Capsule Summary
In an attempt to better classify molecular profiles in chronic rhinosinusitis (CRS)subtypes and promote endotyping, we characterize the expression of relevant type 2 inflammatory markers in several clinical subtypes of CRS.
Keywords: rhinosinusitis, microarray, expression profiling, nasal polyps, allergic fungal sinusitis, type 2 inflammation, IL1RL1, ST2
To the Editor
Chronic rhinosinusitis (CRS) represents a heterogeneous disease comprising several different subtypes which are grouped together by common criteria lasting at least 12 weeks. The classification of CRS subtypes remains largely clinical and carries little prognostic value. At present, clinicians possess limited capability in predicting a patient’s disease course and anticipating response to available therapies. More recently, efforts have focused on categorizing CRS subtypes into endotypes. Endotypes organize disease subtypes according to molecular patterns believed to underlie the expression of different clinical phenotypes1. The push for disease endotyping in CRS derives from the successful application of this method in asthma, a disease sharing many pathophysiologic features with CRS.
Indeed, the presence of type 2 inflammation represents an important point of disease stratification, both in asthma and CRS. In asthma, increased blood and tissue eosinophilia, IgE levels, and expression of type 2 inflammatory biomarkers, such as IL-4, IL-5, IL-13, and periostin, have been leveraged to endotype disease, develop novel biologic therapies and monitor response to treatment2.
In this study, we characterized the expression patterns of several type 2 inflammatory cytokines of interest in different clinical subtypes of CRS. To this end, we employ a top-down approach in the form of lar e-scale microarray ene profilin techniques in 130 patients representin different clinical subtypes of CRS: AERD, AFRS, HC, CRSsNP, and CRSwNP. We believe that a study of this desi n is well-suited to elucidate patterns of differential ene expression between subtypes that may serve to advance disease endotypin in CRS. To date, this is the lar est microarray study of its type in CRS.
Supplementary Table 1 (Online Repository Table E1) summarizes the different disease subtypes enrolled in each group and relevant clinical characteristics.
We first performed hierarchical cluster analysis using microarray data from 130 patients with different subtypes of CRS. This resulted in the identification of 500 distinct gene expression clusters which are illustrated in a heat map (Online Repository Figure E1). Many gene clusters had differential expression patterns characterized by either highly upregulated (red signal) or highly downregulated (green signal) gene expression. Polyp subtypes AERD, AFRS, and CRSwNP demonstrated similar patterns of increased or decreased cluster expression and segregated from CRSsNP and HC based on their differential cluster expression, supporting a clear molecular delineation between polyp and non-polyp CRS phenotypes.
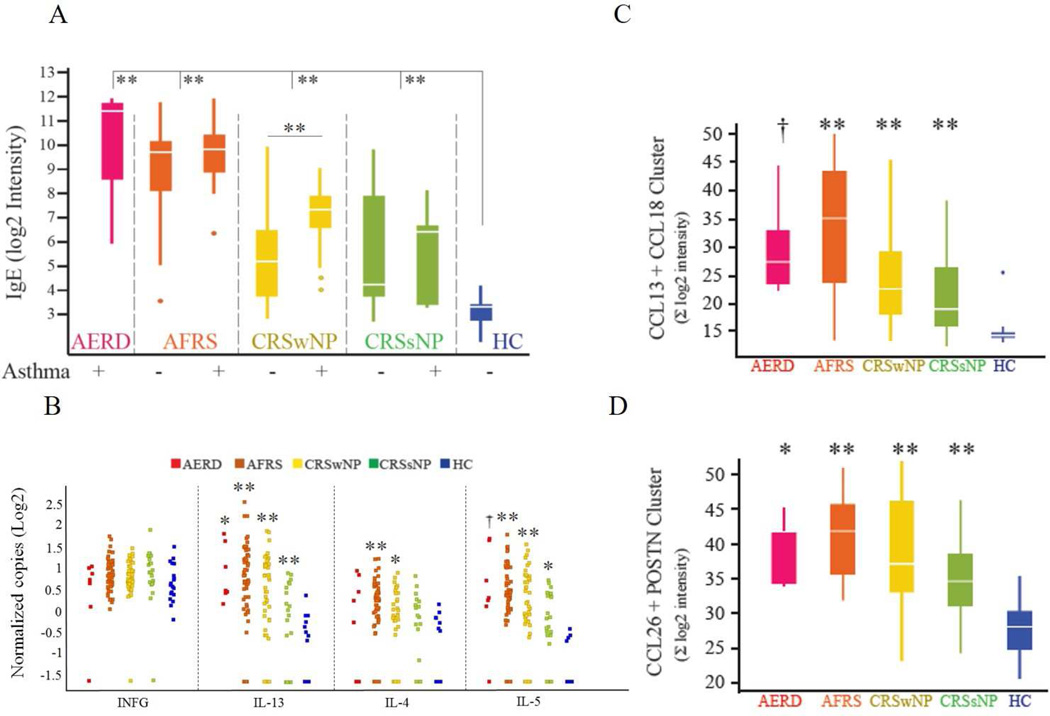
Clusters containing the most differentially expressed sequences between HC and disease, or those of particular interest, were examined for differential expression between subtypes. In addition, data was analyzed to identify correlative and reciprocal relationships between these clusters, and with other biological pathways of interest, especially those involved in type 2 inflammation. Sequences with the highest differential expression between groups were between HC and AFRS or HC and AERD. The single highest differential signal was from a sequence specific to the soluble form of IgE (Figure 1A). This expression was significantly upregulated in all subtypes when compared to HC, with AERD and AFRS demonstrating increased local IgE levels when compared to other disease subtypes. IgE expression was higher in CRSwNP patients with asthma in comparison to CRSwNP patients without asthma (p<0.001), while other subtypes showed no difference in tissue IgE expression when comparing those with and without asthma.
Figure 1. Expression of canonical type 2 inflammatory markers in CRS subtypes.
(A) Expression of IgE. Samples from each disease subtype are stratified according to presence or absence of asthma phenotype. (B) Relative expression of several canonical upstream regulators of type 1 (IFNG) or type 2 inflammation (IL-4, IL-5, IL-13) as measured by droplet digital PCR. (C and D) Relative expression of important downstream type 2 inflammatory gene clusters. †p<0.05, * p<0.01, ** p<0.001 for comparisons of HC vs each subtype. When no annotation is indicated, the comparison was not significant. POSTN - periostin
We next analyzed the expression of several canonical type 2 inflammatory cytokines (IL-4, IL-5, IL-13) and IFNγ by digital droplet quantitative PCR (Figure 1B). We also measured the expression of type 2 cytokine gene clusters that commonly act downstream of these cytokines (CCL13+CCL18 clusters, CCL26+Periostin clusters, Figure 1C and 1D). The cytokines CCL13, -18, -26 and periostin are of particular interest in CRS, as they have been shown to mediate localized eosinophilic inflammation3,4. No subtypes demonstrated increased expression of IFNγ, a canonical type 1 inflammatory cytokine, which is consistent with other studies showing lack of increased IFNγ in CRS subtypes, including CRSsNP, in Japanese, Chinese, and American populations3. All subtypes demonstrated relative increased expression of IL-13. Similarly, all subtypes exhibited increased expression of IL-5 when compared to HC. Only AFRS and CRSwNP subtypes demonstrated significant increases in IL-4 gene expression. Despite no differences detected between the mean copy of IL-13 and IL-4 among CRSwNP, AFRS and CRSsNP samples, CRSwNP and AFRS had more samples expressing higher copies of IL-13 and IL-4 as compared to samples from CRSsNP patients. Gene cluster expression analysis of CCL13+CCL18 and CCL26+periostin revealed overexpression in comparison to HC of these gene clusters in all CRS subtypes. Taken together, with the exception of IL-4, the expression of the above type 2 inflammatory markers did not separate clinical phenotypes but rather highlighted subgroups (possible endotypes) among the clinical phenotypes with different expression levels of these type 2 inflammatory markers when compared to HC.
Because we could not readily delineate subtypes of CRS disease based on the relative expression of markers chosen in Figure 1, we explored additional components of type 2 inflammation that might be driving disease in CRS. The mast cell axis represents a central node of type 2 inflammation, capable of responding to both innate and adaptive immune arms to promote a type 2 response. Studies originating from the work of the senior author have found that mast cells are present at increased levels, independent of atopy, in polyp mucosa of patients with CRSwNP5. We have also shown that inflamed mucosa of CRSwNP patients harbor type 2 innate lymphoid cells (ILC2s) expressing IL1RL1 (also known as ST2), a receptor for IL-33, and that these ILC2s secrete IL-13 in response to stimulation by IL-336. We wished to determine if there existed a correlation between the expression of IL1RL1 and genes associated with mast cell activity, and furthermore, assess if these genes were exclusive to certain disease subtypes.
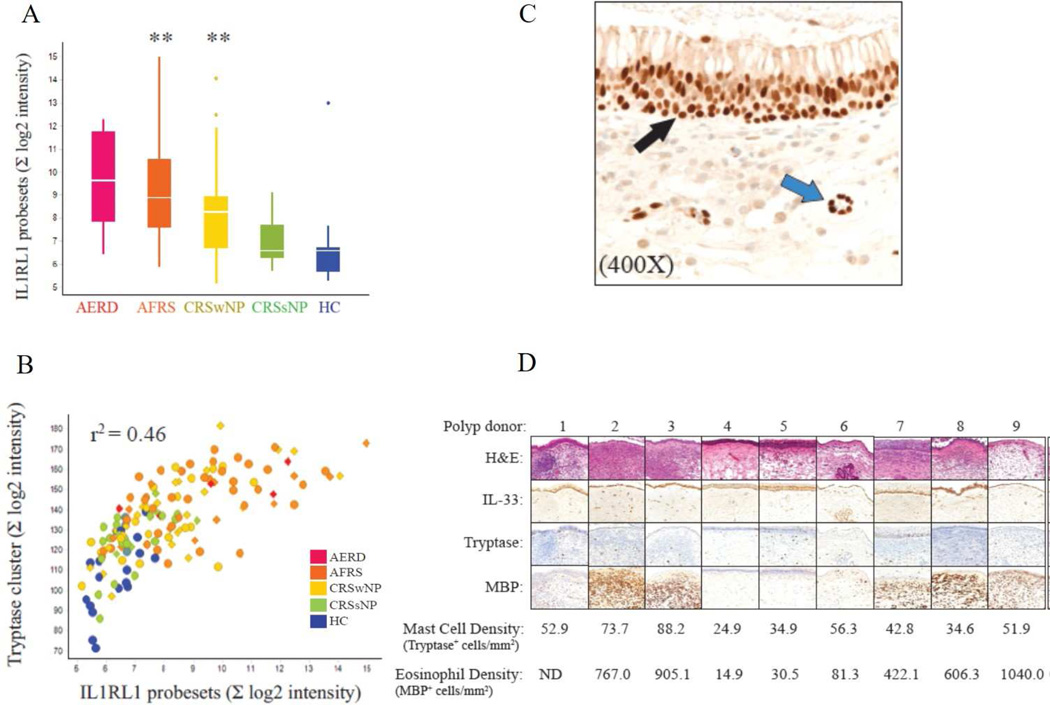
We found that IL1RL1 transcripts were significantly overexpressed in polyp subtypes, AFRS and CRSwNP, when compared to CRSsNP and HC (Figure 2A). Additionally, the expression of IL1RL1 correlated with a cluster enriched for mast cell-related genes (tryptase cluster, Figure 2B). We also found that IL1RL1 expression correlated with the expression of 21 genes associated with mast cell and eosinophil activity (Online Repository, Table E2). We performed an immunohistochemical analysis in a representative collection of surgically removed CRSwNP polyp tissue (Figure 2C and 2D). IL-33 expression was evident in the epithelial cells and around vessels in all polyp samples (Figure 2C). Immunohistochemistry indicated the presence of abundant eosinophils and mast cells, both known IL1RL1-expressing cells (Figure 2D). Taken together, these results implicate an IL1RL1-mast cell signaling axis as a potential endotype marker and target for therapeutic intervention.
Figure 2. ILRL1 expression and mast cell activity in CRS.
(A) Box and whisker plot of differential expression of IL1RL1 by microarray analysis. (B) Correlation of IL1RL1 and tryptase gene cluster. r2 = 0.46, p<0.001. (C and D) IL-33 expression, mast cell and eosinophil infiltration in representative CRSwNP tissue. (C) Representative photomicrograph illustrating IL-33 expression located in the nasal surface epithelium (black arrow) as well as the stroma (blue arrow) containing inflammatory infiltrate. (D) Photomicrographs demonstrating expression of IL-33 associated with varying levels of eosinophil and mast cell infiltration. MBP: major basic protein. ** p<0.001 for comparisons of HC vs each subtype. When no annotation is indicated, the comparison was not significant.
Discussion (See Online Repository)
Molecular pathways that drive disease in different CRS clinical phenotypes – so-called endotypes – need further clarification to facilitate the application of personalized therapies in CRS. Our top down, microarray-based analysis allowed us to characterize the relative expression of several type 2 inflammatory gene clusters of interest in well-defined clinical subtypes of CRS, many of which take into account eosinophilic inflammation, comorbid asthma, aspirin sensitivity, fungal colonization, and atopy. Admittedly, we do not define specific endotypes in this study; however, we evaluated key type 2 inflammatory markers as a means of endotyping CRS patients. We found that there were trends of elevated expression of certain type 2 inflammatory markers in clinical phenotypes but also found notable variation of expression within a given phenotype. One such marker, local IgE expression, was elevated in subgroups of AERD, AFRS, and CRSwNP patients with asthma, clinical subtypes that characteristically exhibit increased local sinonasal eosinophilia. Our observations support previous studies which indicate a correlation between local sinonasal IgE expression and increased tissue eosinophilia.7,8,9,E1
Interestingly, with the exception of IL-4, all CRS subtypes demonstrated increased expression of several canonical type 2 inflammatory markers (Figure 1). The observed increase in these measured type 2 inflammatory markers in CRSsNP may in part be due to the high prevalence of co-morbid allergic rhinitis in this patient population; however, multiple studies have suggested that atopic sinonasal inflammation represents a distinct process from that observed in CRS.E2, E3, E4 Our findings complement those of Tomassen et al., which showed the presence of type 2 inflammation in both polyp and non-polyp clinical subtypes, and highlight the complexity of the inflammatory milieu in CRS mucosa. Together, these findings prompt a need to investigate additional type 2 inflammatory pathways that better separate disease subtypes.
Studies in asthma and CRS have highlighted multiple different mechanisms that can promote type 2 inflammation, including an adaptive immune component and a recently identified innate type 2 inflammatory component. Epithelial-derived cytokines, like IL-25, IL-33, thymic stromal lymphopoietin and mast cells may play important roles in mediating type 2 inflammatory disease independent of adaptive immunity. Indeed, elevation of the IL-33 receptor, IL1RL1, in CRSwNP is not a novel finding; nonetheless, our results add credence to preceding studies which have shown the importance of this pathway in the pathogenesis of CRSwNP6,E4,E5,E6. Additionally, to the best of our knowledge, we present novel findings correlating IL1RL1 expression with mast cell activity in polyp subtypes and we extend these findings to the AFRS clinical subtype. We noted a trend towards increased IL1RL1 expression in the AERD clinical subtype (Figure 2A); however, the small sample size of this group likely precluded achieving statistical significance. Taken together, our results implicate an IL1RL1-mast cell signaling axis as a potential disease marker, a mediator of type 2 inflammation and a potential target for therapeutic intervention.
The study presented herein illustrates the complexity of the inflammatory makeup in CRS. While features of type 2 inflammation appear active in many clinical subtypes of CRS, they may culminate in a type 2 inflammatory response via different mechanisms, including infection, barrier disruption, allergy, etc. Additionally, the degree of type 2 inflammatory activation may be an important factor in differentiating disease endotypes. As clinical trials that test the efficacy of novel therapies in CRS progress, a better understanding of the specific molecular pathways that drive a patient’s disease will aid in identifying those who will benefit most from these novel treatment strategies.
Supplementary Material
Acknowledgments
Financial Support: The cost of the microarray processing was partially supported by Amgen. AL is supported by the Center for Clinical and Translational Sciences, which is funded by National Institutes of Health Clinical and Translational Award UL1 TR000371 and KL2 TR000370 from the National Center for Advancing Translational Science. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Advancing Translational Science or the National Institutes of Health.
We would like to acknowledge Kim Merriam and Ken Ganley for their help with immunohistochemistry.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: The Department of Otorhinolaryngology at the University of Texas McGovern Medical School received industry research funding from Allakos, Amgen and IntersectENT. AL received consulting fees from 480 Biomedical, Aerin Medical, ENTvantagm Laurimed, and Medtronic. MJC received consulting fees from JNJ/Acclarent. SF received consulting fees from IntersectENT.
Authorship: CBR, DES, and AL jointly conceived the study, its design and implemented the analysis. MAT, XH and SA assisted with the analysis. MAT, CBR, DES, and AL wrote the manuscript, and CJPD, MJC, and SF edited the manuscript and provided conceptual advice. We would like to acknowledge Kim Merriam and Ken Ganley for their help with immunohistochemistry.
References
- 1.Akdis CA, Bachert C, Cingi C, Dykewicz MS, Hellings PW, Naclerio RM, et al. Endotypes and phenotypes of chronic rhinosinusitis: a PRACTALL document of the European Academy of Allergy and Clinical Immunology and the American Academy of Allergy, Asthma & Immunology. J Allergy Clin Immunol. 2013 Jun;131(6):1479–1490. doi: 10.1016/j.jaci.2013.02.036. [PubMed: 23587334] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gauthier M, Ray A, Wenzel SE. Evolving Concepts of Asthma. Am J Respir Crit Care Med. 2015 Sep 15;192(6):660–668. doi: 10.1164/rccm.201504-0763PP. [PubMed: 23587334] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kato A. Immunopathology of chronic rhinosinusitis. Allergol Int. 2015 Apr;64(2):121–130. doi: 10.1016/j.alit.2014.12.006. [PubMed: 2583808] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang M, Wang X, Zhang N, Wang N, Wang H, Li Y, et al. Association of periostin expression with eosinophilic inflammation in nasal polyps. J Allergy Clin Immunol. 2015 Dec;136(6):1700–1703. doi: 10.1016/j.jaci.2015.09.005. [PubMed: 26521039] [DOI] [PubMed] [Google Scholar]
- 5.Shaw JL, Ashoori F, Fakhri S, Citardi MJ, Luong A. Increased percentage of mast cells within sinonasal mucosa of chronic rhinosinusitis with nasal polyp patients independent of atopy. Int Forum Allergy Rhinol. 2012 May-Jun;2(3):233–240. doi: 10.1002/alr.21021. [PubMed: 22344928] [DOI] [PubMed] [Google Scholar]
- 6.Shaw JL, Fakhri S, Citardi MJ, Porter PC, Corry DB, Kheradmand F, et al. IL-33-responsive innate lymphoid cells are an important source of IL-13 in chronic rhinosinusitis with nasal polyps. Am J Respir Crit Care Med. 2013 Aug 15;188(4):432–439. doi: 10.1164/rccm.201212-2227OC. [PubMed: 23805875] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bachert C, Zhang N, Holtappels G, De Lobel L, van Cauwenberge P, Liu S, et al. Presence of IL-5 protein and IgE antibodies to staphylococcal enterotoxins in nasal polyps is associated with comorbid asthma. J Allergy Clin Immunol. 2010;126:962–968. doi: 10.1016/j.jaci.2010.07.007. [PubMed: 20810157] [DOI] [PubMed] [Google Scholar]
- 8.Tomassen P, Vandeplas G, Van Zele T, Cardell LO, Arebro J, Olze H, et al. Inflammatory endotypes of chronic rhinosinusitis based on cluster analysis of biomarkers. J Allergy Clin Immunol. 2016 May;137(5):1449–1456. doi: 10.1016/j.jaci.2015.12.1324. [PubMed: 26949058] [DOI] [PubMed] [Google Scholar]
- 9.Bachert C, van Steen K, Zhang N, Holtappels G, Cattaert T, Maus B, et al. Specific IgE against Staphylococcus aureus enterotoxins: an independent risk factor for asthma. J Allergy Clin Immunol. 2012;130:376–381. doi: 10.1016/j.jaci.2012.05.012. [PubMed: 22738677] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.