Abstract
Age spots, also called solar lentigines and lentigo senilis, are light brown to black pigmented lesions of various sizes that typically develop in chronically sun-exposed skin. It is well known that age spots are strongly related to chronic sun exposure and are associated with photodamage and an increased risk for skin cancer, however, the mechanism(s) underlying their development remain poorly understood. We used immunohistochemical analysis and microarray analysis to investigate the processes involved in their formation, focusing on specific markers associated with the functions and proliferation of melanocytes and keratinocytes. A total of 193 genes were differentially expressed in age spots but melanocyte pigment genes were not among them. The increased expression of keratins 5 and 10, markers of basal and suprabasal keratinocytes, respectively, in age spots suggests that the increased proliferation of basal keratinocytes combined with the decreased turnover of suprabasal keratinocytes leads to the exaggerated formation of rete ridges in lesional epidermis which in turn disrupts the normal processing of melanin upwards from the basal layer. Based on our results, we propose a model for the development of age spots that explains the accumulation of melanin and the development of extensive rete ridges in those hyperpigmented lesions.
Keywords: solar lentigo, lentigo senilis, melanin, skin, pigmentation
Introduction
Age spots, also called solar lentigines and lentigo senilis, are light brown to black pigmented lesions of various sizes (several mm to a few cm) that typically develop in chronically sun-exposed skin. Histologically, age spots display elongated, club-shaped rete ridges containing numerous melanocytes and increased melanin production without cellular atypia (1). It is well known that age spots are strongly related to chronic sun exposure and are associated with photodamage and an increased risk for skin cancer (2), however, the mechanism(s) underlying their development remain poorly understood.
Some studies have reported various melanogenic paracrine factors, such as keratinocyte growth factor (KGF/FGF7), hepatocyte growth factor (HGF), stem cell factor (SCF/KITL) and endothelin 1 (EDN1), that might be involved in the development of age spots (3–6). Other studies have suggested an inflammation model for the hyperpigmentation of age spots and the involvement of factors related to inflammation (7, 8). Recently developed imaging techniques and the ultrastructural analysis of age spots have provided insights about how age spots develop and suggested that perturbations in the epidermal-dermal junction might be involved (9, 10). Three studies have characterized the expression of pigment-related genes in the hyperpigmented skin of age spots on the face and legs compared with perilesional skin. One of those studies (11) used PCR and in situ hybridization analysis to demonstrate that several melanocyte-specific genes were increased in expression in lesional skin and they proposed that the increased expression of POMC by keratinocytes (the ligand that activates MC1R) is involved in the stimulation of pigmentation. A recent GWAS study (12) concluded that 4 genes already known to be associated with the regulation of skin color and the risk of skin cancers (IRF4, MC1R, ASIP and BNC2) contribute to the presence of age spots on facial skin, and that study concluded that the mechanism involved is independent of the melanin biosynthetic pathway. A later study (13) reported that 94 genes were significantly up-regulated in the lesional skin of age spots on the back, including 7 that are melanocyte-related as well as a number of inflammation-related genes. They also noted that significantly higher numbers of cell layers in the stratum corneum were present in age spots.
Those previous studies mostly focused on changes in the epidermis but it is becoming increasingly clear that factors in the dermis, principally those derived from dermal fibroblasts, also play important roles in determining the phenotype, including pigmentation, of the overlying epidermis. For example, DKK1 and NRG1 are produced by fibroblasts and affect the functions of melanocytes in palmoplantar skin and in different skin phototypes, respectively (14–17).
In this study, we used immunohistochemical analysis and microarray analysis to investigate the process underlying the development of age spots, focusing on the expression of specific markers associated with the functions and proliferation of melanocytes and keratinocytes, including factors derived from the dermis. Based on our results, we suggest a model for the development of age spots that might explain the long-term accumulation of melanin and the development of rete ridges in those hyperpigmented lesions.
Materials and Methods
Subjects and age spot characteristics
Twenty European Caucasian subjects of phototypes I or II (age range = 55–73 years) with various sizes and colors of age spots on their forearms as well as on their hands were recruited for this study (subject characteristics are summarized in Table S1). We developed a simple age spot grading system that assessed the distribution pattern, size and color intensity of age spots in this study, as detailed below. The protocol was approved by the local Ethical Committee (Hamburg, ID No 2821) and followed all the principles of the Helsinki Declaration; written informed consent was obtained from each volunteer prior to the study. There were 2 phases of this study: Phase 1 (Subjects 1–8) was designed to assess the gene expression patterns of fibroblasts in the dermis underlying age spots. The aim was to generate stable fibroblast cultures from the biopsies as detailed in (15, 16) and then assess gene expression patterns in fibroblasts derived from age spots with fibroblasts derived from perilesional skin. However, the derivation of sufficient numbers of fibroblasts to allow mRNAs to be purified took several months and the gene expression patterns over that time frame were not stable, hence this phase of the study was not pursued. Phase 2 (Subjects 9 – 20) was designed to assess the gene expression patterns of whole skin biopsies (epidermis and dermis) from age spots and perilesional skin, and this part of the study was completed as designed. Four mm whole skin punch biopsies were taken from two different age spots on the forearm of each subject and from a perilesional control area in the vicinity of each age spot. Individual age spots at least 4 mm in diameter (the size of the skin punch) were biopsied along with nearby normal perilesional skin. For microarray analysis, one pair of biopsies from an age spot and its perilesional control was used. The remaining pair of age spot and perilesional control biopsies was used for histology and immunohistochemistry. All age spots that were designated for closer examination were dermatoscopically verified by dermatologists. To quantitate the visual characteristics of age spots, images were taken with an Epi-Flash camera. To assess all highly concentrated colored molecules such as melanin and hemoglobin, reemission spectra were gathered from age spots and from perilesional skin using an MCS 501 Zeiss spectrometer.
Reemission spectrometry
Differences in the color of age spots and perilesional skin were determined using an MCS 501 Zeiss spectrometer. During measurement, the gauge head of the device (with a 4 cm diameter) was precisely positioned on the skin. In the center of the gauge head, two different types of optical fibers are located in a mixed manner. The first type is responsible for the even illumination of the skin, while the second type collects all signals of the reemitted light from the skin. For skin measurements, it is important that the light quantity is precisely controlled. Simultaneously with the illumination, the reemitted light is collected, absorbed by the photomultiplier and quantified (Bjerring and Andersen, 1987; Gillies et al., 1996). During color measurements, 4 single reemission spectra were obtained from the skin (time of integration: individual, from 25 to 55 msec; number of accumulation: 30, wavelength range: 250 to 800 nm), which were used to calculate the mean reemission spectrum. To depict color differences between age spots and the perilesional areas, the integrals of both reemission spectra in the wavelength range from 320 to 780 nm were formed. Subsequently, the ratio between both integrals were determined and are expressed as a percentage change. 100% means the color of the age spot and the surrounding area is identical, and no spot is visible. The greater the decrease of percentage change, the more visible is the age spot.
Epi-Flash photography
This technique is based on digital photography with a standard camera (Canon, EOS 1 with Compact Macro Lens, EF 50 mm) equipped with a circular flash (Macro Ring Lite, MR-14EX) fixed to a macro objective. By the use of a polarizing filter it is possible to eliminate the influence of the skin surface with the result that the skin color can be quantitatively determined. This means the visual color impression of the age spot is assessed and not the exact quantification of skin pigments such as melanin.
Melanin staining
Paraffin-embedded tissues were stain with the Fontana-Masson silver stain to observe melanin distribution in the specimens (18). Stained samples were observed and photographed using a Leica DMRB/DMLD microscope (Leica, Wetzlar, Germany) with visible light and images analyzed using Scion Image software, as previously detailed (19).
Immunohistochemistry
Paraffin-embedded skin biopsies were examined for the expression of various melanocyte proteins and secreted factors/receptors using indirect immunofluorescence. Melanocytes were detected using anti-human MART-1 mouse monoclonal Ab-3 (1:200 dilution; NeoMarkers, Fremont, CA). For the secreted paracrine factor receptors, anti-human c-KIT goat IgG (1 µg/ml; R&D Systems, Minneapolis, MN) and anti-FGFR1 rabbit polyclonal antibody (1:400 dilution; Abcam, Cambridge, MA) were used. After incubation with the primary antibody in the presence of 5% serum overnight at 4°C, sections were incubated with appropriate secondary antibodies, Alexa Fluor® 594 goat anti-rabbit IgG (H+L), Alexa Fluor® 488 goat anti-mouse IgG (H+L), Alexa Fluor® 594 donkey anti-goat IgG (H+L), (1:400 dilution; all from Molecular Probes, Inc., Eugene, OR) or with fluorescein horse anti-mouse IgG (1:100 dilution; Vector Laboratories, Inc., Burlingame, CA) with 5% serum for 1 hr at room temperature. Nuclei were counterstained with DAPI (Vector). Fluorescence was observed and photographed using the Leica DMRB/DMLD microscope with a 3CCD 3-chip color video camera (Dage-MTI, Michigan City, IN), and images were processed using Scion Image software (Scion, Frederick, MD). Paraffin-embedded skin biopsies were examined for expression of KRT5 and KRT10 using the immunoperoxidase method with cytokeratin 5 (1:2,000 dilution; Vector) and cytokeratin 10 (1:800 dilution; Santa Cruz Biotechnology), respectively. Stained samples were observed and photographed using a Leica DMRB/DMLD microscope with visible light.
Microarray analysis, data evaluation, bioinformatics and statistical methods
mRNAs from whole skin biopsies from 10 age spot specimens and their perilesional controls (subjects 9, 11–15 and 17–20 in Table S1) were used for the microarray analysis. The mRNAs from subjects 10 and 16 were partially degraded and were excluded from the study. The paired samples of subject 16987 were excluded from the analysis due to dramatic differences in the expression pattern discovered by PCA analysis as detailed below. A single-color hybridization on Agilent Whole Human Genome Oligo Microarrays and bioinformational analysis was performed for each skin biopsy sample by ImaGenes (Berlin, Germany). The processed signal intensities from FeatureExtraction were loaded into R. After log base 2 transformation, quantile-normalization was performed on the dataset using the Bioconductor R-Project package LinearModels for Microarray Data (“limma”), which enabled the comparison of samples. R package SRA was applied on normalized data to correct for batch effect, and. To detect differentially expressed genes (DEGs) between age spots and the matched perilesional control specimens, two-sided paired T tests were applied. To control the false discovery rate (FDR), the resulting p values from the paired T tests were adjusted by the method of Benjamini & Hochberg correction (20). An adjusted p value <0.05 is considered statistically significant. The top 500 DEGs (sorted by absolute -fold change) with average transformed intensities >5 in at least one of the biological conditions were loaded into DAVID Bioinformatics Resources 6.7 to identify the enriched biological process, cellular component and molecular function GO terms.
Results
Subjects, age spot grading and colorimetry of age spots
The subjects in this study ranged in age from 55 to 73 years with skin phototypes I – II (Table S1). Only subjects with age spots on their forearms classified as types 1 or 2 (as discussed below) were selected for the study. The differential assessment of age spots on chronic sun-damaged skin is challenging. To arrange for the screening of subjects with age spots in a direct way, we developed an age spot grading system for the discrimination and characterization of distribution types that is based on age spot patterns. This system considers 3 basic criteria: i) distribution, ii) spot size, and iii) spot color. The grading system is fast and no special facilities are required. The age spot grading system has 4 different types (examples are shown in Fig. S1A). I. Type 1a – freckle-like spots: few to many light, freckle-like age spots. II. Type 1b – individually distinguishable large, dark age spots. III. Type 2 – multiple, distinguishable large, dark age spots. IV. Type 3 - Many large, dark age spots, which are partly merged.
Differences in the pigmentation of age spots and perilesional skin were determined using remission spectrometry and Epi-Flash photography as detailed in the Methods. Quantitative reemission and Epi-Flash data of the age spots analyzed are shown in Table S2.
Histology and melanin content in age spots
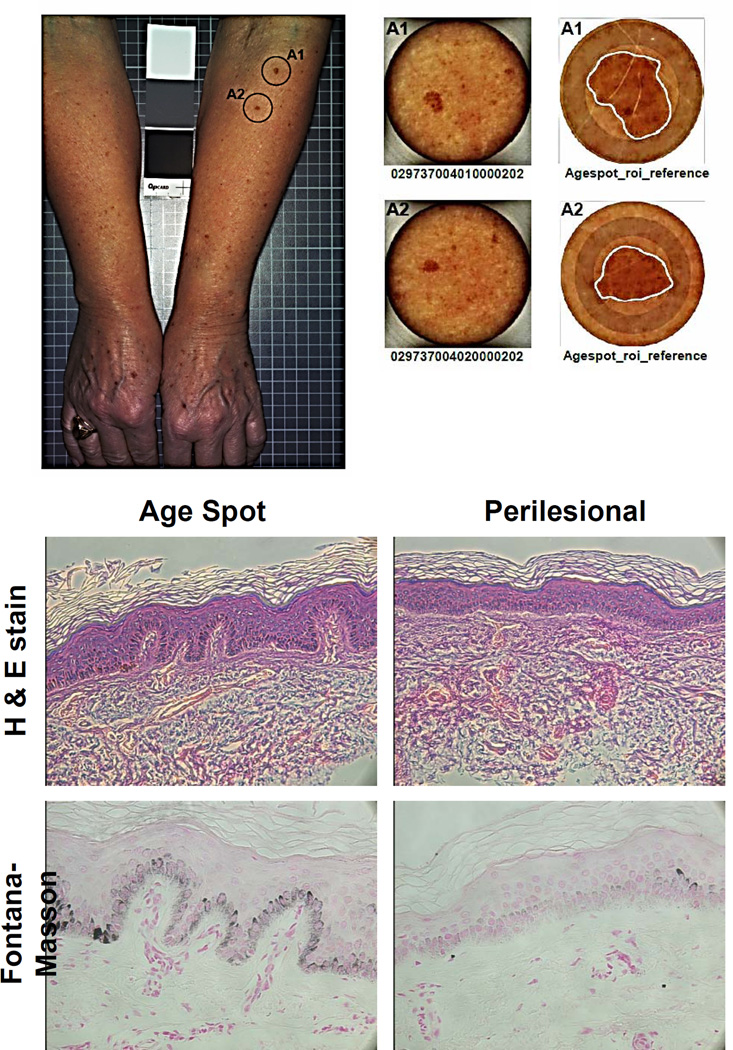
Biopsy specimens (2 per subject) were taken from representative age spots (each ≥4 mm diameter) and perilesional skin and their histological characteristics were investigated with H&E staining and Fontana-Masson silver staining (Fig. 1). The specimens showed the characteristic features of age spots, such as epidermal hyperplasia, elongated epidermal rete ridges with club-shaped or bud-like extensions, and thinned and atrophic epidermis between rete ridges. Quantitative analysis of the Fontana-Masson silver staining revealed that age spot specimens on average had about a 2-fold increase of melanin content compared to the perilesional control skin (Fig. S1B).
Figure 1.
Characteristics of age spots and perilesional skin used in this study. Top. Example of 2 age spots (type 1b) on the left arm of subject 1 (circled, left) and the biopsies taken from them (center). Higher magnification images (right) with the age spots outlined in white. Bottom. Microscopy of a representative age spot and perilesional skin from subject 10; Top, H&E staining; Bottom, Fontana-Masson staining
Melanocyte density and the expression of paracrine factor receptors in age spots
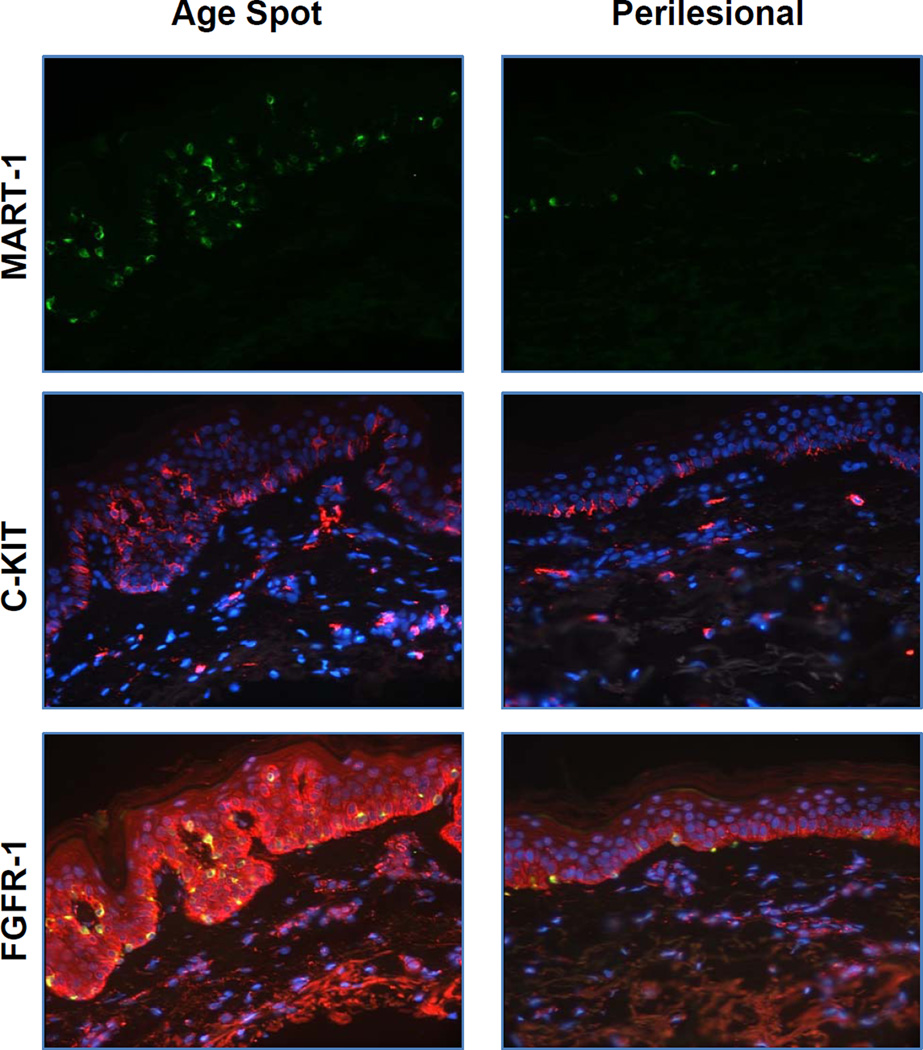
We characterized the melanocyte density in age spots vs. perilesional control skin using immunohistochemical staining with the melanocyte specific marker, MART-1 (Fig. 2). We observed significantly higher numbers of melanocytes in age spots compared to perilesional skin. However, the thickness of the skin and the ratio of the length of the basal layer to the epidermis is significantly higher (p<0.01) in age spots compared to perilesional skin (Fig. S1C), so the actual density of melanocytes along the dermal:epidermal border in age spots is similar to that in perilesional control skin.
Figure 2.
Immunohistochemical staining of MART-1 (green), c-KIT (red) and FGFR-1 (red, co-stained with MART-1 (green)) in age spots and in perilesional skin. Representative images of specimens from subject 4 are shown.
It is well-established that melanocyte functions are regulated by various paracrine factors produced by keratinocytes in the epidermis and by fibroblasts in the dermis (reviewed in (21, 22)). We previously reported that levels of some receptors for well-known melanogenic paracrine factors and their receptors are significantly increased in human skin following repetitive exposure to suberythemal doses of UV (23). Given the fact that age spots typically develop in sun-exposed areas of human skin, we hypothesized that some of those paracrine factors might also be involved in the development of age spots. As shown in Fig. 2, there was a dramatic increase in the expression of c-KIT and FGFR-1 (receptors for SCF and FGF, respectively) in age spots compared to perilesional control skin, but again, this was at least in part offset by the significantly increased length of the dermal:epidermal border in age spots.
Microarray analyses
In order to characterize the process underlying the development of age spots, we used microarray analysis to compare gene expression patterns in age spots vs. perilesional control skin. Two-sided paired T tests were applied on normalized and log2 scaled array data. There were 9,863 probes with a p value <0.05, among which 4,350 had a p value <0.01 (the microarray database has been deposited in GEO, #GSE57103). To control the FDR, the resulting p values from the paired T tests were adjusted by the method of Benjamini & Hochberg correction. A total of 2,003 probes were identified as DEGs at a significance level of 5% (adjusted p value <0.05), and 193 out of those 2,003 DEGs showed a strong difference (>2 fold) between the age spots and the matching perilesional controls.
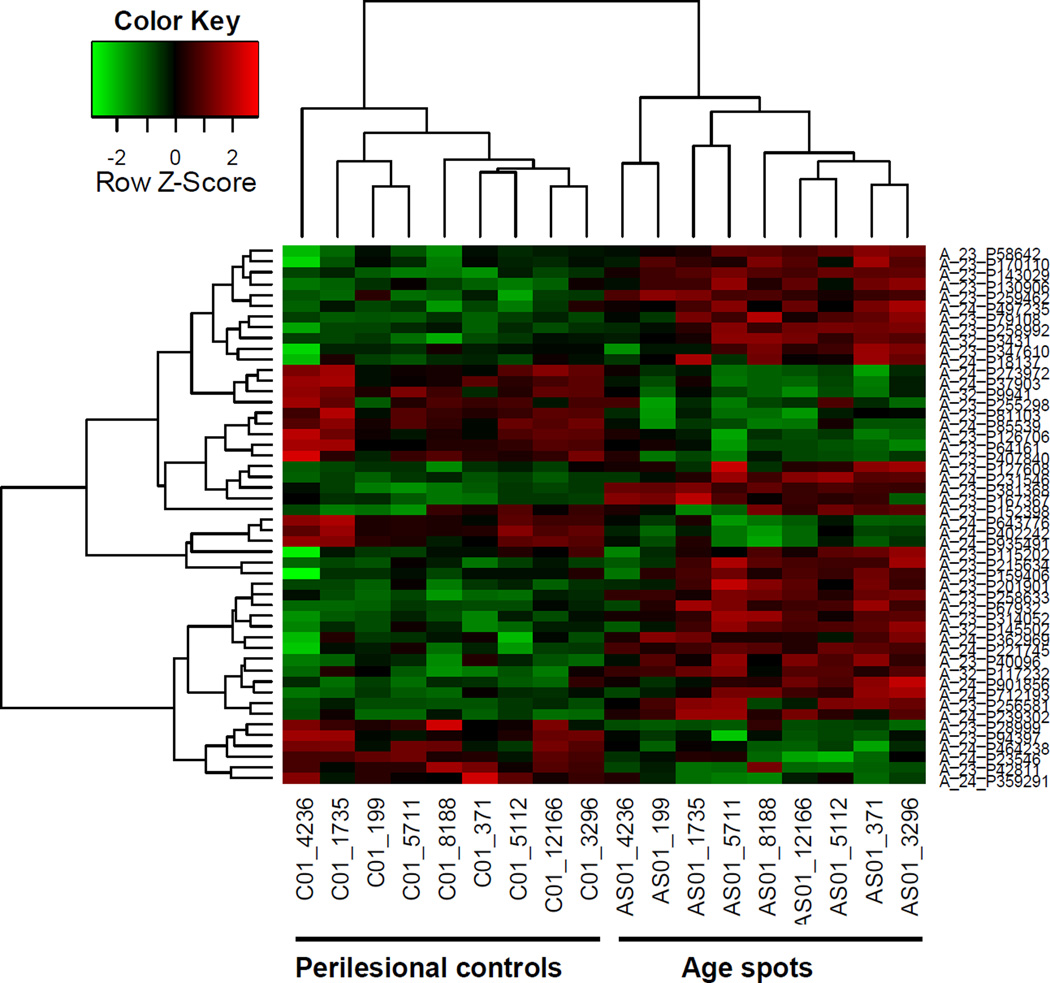
Figure 3 shows a heat map of the two-dimensional clustering of the top 50 genes retrieved by ordering DEGs according to their absolute log -fold change. Probes that fall into one cluster (row labels) have similar behaviors in the experiments. Samples with similar behavior fall into one cluster (column labels). Focusing on those top 50 genes displaying significantly different expression patterns between the 2 types of tissues, all age spots and all perilesional control skins clustered into discrete groups. Expression levels are represented by red and green for high and low intensities, respectively, with black indicating a medium intensity. In order to balance the display, the data are scaled row-wise and symmetrically between −3 and +3.
Figure 3.
Microarray analysis of age spots compared to perilesional skin. A. Heat map of the top 50 DEGs; B. Volcano plot of the 193 DEGs with >2-fold change in expression level; C. Principle component analysis plot of age spot and perilesional controls; Top 50 differential probes.
Next, we checked the DEGs that are both statistically significant and of biological interest. This objective can be achieved by ranking the -fold change value for DEGs and retrieving those with at least a 2-fold change in their expression levels. The volcano plot in Fig. S2A indicates that within the 2,003 significant ProbeIDs, 193 have a -fold change of >2 (log -fold change larger than 1= log(2,2)).
Principal component analysis (PCA) between the microarrays based on the 50 top-ranked DEGs led to a clear clustering of age spot and perilesional controls according to biological conditions (Fig. S2B). The relative variance in gene expression (relative to total variance) comprised in PC1 and PC2 is indicated. Gene set enrichment analysis revealed that “extracellular matrix organization”, “extracellular structure organization”, “epidermis development”, “wound healing” and “ectoderm development” are the top 5 Go term gene sets (Table S3). Most top enriched cellular components are extracellular components (Table S4). Top molecular functions include extracellular matrix structural constituent, peptidase inhibitor activity and polysaccharide binding (Suppl. Table S5).
Interestingly, none of the melanocyte-specific genes showed a significantly different gene expression level in age spots compared to the perilesional control skin. Table S6 shows the list of the top 20 DEGs where the -fold change in age spots compared to the perilesional controls was >2 and was increased or decreased at statistically significant levels (adjusted p<0.05). Since melanocyte density along the dermal:epidermal border in age spots was comparable to the normal perilesional skin, and the expression of melanocyte-specific genes was similar (Table S7), we hypothesized that the hyperpigmentation in age spots reflects decreased melanin removal rather than increased melanin production.
Changes in the expression of keratins in age spots
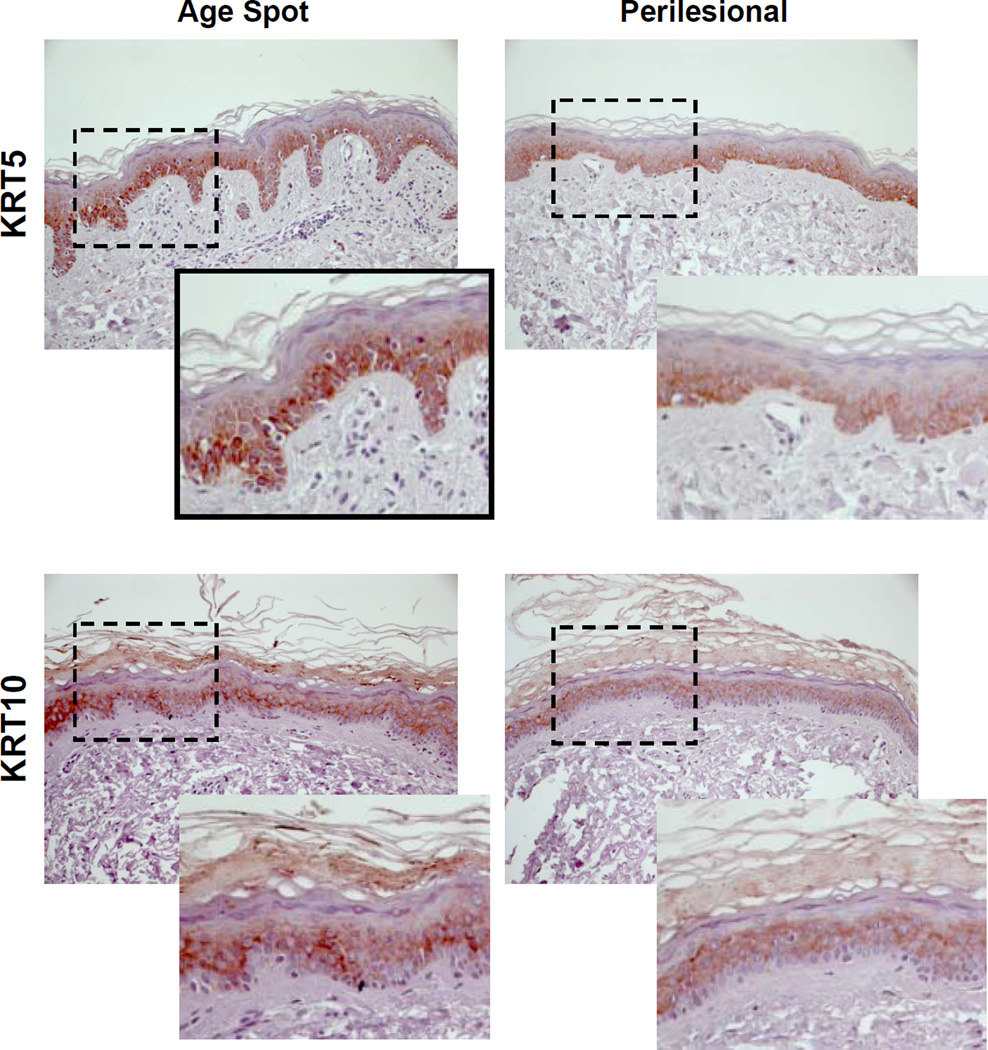
To test the hypothesis that age spots might result from decreased melanin removal (and thus the accumulation of melanin in basal skin), we investigated the expression of various keratins in age spot specimens compared with perilesional control skin. As noted above, keratin 5 (KRT5) is normally expressed in basal keratinocytes whereas keratin 10 (KRT10) is expressed in suprabasal keratinocytes. Figure 4 shows that the staining of KRT5 is more intense and more widely present in age spots than in the perilesional control skin. In particular, keratinocytes in rete ridges expressed KRT5 (see the expanded view in the black squares), which suggests that keratinocytes in rete ridges are actively proliferating and have the characteristics of basal keratinocytes. The staining of KRT10 was also more intense in age spots than in the perilesional skin, which suggests that cells in the suprabasal layer in age spots have a slower turnover than normal keratinocytes.
Figure 4.
Expression of keratins 5 and 10 in age spots and perilesional skin. Left) Expression of KRT5 (subject 14) and KRT10 (subject 15). The area outlined in black shows rete ridges at a higher magnification below right. Right) Scheme proposed for the development of elongated rete ridges in age spots. Normal skin is shown at the top; the bracket indicates suprabasal keratinocytes, with the layer of basal keratinocytes and scattered melanocytes immediately below that. The arrow indicates the basement membrane. In early stage age spots, the increased expression of KRT10 indicates the slower desquamation of suprabasal keratinocytes while the increased expression of KRT5 indicates the more actively proliferating basal keratinocytes, which pressures the basement membrane downwards and weakens the dermal extracellular matrix in conjunction with increased MMP activity. Over time, that downward pressure results in the increased rete ridges and the accumulation of melanin seen in fully developed age spots.
Discussion
In this study, we analyzed age spots and perilesional control skin specimens using immunohistochemical staining and microarray approaches. Immunohistochemical analysis using the melanocyte-specific marker MART-1 revealed an increased number of melanocytes in age spots but a relatively normal density when corrected for the increased length of the dermal:epidermal border in those lesions. It has been suggested that various melanogenic paracrine factors (SCF, KGF/FGF7 and/or ET1) and their receptors (cKIT, FGFR1 and EDNRB, respectively) might be involved in the development of age spots (3–6). SCF/c-KIT is a key regulator in the development of melanocytes as well as in the proliferation and differentiation of mast cells. It was originally described as mast cell growth factor, and it has been shown that mast cells play important roles in many processes that lead to the reorganization/remodeling of skin tissue after injury. Two earlier studies reported that the expression of several genes encoding melanocyte-specific proteins in age spots were increased compared to control skin (11, 13). The numbers of DEGs identified in our study (193) was very similar to the study by Aoki et al. (2007) that found 135 DEGs, but surprisingly, our microarray analysis showed that none of the melanocyte-specific genes showed a significantly different expression pattern in age spots compared to the perilesional skin, including factors secreted by keratinocytes (e.g. EDN1, SCF, KGF and POMC) and fibroblasts (e.g. NRG1 and DKK1) that are known to regulate pigmentation in normal skin (see Tables S6 and S7). The discrepancy of our results from those other 2 previous studies might be due to differences in the ethnic groups of the subjects studied (Caucasian vs. Asian), the locations of age spots studied (forearm vs. back or face/leg), the ages of the study subjects (40–55 yr vs 55–73 yr), the limited sample sizes and/or other factors. It should be noted that while Asian and Caucasian skin is very similar, there are some significant differences between them, especially regarding melanocyte function and responses to UV exposure (19). Further investigation will be needed to provide further insights into the mechanism of age spot development in different phototypes of skin and in different locations on the body.
Interestingly, several other recent studies of hyperpigmented skin, the UV-induced long-lasting pigmentation LLP (24) and ethnic skin (25), also showed that genes and mechanisms other than pigment-related processes correlated with the increased pigmentation. A meta-analysis study of 5 different models of hyperpigmented skin (26), including age spots, demonstrated that the expression of many genes that were not significantly different in any one of those studies, but did reach statistical significance when multiple studies were considered together (primarily due to the limited sample size in any individual study). Although none of the melanocyte-specific genes in our age spot specimens had significantly higher expression levels than the perilesional controls, our study showed that the melanin content in age spots was about 2-fold higher than the surrounding skin, which is consistent with the hyperpigmented phenotype. Those results led us to hypothesize that the hyperpigmentation in age spots results from decreased melanin removal rather than increased melanin production. In fact, the impaired removal of melanin in age spots had been suggested several decades ago (1), and another group had reported that genes involved in keratinization were down-regulated in age spots (8). That latter study also showed that several genes related to the basement membrane and extracellular matrix were down-regulated while the expression of genes encoding some MMPs was increased as was found in our study. Another recent study demonstrated that changes in the dermal papilla are characteristic features of aging skin, at least in facial skin (27). Noblesse et al. (10) showed that at the ultrastructural level, the basement membrane in age spots is thinner and disrupted compared to perilesional skin. In fact, the more extensive rete ridges in age spots, a hallmark feature of those lesions, suggest that the morphological changes in age spots are as important as the pigmentary changes.
Keratins are the major structural proteins of vertebrate epidermis, and various keratins are expressed in different areas of human skin (28). KRT5 and KRT14 are expressed in basal keratinocytes, whereas KRT1, KRT10 and KRT11 are expressed in suprabasal keratinocytes. KRT10 inhibits cell cycle progression and promotes differentiation, and the loss of expression of KRT10 leads to increased keratinocyte turnover (29). Consistent with its inhibitory effect on growth, the ectopic expression of KRT10 suppresses keratinocyte proliferation (30).
Our study shows that age spots have abnormally high levels of expression of KRT5 and KRT10. Therefore, we propose a model for how rete ridges develop and melanin pigment accumulates over time in age spots (shown schematically in Fig. S3). In age spots, early events in UV-exposed skin might involve pigment-related genes, as identified in previous studies (19,24), that initially increase melanocyte growth and differentiation, leading to increased skin pigmentation. Over time, in older subjects, the basal keratinocytes (KRT5-positive) might proliferate more actively while suprabasal keratinocytes (KRT10-positive) desquamate more slowly than in normal skin putting pressure on the basement membrane. The fragile basement membrane and weakened dermal extracellular matrix due to increased MMP activity would allow proliferating basal keratinocytes to push down towards the dermis to form rete ridges instead of moving upwards towards the stratum corneum as in normal skin. The combined effects of these events leads to more extensive rete ridges which would interfere with the normal removal of melanin via desquamation, causing its accumulation in the basal layer and resulting in the hyperpigmentation in those areas. Further investigation will be required to verify this hypothesis.
Supplementary Material
Acknowledgments
This work was supported in part by the Intramural Research Program of the National Cancer Institute. The authors wish to thank Prof. Yasushi Tomita and Dr. Tomonori Motokawa for unpublished information from their study (13).
Abbreviations
- DEG
differentially expressed gene
- FDR
false discovery rate
- KRT5
keratin 5
- KRT10
keratin 10
Footnotes
Conflict of interest statement
None declared.
Author contributions
LY, CS, VJH and LK wrote the manuscript; WC, LY, CS and JB performed the research; WC, LY, CS, VJH and LK analyzed the data; VJH and LK designed the study.
References
- 1.Braun-Falco O, Schoefinius HH. Hautarzt. 1971;22:277–283. [PubMed] [Google Scholar]
- 2.Bastiaens M, Hoefnagel J, Westendorp R, et al. Pigment Cell Res. 2004;17:225–229. doi: 10.1111/j.1600-0749.2004.00131.x. [DOI] [PubMed] [Google Scholar]
- 3.Chen N, Hu Y, Li WH, et al. Exp Dermatol. 2010;19:865–872. doi: 10.1111/j.1600-0625.2009.00957.x. [DOI] [PubMed] [Google Scholar]
- 4.Hattori H, Kawashima M, Ichikawa Y, et al. J Invest Dermatol. 2004;122:1256–1265. doi: 10.1111/j.0022-202X.2004.22503.x. [DOI] [PubMed] [Google Scholar]
- 5.Kadono S, Manaka I, Kawashima M, et al. J Invest Dermatol. 2001;116:571–577. doi: 10.1046/j.1523-1747.2001.01296.x. [DOI] [PubMed] [Google Scholar]
- 6.Kovacs D, Cardinali G, Aspite N, et al. Br J Dermatol. 2010;163:1020–1027. doi: 10.1111/j.1365-2133.2010.09946.x. [DOI] [PubMed] [Google Scholar]
- 7.Iriyama S, Ono T, Aoki H, et al. J Dermatol Sci. 2011;64:223–228. doi: 10.1016/j.jdermsci.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 8.Goyarts E, Muizzuddin N, Maes D, et al. Ann N Y Acad Sci. 2007;1119:32–39. doi: 10.1196/annals.1404.006. [DOI] [PubMed] [Google Scholar]
- 9.Nakajima A, Funasaka Y, Kawana S. Exp Dermatol. 2012;21(Suppl 1):18–21. doi: 10.1111/j.1600-0625.2012.01497.x. [DOI] [PubMed] [Google Scholar]
- 10.Noblesse E, Nizard C, Cario-Andre M, et al. Skin Pharmacol Physiol. 2006;19:95–100. doi: 10.1159/000091976. [DOI] [PubMed] [Google Scholar]
- 11.Motokawa T, Kato T, Katagiri T, et al. J Dermatol Sci. 2005;37:120–123. doi: 10.1016/j.jdermsci.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 12.Jacobs LC, Hamer MA, Gunn DA, et al. J Invest Dermatol. 2015;135:1735–1742. doi: 10.1038/jid.2015.62. [DOI] [PubMed] [Google Scholar]
- 13.Aoki H, Moro O, Tagami H, et al. Br J Dermatol. 2007;156:1214–1223. doi: 10.1111/j.1365-2133.2007.07830.x. [DOI] [PubMed] [Google Scholar]
- 14.Yamaguchi Y, Passeron T, Hoashi T, et al. FASEB J. 2008;22:1009–1020. doi: 10.1096/fj.07-9475com. [DOI] [PubMed] [Google Scholar]
- 15.Yamaguchi Y, Itami S, Watabe H, et al. J Cell Biol. 2004;165:275–285. doi: 10.1083/jcb.200311122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi W, Wolber R, Gerwat W, et al. J Cell Sci. 2010;123:3102–3111. doi: 10.1242/jcs.064774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi W, Kolbe L, Hearing VJ. Pigment Cell Melanoma Res. 2012;25:477–481. doi: 10.1111/j.1755-148X.2012.01002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bancroft JD, Stevens A. Theory and Practice of Histological Techniques. New York: Churchill Livingstone; 1982. [Google Scholar]
- 19.Tadokoro T, Kobayashi N, Zmudzka BZ, et al. FASEB J. 2003;17:1177–1179. doi: 10.1096/fj.02-0865fje. [DOI] [PubMed] [Google Scholar]
- 20.Hochberg Y, Benjamini Y. Stat Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 21.Imokawa G. Pigment Cell Res. 2004;17:96–110. doi: 10.1111/j.1600-0749.2003.00126.x. [DOI] [PubMed] [Google Scholar]
- 22.Yamaguchi Y, Brenner M, Hearing VJ. J Biol Chem. 2007;282:27557–27561. doi: 10.1074/jbc.R700026200. [DOI] [PubMed] [Google Scholar]
- 23.Choi W, Miyamura Y, Wolber R, et al. J Invest Dermatol. 2010;130:1685–1696. doi: 10.1038/jid.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coelho SG, Yin L, Valencia JC, et al. J Pathol. 2015;236:17–29. doi: 10.1002/path.4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yin L, Coelho SG, Ebsen D, et al. Exp Dermatol. 2014;23:731–735. doi: 10.1111/exd.12518. [DOI] [PubMed] [Google Scholar]
- 26.Yin L, Coelho SG, Valencia JC, et al. J Invest Dermatol. 2015;135:2455–2463. doi: 10.1038/jid.2015.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizukoshi K, Yonekura K, Futagawa M, et al. Skin Res Technol. 2015;21:224–231. doi: 10.1111/srt.12180. [DOI] [PubMed] [Google Scholar]
- 28.Fuchs E. Annu Rev Cell Dev Biol. 1995;11:123–153. doi: 10.1146/annurev.cb.11.110195.001011. [DOI] [PubMed] [Google Scholar]
- 29.Reichelt J, Furstenberger G, Magin TM. J Invest Dermatol. 2004;123:973–981. doi: 10.1111/j.0022-202X.2004.23426.x. [DOI] [PubMed] [Google Scholar]
- 30.Santos M, Paramio JM, Bravo A, et al. J Biol Chem. 2002;277:19122–19130. doi: 10.1074/jbc.M201001200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.