Abstract
Objective
Untreated advanced HIV infection alters the gut microbiota, but it is unclear whether antiretroviral therapy (ART) reverses these changes. We compared the composition of the rectal microbiota among three groups of men who have sex with men (MSM): HIV-uninfected, untreated HIV, and ART-treated HIV-infected.
Design
A cross-sectional study was conducted among 130 MSM (55 HIV-uninfected, 41 untreated HIV, 34ART-treated HIV) in Abuja, Nigeria.
Methods
Bacterial 16S rRNA genes were amplified from rectal swabs, sequenced and clustered into Genera-level operational taxonomic units. Alpha diversity was quantified using the Shannon Index and compared among groups using the Kruskal-Wallis test; Associations with other scale variables were quantified using Spearman's Rank Correlation (Rs). The relative abundance of the top 15 taxa was compared according to HIV infection/treatment status using the Wilcoxon-Rank sum test.
Results
HIV treated MSM had a decrease in a commensal phylum, Bacteroidetes (p<0.01). Alpha diversity was positively correlated with viral loads (Rs=0.32, p<0.01). Statistically significant shifts in relative abundance of rectal microbiota for the HIV treated group included a decrease in the most abundant bacteria, Prevotella (p=0.02) and an increase in pathogenic bacteria, Peptoniphilus (p=0.04), Finegoldia (p=0.01), Anaerococcus (p=0.03), and Campylobacter (p=0.03) as compared to the other groups.
Conclusions
Untreated HIV infection does not significantly alter the rectal microbiota, whereas prior treatment is associated with a shift towards a more pathogenic pattern of microbiota. Treatment with an antibiotic, co-trimoxazole, in conjunction with ART may have contributed to this shift.
Keywords: antiretroviral therapy, co-trimoxazole, Prevotella, alpha diversity, relative abundance
Introduction
The rectal microbiota may be important in driving mucosal immune responses that defend against infectious pathogens and regulate local inflammation during receptive anal intercourse [1]. HIV has been implicated in reducing rectal microbiota diversity, shifting the composition of commensals, enhancing microbial translocation, and disrupting local immunity [2-8]. While several studies have focused on the upper gastrointestinal tract microbiota, virtually no data are available from sub-Saharan Africa and only a few studies [9, 10] have begun to characterize the rectal microbiota.
The rectal microbiota is dominated by 5 phyla: Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, and Fusobacteria, ranked in order of abundance. Early studies suggested a decline in species richness and increase in a more pathogenic phyla, Fusobacteria, among individuals with advanced HIV infection who are not on antiretroviral therapy (ART) [9, 10]. Fusobacteria harbor virulence factors that potentially disrupt local immunity [11-14], which may contribute to the pathogenesis of diseases such as inflammatory bowel disease and gastrointestinal malignancies [15-19].
The impact of ART on the microbiota is unclear. Some studies suggest that long-term ART partially restores the microbiota to a level between untreated HIV and HIV-uninfected [4, 5, 7]. Others suggest ART has little impact [6, 8] or even disrupts the microbiota [7]. Taken together, the stage of HIV disease and the timing and duration of ART may impact the microbiota differently.
The objective of this study was to evaluate the rectal microbiota among three groups: HIV-uninfected, untreated HIV, and ART-treated HIV.
Materials and Methods
Study design and Population
Our cross-sectional rectal microbiota study was nested in the Abuja, Nigeria, site of the parent cohort, TRUST/RV368. Data on demographics, sexual behaviors, HIV and sexually transmitted infections (STIs) among Nigerian men who have sex with men (MSM) were previously collected [20, 21]. The prevalence of high-risk human papillomavirus (HR-HPV) was characterized in a subset of 165 participants [22]. After HPV testing, seven samples had insufficient volume and 28 failed the 16S rRNA amplification criteria of 1000 reads, leaving a final sample size of 130 for the rectal microbiota analysis.
The study was approved by the Federal Capital Territory Health Research Ethics Committee in Nigeria and the University of Maryland Baltimore Institutional Review Board.
Laboratory Procedures
DNA was extracted and processed as previously described [10]. Sequencing targeted an amplicon size of ∼469bp within the V3 and V4 hypervariable regions using primers 319F and 806R. The paired-end reads (300bp each) were sequenced on the Illumina MiSeq platform with an overlap of ∼90bp, resulting in entire coverage of the amplicons. Paired-end reads were assembled using FLASH [23] with error correction. Dual barcoding information was used to index individual samples [24], and the sequences were de-multiplexed by binning sequences with the same barcodes in QIIME (version 1.8.0) [25]. QIIME quality trimming was performed. Sequences with less than 3% dissimilarity were clustered together using USEARCH (v5.2.32) [26] and de novo chimera detection was conducted in UCHIME v5.1 [27]. Genus-level taxonomic ranks were assigned for a representative sequence of each operational taxonomic unit using Ribosomal Database Project (RDP) Naïve Bayes Classifier v.2.2 [28], using 0.8 confidence values as the cutoff to greengenes 16S rRNA gene sequences (Aug, 2013 vers.) [29].
Statistical Analyses
Baseline characteristics were compared between groups using Pearson's chi square test, Kruskal-Wallis test or Wilcoxon rank sum test.
The Shannon index, a measure of alpha diversity, was calculated as the negative of the sum of the relative abundance multiplied by the natural log of the relative abundance for each genus and further stratified by phyla. Shannon indices were compared using the Kruskal-Wallis test. Associations with CD4+ T cell counts and viral loads were quantified using Spearman's Rank Correlation (Rs).
Shifts in composition were estimated from the relative abundance of the 15 most abundant genera within the top 5 phyla. The remaining genera were pooled within their phyla category prior to calculating a relative abundance. This resulted in relative abundances for 21 taxa categories within the 5 phyla that were compared between the three HIV infection/treatment groups. Age, rectal gonorrhea, and HR-HPV were tested separately for associations with relative abundance using Rs and Wilcoxon rank sum test in order to determine if these could be confounding any associations with HIV/ART status.
Results
Participant characteristics
Of the 130 participants, 55 were HIV-uninfected, 41 had untreated HIV, and 34 had ART-treated HIV. HIV-uninfected were younger than untreated and ART-treated HIV-infected men [median (interquartile range, IQR) 23 (20-28), 25 (21-27) and 27 (24-32), respectively, p<0.01]. The HIV treated group had the lowest prevalence of rectal gonorrhea than untreated and HIV-uninfected men [11.8%, 39%, 27.8%, respectively, p=0.03] and more HR-HPV [median (IQR) 3 (1-4), 2 (1-4), 0 (0-1), respectively, p<0.01]. The three groups did not differ significantly on rectal chlamydia, treatment for symptomatic STIs, number of male partners with whom they engaged in receptive sex, condomless receptive sex, and concurrent partnerships. The ART-treated HIV group had higher CD4 counts (p=0.05) and lower log10 transformed viral loads (p<0.01) than the HIV-untreated group. Years since HIV diagnosis was longer for the ART-treated vs. untreated group [median (IQR) 2 (1-4) and 1(0-2), respectively, p=0.02].
Alpha Diversity
Overall, there was no significant difference in the alpha diversity between the HIV-uninfected, untreated and ART-treated HIV-infected men [median (IQR) 2.68 (2.43-2.89), 2.58 (2.27-2.94), 2.68 (2.34-2.87), respectively, p=0.73]. When the Shannon index was stratified by phyla, the treated HIV group had a decrease in species richness and evenness in the Bacteroidetes phylum (p=0.002) and a non-significant increase in the Fusobacteria phylum (p=0.12). For men who were HIV-infected, the alpha diversity of the Bacteroidetes phylum did not differ by CD4 counts (Rs=-0.10, p=0.39) but was positively correlated with viral load (Rs=0.32, p<0.01).
Shifts in microbial composition
The HIV-uninfected and untreated HIV groups did not differ in their relative abundances of the 21 different taxa categorizations (Table 1, Figure 1). Compared to the HIV-uninfected group, the ART-treated had significant shifts in the composition of their rectal microbiota (Table 1). Within the Firmicutes phylum, there was an increase in the Peptoniphilus, Finegoldia and Anaerococcus genera. Within the Bacteroidetes phylum, there was a decrease in Prevotella. Within the Proteobacteria phylum, there was an increase in Campylobacter. Within the Fusobacteria phylum, there was a non-significant increase in Fusobacterium (p=0.11). These shifts were not significantly associated with age or number of HR-HPV. Participants with rectal gonorrhea had increased abundance of Prevotella compared to those without gonorrhea [median (IQR) 29.3 (16.2-43.1) vs. 22.7 (14.2-33.4), p=0.05].
Table 1. Relative abundance of most common rectal genera within 5 phyla, stratified by HIV and ART status.
| HIV-Uninfected | Untreated HIV | HIV- vs. Untreated | Treated HIV | HIV- vs. Treated | ||
|---|---|---|---|---|---|---|
|
| ||||||
| Median % (IQR) n=55 | Median % (IQR) n=41 | P* | Median % (IQR) n=34 | P* | Abundance shift | |
| Firmicutes | ||||||
| g_faecalibacterium | 5.2 (2.3-8.1) | 6.6 (4.1-9.0) | 0.15 | 3.4 (1.1-6.1) | 0.07 | |
| g_peptoniphilus | 2.1 (0.6-6.7) | 2.1 (0.5-5.4) | 0.54 | 4.8 (1.3-7.0) | 0.04 | ↑ |
| g_wal_1855D | 0.7 (0.0-5.0) | 0.0 (0.0-1.4) | 0.05 | 1.3 (0.0-4.8) | 0.90 | |
| g_finegoldia | 2.6 (0.7-5.9) | 1.7 (0.9-5.8) | 0.91 | 5.1 (1.9-14.1) | 0.01 | ↑ |
| g_roseburia | 3.5 (1.0-6.9) | 3.0 (1.7-6.6) | 0.86 | 1.7 (0.8-3.7) | 0.09 | |
| g_anaerococcus | 1.7 (0.6-4.5) | 1.5 (0.4-2.9) | 0.47 | 3.0 (1.3-9.5) | 0.03 | ↑ |
| f_ruminococcaceae | 1.8 (1.0-2.9) | 1.5 (0.8-3.3) | 0.90 | 1.2 (0.7-2.1) | 0.07 | |
| g_blautia | 2.2 (1.0-3.7) | 1.6 (0.9-3.0) | 0.24 | 1.1 (0.5-3.1) | 0.08 | |
| g_ruminococcus | 1.2 (0.5-2.7) | 1.4 (0.4-3.4) | 0.62 | 2.1 (0.6-3.8) | 0.38 | |
| g_other | 20.6 (16.5-26.6) | 21.9 (15.7-25.6) | 0.92 | 19.6 (12.7-25.7) | 0.26 | |
| Bacteroidetes | ||||||
| g_prevotella | 26.4 (15.6-38.2) | 27.8 (16.2-39.7) | 0.53 | 18.7 (10.8-27.2) | 0.02 | ↓ |
| g_bacteroides | 1.2 (0.4-3.0) | 2.1 (0.3-4.9) | 0.44 | 0.5 (0.3-2.4) | 0.24 | |
| g_porphyromonas | 0.5 (0.0-2.2) | 0.3 (0.1-2.1) | 0.82 | 0.8 (0.2-3.4) | 0.27 | |
| g_other | 1.3 (0.5-2.9) | 2.0 (0.5-2.9) | 0.78 | 0.7 (0.2-1.4) | <0.01 | ↓ |
| Proteobacteria | ||||||
| g_campylobacter | 0.5 (0.0-3.2) | 0.6 (0.0-1.4) | 0.53 | 1.7 (0.4-4.1) | 0.03 | ↑ |
| g_other | 2.0 (0.7-4.7) | 1.7 (0.8-4.5) | 0.88 | 0.5 (0.3-1.9) | 0.01 | ↓ |
| Actinobacteria | ||||||
| g_all | 0.4 (0.1-0.9) | 0.4 (0.2-1.2) | 0.71 | 0.6 (0.2-1.3) | 0.21 | |
| Fusobacteria | ||||||
| g_sneathia | 0.0 (0.00-0.2) | 0.0 (0.0-0.0) | 0.32 | 0.0 (0.0-4.2) | 0.32 | |
| g_fusobacterium | 0.2 (0.00-2.2) | 0.1 (0.0-1.6) | 0.44 | 1.2 (0.0-3.6) | 0.11 | ↑ |
| g_other | 0.0 (0.00-0.0) | 0.0 (0.0-0.0) | 0.29 | 0.0 (0.0-0.0) | 0.49 | |
| Other phyla | ||||||
| g_all | 0.2 (0.0-0.6) | 0.1 (0.0-0.3) | 0.1 (0.0-0.3) | 0.02 | ↓ | |
Abbreviations:ART, antiretroviral therapy; IQR, interquartile range; g_, Genus; f_, Family
Wilcoxon rank sum test
Figure 1.
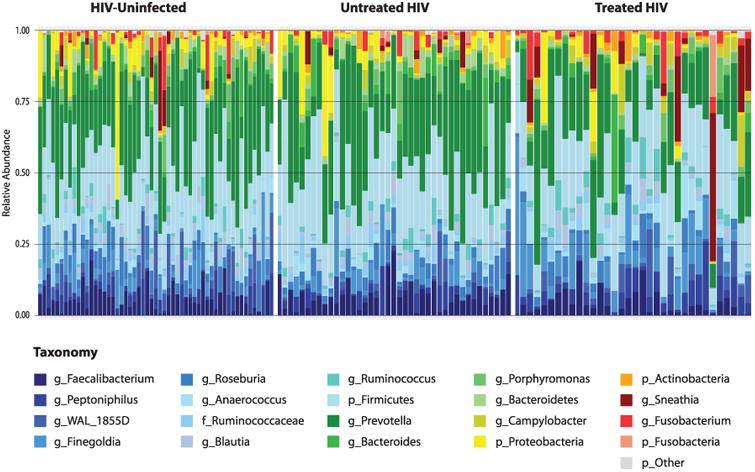
Relative Abundance of Rectal Taxa, by HIV and ART Status.
Color shades of phyla (Firmicutes: blue; Bacteroidetes: green; Proteobacteria: yellow; Actinobacteria: orange; Fusobacteria: red; other: gray)
Discussion
Nigerian MSM with untreated HIV did not differ in alpha diversity or composition of their rectal microbiota as compared to HIV-uninfected MSM. Treated HIV was associated with a decreased diversity of the Bacteroidetes phylum and compositional shifts towards more pathogenic bacteria. Lower prevalence of rectal gonorrhea may have contributed to a lower abundance of Prevotella in the ART-treated group. The alpha diversity and relative abundance of Fusobacteria was similar across the groups.
Interestingly, participants on ART had a shift in abundance of common bacteria, which may have been confounded by antibiotic therapy. WHO guidelines recommend long-term co-trimoxazole prophylaxis for adults receiving ART since it has been shown to reduce early mortality by 58% [30-33]. The extent of the confounding is unclear since none of the treated HIV-infected men (n=34) reported using co-trimoxazole and 44% (15/34) reported either an allergy or a CD4+ T cell count of >350 cells/mm3 that contraindicated them from receiving co-trimoxazole [34]. This suggests only a fraction of the men were on antibiotic prophylaxis.
Other antibiotics have been associated with reduced gut and fecal microbial diversity [35-37] but the impact of co-trimoxazole has not been well investigated. Yu et al. found co-trimoxazole was only significantly associated with an altered rectal microbiota in one of six alpha and beta diversity measures [10]. Another study among HIV-exposed uninfected infants found co-trimoxazole was associated with an increase in blood levels of lipopolysaccharide, a marker of microbial translocation and poor gut integrity, but not a direct measure of rectal microbiota changes [38].
Alternatively, HIV treatment itself may negatively impact the rectal microbiota. A decrease in species diversity after ART initiation was observed in two longitudinal studies, one in humans [7] and the other in macaques [39]. Other studies suggest that ART impacts the phylum, Bacteroidetes [4, 6], and in particular the genus Prevotella, as opposed to Bacteroides [5, 7]. Bacteroidetes may be an important commensal because it is preserved in HIV-positive elite controllers [7] and is depleted among those with inflammatory bowel disease [40]. The dominance of Prevotella may be attributed to sexual behavior [8] or a diet comprised mostly of simple carbohydrates [41].
There was also an increase in more Gram-positive anaerobic bacteria such as Peptoniphilus, Finegoldia, and Anaerococcus, all of which have been previously found to be associated with vaginal or genitourinary tract infections [42] and were reduced in the penile microbiota after circumcising HIV-uninfected men [43]. Additionally there was an increase in Campylobacter in the gut microbiota of ART-treated HIV-infected individuals [6]. Further studies of the influence of anaerobic species on local immunity and long term susceptibility to STIs are needed.
The rectal microbiota did not differ between untreated HIV and HIV-uninfected participants, but it is possible that HIV infection was too recent within this cohort to observe long-term shifts. Simian studies have found that SIV does not immediately impact gut microbial diversity [44, 45]. In a human study, diversity was not different for those with untreated HIV and >500 CD4+ T cell counts as compared to HIV-uninfected [8]. Furthermore, diversity and number of taxa were not altered among men who had been living with HIV for seven to eleven years [10]. Alterations may only become apparent during later stages of HIV disease.
A few limitations should be acknowledged. First, this cross-sectional study cannot assess directionality, but is one of the first studies to describe the rectal microbiota in sub-Saharan Africa. Second, self-reported data on co-trimoxazole prophylaxis were incomplete and the component of treatment regimens contributing to observed shifts could not be determined. Third, co-infection with rectal gonorrhea may have confounded the abundance differences in one genus, Prevotella, but a larger sample size would be needed to account for this effect. Lastly, diet, antibiotic use and smoking status were not collected and may have confounded our main association.
In summary, there were subtle shifts in the rectal microbiota among HIV-positive individuals receiving treatment, including a decrease in diversity of the Bacteroidetes phylum driven primarily by a loss in Prevotella. There was a shift towards more pathogenic bacteria. Prospective studies are needed to evaluate the long-term consequences of alterations of the microbiota with HIV infection and its treatment.
Acknowledgments
The authors thank Mike Humphrey for his assistance in the sequencing, the clients who willingly engaged in the TRUST/RV368 study regardless of the difficult contexts, and the ceaseless commitment of the study staff to their clients and the study team.
Funding Sources: The research reported in this publication was supported by seed funding from MPower and salary support from UMGCC P30 grant (P30 CA 134274-04) from the National Cancer Institute. Additional support has been provided by the U.S National Institutes of Health under award number R01MH099001-01 and R01AI120913, the U.S. Military HIV Research Program (Grant No. W81XWH-07-2-0067), Fogarty AITRP (D43TW01041), and the President's Emergency Plan for AIDS Relief through cooperative agreement U2G IPS000651 from the HHS/Centers for Disease Control and Prevention (CDC), Global AIDS Program with IHVN. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Department of Defense, National Institutes of Health or other funders.
Footnotes
The TRUST Study Group is constituted as follows: Principal investigators: William Blattner and Man Charurat (IHV, University of Maryland, Baltimore, MD, USA)
Co-investigators: Alash'le Abimiku, Sylvia Adebajo, Julie Ake, Stefan Baral, Trevor Crowell, Charlotte Gaydos, Lindsay Hughes, Babajide Keshinro, Jerome Kim, Hongjie Liu, Jennifer Malia, Nelson Michael, Ogbonnaya Njoku, Rebecca Nowak, Helen Omuh, Ifeanyi Orazulike, Sheila Peel, Merlin Robb, Cristina Rodriguez-Hart, Sheree Schwartz
Institutions: Institute of Human Virology at the University of Maryland School of Medicine (IHV), Johns Hopkins Bloomberg School of Public Health (JHSPH), Walter Reed Army Institute of Research, U.S. Military HIV Research Program (MHRP), Department of Defense, Walter Reed Program, Nigeria (WRP), Institute of Human Virology Nigeria (IHVN), International Centre for Advocacy for the Right to Health (ICARH), Population Council (Pop Council)
Conflicts of Interest and Source of Funding: TC has received a speaker fee from Gilead Sciences.
Contributors: WB and MC designed the TRUST study. RN, WB, and MC conceived the analysis for the manuscript. Data collection and management was facilitated by RN, WD, JR, BM. RN conducted the data analysis with input from SMB, JR, HL, and BM. RN drafted the manuscript and SMB, JR, BM, TC, SB, HL, MC, and WB provided critical review and editing. All authors have seen and approved the paper; the corresponding author had full access to the data and had final responsibility for the decision to submit for publication.
References
- 1.Denny JE, Powell WL, Schmidt NW. Local and Long-Distance Calling: Conversations between the Gut Microbiota and Intra- and Extra-Gastrointestinal Tract Infections. Front Cell Infect Microbiol. 2016;6:41. doi: 10.3389/fcimb.2016.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12:1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 3.Klatt NR, Funderburg NT, Brenchley JM. Microbial translocation, immune activation, and HIV disease. Trends Microbiol. 2013;21:6–13. doi: 10.1016/j.tim.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vujkovic-Cvijin I, Dunham RM, Iwai S, Maher MC, Albright RG, Broadhurst MJ, et al. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci Transl Med. 2013;5:193ra91. doi: 10.1126/scitranslmed.3006438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lozupone CA, Rhodes ME, Neff CP, Fontenot AP, Campbell TB, Palmer BE. HIV-induced alteration in gut microbiota: driving factors, consequences, and effects of antiretroviral therapy. Gut Microbes. 2014;5:562–570. doi: 10.4161/gmic.32132. [DOI] [PubMed] [Google Scholar]
- 6.Mutlu EA, Keshavarzian A, Losurdo J, Swanson G, Siewe B, Forsyth C, et al. A compositional look at the human gastrointestinal microbiome and immune activation parameters in HIV infected subjects. PLoS Pathog. 2014;10:e1003829. doi: 10.1371/journal.ppat.1003829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nowak P, Troseid M, Avershina E, Barqasho B, Neogi U, Holm K, et al. Gut microbiota diversity predicts immune status in HIV-1 infection. AIDS. 2015;29:2409–2418. doi: 10.1097/QAD.0000000000000869. [DOI] [PubMed] [Google Scholar]
- 8.Noguera-Julian M, Rocafort M, Guillen Y, Rivera J, Casadella M, Nowak P, et al. Gut Microbiota Linked to Sexual Preference and HIV Infection. EBioMedicine. 2016;5:135–146. doi: 10.1016/j.ebiom.2016.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McHardy IH, Li X, Tong M, Ruegger P, Jacobs J, Borneman J, et al. HIV Infection is associated with compositional and functional shifts in the rectal mucosal microbiota. Microbiome. 2013;1:26. doi: 10.1186/2049-2618-1-26. 26-2618-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu G, Fadrosh D, Ma B, Ravel J, Goedert JJ. Anal microbiota profiles in HIV-positive and HIV-negative MSM. AIDS. 2014;28:753–760. doi: 10.1097/QAD.0000000000000154. [DOI] [PubMed] [Google Scholar]
- 11.Allen-Vercoe E, Jobin C. Fusobacterium and Enterobacteriaceae: important players for CRC? Immunol Lett. 2014;162:54–61. doi: 10.1016/j.imlet.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohkusa T, Yoshida T, Sato N, Watanabe S, Tajiri H, Okayasu I. Commensal bacteria can enter colonic epithelial cells and induce proinflammatory cytokine secretion: a possible pathogenic mechanism of ulcerative colitis. J Med Microbiol. 2009;58:535–545. doi: 10.1099/jmm.0.005801-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han YW, Shi W, Huang GT, Kinder Haake S, Park NH, Kuramitsu H, et al. Interactions between periodontal bacteria and human oral epithelial cells: Fusobacterium nucleatum adheres to and invades epithelial cells. Infect Immun. 2000;68:3140–3146. doi: 10.1128/iai.68.6.3140-3146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jewett A, Hume WR, Le H, Huynh TN, Han YW, Cheng G, et al. Induction of apoptotic cell death in peripheral blood mononuclear and polymorphonuclear cells by an oral bacterium, Fusobacterium nucleatum. Infect Immun. 2000;68:1893–1898. doi: 10.1128/iai.68.4.1893-1898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strauss J, Kaplan GG, Beck PL, Rioux K, Panaccione R, Devinney R, et al. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm Bowel Dis. 2011;17:1971–1978. doi: 10.1002/ibd.21606. [DOI] [PubMed] [Google Scholar]
- 16.Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol. 2012;9:599–608. doi: 10.1038/nrgastro.2012.152. [DOI] [PubMed] [Google Scholar]
- 17.Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahn J, Sinha R, Pei Z, Dominianni C, Wu J, Shi J, et al. Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst. 2013;105:1907–1911. doi: 10.1093/jnci/djt300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flanagan L, Schmid J, Ebert M, Soucek P, Kunicka T, Liska V, et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis. 2014;33:1381–1390. doi: 10.1007/s10096-014-2081-3. [DOI] [PubMed] [Google Scholar]
- 20.Charurat ME, Emmanuel B, Akolo C, Keshinro B, Nowak RG, Kennedy S, et al. Uptake of treatment as prevention for HIV and continuum of care among HIV-positive men who have sex with men in Nigeria. J Acquir Immune Defic Syndr. 2015;68 Suppl 2:S114–23. doi: 10.1097/QAI.0000000000000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baral SD, Ketende S, Schwartz S, Orazulike I, Ugoh K, Peel SA, et al. Evaluating respondent-driven sampling as an implementation tool for universal coverage of antiretroviral studies among men who have sex with men living with HIV. J Acquir Immune Defic Syndr. 2015;68 Suppl 2:S107–13. doi: 10.1097/QAI.0000000000000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nowak RG, Gravitt PE, He X, Ketende S, Dauda W, Omuh H, et al. Prevalence of Anal High-Risk Human Papillomavirus Infections Among HIV-Positive and HIV-Negative Men Who Have Sex With Men in Nigeria. Sex Transm Dis. 2016;43:243–248. doi: 10.1097/OLQ.0000000000000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fadrosh DW, Ma B, Gajer P, Sengamalay N, Ott S, Brotman RM, et al. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome. 2014;2 doi: 10.1186/2049-2618-2-6. 6-2618-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 27.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Church JA, Fitzgerald F, Walker AS, Gibb DM, Prendergast AJ. The expanding role of co-trimoxazole in developing countries. Lancet Infect Dis. 2015;15:327–339. doi: 10.1016/S1473-3099(14)71011-4. [DOI] [PubMed] [Google Scholar]
- 31.Harries AD, Lawn SD, Suthar AB, Granich R. Benefits of combined preventive therapy with co-trimoxazole and isoniazid in adults living with HIV: time to consider a fixed-dose, single tablet coformulation. Lancet Infect Dis. 2015;15:1492–1496. doi: 10.1016/S1473-3099(15)00242-X. [DOI] [PubMed] [Google Scholar]
- 32.WHO 2014 Supplement to the 2013 consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection. 2014 Mar; [PubMed] [Google Scholar]
- 33.Suthar AB, Granich R, Mermin J, Van Rie A. Effect of cotrimoxazole on mortality in HIV-infected adults on antiretroviral therapy: a systematic review and meta-analysis. Bull World Health Organ. 2012;90:128C–138C. doi: 10.2471/BLT.11.093260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Federal Ministry of Health. National Guidelines for HIV and AIDS Treatment and Care in Adolescents and Adults; Abuja, Nigeria: Oct, 2010. [Google Scholar]
- 35.Jakobsson HE, Jernberg C, Andersson AF, Sjolund-Karlsson M, Jansson JK, Engstrand L. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One. 2010;5:e9836. doi: 10.1371/journal.pone.0009836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vrieze A, Out C, Fuentes S, Jonker L, Reuling I, Kootte RS, et al. Impact of oral vancomycin on gut microbiota, bile acid metabolism, and insulin sensitivity. J Hepatol. 2014;60:824–831. doi: 10.1016/j.jhep.2013.11.034. [DOI] [PubMed] [Google Scholar]
- 37.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011;108 Suppl 1:4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kourtis AP, Ibegbu CC, Wiener J, King CC, Tegha G, Kamwendo D, et al. Role of intestinal mucosal integrity in HIV transmission to infants through breast-feeding: the BAN study. J Infect Dis. 2013;208:653–661. doi: 10.1093/infdis/jit221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klase Z, Ortiz A, Deleage C, Mudd JC, Quinones M, Schwartzman E, et al. Dysbiotic bacteria translocate in progressive SIV infection. Mucosal Immunol. 2015;8:1009–1020. doi: 10.1038/mi.2014.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu GD, Bushmanc FD, Lewis JD. Diet, the human gut microbiota, and IBD. Anaerobe. 2013;24:117–120. doi: 10.1016/j.anaerobe.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 42.Murphy EC, Frick IM. Gram-positive anaerobic cocci--commensals and opportunistic pathogens. FEMS Microbiol Rev. 2013;37:520–553. doi: 10.1111/1574-6976.12005. [DOI] [PubMed] [Google Scholar]
- 43.Price LB, Liu CM, Johnson KE, Aziz M, Lau MK, Bowers J, et al. The effects of circumcision on the penis microbiome. PLoS One. 2010;5:e8422. doi: 10.1371/journal.pone.0008422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Handley SA, Thackray LB, Zhao G, Presti R, Miller AD, Droit L, et al. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell. 2012;151:253–266. doi: 10.1016/j.cell.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McKenna P, Hoffmann C, Minkah N, Aye PP, Lackner A, Liu Z, et al. The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis. PLoS Pathog. 2008;4:e20. doi: 10.1371/journal.ppat.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]


