Abstract
MicroRNAs (miRNAs) are a central player in post-transcriptional regulation of gene expression and are involved in numerous biological processes in eukaryotes. Knowledge of the origins and divergence of miRNAs paves the way for a better understanding of the complexity of the regulatory networks that they participate in. The biogenesis, degradation, and regulatory activities of miRNAs are relatively better understood, but the evolutionary history of miRNAs still needs more exploration. Inverted duplication of target genes, random hairpin sequences and small transposable elements constitute three main models that explain the origination of miRNA genes (MIR). Both inter- and intra-species divergence of miRNAs exhibits functional adaptation and adaptation to changing environments in evolution. Here we summarize recent progress in studies on the evolution of MIR and related genes.
1. Introduction
MicroRNAs (miRNAs) and small interfering RNAs (siRNAs) are central players in RNA silencing. SiRNAs are derived en mass from long double-stranded RNAs, and silence transgenes, viruses, and transposable elements [1]. MiRNAs are processed from short hairpin precursors encoded by MIR genes and regulate endogenous genes to impact development and stress responses [2,3]. RNA silencing as a mechanism to silence foreign genetic material (such as viruses) is conserved in all domains of life in eukaryotes, thus it is possible that it existed in the common eukaryotic ancestor. MiRNAs are found in all plant and animal lineages [4,5], and share similar biogenesis, function and turnover pathways in diverse lineages [2,6,7]. In this review, we summarize findings from studies on the origin and evolution of MIR genes and genes related to miRNA biogenesis and function, with a focus on plants.
1. The evolution of genes related to small RNA biogenesis and function
In miRNA biogenesis, Drosha generates pre-miRNAs from pri-miRNAs and Dicer then processes pre-miRNAs into mature miRNAs [7]. In plants, DICER-LIKE proteins (DCLs) perform the functions of both Drosha and Dicer [7,8]. A comprehensive phylogenetic study on DCLs implicated the same origin but independent evolution of DCLs in plants and animals, and suggested that antiviral immunity served as a driving force in the evolution of plant DCLs [6]. In the green alga Chlamydomonas reinhardtii, which separated early from higher plants, miRNA biogenesis is likely conducted by CrDCL3 [9]. Intriguingly, CrDCL3 shows some Drosha-like features, possessing a proline-rich domain and lacking the PAZ domain. This is in contrast to the absence of Drosha-like proteins in higher plants; these differences in algal and higher plant DCLs indicate parallel evolution of the miRNA machinery in different plant lineages. On the other hand, the presence of a Drosha-like protein in Chlamydomonas suggests a single origin of DCL genes in animals and plants [9]. Echoing the similarity in miRNA biogenesis between Chlamydomonas and animals, a recent study found that Chlamydomonas miRNAs have similar features to animal miRNAs in terms of target recognition [10]. In animals, a miRNA recognizes its target transcripts via complementarity between the target and nucleotides 2 to 8 of the miRNA, namely the seed region. The seed region of a Chlamydomonas miRNA is sufficient to render miRNA-based repression of a target transcript [10]. This is different from the requirement for near-perfect base pairing for target recognition by higher plant miRNAs [11].
ARGONAUTE (AGO) family proteins associate with small RNAs and mediate their activities. The plant AGO family can be divided into 3 major higher plant lineages, which are AGO1/5/10, AGO2/3/7, and AGO4/6/8/9, plus an algal lineage [12]. There exists a grass-specific AGO subfamily, designated AGO18, which is close to the AGO1/5/10 lineage, but its origination is unclear [13]. AGO18 in rice sequesters miR168 and acts as the decoy of AGO1, which is the target of miR168 [14]. In maize, however, ZmAGO18b binds 24-nt phased secondary siRNAs in male reproductive development [15]. In Arabidopsis, AGO10 serves as the decoy of AGO1 in binding miR165/166 [16,17]. Although AGO18 is grass-specific, its miRNA partner (i.e. miR168) and the miRNA target gene (i.e. AGO1) are conserved in land plants [4,18], implying a later origin of AGO18. Considering their similar functions in blocking AGO1-miRNA interactions but their different miRNA partners as well as different biological functions, AGO18 in cereals and AGO10 in Arabidopsis may have resulted from parallel evolution. The earlier emergence of miRNAs that are currently specifically bound by AGO proteins that emerged later in evolution, such as miR165/166, miR168 and miR390 [4,13], implies that the miRNAs initially perform some vital regulatory functions common in all plant lineages and the expansion in the AGOs adds additional layers to the regulatory networks or endow new biological functions to the miRNAs in specific lineages. These findings also suggest that the enormous variety of small RNAs contributes to the expansion and functional specialization of AGO proteins.
In summary, the studies on the evolution of DCLs and AGOs suggest a single and ancient origin of the small RNA machinery in plants as well as in eukaryotes. The ancient DCLs and AGOs could be working with siRNAs in immunity, and those cooperating with miRNAs might have evolved later from them. It is unclear whether the miRNA machinery evolved prior to or after the divergence of plants and animals. The lack of conservation of miRNAs between plants and animals or between algae and higher plants indicates multiple independent origins of MIR genes themselves.
1. Identification of miRNAs in plants and insights into miRNA evolution
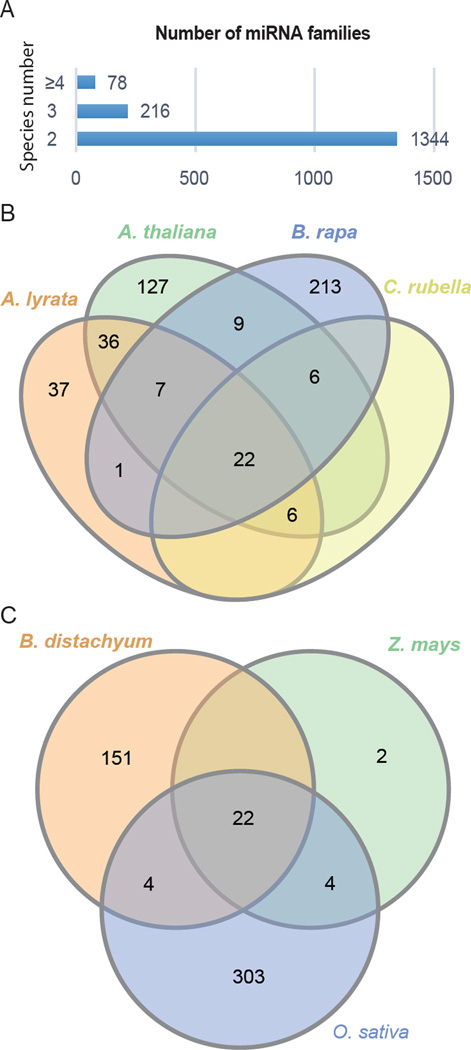
During the past decade, next-generation sequencing boosted the yield of genome and transcriptome sequences from a tremendous number of plants. Efforts to identify miRNAs from the ever-increasing databases of plant genomes and transcriptomes shed light on the overall features of MIR gene evolution. The numbers of miRNAs identified by these efforts are not related to the phylogeny but rather to the tissue type and developmental stages of the samples [19–21]. For example, 428 miRNAs were found in the liverwort Pellia endiviifolia [19] but only 129 were found in another liverwort Marchantia polymorpha [20]. The numbers of MIR genes in species that underwent extra whole genome duplications are much higher than those in related species without such duplications. For example, currently, there are 2787 annotated MIR genes in Glycine max [22] vs. 216 in Phaseolus vulgaris [23] (two species in the Fabaceae family), and 680 in Brassica rapa [24,25] vs. 80 in Capsella rubella [21] (two species in the Brassicaceae family). The identification of miRNAs from various plants also provided an opportunity to understand the conservation of miRNAs over both large and small evolutionary distances. By comparing miRNA families identified in select angiosperms, it is evident that only a few miRNA families are conserved in land plants or even angiosperms (Figure 1A). Even in one family, such as Poaceae and Brassicaceae (Figure 1B,C), many more miRNAs are species-specific rather than conserved, suggesting rapid origination and divergence of MIR genes.
Figure 1. miRNA families identified in selected plant species.
(A) miRNA family distribution in selected plant species. Data are from miRBase v21 and [19–24,42,49]. The selected species are basal land plants Physcomitrella patens, Marchantia polymorpha and Pellia endiviifolia, the basal vascular plant Selaginella moellendorffii, the basal angiosperm Amborella trichopoda, monocots Brachypodium distachyum, Oryza sativa and Zea mays, and eudicots Glycine max, Phaseolus vulgaris, Populus trichocarpa, Moringa oleifera, Brassica rapa, Capsella rubella, Arabidopsis lyrata, and Arabidopsis thaliana. Only 78 miRNA families are found in more than 4 species, most miRNA families are present in a smaller number of species. (B, C) Venn diagrams for shared miRNA families in closely related species in Brassicaceae (B) and Poaceae (C). The data sources are the same as those in (A). The number of shared miRNA families in closely related species is smaller than that of species-specific ones, indicating rapid and independent origination of miRNA families.
1. Origin of miRNAs
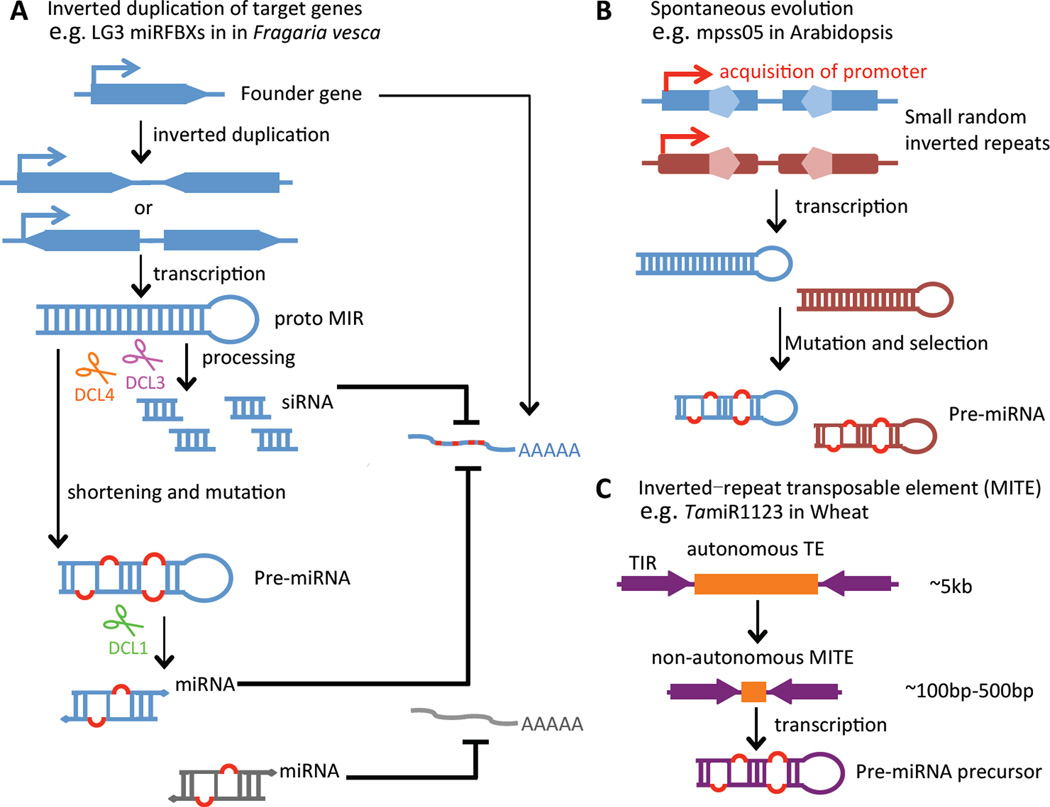
Three possible de novo origins of MIR genes have been uncovered by previous studies (reviewed by [8], Figure 2).
Figure 2. Origins of plant MIR genes.
Three models that explain the de novo origination of miRNAs. (A) MIR genes stem from inverted duplication of target genes. The transcription of inverted duplicated founder genes leads to the emergence of proto MIRs. Proto MIRs often exhibit a long stem-loop structure fitting for the processing by DCL3 and DCL4 to generate siRNAs to suppress target gene expression. Shortening and mutations occur to the inverted repeats during evolution, resulting in the formation of pre-miRNA-like hairpins, which are compatible with processing by DCL1 to generate mature miRNAs. The transcript of the founder gene serves as the target of the mature miRNA. Red spots on the transcript indicate mutations that also accumulate when the founder gene evolves. Occasionally, the target of the miRNA is not the founder gene, which implies divergence in the course of evolution. (B) Small random inverted sequences scattered in the genome provide a source of MIR genes. First, the random inverted repeats obtain a promoter element to enable transcription; then mutations and selections occur during evolution to form the precursor of a miRNA. (C) MITEs contribute to the origination of miRNAs. MITEs, a type of non-autonomous transposons, are composed of terminal inverted repeats (TIRs) and a short open reading frame (ORF), and are roughly 100~500bp in size. MITEs may serve as templates to produce miRNA precursors.
1.1. Gene inverted duplication
Studies on some young MIR genes suggest their evolution from inverted duplication of target genes, as sequences flanking the mature miRNAs in MIR genes show similarity to those flanking the miRNA-binding sites in target genes [26]. Such inverted repeats could generate hairpin structures that initially produce many small RNAs (i.e. siRNAs). During evolution, only a key portion of the hairpin is retained to result in an MIR gene that encodes only one predominant small RNA species - the miRNA (Figure 2A). There are many examples showing sequence homology between MIR genes and target genes, thus supporting this model [26–29]. The original target inverted duplication hypothesis calls for neutral evolution of the sequences other than those of the mature miRNAs and miRNA stars in MIR genes [26]. However, this is not the case for some young miRNAs. For example, for AtmiR824, selection also affected the fold-back structure of the pre-miRNA rather than only the miR824/miR824* sequences in different Arabidopsis thaliana ecotypes [30].
1.1. Spontaneous evolution
Only half of Arabidopsis lyrata MIR gene families that can align to protein-coding genes yield miRNAs targeting the homologous genes [27]. This cannot be explained by the target inverted duplication hypothesis and implies that there must be other origins for MIR genes in plants. A source of miRNAs may be hairpin regions scattered in the genome, which may occasionally give rise to precursors of miRNAs upon acquiring promoters enabling transcription [31] (Figure 2B). For example, mpss05, a candidate pre-miRNA foldback found in Arabidopsis thaliana, can align to two regions that may have originated by a duplication of a chromosomal fragment [31].
1.1. Miniature Inverted-repeat Transposable Elements (MITE)
Another potential source of miRNAs is MITEs, as MITE RNAs may be able to fold into stem loops that resemble miRNA precursors. 10 miRNAs in Arabidopsis and 38 in rice were reported to be derived from MITEs [32,33] (Figure 2C). However, MITEs tend to generate many small RNAs, which maybe better classified as siRNAs [32]. Thus, the MITE-origin hypothesis is still controversial. Recently, TamiR1123 that functions in vernalization in wheat was found to be produced from a MITE locus and is an example that supports this model [34].
1. Functional divergence of miRNAs
Besides the de novo origination of MIR genes, duplications of existing MIR genes produce paralogous genes, enlarging the MIR gene family and potentially expanding the functions of the family. One clear case of functional divergence of paralogous miRNAs is Arabidopsis MIR159/319, which originated from the same ancestor but separated in the common ancestors of land plants and evolved to target two distinct gene families [4,35]. Despite few examples in the divergence of miRNA target specificity as in the case of miR159/319, plenty of evidence supports tissue- and/or taxonomy-specific divergence in miRNA evolution. Two related genes, MIR156 and MIR529, encode highly similar miRNAs but exhibit differences in their distribution among plant species, tissue expression patterns and evolutionary rates [36]. Intriguingly, even though there still exists some potentially functional miR529-responsive elements in the Arabidopsis thaliana genome, miR529 was lost in Arabidopsis. Members of the MIR156 family may compensate for the regulatory functions of miR529 in Arabidopsis [36]. The divergent expression patterns of paraloguos miRNAs may have also contributed to plant adaptation to extreme environments, as suggested by a recent study on the small RNA transcriptome of mangroves, which grow under high salinity, poor oxygen and low nutrient conditions [37]. A study in Brassica rapa found that MIR genes are over retained compared with protein-coding genes after a species-specific whole genome triplication (WGT) event [24]. The retained multiple-copy MIR genes are under increased purifying selection as compared to protein-coding genes, suggesting functional importance of these MIR genes. A recent study on 22-nt miRNAs raised a new hypothesis that some miRNA (super) families are derived from existing miRNA families in a target-driven manner. Rapidly evolving target genes drive the evolution of miRNAs to maintain the miRNA-target relationship. For example, the MIR7122 family targeting TAS-LIKE (TASL) genes probably evolved from the conserved MIR390 family targeting TAS3 [38]. Divergence in miRNAs or targets may have played a role in crop domestication. Studies comparing cultivated rice with wild rice (Oryza rufipogon) found positively selected miRNAs and target genes in cultivated rice, suggesting that miRNAs were involved in or even drove rice domestication [39,40]. Another study suggested that a loss-of-function mutation in MIR172p improved fruit size during apple domestication [41].
1. Co-evolution of miRNAs and miRNA target genes
A mutual selection between miRNAs and their target genes probably occurs in evolution. A recent report shows that during the domestication of soybean, many factors influenced the evolution of MIR genes and miRNA targets, including the duplication status, expression level, and miRNA-target interactions [22]. A similar case is a study on Populus trichocarpa that experienced a recent whole genome duplication (WGD) event compared to other related species. The authors found the de novo emergence of new Populus-specific miRNAs after WGD as regulators of the newly formed duplicated genes in salicoids [42].
Some miRNA-target interactions result in small RNA amplification. Certain miRNA targets, such as PPR, NBS-LRR, MYB, and noncoding TAS loci, generate secondary, phased siRNAs (phasiRNAs) upon miRNA-guided RNA cleavage [43]. Such small RNA amplification is probably an economic way to suppress a large number of similar genes with only one initial miRNA. This is a broadly adopted regulatory strategy during plant evolution [4,38,44]. The miRNA-target pairs exhibit patterns of co-evolution. In soybean, the miRNA targeting sites in NBS-LRR genes exhibit evolutionary patterns different from those of flanking sequences, suggesting that miRNAs influence the evolution of the targets [45]. On the other hand, a newly published report shows that NBS-LRR genes keep giving birth to new miRNAs targeting themselves in a convergent manner in diverse lineages. The evolution of these miRNAs appears to be driven by the amino acid diversity of their target sites, as diversity in miRNAs lies mainly in nucleotides corresponding to the third codon positions of the target site sequences [46].
Occasionally, conserved miRNA-target pairs could be lost in specific plant groups [4], and in some cases, re-construction of the regulation of the target genes is vital for their survival. For instance, DCL1 is targeted by miR162 to form a feedback loop to achieve homeostasis of miRNA biogenesis. However, in some plants including Physcomitrella patens, Selaginella moellendorffii, and Salvia miltiorrhiza, either the binding site for miR162 is highly diverged or miR162 is poorly expressed. In Salvia miltiorrhiza, miR397 replaces miR162 to maintain the regulation [47]. Similarly, miR398 that targets CSD2 is missing in Physcomitrella patens, but miR1073 targets CSD2 in this species and this miRNA shares a similar expression pattern with Arabidopsis miR398 [48].
1. Perspectives
MiRNAs and their target genes constitute a complex and dynamic gene regulatory network. From an evolutionary perspective, this network underwent parallel evolution in animals and plants, and in eudicots and monocots after the divergence of these lineages, leading to similar logistics but different flavors of the network in different lineages. When a new small RNA is produced, it may be retained and rewired into an existing regulatory network, making up a new miRNA and a new component of the network. The retained miRNA could also shape the target sequences, introducing even more divergence between lineages. With advanced sequencing technology, more and more lineage-specific miRNAs will be discovered from different plants and tissues, thus revealing more patterns of evolution, either supporting or opposing the current hypotheses.
Highlights.
-
➢
Most miRNAs are lineage-specific.
-
➢
MIR genes evolve rapidly and show much divergence.
-
➢
MIR genes may keep originating from target genes.
-
➢
miRNA-target pairs may co-evolve.
Acknowledgments
The authors' research is supported by Betty and Gordon Moore Foundation (GBMF3046), National Institutes of Health (GM061146), National Science Foundation (IOS-1340001), National Science Foundation of China (91440105), and Guangdong Innovation Research Team Fund (2014ZT05S078).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Martínez de Alba AE, Elvira-Matelot E, Vaucheret H. Gene silencing in plants: a diversity of pathways. Biochim Biophys Acta. 2013;1829:1300–1308. doi: 10.1016/j.bbagrm.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol. 2013;14:475–488. doi: 10.1038/nrm3611. [DOI] [PubMed] [Google Scholar]
- 3.Sunkar R, Li Y-F, Jagadeeswaran G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 2012;17:196–203. doi: 10.1016/j.tplants.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 4. Chávez Montes RA, de Fátima Rosas-Cárdenas F, De Paoli E, Accerbi M, Rymarquis LA, Mahalingam G, Marsch-Martínez N, Meyers BC, Green PJ, de Folter S. Sample sequencing of vascular plants demonstrates widespread conservation and divergence of microRNAs. Nat Commun. 2014;5:3722. doi: 10.1038/ncomms4722. ** In this study, the authors sequenced small RNAs from algae and from representative species across vascular plants, identified miRNAs, and characterized the 5 main features of these miRNAs. This study provided an overview of miRNAs across the plant kingdom for the first time and set the foundation for future studies on miRNA evolution.
- 5.Meunier J, Lemoine F, Soumillon M, Liechti A, Weier M, Guschanski K, Hu H, Khaitovich P, Kaessmann H. Birth and expression evolution of mammalian microRNA genes. Genome Res. 2013;23:34–45. doi: 10.1101/gr.140269.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mukherjee K, Campos H, Kolaczkowski B. Evolution of animal and plant dicers: early parallel duplications and recurrent adaptation of antiviral RNA binding in plants. Mol Biol Evol. 2013;30:627–641. doi: 10.1093/molbev/mss263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 8.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 9. Valli AA, Santos BACM, Hnatova S, Bassett AR, Molnar A, Chung BY, Baulcombe DC. Most microRNAs in the single-cell alga Chlamydomonas reinhardtii are produced by Dicer-like 3-mediated cleavage of introns and untranslated regions of coding RNAs. Genome Res. 2016;26:519–529. doi: 10.1101/gr.199703.115. ** This study provided new evidence supporting that miRNA pathways in plants and animals might have had a common origin but underwent parallel evolution. The authors employed a forward genetic screen and identified DCL3 as an important miRNA biogenesis factor for the unicellular model plant, Chlamydomonas reinhardtii. The Chlamydomonas DCL3 protein shows similarity to Drosha in animals. Other features of Chlamydonomas miRNAs, such as miRNA gene distribution within introns of protein coding genes and the location of miRNA target sites in the 3' UTR of target genes, are similar to those in animals rather than higher plants.
- 10. Yamasaki T, Voshall A, Kim EJ, Moriyama E, Cerutti H, Ohama T. Complementarity to an miRNA seed region is sufficient to induce moderate repression of a target transcript in the unicellular green alga Chlamydomonas reinhardtii. Plant J. 2013;76:1045–1056. doi: 10.1111/tpj.12354. * This work revealed that complementarity to the seed region of a miRNA is sufficient for target recognition by this miRNA in the green alga Chlamydomonas reinhardtii. The findings in this study, along with those in [9], suggest that miRNA biogenesis and action in Chlamydomonas resemble those in metazoans.
- 11.Tang G, Reinhart BJ, Bartel DP, Zamore PD. A biochemical framework for RNA silencing in plants. Genes Dev. 2003;17:49–63. doi: 10.1101/gad.1048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh RK, Gase K, Baldwin IT, Pandey SP. Molecular evolution and diversification of the Argonaute family of proteins in plants. BMC Plant Biol. 2015;15:23. doi: 10.1186/s12870-014-0364-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang H, Xia R, Meyers BC, Walbot V. Evolution, functions, and mysteries of plant ARGONAUTE proteins. Curr Opin Plant Biol. 2015;27:84–90. doi: 10.1016/j.pbi.2015.06.011. * This review article summarizes recent progress in studies of AGO proteins in both eudicots and monocots, especially in Arabidopsis and rice, and designates a new nomenclature for plant AGO genes. Moreover, this paper shows that the AGO18 subfamily that functions in reproductive development and viral defense is phylogenetically grass-specific.
- 14.Wu J, Yang Z, Wang Y, Zheng L, Ye R, Ji Y, Zhao S, Ji S, Liu R, Xu L, et al. Viral-inducible Argonaute18 confers broad-spectrum virus resistance in rice by sequestering a host microRNA. eLife. 2015;4:356. doi: 10.7554/eLife.05733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhai L, Sun W, Zhang K, Jia H, Liu L, Liu Z, Teng F, Zhang Z. Identification and characterization of Argonaute gene family and meiosis-enriched Argonaute during sporogenesis in maize. J Integr Plant Biol. 2014;56:1042–1052. doi: 10.1111/jipb.12205. [DOI] [PubMed] [Google Scholar]
- 16.Zhu H, Hu F, Wang R, Zhou X, Sze S-H, Liou LW, Barefoot A, Dickman M, Zhang X. Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell. 2011;145:242–256. doi: 10.1016/j.cell.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ji L, Liu X, Yan J, Wang W, Yumul RE, Kim YJ, Dinh TT, Liu J, Cui X, Zheng B, et al. ARGONAUTE10 and ARGONAUTE1 regulate the termination of floral stem cells through two microRNAs in Arabidopsis. PLoS Genet. 2011;7:e1001358. doi: 10.1371/journal.pgen.1001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang X, Qi Y. RNAi in plants: an Argonaute-centered view. Plant Cell. 2016;28:272–285. doi: 10.1105/tpc.15.00920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alaba S, Piszczalka P, Pietrykowska H, Pacak AM, Sierocka I, Nuc PW, Singh K, Plewka P, Sulkowska A, Jarmolowski A, et al. The liverwort Pellia endiviifolia shares microtranscriptomic traits that are common to green algae and land plants. New Phytol. 2015;206:352–367. doi: 10.1111/nph.13220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin P-C, Lu C-W, Shen B-N, Lee G-Z, Bowman JL, Arteaga-Vazquez MA, Liu L-YD, Hong S-F, Lo C-F, Su G-M, et al. Identification of miRNAs and their targets in the liverwort Marchantia polymorpha by integrating RNA-Seq and degradome analyses. Plant Cell Physiol. 2016;57:339–358. doi: 10.1093/pcp/pcw020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith LM, Burbano HA, Wang X, Fitz J, Wang G, Ural-Blimke Y, Weigel D. Rapid divergence and high diversity of miRNAs and miRNA targets in the Camelineae. Plant J. 2015;81:597–610. doi: 10.1111/tpj.12754. [DOI] [PubMed] [Google Scholar]
- 22.Liu T, Fang C, Ma Y, Shen Y, Li C, Li Q, Wang M, Liu S, Zhang J, Zhou Z, et al. Global investigation of the co-evolution of MIRNA genes and microRNA targets during soybean domestication. Plant J. 2016;85:396–409. doi: 10.1111/tpj.13113. [DOI] [PubMed] [Google Scholar]
- 23.Nithin C, Patwa N, Thomas A, Bahadur RP, Basak J. Computational prediction of miRNAs and their targets in Phaseolus vulgaris using simple sequence repeat signatures. BMC Plant Biol. 2015;15:140. doi: 10.1186/s12870-015-0516-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun C, Wu J, Liang J, Schnable JC, Yang W, Cheng F, Wang X. Impacts of whole-genome triplication on MIRNA evolution in Brassica rapa. Genome Biol Evol. 2015;7:3085–3096. doi: 10.1093/gbe/evv206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woodhouse MR, Cheng F, Pires JC, Lisch D, Freeling M, Wang X. Origin, inheritance, and gene regulatory consequences of genome dominance in polyploids. Proc Natl Acad Sci USA. 2014;111:5283–5288. doi: 10.1073/pnas.1402475111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allen E, Xie Z, Gustafson AM, Sung G-H, Spatafora JW, Carrington JC. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat Genet. 2004;36:1282–1290. doi: 10.1038/ng1478. [DOI] [PubMed] [Google Scholar]
- 27.Fahlgren N, Jogdeo S, Kasschau KD, Sullivan CM, Chapman EJ, Laubinger S, Smith LM, Dasenko M, Givan SA, Weigel D, et al. MicroRNA gene evolution in Arabidopsis lyrata and Arabidopsis thaliana. Plant Cell. 2010;22:1074–1089. doi: 10.1105/tpc.110.073999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He H, Liang G, Li Y, Wang F, Yu D. Two young MicroRNAs originating from target duplication mediate nitrogen starvation adaptation via regulation of glucosinolate synthesis in Arabidopsis thaliana. Plant Physiol. 2014;164:853–865. doi: 10.1104/pp.113.228635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xia R, Ye S, Liu Z, Meyers BC, Liu Z. Novel and recently evolved microRNA clusters regulate expansive F-BOX gene networks through phased small interfering RNAs in wild diploid strawberry. Plant Physiol. 2015;169:594–610. doi: 10.1104/pp.15.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Meaux J, Hu J-Y, Tartler U, Goebel U. Structurally different alleles of the ath-MIR824 microRNA precursor are maintained at high frequency in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2008;105:8994–8999. doi: 10.1073/pnas.0803218105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fenselau de Felippes F, Schneeberger K, Dezulian T, Huson DH, Weigel D. Evolution of Arabidopsis thaliana microRNAs from random sequences. RNA. 2008;14:2455–2459. doi: 10.1261/rna.1149408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piriyapongsa J, Jordan IK. Dual coding of siRNAs and miRNAs by plant transposable elements. RNA. 2008;14:814–821. doi: 10.1261/rna.916708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Li C, Xia J, Jin Y. Domestication of transposable elements into microRNA genes in plants. PLoS ONE. 2011;6:e19212. doi: 10.1371/journal.pone.0019212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu M, Carver BF, Yan L. TamiR1123 originated from a family of miniature inverted-repeat transposable elements (MITE) including one inserted in the Vrn-A1a promoter in wheat. Plant Sci. 2014;215–216:117–123. doi: 10.1016/j.plantsci.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 35.Palatnik JF, Wollmann H, Schommer C, Schwab R, Boisbouvier J, Rodriguez R, Warthmann N, Allen E, Dezulian T, Huson D, et al. Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319. Dev Cell. 2007;13:115–125. doi: 10.1016/j.devcel.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 36. Morea EGO, da Silva EM, e Silva GFF, Valente GT, Barrera Rojas CH, Vincentz M, Nogueira FTS. Functional and evolutionary analyses of the miR156 and miR529 families in land plants. BMC Plant Biol. 2016;16:40. doi: 10.1186/s12870-016-0716-5. * This study provided a clear example for the birth, divergence and death of MIR genes, which will provoke further thoughts on MIR gene evolution. Phylogenetic analysis revealed the distinct evolutionary patterns of miR156, miR529 and their target sites. Further genetic analysis showed that the miR529 target sites in several SPL genes are still functional in Arabidopsis in which miR529 has been lost, suggesting asymmetric evolution for miRNAs and targets. The study also revealed the partially redundant functions of the evolutionarily related miR156 and miR529.
- 37.Wen M, Lin X, Xie M, Wang Y, Shen X, Liufu Z, Wu C-I, Shi S, Tang T. Small RNA transcriptomes of mangroves evolve adaptively in extreme environments. Sci Rep. 2016;6:27551. doi: 10.1038/srep27551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia R, Meyers BC, Liu Z, Beers EP, Ye S, Liu Z. MicroRNA superfamilies descended from miR390 and their roles in secondary small interfering RNA biogenesis in eudicots. Plant Cell. 2013;25:1555–1572. doi: 10.1105/tpc.113.110957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Q, Wang H, Hu H, Zhang H. Genome-wide identification and evolutionary analysis of positively selected miRNA genes in domesticated rice. Mol Genet Genomics. 2015;290:593–602. doi: 10.1007/s00438-014-0943-0. [DOI] [PubMed] [Google Scholar]
- 40.Liu Y, Wang Y, Zhu Q-H, Fan L. Identification of phasiRNAs in wild rice (Oryza rufipogon) Plant Signal Behav. 2013;8 doi: 10.4161/psb.25079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yao J-L, Xu J, Cornille A, Tomes S, Karunairetnam S, Luo Z, Bassett H, Whitworth C, Rees-George J, Ranatunga C, et al. A microRNA allele that emerged prior to apple domestication may underlie fruit size evolution. Plant J. 2015;84:417–427. doi: 10.1111/tpj.13021. [DOI] [PubMed] [Google Scholar]
- 42.Xie J, Yang X, Song Y, Du Q, Li Y, Chen J, Zhang D. Adaptive evolution and functional innovation of Populus-specific recently evolved microRNAs. New Phytol. 2016 doi: 10.1111/nph.14046. [DOI] [PubMed] [Google Scholar]
- 43.Fei Q, Xia R, Meyers BC. Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell. 2013;25:2400–2415. doi: 10.1105/tpc.113.114652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Axtell MJ, Jan C, Rajagopalan R, Bartel DP. A two-hit trigger for siRNA biogenesis in plants. Cell. 2006;127:565–577. doi: 10.1016/j.cell.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 45. Zhao M, Meyers BC, Cai C, Xu W, Ma J. Evolutionary patterns and coevolutionary consequences of MIRNA genes and microRNA targets triggered by multiple mechanisms of genomic duplications in soybean. Plant Cell. 2015;27:546–562. doi: 10.1105/tpc.15.00048. ** Through globally analyzing MIR genes in the soybean genome, the authors found that the retention of duplicated MIR genes is correlated with that of adjacent protein-coding genes, and with the functions of miRNAs. Also, the retention of target genes contributes to the retention of MIR genes, which reflects a new aspect of miRNA-target relationship and co-evolution.
- 46.Zhang Y, Xia R, Kuang H, Meyers BC. The diversification of plant NBS-LRR defense genes directs the evolution of MicroRNAs that target them. Mol Biol Evol. 2016;33:2692–2705. doi: 10.1093/molbev/msw154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shao F, Qiu D, Lu S. Comparative analysis of the Dicer-like gene family reveals loss of miR162 target site in SmDCL1 from Salvia miltiorrhiza. Sci Rep. 2015;5:9891. doi: 10.1038/srep09891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Higashi Y, Takechi K, Takano H, Takio S. Involvement of microRNA in copper deficiency-induced repression of chloroplastic CuZn-superoxide dismutase genes in the moss Physcomitrella patens. Plant Cell Physiol. 2013;54:1345–1355. doi: 10.1093/pcp/pct084. [DOI] [PubMed] [Google Scholar]
- 49.Pirrò S, Zanella L, Kenzo M, Montesano C, Minutolo A, Potestà M, Sobze MS, Canini A, Cirilli M, Muleo R, et al. MicroRNA from Moringa oleifera: identification by high throughput sequencing and their potential contribution to plant medicinal value. PLoS ONE. 2016;11:e0149495. doi: 10.1371/journal.pone.0149495. [DOI] [PMC free article] [PubMed] [Google Scholar]