aldrich JT, Clauss TRW, Purvine SO, Wang Z, Camp DG, Shabanowitz J, Stanley P, Hart GW, Hunt DF, Yang F, Smith RD. Tandem mass spectrometry identifies many mouse brain O-GlcNAcylated proteins including EGF domain-specific O-GlcNAc transferase targets. Proc Natl Acad Sci. 2012;109(19):7280. doi: 10.1073/pnas.1200425109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nilsson J, Ruetschi U, Halim A, Hesse C, Carlsohn E, Brinkmalm G, Larson G. Enrichment of glycopeptides for glycan structure and attachment site identification. Nat Meth. 2009;6(11):809. doi: 10.1038/nmeth.1392. [DOI] [PubMed] [Google Scholar]
- 26.Steentoft C, Vakhrushev SY, Vester-Christensen MB, Schjoldager KT, Kong Y, Bennett EP, Mandel U, Wandall H, Levery SB, Clausen H. Mining the O-glycoproteome using zinc-finger nuclease-glycoengineered Simple Cell lines. Nat Meth. 2011;8(11):977. doi: 10.1038/nmeth.1731. [DOI] [PubMed] [Google Scholar]
- 27.Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KT, Lavrsen K, Dabelsteen S, Pedersen NB, Marcos-Silva L, Gupta R, Bennett EP, Mandel U, Brunak S, Wandall HH, Levery SB, Clausen H. Precision mapping of the human O-GalNAc glycoproteome through Simple Cell technology. EMBO J. 2013;32(10):1478. doi: 10.1038/emboj.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao P, Viner R, Teo CF, Boons G-J, Horn D, Wells L. Combining High-Energy C-Trap Dissociation and Electron Transfer Dissociation for Protein O-GlcNAc Modification Site Assignment. J Proteome Res. 2011;10(9):4088. doi: 10.1021/pr2002726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu SW, Pu TH, Viner R, Khoo KH. Novel LC-MS(2) product dependent parallel data acquisition function and data analysis workflow for sequencing and identification of intact glycopeptides. Anal Chem. 2014;86(11):5478. doi: 10.1021/ac500945m. [DOI] [PubMed] [Google Scholar]
- 30.Shah P, Wang X, Yang W, Toghi Eshghi S, Sun S, Hoti N, Chen L, Yang S, Pasay J, Rubin A, Zhang H. Integrated Proteomic and Glycoproteomic Analyses of Prostate Cancer Cells Reveal Glycoprotein Alteration in Protein Abundance and Glycosylation. Mol Cell Proteomics. 2015;14(10):2753. doi: 10.1074/mcp.M115.047928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh C, Zampronio CG, Creese AJ, Cooper HJ. Higher Energy Collision Dissociation (HCD) Product Ion-Triggered Electron Transfer Dissociation (ETD) Mass Spectrometry for the Analysis of N-Linked Glycoproteins. J Proteome Res. 2012;11(9):4517. doi: 10.1021/pr300257c. [DOI] [PubMed] [Google Scholar]
- 32.Parker BL, Thaysen-Andersen M, Solis N, Scott NE, Larsen MR, Graham ME, Packer NH, Cordwell SJ. Site-Specific Glycan-Peptide Analysis for Determination of N-Glycoproteome Heterogeneity. J Proteome Res. 2013;12(12):5791. doi: 10.1021/pr400783j. [DOI] [PubMed] [Google Scholar]
- 33.He L, Xin L, Shan B, Lajoie GA, Ma B. GlycoMaster DB: Software To Assist the Automated Identification of N-Linked Glycopeptides by Tandem Mass Spectrometry. J Proteome Res. 2014;13(9):3881. doi: 10.1021/pr401115y. [DOI] [PubMed] [Google Scholar]
- 34.Hua S, Nwosu CC, Strum JS, Seipert RR, An HJ, Zivkovic AM, German JB, Lebrilla CB. Site-specific protein glycosylation analysis with glycan isomer differentiation. Anal Bioanal Chem. 2012;403(5):1291. doi: 10.1007/s00216-011-5109-x. [DOI] [PubMed] [Google Scholar]
- 35.Toghi Eshghi S, Shah P, Yang W, Li X, Zhang H. GPQuest: A Spectral Library Matching Algorithm for Site-Specific Assignment of Tandem Mass Spectra to Intact N-glycopeptides. Anal Chem. 2015;87(10):5181. doi: 10.1021/acs.analchem.5b00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woo CM, Iavarone AT, Spiciarich DR, Palaniappan KK, Bertozzi CR. Isotope-targeted glycoproteomics (IsoTaG): a mass-independent platform for intact N- and O-glycopeptide discovery and analysis. Nat Meth. 2015;12(6):561. doi: 10.1038/nmeth.3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laughlin ST, Baskin JM, Amacher SL, Bertozzi CR. In Vivo Imaging of Membrane-Associated Glycans in Developing Zebrafish. Science. 2008;320:664. doi: 10.1126/science.1155106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang PV, Chen X, Smyrniotis C, Xenakis A, Hu T, Bertozzi CR, Wu P. Metabolic Labeling of Sialic Acids in Living Animals with Alkynyl Sugars. Angew Chem Int Ed. 2009;48(22):4030. doi: 10.1002/anie.200806319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hubbard SC, Boyce M, McVaugh CT, Peehl DM, Bertozzi CR. Cell surface glycoproteomic analysis of prostate cancer-derived PC-3 cells. Bioorg Med Chem Lett. 2011;21:4945. doi: 10.1016/j.bmcl.2011.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsu TL, Hanson SR, Kishikawa K, Wang SK, Sawa M, Wong CH. Alkynyl sugar analogs for the labeling and visualization of glycoconjugates in cells. Proc Natl Acad Sci. 2007;104(8):2614. doi: 10.1073/pnas.0611307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chuh KN, Zaro BW, Piller F, Piller V, Pratt MR. Changes in Metabolic Chemical Reporter Structure Yield a Selective Probe of O-GlcNAc Modification. J Am Chem Soc. 2014;136(35):12283. doi: 10.1021/ja504063c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pangborn AB, Giardello MA, Grubbs RH, Rosen RK, Timmers FJ. Safe and Convenient Procedure for Solvent Purification. Organometallics. 1996;15(5):1518. [Google Scholar]
- 43.Lee PJJ, Compton BJ. Desctructible surfactants and uses thereof. USA Patent. 2007 [Google Scholar]
- 44.Wang W, Hong S, Tran A, Jiang H, Triano R, Liu Y, Chen X, Wu P. Sulfated Ligands for the Copper(I)-Catalyzed Azide–Alkyne Cycloaddition. Chem Asian J. 2011;6(10):2796. doi: 10.1002/asia.201100385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prescher JA, Dube DH, Bertozzi CR. Chemical remodelling of cell surfaces in living animals. Nature. 2004;430:873. doi: 10.1038/nature02791. [DOI] [PubMed] [Google Scholar]
- 46.Bern M, Kil YJ, Becker C. Byonic: advanced peptide and protein identification software. Curr Protoc Bioinformatics. 2012 doi: 10.1002/0471250953.bi1320s40. Chapter 13:Unit13 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vizcaino JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, Xu QW, Wang R, Hermjakob H. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44(D1):D447. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Szychowski J, Mahdavi A, Hodas JJL, Bagert JD, Ngo JT, Landgraf P, Dieterich DC, Schuman EM, Tirrell DA. Cleavable Biotin Probes for Labeling of Biomolecules via Azide–Alkyne Cycloaddition. J Am Chem Soc. 2010;132:18351. doi: 10.1021/ja1083909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palaniappan KK, Pitcher AA, Smart BP, Spiciarich DR, Iavarone AT, Bertozzi CR. Isotopic Signature Transfer and Mass Pattern Prediction (IsoStamp): An Enabling Technique for Chemically-Directed Proteomics. ACS Chem Biol. 2011;6(8):829. doi: 10.1021/cb100338x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Woo CM, Bertozzi CR. Isotope Targeted Glycoproteomics (IsoTaG) to Characterize Intact, Metabolically Labeled Glycopeptides from Complex Proteomes. Curr Protoc Chem Biol. 2016;8(1):59. doi: 10.1002/9780470559277.ch150185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.