SUMMARY
Ca2+ signals were reported to control lipid homeostasis but the Ca2+ channels and pathways involved are largely unknown. Store-operated Ca2+ entry (SOCE) is a ubiquitous Ca2+ influx pathway regulated by stromal interaction molecule 1 (STIM1) and STIM2, and by the Ca2+ channel ORAI1. We show that SOCE-deficient mice accumulate pathological amounts of lipid droplets in liver, heart and skeletal muscle. Cells from patients with loss-of-function mutations in STIM1 or ORAI1 show a similar phenotype, suggesting a cell intrinsic role for SOCE in the regulation of lipid metabolism. SOCE is crucial to induce mobilization of fatty acids from lipid droplets, lipolysis and mitochondrial fatty acid oxidation. SOCE regulates cAMP production and the expression of neutral lipases as well as the transcriptional regulators of lipid metabolism, PGC-1α and PPARα. SOCE-deficient cells upregulate lipophagy that protects them against lipotoxicity. Our data provide evidence for an important role of SOCE in lipid metabolism.
eTOC
Calcium signaling is fundamental to many cellular processes and alterations in cellular Ca2+ homeostasis occur in metabolic disorders. Maus et al delineate the role of store-operated calcium entry (SOCE), an important pathway for increasing intracellular Ca2+ levels, in lipid metabolism in SOCE-deficient mice and cells from human patients.
INTRODUCTION
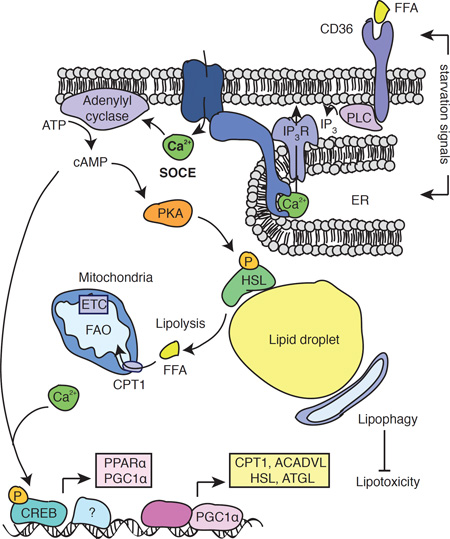
Ca2+ is a versatile signaling molecule regulating a wide variety of cellular processes including muscle contraction, cellular motility, vesicle fusion, gene transcription and cell metabolism (Berridge et al., 2000). The cytosolic Ca2+ concentration is tightly regulated by an array of Ca2+ channels, transporters, exchangers and pumps (Berridge, 2012). An important pathway for increasing intracellular Ca2+ levels in many different cell types is store-operated Ca2+ entry (SOCE). SOCE is mediated by two families of proteins: ORAI proteins in the plasma membrane form the Ca2+ release-activated Ca2+ (CRAC) channel that conducts Ca2+ influx from the extracellular space (Feske et al., 2006; Vig et al., 2006; Zhang et al., 2006), and stromal interaction molecules (STIM) 1 and 2 in the endoplasmic reticulum (ER) membrane that bind to ORAI proteins resulting in the opening of CRAC channels (Liou et al., 2005; Zhang et al., 2005). STIM proteins are activated following a reduction in the ER Ca2+ concentration in response to stimulation of cell surface receptors that induce production of inositol 1,4,5-trisphosphate (IP3) and opening of Ca2+ release channels in the ER such as the IP3 receptor (IP3R) (Feske, 2007). The subsequent reduction in the ER Ca2+ concentration results in the dissociation of Ca2+ from an ER luminal EF-hand domain in STIM1 (Liou et al., 2005; Zhang et al., 2005) and allow the cytoplasmic tail of STIM1 to bind to ORAI1 (Maus et al., 2015; Soboloff et al., 2012). ORAI1 proteins form hexameric complexes in the plasma membrane that constitute the Ca2+ permeant pore of the CRAC channel (Prakriya et al., 2006). Patients with loss-of-function mutations in ORAI1, the best characterized member of the ORAI family, or in STIM1 suffer from an autosomal-recessive disease syndrome named CRAC channelopathy that is characterized by severe immunodeficiency, muscular hypotonia and anhidrotic ectodermal dysplasia (Lacruz and Feske, 2015). The cellular mechanisms underlying disease pathogenesis in different tissues are incompletely understood.
Previous studies have suggested a role for Ca2+ in controlling cell metabolism (Arruda and Hotamisligil, 2015). Allosteric regulation of metabolic enzymes by Ca2+ controls metabolic pathways, such as the tricarboxylic acid (TCA) cycle (Hajnoczky et al., 1995; McCormack et al., 1990). Transcriptional control of metabolism by Ca2+ is exerted indirectly via Ca2+ dependent kinases and phosphatases, such as calmodulin-regulated kinases (CAMK) and calcineurin that control expression of the peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PGC-1α) (Czubryt et al., 2003; Handschin et al., 2003; Schaeffer et al., 2004). However, little is known about the Ca2+ channels involved in these regulatory processes, which could be important targets to therapeutically modulate Ca2+ responsive metabolic pathways.
Alterations in cellular Ca2+ homeostasis have been observed in metabolic disorders, such as obesity and diabetes (Arruda and Hotamisligil, 2015). Genome wide association studies (GWAS) showed that single nucleotide polymorphisms (SNP) in genes regulating intracellular Ca2+ homeostasis such as sarco/endoplasmic reticulum Ca2+ ATPase (SERCA) (Varadi et al., 1999) and the inositol trisphosphate receptor (IP3R) (Shungin et al., 2015) are associated with changes in body mass index and susceptibility to diabetes. Recent functional genetics screens in Drosophila demonstrated the importance of dSERCA and the ryanodine receptor (dRyR) (Bi et al., 2014), dIP3R (Subramanian et al., 2013) and dStim (Baumbach et al., 2014) in lipid homeostasis, implicating both Ca2+ release from the ER and Ca2+ influx across the plasma membrane in lipid metabolism.
Here we identify CRAC channels and SOCE as a critical Ca2+ signaling pathway that controls lipid metabolism in mouse and human cells. We find that ORAI1- or STIM1/STIM2-deficient mice that lack SOCE accumulate pathological amounts of lipid droplets (LD) in skeletal and heart muscles and in liver. Isolated fibroblasts from human patients with loss-of-function mutations in STIM1 or ORAI1 also show increased LD levels. In the absence of SOCE, cells display impaired lipolysis, and upregulate lipophagy that protects them from lipotoxicity. We find that SOCE is crucial for the basal and starvation-induced transcription of peroxisome proliferator-activated receptor α (PPARα) and PGC-1α genes, and of neutral lipases and other downstream genes involved in fatty acid metabolism.
RESULTS
Impaired SOCE results in LD accumulation in cells and tissues
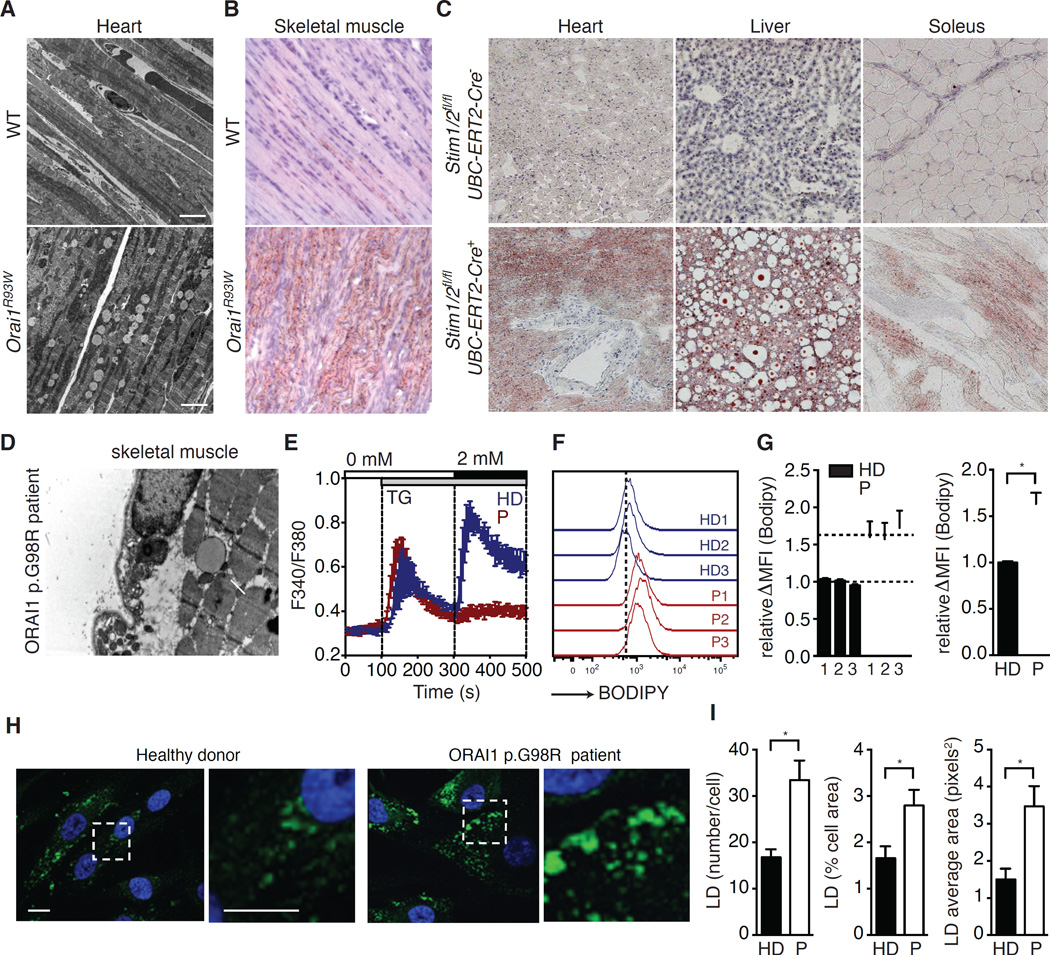
To investigate the role of SOCE in the regulation of lipid metabolism we used Orai1R93W knock-in mice that express a non-functional ORAI1 channel protein (McCarl et al., 2010), which abolishes SOCE in all tissues and is equivalent to the human ORAI1 p.R91W mutation found in a patient with CRAC channelopathy (Feske et al., 2006). As reported previously, only a small fraction of skeletal myofibers in Orai1R93W mice contain markedly swollen mitochondria with abnormal cristae structure (McCarl et al., 2010). However, transmission electron microscopy (TEM) and Oil Red O staining both revealed that Orai1R93W heart and skeletal muscle accumulate abnormal amounts of LDs (Figure 1A and 1B). Because Orai1R93W mice die perinatally, we established a tamoxifen-inducible mouse model in which Stim1 and Stim2 genes can be deleted in all tissues of adult animals to abolish SOCE. Treatment of Stim1fl/flStim2fl/fl UBC-ERT2-Cre mice with tamoxifen resulted in rapid development of muscular dystonia and myoclonus-like symptoms (Video S2–S3) and death within 5–10 days. Histologies of their heart muscle, skeletal muscle and liver revealed the accumulation of pathological amounts of LDs when compared to tamoxifen injected Cre-negative littermates (Figure 1C). TEM of a muscle biopsy from a ORAI1 p.G98R patient also showed LD deposition (Figure 1D). LD accumulation in the absence of SOCE was confirmed in fibroblasts from patients with loss-of-function mutations in ORAI1 or STIM1 that abolish SOCE (Figure 1E), which showed a significantly higher LD content compared to healthy donors when cultured in high-glucose media (Figure 1F and 1G) and when challenged with oleic acid (OA) (Figure 1H and 1I). Collectively these findings suggest that SOCE has a conserved role in controlling lipid homeostasis in mice and humans both in vivo and vitro.
Figure 1. SOCE regulates lipid balance in cells and tissues.
(A–C) Lipid droplet (LD) accumulation in SOCE-deficient mice. EM images of heart (A) and Oil Red O stain of skeletal muscle (B) from wild-type (WT) and Orai1R93W knock-in mice 5 days postpartum. (C) Oil Red O stain of heart, liver and soleus sections (10x) from 16-week-old Stim1fl/flStim2fl/fl UBC-ERT2-Cre mice and Cre-negative littermates 5 days after the first dose of tamoxifen injection. (D) EM image (20,000×) of skeletal muscle from a patient with ORAI1 p.G98R mutation shows LD deposition (arrow). (E) SOCE in fibroblasts from healthy donors (HD, blue) and patients (P, red) with loss-of-function mutations in STIM1 or ORAI1 measured by Ca2+ imaging. ER Ca2+ stores were depleted with thapsigargin (TG) and SOCE was measured after re-addition of 2 mM Ca2+. Traces represent the averages of 3 HDs and 3 Ps (≥ 20 cells per donor analyzed). (F and G) Neutral lipid staining of fibroblasts from HDs and Ps cultured in high glucose medium. Cells were stained with BODIPY 493/503, and analyzed by flow cytometry. Representative histograms (F) and average mean fluorescence intensities (MFI) (G) of BODIPY 493/503 after subtraction of autofluorescence (ΔMFI) in cells from 3 HDs and 3 Ps. ΔMFI values of patients were normalized to the mean ΔMFI in HD cells. (H–I) HD and P fibroblasts were challenged with oleic acid (OA, 0.125 mM, 24 hours) and stained with BODIPY 493/503 (green) to identify neutral lipid stores. (H) Dashed boxes indicate regions shown at higher magnification in the right panels. Nuclei were counterstained with DAPI. (I) Average numbers of LDs per cell (left), percentage of cellular area occupied by LDs (middle) and average LD area (right). All values are mean ± SEM. Statistical analysis was performed using Student’s t-test. * p<0.05.
SOCE is required for mitochondrial gene expression, function and fatty acid oxidation
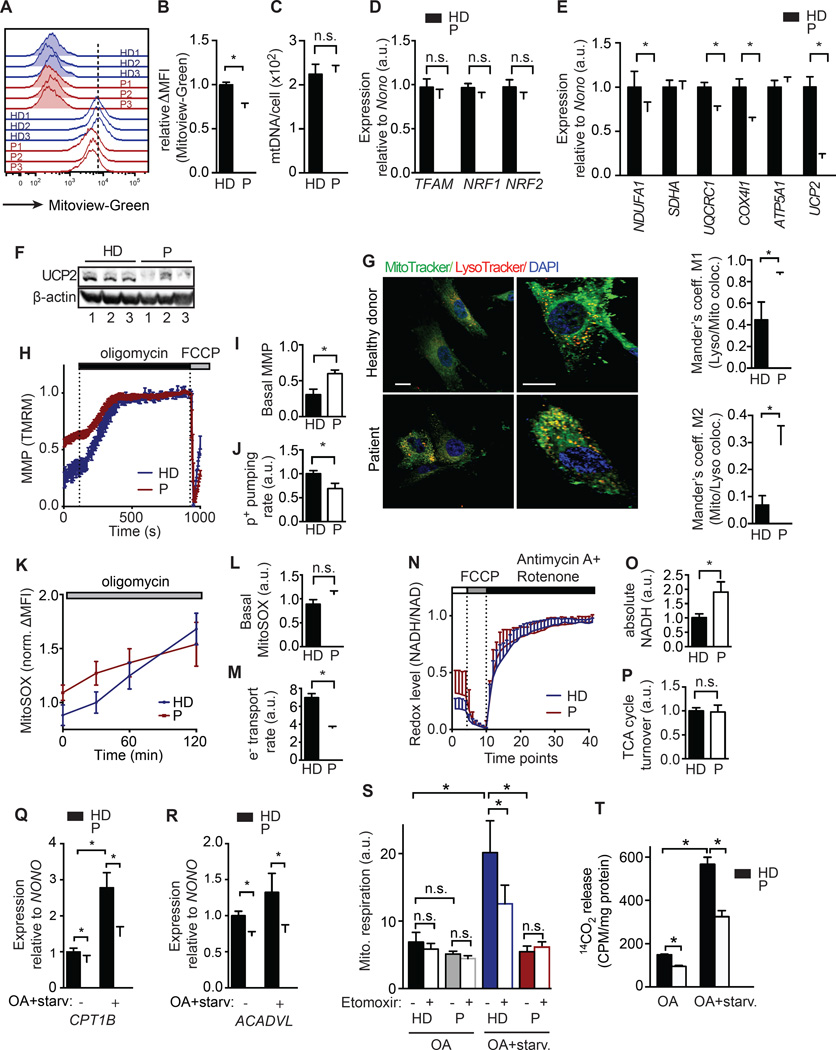
Lipid catabolism in cells depends on mitochondria, and reduced mitochondrial function leads to triglyceride accumulation in muscle and liver (Petersen et al., 2003). Since intracellular Ca2+ regulates mitochondrial function (Brookes et al., 2004), we tested if a specific role of SOCE in mitochondrial function could account for the higher lipid content in SOCE-deficient cells. Using fibroblasts from patients with loss-of-function mutations in ORAI1 or STIM1, we found that their mitochondrial volume was decreased when measured using MitoView Green, a membrane potential-insensitive mitochondrial dye (Figure 2A and 2B) and by TEM (Figure S1). By contrast, the mitochondrial DNA copy numbers assessed by quantitative PCR were normal in patient fibroblasts (Figure 2C), which is consistent with the observed normal expression of transcription factors responsible for mitochondrial genome replication, Transcription Factor A Mitochondrial (TFAM) and Nuclear Respiratory Factor (NRF) 1 and 2 (Figure 2D). However, the expression of components of the electron transport chain (ETC) complexes CI, CIII and CIV in mitochondria, which are critical for oxidative phosphorylation was reduced in SOCE-deficient fibroblasts, whereas expression of complex CII and CV was normal (Figure 2E and S2A). Analysis of isolated mitochondria by blue native polyacrylamide gel electrophoresis (BN-PAGE) confirmed reduced expression of complexes CI and CIV and of supercomplex CICIII2 in patient fibroblasts (Figure S2B). SOCE-deficient cells also showed a severe defect in mRNA (Figure 2E) and protein (Figure 2F) expression of uncoupler protein 2 (UCP2), a proton transporter in the inner mitochondrial membrane. Opening of UCP2, which is controlled by fatty acids and other factors, results in the reduction of the mitochondrial membrane potential (MMP) and of ROS production and thus prevents mitochondrial damage (Brand and Esteves, 2005). Consistent with reduced UCP2 expression, we observed higher numbers of damaged mitochondria as suggested by increased colocalization of mitochondria and acidic vesicular compartments (lysosomes) in fibroblasts of SOCE-deficient patients (Figure 2G). Similar results were found using the pH sensitive MitoKeima reporter (Katayama et al., 2011), which detects mitochondria undergoing lysosomal degradation (mitophagy) (Figure S2D). We observed a similar higher number of damaged mitochondria in the skeletal muscle and hearts of Orai1R93W knock-in mice by TEM (Figure S1) (McCarl et al., 2010). Also consistent with reduced UCP2 expression, the MMP-sensitive dye tetramethylrohodamine demonstrated that SOCE-deficient patient cells had significantly higher basal MMP levels than control cells (Figure 2H and 2I).
Figure 2. SOCE is required for mitochondrial function and FAO.
(A and B) Mitochondrial volume in fibroblasts from healthy donors (HD, blue) and patients (P) with loss-of-function mutations in STIM1 or ORAI1 (P, red) analyzed with MitoView Green and flow cytometry. Representative histograms (A) and average MFI of MitoView Green after subtraction of autofluorescence (ΔMFI) in HD and P fibroblasts (B). (C) Mitochondrial DNA copy number in fibroblasts from 3 HDs and 3 Ps assessed by real time PCR. (D and E) Relative mRNA levels of transcription factors involved in mitochondrial DNA replication (D) and electron transport chain (ETC) components and uncoupler protein 2 (UCP2) (E) in fibroblasts from 3 HDs and 3 Ps assessed by real time PCR and normalized to the house keeping gene NONO (Non-POU domain-containing octamer-binding protein). (F) Western blot for UCP2 and β-actin in fibroblasts from 3 HDs and 3 Ps. (G and H) Analysis of mitochondrial degradation (mitophagy) by co-staining of fibroblasts from HD and P with MitoTracker (green), LysoTracker DND-99 (red) and DAPI (blue). Images show cells at low (left) and high (right) magnification. Bar graphs show Mander’s coefficients of colocalization of MitoTracker and LysoTracker in cells from 3 HDs and 3 Ps (right). (H–J) Analysis of the mitochondrial membrane potential (MMP) in TMRM-loaded HD (blue) and P (red) fibroblasts by single cell imaging. Averaged traces (H), and averaged basal MMP (I) before and after oligomycin treatment (0 and 1 represent minimum and maximum MMP after de- and hyperpolarization with 1 µM FCCP and 1 µM oligomycin, respectively). (J) Proton pumping rate of the ETC measured as the MMP hyperpolarization rate after oligomycin. (K–M) Mitochondrial superoxide production and ETC electron transport rate. Fibroblasts from 3 HDs (blue) and 3 Ps (red) were loaded with MitoSOX and analyzed by flow cytometry before and after 1 µM oligomycin. Normalized MitoSOX signal (ΔMFI) over time (K), basal MitoSOX signal (ΔMFI) before oligomycin (L) and electron transport rate as the rate of superoxide production after oligomycin (M). (N–P) NADH levels and TCA cycle turnover. The redox state of cells was measured by NADH autofluorescence in fibroblasts from 3 HDs (blue) and 3 Ps (red) by time-lapse microscopy. Relative minimum (0) and maximum (1) NADH fluorescence was determined by treating cells with 1 µM FCCP to deplete NADH and 1 µM antimycin A / 100 nM rotenone to saturate NADH levels. Averaged relative NADH autofluorescence over time on a scale from 0 to 1 (N), basal NADH levels measured as total NADH autofluorescence per cell after background subtraction (O) and TCA cycle turnover (P) calculated as the rate of NADH increase after antimycin A / rotenone. (Q, R) Relative mRNA levels of CPT1B (Q) and ACADVL (R) in fibroblasts after 24h culture in high-glucose or oleic acid (OA) medium followed by 6h starvation. mRNA levels in 3 Ps were normalized to those in 3 HDs. (S) Mitochondrial respiration in fibroblasts cultured in OA medium only (HD, black and P, grey) or with subsequent starvation (HD, blue and P, red) in the presence or absence of the CPT1 inhibitor etomoxir (40 µM). (T) 14C-OA oxidation measured in cells (HD, black and P, white) cultured in OA alone or OA plus starvation. Data are mean ± SEM and normalized to HD fibroblasts (unless indicated otherwise). Statistical analysis was performed using Student’s t-test. * p<0.05.
To assess the function of the ETC in mitochondria of SOCE-deficient cells, we measured the hyperpolarization rate of the MMP after treatment of cells with oligomycin, which inhibits the ATP synthase and thus traps protons in the intermembrane space. We found a reduced proton pumping rate by the ETC in patient compared to control cells (Figure 2H and 2J). We next analyzed the electron transport rate of the ETC by measuring the formation of O2− using the superoxide-sensitive mitochondria-targeted dye MitoSOX. Whereas basal mitochondrial superoxide levels were slightly, but not statistically significantly increased in SOCE-deficient cells (Figure 2K and 2L), the rate of superoxide production after ATP synthase inhibition with oligomycin was significantly reduced in the absence of SOCE, indicative of lower electron-transport rate by the ETC (Figure 2K and 2M). Direct analysis of O2 consumption rate revealed normal or moderately reduced basal respiration in patient fibroblasts, but a significant decrease in the maximal respiration rate in SOCE-deficient patient cells compared to controls (Figure S2C). Reduced ETC function was not limited to fibroblasts because a functional analysis of individual ETC complexes in skeletal muscle biopsies of ORAI1-deficient patients revealed decreased activity of complexes I and IV in two out of three patients (Table S1).
Ca2+ is a co-factor of three enzymes in the TCA cycle (Hajnoczky et al., 1995; McCormack et al., 1990), and impaired ETC function in SOCE-deficient cells might therefore be due to decreased levels of reduced co-enzymes nicotinamide adenine dinucleotide (NADH) and flavin adenine dinucleotide (FADH2). However, SOCE-deficient cells showed significantly increased absolute levels of NADH in high glucose-containing culture (Figure 2N and 2O). Furthermore, they had no defect in replenishing the NADH pool after its depletion with FCCP and subsequent blockade of the electron acceptor complexes I and III with antimycin A and rotenone (Figure 2N and 2P). These findings indicate that the TCA cycle is not dependent on SOCE, at least under the experimental conditions used here, and does not account for impaired ETC function in SOCE-deficient cells.
An important function of the mitochondria is to break down fatty acids into acetyl-CoA by β-oxidation. Several enzymes are critical for this process including acyl-CoA dehydrogenase very long-chain (ACADVL), an enzyme that catalyzes the first step of β-oxidation, and the long-chain fatty acid transporter carnitine palmitoyltransferase 1B (CPT1B) that facilitates the transport of fatty acids into the mitochondria (Houten and Wanders, 2010). Expression of both genes was reduced in patient fibroblasts when cultured in high-glucose medium, and this difference became more pronounced when cells were cultured in OA-containing medium followed by starvation in 2 mM glucose medium (Figure 2Q and 2R). To assess mitochondrial fatty acid oxidation (FAO), we measured cellular O2 utilization of fibroblasts cultured in OA-containing medium with or without subsequent starvation. The CPT1 inhibitor etomoxir, which inhibits fatty acid transport into the mitochondria, was used to estimate the contribution of FAO to total mitochondrial respiration. Healthy donor and SOCE-deficient patient fibroblasts showed comparable low levels of O2 consumption in OA culture (Figure 2S). Starvation induced a drastic increase in etomoxir-sensitive mitochondrial respiration in control, but not in SOCE-deficient patient cells (Figure 2S). To directly quantify FAO, we measured 14CO2 release from cells cultured in 14C-OA-containing medium, with or without subsequent starvation. SOCE-deficient patient fibroblasts had reduced rates of FAO compared to healthy donor fibroblasts when cultured in OA or cultured in OA and then starved (Figure 2T). As observed previously, FAO was more pronounced in starved cells (Figure 2T). We conclude that SOCE is critical for mitochondrial FAO. Taken together, our data show that SOCE controls expression of several key mitochondrial enzymes including uncoupler proteins, ETC components and enzymes involved in fatty acid oxidation and thereby regulates mitochondrial function and FAO.
SOCE regulates lipolysis by controlling lipase expression and adenylyl cyclase dependent signaling
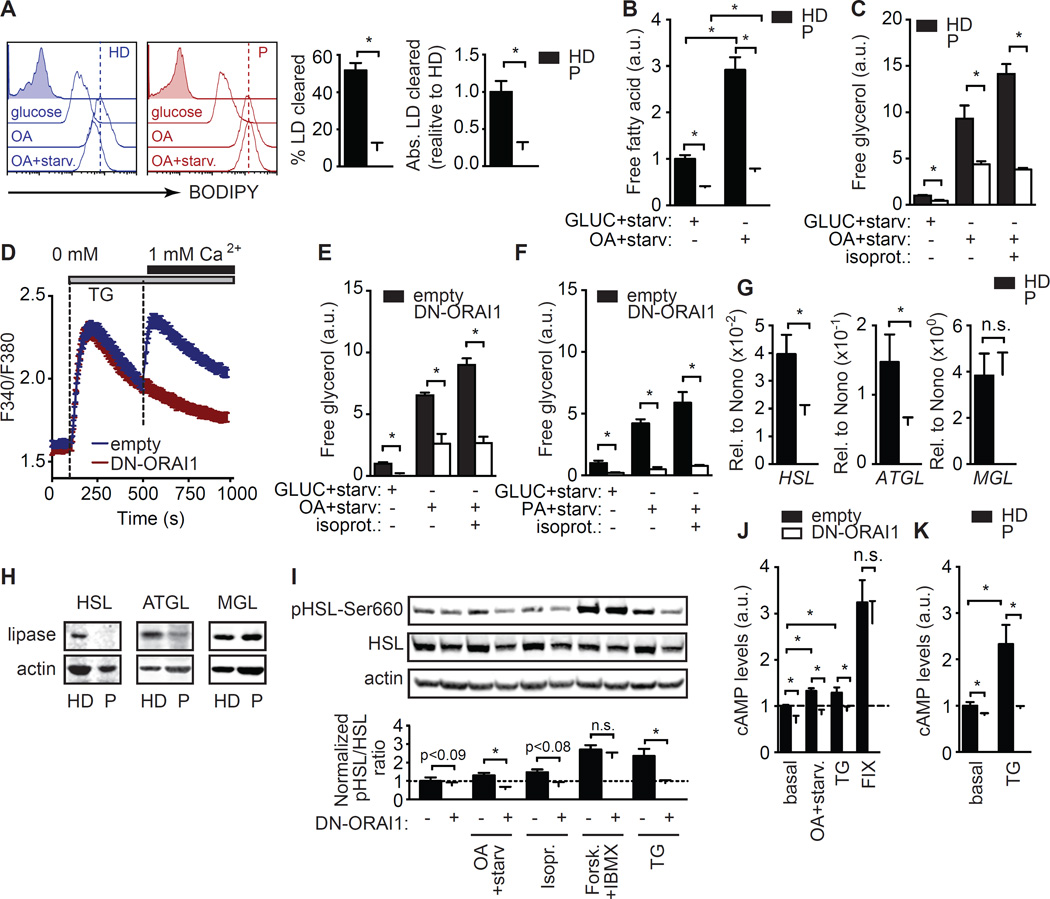
Next, we determined if changes in triacylglyceride (TAG) mobilization from LDs could also account for lipid accumulation in SOCE-deficient cells. BODIPY staining of fibroblasts cultured in glucose medium, OA medium or OA culture followed by by 24 hour starvation revealed that whereas HD fibroblasts readily mobilized about half of their stored LDs in response to starvation, SOCE-deficient cells showed a severe defect in LD mobilization (Figure 3A). Lipolysis, measured by free fatty acid (FFA) (Figure 3B) and glycerol (Figure 3C) release upon starvation after high glucose or OA culture was significantly reduced in SOCE-deficient compared to control cells. This defect could not be rescued by inducing lipolysis with the β-adrenergic receptor agonist isoproterenol, which only increased glycerol release in control cells (Anthonsen et al., 1998) (Figure 3C). Similarly, mouse NIH3T3-L1 cells expressing a dominant negative form of ORAI1 (DN-ORAI1) that abolishes SOCE (Figure 3D) showed significantly reduced lipolysis upon starvation after culture in high-glucose medium or OA (Figure 3E). Isoproterenol was again unable to rescue this defect (Figure 3E). The lipolysis defect was not dependent on the saturation of the fatty acid nutrient, as SOCE-deficient cells cultured with palmitic acid (PA) instead of OA also showed impaired free glycerol release upon starvation when compared to control cells (Figure 3F).
Figure 3. SOCE regulates starvation-induced lipolysis.
(A) Analysis of lipid droplet (LD) levels in fibroblasts from 3 healthy donors (HD) and 3 patients (P) with loss-of-function mutations in STIM1 or ORAI1. Cells were cultured in high glucose or oleic acid (OA) medium for 12 h without or with subsequent starvation (OA+starv) in low glucose (2 mM) medium for 24 h and stained with BODIPY (open traces) or left unstained (filled gray traces) to measure LD content. Representative histograms (left) and quantification of 3 repeat experiments (right). ΔMFI, mean fluorescence intensity (MFI) BODIPY - MFI unstained cells. % LD clearance was calculated as described in Methods. (B–C) Free fatty acid (B) and free glycerol (C) release by fibroblasts from 3 HDs and 3 Ps after 24 h culture in high glucose (11 mM) or OA-containing medium followed by 24 h of starvation in 2 mM glucose in the presence or absence of isoproterenol (10 µM). a.u., arbitrary units. (D–F) Lipolysis in NIH3T3-L1 cells transduced with dominant negative ORAI1-E106Q (red, DN-ORAI1) or empty vector control (blue, empty). (D) SOCE following stimulation with thapsigargin (TG) and re-addition of 1 µM Ca2+ Ringer’s solution. (E and F) Free glycerol release by NIH3T3-L1 cells cultured for 24 h in medium containing high glucose (E and F), OA (E) or PA (F) followed by starvation for 6 h in 1 mM glucose in the presence or absence of isoproterenol (10 µM). (G and H) Expression of HSL, ATGL and MGL mRNA (normalized to the housekeeping gene NONO) (G) and protein (H) determined in fibroblasts from 3 HDs and 3 Ps. Data are representative of 3 repeat experiments. (I) HSL phosphorylation on the activating Ser660 residue in NIH3T3-L1 cells transduced with empty vector or DN-ORAI1. Cells were cultured for 30 min in high glucose (24 mM) medium alone or with forskolin (10 µM, Forsk.) + IBMX (1 mM) or with thapsigargin (1 µM, TG), or in medium containing 0.5 mM OA followed by 30 min starvation (OA+starv) in the presence or absence of isoproterenol (10 µM). Representative Western blots (top) and normalized ratios of phospho-HSL to total HSL expression (bottom) from 5 repeat experiments (see Methods for details). (J and K) cAMP levels in NIH3T3-L1 cells (J) cultured as described in (I) and in fibroblasts from 3 HDs and 3 Ps (K) cultured in high glucose (11 mM) RPMI 1640 medium alone (basal) or stimulated for 30 min with 1 µM TG. All values represented as bar graphs in A, B, C, E, F, G, I, J and K were normalized to mean values obtained in HD fibroblasts or empty vector-transduced NIH3T3-L1 cells after culture in glucose medium. All values are mean ± SEM of at least 3 repeat experiments. Statistical analysis was performed using Student’s t-test. * p<0.05.
To understand the mechanisms underlying impaired lipolysis in the absence of SOCE, we analyzed the expression of the neutral lipases hormone sensitive lipase (HSL), adipose triglyceride lipase (ATGL) and monoacylglycerol lipase (MGL). The expression of HSL and ATGL in SOCE-deficient fibroblasts was reduced both at the mRNA (Figure 3G) and protein (Figure 3H) level, whereas MGL expression was unchanged. HSL function is activated by phosphorylation on serine residues by protein kinase A (PKA). PKA activity in turn depends on the cytosolic levels of cyclic adenosine monophosphate (cAMP) generated by plasma membrane-associated adenylyl cyclases (AC) whose activity was shown to be regulated by ORAI1-mediated SOCE (Willoughby and Cooper, 2006; Willoughby et al., 2012). We therefore tested the hypothesis that SOCE regulates HSL phosphorylation in a cAMP dependent manner. We observed a small, but visible reduction in HSL phosphorylation on Ser660 in SOCE-deficient NIH3T3-L1 cells when cultured in high-glucose medium, which became more pronounced when cells were cultured in OA followed by starvation in the presence or absence of isoproterenol. The induction of SOCE by thapsigargin stimulation (to passively deplete ER Ca2+ stores) for 30 min was sufficient to increase HSL phosphorylation in control cells but to a significantly lower extend in SOCE-deficient fibroblasts (Figure 3I). Treatment of SOCE-deficient NIH3T3-L1 cells with the AC activator Forskolin and the phosphodiesterase inhibitor 3-isobutyl-1-methylxanthine (IBMX) to elevate cAMP levels while bypassing the SOCE requirement for cAMP production restored HSL phosphorylation (Figure 3I). These findings suggest that SOCE acts upstream of ACs to increase cAMP levels and induce HSL phosphorylation. To confirm that SOCE regulates AC function, we analyzed cAMP production and found that cAMP levels were significantly reduced in SOCE-deficient NIH3T3-L1 cells when cultured in high glucose or OA medium followed by starvation (Figure 3J). cAMP levels increased significantly in control cells after thapsigargin stimulation, but this increase was absent in SOCE-deficient NIH3T3-L1 cells (Figure 3J). Treatment of SOCE-deficient fibroblasts with forskolin + IBMX restored cAMP to WT levels (Figure 3J). We observed a similar defect in cAMP production in fibroblasts from SOCE-deficient patients (Figure 3K). Taken together, we conclude that SOCE regulates the expression of the neutral lipases HSL and ATGL as well as the cAMP-dependent activation of HSL and thereby controls lipolysis.
Protective upregulation of lipophagy in SOCE-deficient cells
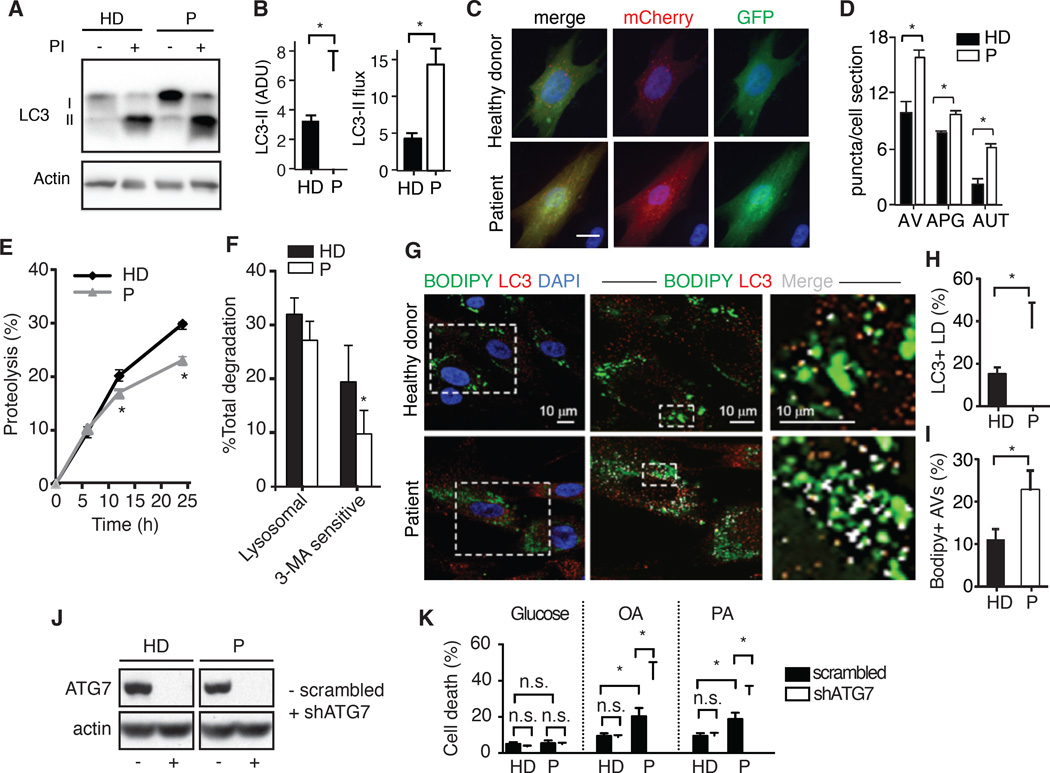
In addition to conventional lipolysis, LDs can also be targeted for lysosomal hydrolysis through lipophagy (Singh et al., 2009). We investigated if SOCE deficiency in patient fibroblasts affects the status of the autophagy/lysosomal system. Immunoblots for the autophagosome marker microtubule-associated protein light chain 3 (LC3) in cells maintained in the presence or absence of lysosomal protease inhibitors revealed consistently higher steady-state levels of LC3-II and increased autophagic flux (measured as the amount of LC3-II degraded in lysosomes) in patient cells, suggesting higher rates of autophagy in the absence of SOCE (Figure 4A and 4B). We confirmed this finding using a tandem fluorescent LC3 construct that allows for measuring autophagosome content (as mCherry+ and GFP+ fluorescent puncta) and autophagosome maturation into autolysosomes (as mCherry+ only puncta). Consistent with the immunoblot data, SOCE-deficient cells had significantly higher content of autophagosomes and autolysosomes (Figure 4C and 4D). Levels of other autophagy-related proteins (Atg5/12, p62, LAMP1) and of the transcription factor EB (TFEB) that controls the autophagic and lysosomal transcriptional programs, were also significantly higher in SOCE-deficient patient cells (Figure S3A–E). We found a similar upregulation of autophagy in the absence of SOCE in the skeletal muscle of Orai1R93W knock-in mice. Morphometric analysis of TEM images revealed a significant increase in the number of autophagic vacuoles in knock-in compared to WT mice (Figure S3F and S3G). As in human fibroblasts, the higher content of autophagic compartments in muscles of knock-in mice was a result of higher number of autolysosomes, further supporting an increase in autophagic flux in these animals (Figure S3H).
Figure 4. Increased lipophagy in SOCE-deficient cells.
(A) Immunoblot for LC3 in fibroblasts from healthy donors (HD) and patients (P) with loss-of-function mutations in STIM1 or ORAI1 treated or not with lysosomal protease inhibitors (PI). (B) LC3-II steady-state levels (left) and autophagic flux (right) calculated by densitometric analysis of immunoblots. (C and D) Autophagy in HD and P fibroblasts lentivirally transduced with the LC3 autophagy reporter mCherry-GFP-LC3 and stained with DAPI. (C) Images of representative cells. (D) Average number of autophagic vacuoles (AV), autophagosomes (APG) and autolysosomes (AUT) per cell section calculated by imaging >1,200 cells in 9 fields from 3 HDs and 3 Ps in duplicate by high content microscopy. (E and F) Long-lived protein degradation in 3 HD and 3 P fibroblasts. Total protein degradation rates (E) and percentage of lysosomal protein degradation and macroautophagy-dependent degradation (3-methyladenine (3-MA) sensitive) in cells treated without or with 10 mM 3-MA for the duration of the chase. (F). (G) HD and P cells were cultured in oleic acid (OA) medium, transferred to medium without OA for 12 h and stained with BODIPY (green, to detect neutral lipid stores), LC3 antibody (red) and DAPI. Representative images of merged channels. Boxed area at higher magnification (middle) and detail of lipid droplets (right) show colocalized pixels in white. (H and I) Percentages of LC3+ lipid droplets (LD) (H) and BODIPY+ autophagic vacuoles (AV) (I) calculated from images similar those in (G). (J and K) ATG7 knockdown causes lipotoxicity. (J) ATG7 protein expression in HD and P fibroblasts lentivirally transduced with scrambled shRNA (−) or shRNA targeting ATG7 (+) analyzed by immunoblotting with anti-ATG7 antibody. (K) Viability of cells after culture in 10 mM glucose, 1 mM OA or 0.5 mM palmitic acid (PA) for 24 hours. All values are mean ± SEM. Statistical analysis was performed using Student’s t-test. * p<0.05.
Despite marked upregulation of autophagy in SOCE-deficient cells, we found that protein is not favored as targeted cargo, as proteolysis of long-lived proteins was reduced in the absence of SOCE (Figure 4E) and this reduction was preferentially due to decreased 3-Methyladenine (3-MA)-sensitive proteolysis (macroautophagy) (Figure 4F). We next investigated lipophagy upon OA challenge by measuring the co-localization of LDs with LC3. Both the percentage of LDs decorated with LC3 (Figure 4G and 4H) and the percentage of autophagic vacuoles containing lipid as cargo (Figure 4G and 4I) were significantly higher in SOCE-deficient cells, suggesting increased lipophagy. Analysis of the LDs that accumulate in the skeletal muscle from Orai1R93W knock-in mice also revealed that a significantly higher percentage of LDs in these animals had morphological features previously attributed to active lipophagy (Figure S3I–S3K) (Singh et al., 2009). To assess the functional importance of increased lipophagy in SOCE-deficient cells, we ablated expression of autophagy-related protein 7 (ATG7), which is essential for autophagosome formation, using specific short-hairpin RNA. Deletion of ATG7 (Figure 4J) caused a significant decrease in the viability of SOCE-deficient cells cultured in OA or PA, whereas it had no effect on the viability of lipid-challenged WT cells or the sensitivity of patient cells to high glucose levels (Figure 4K). Taken together, these results suggest that the increase in lipophagy in SOCE-deficient cells may compensate for the loss of conventional lipolysis and serve as a protective mechanism against lipotoxicity.
Transcriptional regulation of lipid metabolism by PGC-1α and PPARα depends on SOCE
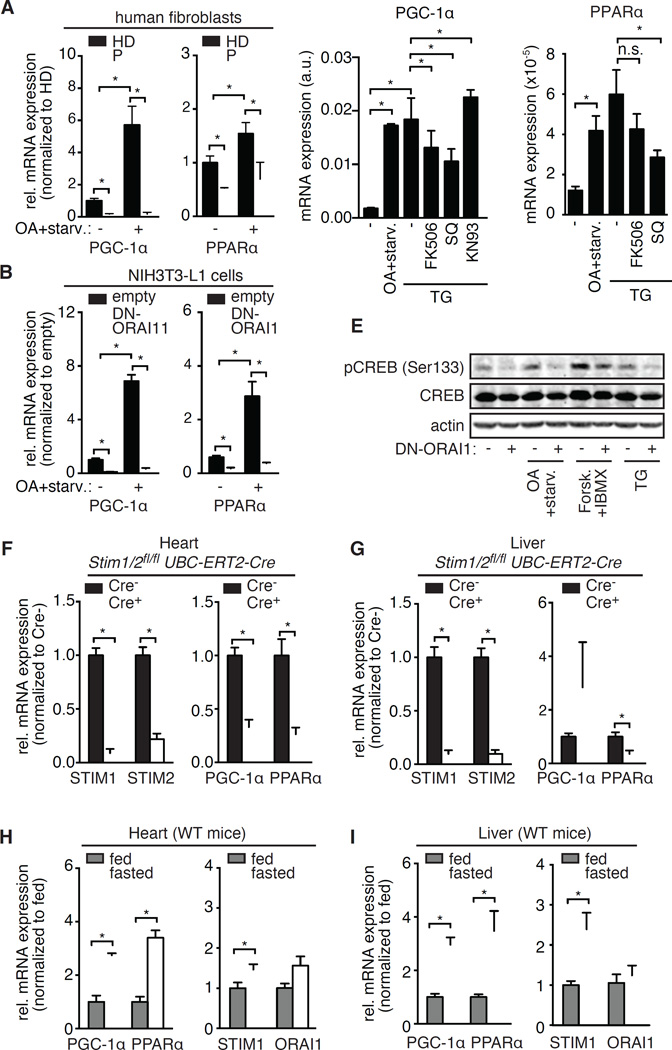
Mammals adapt to prolonged fasting by activating lipolysis in their adipose tissues, thereby releasing fatty acids into the circulation (Finn and Dice, 2006). To adjust to the abundance of fatty acids, the liver and other organs reprogram their metabolism by transcribing genes involved in fatty acid utilization. PPARα (Leone et al., 1999) and its co-regulator PGC-1α (Rhee et al., 2003) were identified as transcriptional regulators of the fasting response in the liver and heart; however, the upstream signaling events leading to metabolic reprogramming are still incompletely understood. Our finding of reduced levels of Cox4l1, UCP2 (Figure 2E), CPT1b, ACADVL (Figure 2Q and 2R), HSL and ATGL (Figure 3G and 3H), all of which are PGC-1α and PPARα target genes (Nikolic et al., 2012; Rakhshandehroo et al., 2010; Rakhshandehroo et al., 2007) suggests a role for SOCE in the transcriptional regulation of the fasting response. We observed a significant reduction in basal levels of PGC-1α and PPARα in SOCE-deficient patient cells compared to controls, and an impaired induction of their expression after culture in OA followed by starvation (Figure 5A). Similar results were obtained analyzing SOCE-deficient NIH3T3-L1 cells (Figure 5B) suggesting a conserved role for SOCE in the regulation of PGC-1α and PPARα expression in mice and humans.
Figure 5. SOCE controls PGC-1α and PPARα expression.
(A and B) Relative mRNA levels of PGC-1α and PPARα in fibroblasts from 3 patients (P) with loss-of-function mutations in STIM1 or ORAI1 normalized to 3 healthy donors (HD, in A) and in NIH3T3-L1 cells expressing dominant negative ORAI1-E106Q (DN-ORAI1) normalized to cells expressing an empty vector (B). Cells were cultured in high glucose (−) or oleic acid (OA)-containing medium followed by 6 hours of starvation (OA+starv.). mRNA expression was normalized to the housekeeping genes NONO (Non-POU domain-containing octamer-binding protein, in A) and ACTB (Actin beta, in B). (C–D) Relative mRNA levels of PGC-1α (C) and PPARα (D) in NIH3T3-L1 cells cultured in high glucose medium alone or stimulated with 1 µM thapsigargin (TG) in the presence or absence of calcineurin inhibitor FK506 (1 µM), adenylyl cyclase inhibitor SQ22,536 (100 µM) or CAMK inhibitor KN93 (10 µM) for 6 h. Alternatively, cells were cultured in medium containing 0.5 mM OA followed by starvation for 6 h. mRNA expression was normalized to ACTB. (E) Phosphorylation of CREB on the activating Ser133 residue assessed by Western blotting in NIH3T3-L1 cells transduced with empty vector (−) or DN-ORAI1 (+). Cells were cultured in high glucose with or without 10 mM isoproterenol, 10 µM forskolin (Forsk.) + 1 mM IBMX or 1 µM thapsigargin (TG) for 30 minutes. Alternatively, cells were cultured in 0.5 mM OA followed by starvation for 6 h. (F and G) Relative mRNA levels of PGC-1α, PPARα, STIM1 and STIM2 in the heart (F) and liver (G) of Stim1fl/flStim2fl/fl UBC-ERT2-Cre mice and Cre-negative littermates 5 days after the first injection of tamoxifen. mRNA levels were determined by real time PCR and normalized to ACTB. (H and I) Relative mRNA levels of PGC-1α, PPARα, STIM1 and ORAI1 in the heart (H) and liver (I) of WT mice fed a normal diet or fasted for 24 h. mRNA expression was normalized to ACTB (PGC1α, PPARα) or HPRT1 (STIM1, ORAI1). Bar graphs in A, B and F-I show data normalized to mean values obtained from the respective control samples and represent the mean ± SEM from at least 3 repeat experiments. Statistical analysis was performed using Student’s t-test. * p<0.05.
Interestingly, low PPARα expression may also contribute to the induction of autophagy observed in SOCE-deficient cells as PPAR agonists pioglitazone and WY14643 suppressed the increased autophagy flux in SOCE-deficient patient fibroblasts (Figure S4A) and in NIH3T3-L1 cells expressing DN-ORAI1 (Figure S4B) to levels comparable to their respective control cells. Furthermore, treatment of control human fibroblasts with the PPARα antagonist GW6471 (to mimic reduced PPARα expression in patient cells) was sufficient to elicit a marked upregulation of autophagy (Figure S5A and S5B). Although in certain settings PPARα stimulates lipophagy (Lee et al., 2014), upregulation of autophagy in SOCE-deficient cells occurs by mechanisms independent of PPARα. Thus, despite blunted PPARα signaling, SOCE-deficient cells had significantly elevated mRNA and protein levels of TFEB (Figure S5C and S3), the transcription factor shown to mediate PPARα-induced autophagy (Lee et al., 2014). This transcriptional upregulation of TFEB in SOCE-deficient cells was insensitive to modulation of PPARα signaling with agonists or antagonists (Figure S5C). Overall these findings suggest that upregulation of autophagy in SOCE-defective cells is a response to the loss of PPARα signaling but not directly mediated by this pathway.
SOCE appears to be not only necessary but also sufficient to induce PGC-1α expression because stimulation of WT fibroblasts with thapsigargin to induce SOCE resulted in levels of PGC-1α expression that were comparable to those observed in cells following OA and subsequent starvation (Figure 5C). Induction of PGC-1α expression by thapsigargin was partially dependent on the activity of calcineurin and ACs, because it was inhibited by FK506 and SQ22,536, respectively, whereas CAMK inhibition with KN93 had no inhibitory effect on PGC-1α expression (Figure 5C). Like PGC-1α, PPARα expression was also induced by thapsigargin in an AC-dependent manner (Figure 5D).
To better understand the mechanisms by which SOCE regulates PGC-1α and PPARα expression, we analyzed the phosphorylation of the cAMP response element-binding protein (CREB), a transcription factor reported to regulate expression of PGC-1α (Herzig et al., 2001) and PPARγ (Herzig et al., 2003). We found impaired CREB phosphorylation in SOCE-deficient cells that could be rescued by elevating cAMP levels with forskolin and IBMX treatment, suggesting that SOCE regulates CREB phosphorylation upstream of ACs (Figure 5E). We observed a similar dependence of PPARα expression on SOCE in vivo. After abolishing SOCE in all tissues of Stim1fl/flStim2fl/fl UBC-ERT2-Cre mice by tamoxifen injection, we observed significantly reduced expression of PPARα in the heart and liver of mice 5 days after the first injection (Figure 5F and 5G). A similar reduction of PGC-1α was observed in the heart, but not the liver of these mice. Fasting results in increased fatty acid levels in the serum and induces reprogramming of tissue metabolism (Cahill, 2006). To test if this switch is associated with altered expression of CRAC channel genes, we measured expression of STIM1 and ORAI1 in mice fed a normal diet and mice fasted for 24 hours. We observed increased expression of PGC-1α and PPARα (Figure 5H and 5I) and of STIM1, but not ORAI1 (Figure 5H and 5I) in the heart and liver of fasted WT animals, potentially in an effort to adapt to the abundance of fatty acids.
Given the dependence of the starvation response on SOCE, we asked which signal activates CRAC channels under starvation conditions. Consistent with previous reports showing that OA induces SOCE (Dramane et al., 2012; El-Yassimi et al., 2008), we found that control fibroblasts responded to OA with a sustained elevation in intracellular Ca2+ levels but that this response was almost completely abolished in SOCE-deficient patient fibroblasts (Figure S5D). The OA-induced SOCE was dependent on PLCγ function and the fatty acid transporter CD36, because the PLCγ inhibitor U73122 and the CD36 inhibitor sulfo-N-succinimidyl oleate (SSO) abolished OA-induced SOCE in HD fibroblasts (Figure S5E). These data show that fatty acids can induce SOCE by binding to PLCγ-coupled cell surface receptors such as CD36 consistent with previous reports. Taken together, these data demonstrate that SOCE directs the transcriptional reprogramming of cells to fatty acid utilization by controlling the expression of PGC-1α and PPARα.
DISCUSSION
In this study, we provide evidence for an important role of SOCE in the lipid metabolism of mammalian cells. We show that SOCE regulates lipolysis by controlling lipase expression and AC-dependent signaling. SOCE-deficient cells showed accumulation of LDs and impaired mobilization of LDs upon starvation. At least part of this defect was due to impaired lipolysis caused by strongly reduced expression of two neutral lipases, ATGL and HSL. In addition, SOCE-deficient cells showed reduced HSL phosphorylation on the activating Ser660 residue (Zechner et al., 2009) by protein kinase A (PKA). Since PKA is activated by cAMP (Zechner et al., 2009), these data suggested that SOCE regulates activation of plasma membrane ACs. This interpretation is supported by our finding that impaired HSL phosphorylation in the absence of SOCE could be rescued by increasing cAMP levels with the AC agonist Forskolin and the phosphodiesterase inhibitor IBMX, indicating that SOCE regulates HSL lipase function upstream of ACs. AC function is known to be Ca2+ dependent (Cooper et al., 1995) and is either positively or negatively regulated by intracellular Ca2+ levels depending on the AC isoforms expressed. Whereas AC1 and AC8 are activated by Ca2+, AC5 and AC6 are inhibited (Willoughby and Cooper, 2007). Intriguingly, ORAI1 directly interacts with AC8, thus allowing Ca2+ influx via ORAI1 channels to modulate cAMP levels (Willoughby 2012). In addition, a recent report identified an ER store depletion- and STIM1-dependent, but Ca2+ influx-independent pathway leading to AC activation and cAMP production (Lefkimmiatis et al., 2009). Our data, however, suggest an important role for SOCE in AC activation and cAMP production as ORAI1- and SOCE-deficient fibroblasts showed impaired cAMP production and cAMP-dependent signaling. In summary, we delineate a pathway that connects SOCE to activation of ACs, cAMP production, HSL phosphorylation and the induction of lipolysis. The physiological signal initiating SOCE and lipolysis at the cellular level and in vivo remains to be investigated. We show that fatty acids such as OA can induce SOCE via binding to the fatty acid transporter CD36 and activation of PLCγ. In addition, it is possible that metabolites or cytosolic factors produced by cells during starvation also initiate SOCE, for instance by releasing Ca2+ from ER stores.
In addition to HSL and ATGL, the expression of the fatty acid transporter CPT1b and the beta-oxidation enzyme ACADVL were also impaired in SOCE-deficient cells, suggesting an important role of SOCE in the transcriptional regulation of lipolysis and lipid utilization. We here show that SOCE controls the transcriptional reprogramming to lipid metabolism, which is dependent on the transcription factors PGC-1α and PPARα (Leone et al., 1999; Rhee et al., 2003; Vega et al., 2000). We found that SOCE on its own was sufficient to induce both PGC-1α and PPARα expression and that this effect was dependent on AC function. Consistent with the role of SOCE and ACs in PGC-1α and PPARα transcription, we found strongly impaired phosphorylation of the transcription factor CREB, which is activated by cAMP and a known regulator of PGC-1α and PPAR expression (Herzig et al., 2003; Herzig et al., 2001). We confirmed blunted PGC-1α and PPARα levels in the heart of Stim1/Stim2-deficient mice in vivo, and found that CRAC channel genes are induced in WT mice upon fasting. Collectively, our data support an important role of SOCE in orchestrating the fasting response at the transcriptional level to promote lipid utilization. Not all Ca2+ channels seem to have the same effect on PGC-1α expression. TRPV4, a non-selective Ca2+ channel, has recently been shown to suppress PGC-1α expression and thereby browning of white adipocytes (Ye et al., 2012), suggesting that different Ca2+ channels may mediate distinct Ca2+ signals that shape alternative metabolic programs of cells.
SOCE is furthermore required for mitochondrial function and FAO. Ca2+ is a known regulator of several mitochondrial functions including energy production by controlling the activity of several dehydrogenases in the TCA cycle and enzymes in the electron transport chain (Denton and McCormack, 1980; Glancy and Balaban, 2012; Hajnoczky et al., 1995; McCormack et al., 1990). Fatty acids are actively transported into the mitochondrial matrix where they are broken down during beta-oxidation to supply the TCA cycle with acetyl-CoA. Clinically, a decline in mitochondrial function is associated with triglyceride accumulation in the skeletal muscle and heart (Petersen et al., 2003). We observed impaired mitochondrial function in SOCE-deficient cells, which showed reduced expression and function of ETC complexes CI and CIV and consequently reduced proton pumping, electron transport, O2 consumption and oleic acid oxidation. Furthermore, SOCE-deficient cells had reduced levels of UCP2, a proton transporter in the inner mitochondrial membrane, whose opening prevents mitochondrial damage (Berardi and Chou, 2014; Brand and Esteves, 2005). Consistent with reduced UCP2 expression, we observed a higher MMP and more mitochondria undergoing degradation in acidic compartments. Many of the genes expressed at lower levels in the absence of SOCE such as CPT1B, ACADVL, UCP2, Cox4i1 and NDUFA1 are regulated by PGC-1α and PPARα (Rakhshandehroo et al., 2010; Rakhshandehroo et al., 2007; Rhee et al., 2003; Vega et al., 2000) providing a likely explanation for the mitochondrial dysfunction in SOCE-deficient cells. It is noteworthy that despite impaired expression of PGC-1α, and its reported role in mitochondrial biogenesis (Wu et al., 1999), we found normal numbers of mitochondria in SOCE deficient cells, possibly due to a compensatory upregulation of PGC-1β (data not shown). Collectively these findings indicate a direct and novel role for SOCE in the regulation of mitochondrial function and FAO.
SOCE-deficient cells showed marked upregulation of autophagy, preferentially of lipid droplets, i.e. lipophagy. The connection between Ca2+ signaling and autophagy is complex and may depend on the context and Ca2+ source. Whereas lysosomal Ca2+ release induces autophagy in a TFEB-dependent manner (Medina et al., 2015), increased cytosolic Ca2+ levels in obese mice interfere with autophagy (Arruda and Hotamisligil, 2015; Park and Lee, 2014) and inhibition of SOCE was recently reported to induce autophagy (Chen et al., 2016). Consistent with this inhibitory effect of cytosolic Ca2+, we observed higher rates of autophagy in SOCE-deficient cells and pronounced expansion of autophagic compartments in skeletal muscle of SOCE-deficient Orai1R93W knock-in mice. Our data indicates that this marked upregulation of autophagy favors lipids as predominant cargo. Lipophagy upregulation occurs during starvation and in response to lipid challenges in multiple cell types and failure to do so contributes to steatosis in common liver disorders (Dolganiuc et al., 2012; Singh et al., 2009). Increased lipophagy appears to be protective in SOCE-deficient cells as blockage of autophagy makes these cells more vulnerable to lipotoxicity. We propose that increased lipophagy in SOCE-deficient cells is an attempt by SOCE-deficient cells to compensate for impaired conventional lipolysis and an attempt to provide the remaining functional mitochondria with FFA for beta oxidation and energy production. The protective effect of increased lipophagy in SOCE-deficient cells may appear counterintuitive if lipophagy were to increase cytosolic FFA levels, especially considering the reduced FAO of SOCE-deficient cells. However, FFA levels in remained markedly low in the absence of SOCE. Protection from lipotoxicity may be explained by slower lysosomal release of FFAs by lipophagy and/or transfer of FFAs by direct lysosome-mitochondria interactions and kiss-and-run events (Daniele and Schiaffino, 2016), thus avoiding the toxicity of cytosolic release of FFAs. Part of the beneficial effect of upregulated autophagy in SOCE-deficient cells could also originate from the role of this process in organelle quality control, in particular of mitochondria.
PPARα was shown to stimulate autophagy (Lee et al., 2014) and since PPARα levels are decreased in SOCE-deficient cells, one might expect autophagy to be suppressed. However, we found that treatment of SOCE-deficient cells with the PPARα agonist WY14643 abolished their increased lipophagy suggesting that (1) the increase in autophagy was reactive to PPARα downregulation in these cells and that (2) upregulation of lipophagy in our setting occurs by mechanisms independent of PPARα. PPARα is not the only regulator of autophagy, and it is likely that alternative regulatory mechanisms are activated in cells to induce autophagy. In fact, even under conditions of blunted PPARα signaling, SOCE-deficient cells had significantly elevated levels of TFEB compared to control cells, which remained unchanged in the presence of PPARα agonists and antagonists. Since TFEB is downstream of the PPARα stimulatory effect on autophagy, it is possible that activation of this master regulator of the autophagic program in SOCE-deficient cells occurs transcriptionally through other pathways (e.g. FOXO, SREBP2, C/EBPb, Erk1/2) and/or posttranscriptionally through TFEB protein phosphorylation/dephosphorylation (Medina et al., 2015).
Ca2+ signals appear to play an important role in systemic lipid homeostasis. Functional screens and studies in Drosophila identified several molecules regulating intracellular Ca2+ homeostasis such as dSERCA, dIP3R, dRyR (Bi et al., 2014; Subramanian et al., 2013) and dSTIM (Baumbach et al., 2014). The effects of deleting these genes on lipid metabolism in Drosophila have been attributed to alterations at the neurological, adipose tissue and cellular level. Drosophila cells lacking dSERCA had increased cytosolic Ca2+ levels and decreased lipid storage (Bi et al., 2014). By contrast, Drosophila cells deficient for the ER release channels dIP3R and dRyR (Bi et al., 2014) or dSTIM (Baumbach et al., 2014) had reduced cytosolic Ca2+ concentrations and presented with increased lipid storage. These findings suggest an inverse correlation between cytosolic Ca2+ levels and cellular lipid load. They are consistent with our findings regarding the role of SOCE in lipid metabolism in mammalian cells as abolishing SOCE in Orai1R93W knock-in mice or Stim1fl/flStim2fl/fl UBC-ERT2-Cre mice resulted in excessive lipid droplet accumulation in the heart, liver and skeletal muscle. In contrast to dSTIM-deficient Drosophila (Baumbach et al., 2014), Stim1/Stim2-deficient mice did not show signs of weight gain or hyperphagia. We cannot exclude that these mice would gain weight over time, but they died 7–10 days after induction of Stim1/Stim2 deletion because complete deletion of SOCE is not compatible with survival.
In this study, we describe a cell intrinsic role for SOCE in the regulation of lipid metabolism at both the functional and transcriptional level. These findings may contribute to a better understanding of the pathogenic mechanisms behind common metabolic diseases and underscore SOCE-dependent signaling as a potential novel therapeutic target for the treatment of disorders of lipid metabolism.
EXPERIMENTAL PROCEDURES
More detailed methods can be found in Supplemental Experimental Procedures.
Mice
Orai1R93W knock-in mice (McCarl et al., 2010) have been described previously. Stim1fl/flStim2fl/fl mice (Oh-Hora et al., 2008) were crossed with B6.Cg-Tg (UBC-Cre/ERT2)1Ejb/J (UBC-ERT2-Cre) mice (The Jackson Laboratory, strain 008085). All animal experiments were conducted in accordance with institutionally approved protocols.
Cells and cell culture
Retroviral (Platinum E) and lentiviral (HEK293 FT) packaging cells, NIH3T3-L1 cells were cultured in DMEM at 37°C, 10% CO2. SOCE-deficient fibroblasts from 3 patients homozygous for missense mutations in STIM1 (p.R429C, described in (Maus et al., 2015), and ORAI1 (p.R91W described in (Feske et al., 2006) and p.G98R (unpublished)) were cultured in RPMI 1640 at 37°C, 5% CO2.
Plasmids and Reagents
Human dsRed-LC3 plasmid (Boland et al., 2008), mtKeima plasmid (Katayama et al., 2011), GFP-dgn/GFP-degFS (Greussing et al., 2012), ORAI1-E106Q (Prakriya et al., 2006) plasmids and other chemicals (Pampliega et al., 2013; Singh et al., 2009) were used as described. Cells were incubated with BODIPY 493/503 (D-3922, 20 mg/ml), LysoTracker® Red DND-99 (L-7528), MitoTracker® Green FM (M-7514, 50 mM) or MitoSOX (50nM, all from Invitrogen) according to the manufacturers’ instructions. Oleic acid (OA) and palmitic acid (PA), both from Sigma Aldrich, were dissolved in chloroform, chloroform was evaporated and fatty acids were conjugated to bovine serum albumin (BSA). Oligomycin, carbonyl cyanide-4-(trifluoromethoxy) phenylhydrazone (FCCP), KN93, SQ22,536 were from Sigma Aldrich, FK506 was purchased from Tocris.
Mitochondrial function analyses
TCA cycle function was measured as described (Abramov et al., 2011). The electron transport rate of the ETC was analyzed using the mitochondrial O2− sensitive dye MitoSOX (Invitrogen) and the mitochondrial membrane potential (MMP) using 20 nM tetramethylrhodamine (TMRM, Biovision) in a non-quenching mode as described (Abramov et al., 2011). Respiration of intact patient cells was measured using a Seahorse XF24 analyzer (Seahorse Bioscience) or the respiratory chain enzyme activity in muscle biopsies from patients (Table S1) was measured as described (Gempel et al., 2007; Kirby et al., 2007). FAO was analyzed using either a Seahorse XF24 analyzer or by incubation of cells with 14C-labeled oleic acid followed by analysis of 14CO2 as previously described (Huynh et al., 2014).
Analysis of lipid storage, lipolysis and lipotoxicity
Lipid droplet (LD) dynamics were measured by BODIPY 493/503 staining of cells. Lipolysis was analyzed with a colorimetric assay (F6428, Sigma) to measure the free glycerol content or a fluorescent quantification kit (MAK044, Sigma) to measure the FFA content in the extracellular medium. To test lipotoxicity, cells were stained with propidium iodide (PI) and analyzed by flow cytometry. Intracellular cAMP levels were measured by ELISA according to the manufacturer’s instructions (ADI-900-066, Enzo Life Sciences).
Intracellular Ca2+ measurements
Cells were loaded with Fura-2 and analyzed as described (Maus et al., 2015) using either a FlexStation 3 multi-mode microplate reader (Molecular Devices) or single cell Ca2+ imaging using an IX81 epifluorescence microscope (Olympus).
Direct fluorescence (DF) and transmission electron microscopy (TEM)
TEM was conducted as described (McCarl et al., 2010). DF was conducted using confocal microscopy or fluorescence microscopy with deconvolution. TEM and DF images were analyzed using ImageJ software.
Autophagy flux analysis
Intracellular protein degradation was measured by pulse-chase after 48h labeling with [3H] leucine (2 µCi/ml) (NENPerkinElmer Life Sciences) (Pampliega et al., 2013). Autophagy activity was estimated by immunoblotting for LC3-II or direct fluorescence in cells expressing mCherry-GFP-LC3.
Other methods
Quantitative real-time PCR was performed after reverse transcription (iScript cDNA synthesis kit (Bio-Rad) using TaqMan probes (Applied Biosystems) or gene specific primers (Table S2) and Maxima SYBR Green qPCR Master Mix (Thermo). For Mitochondrial DNA copy numbers, 3 primers against mtDNA and 2 against genomic DNA (Table S2) were used for amplification. Immunoblot and immunofluorescence procedures and antibodies can be found in Supplemental Experimental Procedures.
Statistical Analyses
All numerical results are mean ± standard error of the mean (SEM). Two-tailed unpaired Student’s t-test was used for statistical analysis upon normalization to control values (if multiple comparisons).
Supplementary Material
Highlights.
SOCE mediated by ORAI1 and STIM1 regulates lipid metabolism
SOCE is required for mitochondrial function and fatty acid oxidation (FAO)
SOCE controls cAMP-dependent induction of PGC-1α/PPARα expression and lipolysis
Increased lipophagy in SOCE-deficient cells prevents lipotoxicity
Acknowledgments
We thank Drs. Y. Deng and M. Cammer (NYUSoM) for help with NADH measurements, Dr. R. W. Taylor (Wellcome Trust Mitochondrial Research Centre, Institute for Ageing and Health) for respiratory chain enzyme activity measurements and Dr. R. Ramasamy for assistance with FAO experiments. The biobank of the MRC Centre for Neuromuscular Disease at Newcastle University provided technical support. This work was funded by NIH grants AI097302 to S.F., AG031782 and AG038072 to A.M.C., and HL119047 to K.J.M., grant 15POST25090199 from the American Heart Association to M.O., Medical Research Council (MRC, G1000848) and European Research Council (ERC, 309548) grants to R.H., Wellcome Trust Strategic Award 096919/Z/11/Z to Z.C.L., a Young Investigator Award by the Alex Lemonade Stand Foundation to M.M. and a postdoctoral fellowship by the Deutsche Forschungsgemeinschaft (DFG) to H.-T.H.-D. The Experimental Pathology Shared Resource at NYUSoM is supported by a Cancer Center Support Grant P30CA016087 at the Laura and Isaac Perlmutter Cancer Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement: S.F. is a cofounder of Calcimedica. The remaining authors declare no conflict of interest.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, eight figures, two tables and three videos that can be found with this article online at xxxxxx
AUTHOR CONTRIBUTIONS
M.M., M.C., B.P., J.L., O.M., R.H., H.-T.H.-D., Z. C.-L., A.M.C. and S.F. designed the research. M.M., B.P., J.L., O.M., U.K., J.Y., R.H., H.-T.H.-D., K.J.M., A.M.C. and S.F. performed experiments and/or analyzed data. M.M., A.M.C. and S.F. wrote the manuscript.
REFERENCES
- Abramov AY, Gegg M, Grunewald A, Wood NW, Klein C, Schapira AH. Bioenergetic consequences of PINK1 mutations in Parkinson disease. PLoS One. 2011;6:e25622. doi: 10.1371/journal.pone.0025622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthonsen MW, Ronnstrand L, Wernstedt C, Degerman E, Holm C. Identification of novel phosphorylation sites in hormone-sensitive lipase that are phosphorylated in response to isoproterenol and govern activation properties in vitro. J Biol Chem. 1998;273:215–221. doi: 10.1074/jbc.273.1.215. [DOI] [PubMed] [Google Scholar]
- Arruda AP, Hotamisligil GS. Calcium Homeostasis and Organelle Function in the Pathogenesis of Obesity and Diabetes. Cell Metab. 2015;22:381–397. doi: 10.1016/j.cmet.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumbach J, Hummel P, Bickmeyer I, Kowalczyk KM, Frank M, Knorr K, Hildebrandt A, Riedel D, Jackle H, Kuhnlein RP. A Drosophila in vivo screen identifies store-operated calcium entry as a key regulator of adiposity. Cell Metab. 2014;19:331–343. doi: 10.1016/j.cmet.2013.12.004. [DOI] [PubMed] [Google Scholar]
- Berardi MJ, Chou JJ. Fatty acid flippase activity of UCP2 is essential for its proton transport in mitochondria. Cell Metab. 2014;20:541–552. doi: 10.1016/j.cmet.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge MJ. Calcium signalling remodelling and disease. Biochem Soc Trans. 2012;40:297–309. doi: 10.1042/BST20110766. [DOI] [PubMed] [Google Scholar]
- Berridge MJ, Lipp P, Bootman MD. The versatility and universality of calcium signalling. Nat Rev Mol Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- Bi J, Wang W, Liu Z, Huang X, Jiang Q, Liu G, Wang Y. Seipin promotes adipose tissue fat storage through the ER Ca(2)(+)-ATPase SERCA. Cell Metab. 2014;19:861–871. doi: 10.1016/j.cmet.2014.03.028. [DOI] [PubMed] [Google Scholar]
- Boland B, Kumar A, Lee S, Platt FM, Wegiel J, Yu WH, Nixon RA. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer’s disease. J Neurosci. 2008;28:6926–6937. doi: 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand MD, Esteves TC. Physiological functions of the mitochondrial uncoupling proteins UCP2 and UCP3. Cell Metab. 2005;2:85–93. doi: 10.1016/j.cmet.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Brookes PS, Yoon Y, Robotham JL, Anders MW, Sheu SS. Calcium, ATP, and ROS: a mitochondrial love-hate triangle. Am J Physiol Cell Physiol. 2004;287:C817–C833. doi: 10.1152/ajpcell.00139.2004. [DOI] [PubMed] [Google Scholar]
- Cahill GF., Jr Fuel metabolism in starvation. Annu Rev Nutr. 2006;26:1–22. doi: 10.1146/annurev.nutr.26.061505.111258. [DOI] [PubMed] [Google Scholar]
- Chen YW, Chen YF, Chen YT, Chiu WT, Shen MR. The STIM1-Orai1 pathway of store-operated Ca2+ entry controls the checkpoint in cell cycle G1/S transition. Sci Rep. 2016;6:22142. doi: 10.1038/srep22142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DM, Mons N, Karpen JW. Adenylyl cyclases and the interaction between calcium and cAMP signalling. Nature. 1995;374:421–424. doi: 10.1038/374421a0. [DOI] [PubMed] [Google Scholar]
- Czubryt MP, McAnally J, Fishman GI, Olson EN. Regulation of peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PGC-1 alpha ) and mitochondrial function by MEF2 and HDAC5. Proc Natl Acad Sci U S A. 2003;100:1711–1716. doi: 10.1073/pnas.0337639100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniele T, Schiaffino MV. Lipid transfer and metabolism across the endolysosomal-mitochondrial boundary. Biochim Biophys Acta. 2016;1861:880–894. doi: 10.1016/j.bbalip.2016.02.001. [DOI] [PubMed] [Google Scholar]
- Denton RM, McCormack JG. On the role of the calcium transport cycle in heart and other mammalian mitochondria. FEBS Lett. 1980;119:1–8. doi: 10.1016/0014-5793(80)80986-0. [DOI] [PubMed] [Google Scholar]
- Dolganiuc A, Thomes PG, Ding WX, Lemasters JJ, Donohue TM., Jr Autophagy in alcohol-induced liver diseases. Alcohol Clin Exp Res. 2012;36:1301–1308. doi: 10.1111/j.1530-0277.2012.01742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dramane G, Abdoul-Azize S, Hichami A, Vogtle T, Akpona S, Chouabe C, Sadou H, Nieswandt B, Besnard P, Khan NA. STIM1 regulates calcium signaling in taste bud cells and preference for fat in mice. J Clin Invest. 2012;122:2267–2282. doi: 10.1172/JCI59953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Yassimi A, Hichami A, Besnard P, Khan NA. Linoleic acid induces calcium signaling, Src kinase phosphorylation, and neurotransmitter release in mouse CD36-positive gustatory cells. J Biol Chem. 2008;283:12949–12959. doi: 10.1074/jbc.M707478200. [DOI] [PubMed] [Google Scholar]
- Feske S. Calcium signalling in lymphocyte activation and disease. Nat Rev Immunol. 2007;7:690–702. doi: 10.1038/nri2152. [DOI] [PubMed] [Google Scholar]
- Feske S, Gwack Y, Prakriya M, Srikanth S, Puppel SH, Tanasa B, Hogan PG, Lewis RS, Daly M, Rao A. A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature. 2006;441:179–185. doi: 10.1038/nature04702. [DOI] [PubMed] [Google Scholar]
- Finn PF, Dice JF. Proteolytic and lipolytic responses to starvation. Nutrition. 2006;22:830–844. doi: 10.1016/j.nut.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Gempel K, Topaloglu H, Talim B, Schneiderat P, Schoser BG, Hans VH, Palmafy B, Kale G, Tokatli A, Quinzii C, et al. The myopathic form of coenzyme Q10 deficiency is caused by mutations in the electron-transferring-flavoprotein dehydrogenase (ETFDH) gene. Brain. 2007;130:2037–2044. doi: 10.1093/brain/awm054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glancy B, Balaban RS. Role of mitochondrial Ca2+ in the regulation of cellular energetics. Biochemistry. 2012;51:2959–2973. doi: 10.1021/bi2018909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greussing R, Unterluggauer H, Koziel R, Maier AB, Jansen-Durr P. Monitoring of ubiquitin-proteasome activity in living cells using a Degron (dgn)-destabilized green fluorescent protein (GFP)-based reporter protein. J Vis Exp. 2012 doi: 10.3791/3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajnoczky G, Robb-Gaspers LD, Seitz MB, Thomas AP. Decoding of cytosolic calcium oscillations in the mitochondria. Cell. 1995;82:415–424. doi: 10.1016/0092-8674(95)90430-1. [DOI] [PubMed] [Google Scholar]
- Handschin C, Rhee J, Lin J, Tarr PT, Spiegelman BM. An autoregulatory loop controls peroxisome proliferator-activated receptor gamma coactivator 1alpha expression in muscle. Proc Natl Acad Sci U S A. 2003;100:7111–7116. doi: 10.1073/pnas.1232352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzig S, Hedrick S, Morantte I, Koo SH, Galimi F, Montminy M. CREB controls hepatic lipid metabolism through nuclear hormone receptor PPAR-gamma. Nature. 2003;426:190–193. doi: 10.1038/nature02110. [DOI] [PubMed] [Google Scholar]
- Herzig S, Long F, Jhala US, Hedrick S, Quinn R, Bauer A, Rudolph D, Schutz G, Yoon C, Puigserver P, et al. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- Houten SM, Wanders RJ. A general introduction to the biochemistry of mitochondrial fatty acid beta-oxidation. J Inherit Metab Dis. 2010;33:469–477. doi: 10.1007/s10545-010-9061-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh FK, Green MF, Koves TR, Hirschey MD. Measurement of fatty acid oxidation rates in animal tissues and cell lines. Methods Enzymol. 2014;542:391–405. doi: 10.1016/B978-0-12-416618-9.00020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama H, Kogure T, Mizushima N, Yoshimori T, Miyawaki A. A sensitive and quantitative technique for detecting autophagic events based on lysosomal delivery. Chem Biol. 2011;18:1042–1052. doi: 10.1016/j.chembiol.2011.05.013. [DOI] [PubMed] [Google Scholar]
- Kirby DM, Thorburn DR, Turnbull DM, Taylor RW. Biochemical assays of respiratory chain complex activity. Methods Cell Biol. 2007;80:93–119. doi: 10.1016/S0091-679X(06)80004-X. [DOI] [PubMed] [Google Scholar]
- Lacruz RS, Feske S. Diseases caused by mutations in ORAI1 and STIM1. Ann N Y Acad Sci. 2015;1356:45–79. doi: 10.1111/nyas.12938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Wagner M, Xiao R, Kim KH, Feng D, Lazar MA, Moore DD. Nutrient-sensing nuclear receptors coordinate autophagy. Nature. 2014;516:112–115. doi: 10.1038/nature13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefkimmiatis K, Srikanthan M, Maiellaro I, Moyer MP, Curci S, Hofer AM. Store-operated cyclic AMP signalling mediated by STIM1. Nat Cell Biol. 2009;11:433–442. doi: 10.1038/ncb1850. [DOI] [PubMed] [Google Scholar]
- Leone TC, Weinheimer CJ, Kelly DP. A critical role for the peroxisome proliferator-activated receptor alpha (PPARalpha) in the cellular fasting response: the PPARalpha-null mouse as a model of fatty acid oxidation disorders. Proc Natl Acad Sci U S A. 1999;96:7473–7478. doi: 10.1073/pnas.96.13.7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou J, Kim ML, Heo WD, Jones JT, Myers JW, Ferrell JE, Jr, Meyer T. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr Biol. 2005;15:1235–1241. doi: 10.1016/j.cub.2005.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maus M, Jairaman A, Stathopulos PB, Muik M, Fahrner M, Weidinger C, Benson M, Fuchs S, Ehl S, Romanin C, et al. Missense mutation in immunodeficient patients shows the multifunctional roles of coiled-coil domain 3 (CC3) in STIM1 activation. Proc Natl Acad Sci U S A. 2015;112:6206–6211. doi: 10.1073/pnas.1418852112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarl CA, Khalil S, Ma J, Oh-hora M, Yamashita M, Roether J, Kawasaki T, Jairaman A, Sasaki Y, Prakriya M, et al. Store-operated Ca2+ entry through ORAI1 is critical for T cell-mediated autoimmunity and allograft rejection. J Immunol. 2010;185:5845–5858. doi: 10.4049/jimmunol.1001796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack JG, Halestrap AP, Denton RM. Role of calcium ions in regulation of mammalian intramitochondrial metabolism. Physiol Rev. 1990;70:391–425. doi: 10.1152/physrev.1990.70.2.391. [DOI] [PubMed] [Google Scholar]
- Medina DL, Di Paola S, Peluso I, Armani A, De Stefani D, Venditti R, Montefusco S, Scotto-Rosato A, Prezioso C, Forrester A, et al. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat Cell Biol. 2015;17:288–299. doi: 10.1038/ncb3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolic N, Rhedin M, Rustan AC, Storlien L, Thoresen GH, Stromstedt M. Overexpression of PGC-1alpha increases fatty acid oxidative capacity of human skeletal muscle cells. Biochem Res Int. 2012;2012:714074. doi: 10.1155/2012/714074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh-Hora M, Yamashita M, Hogan PG, Sharma S, Lamperti E, Chung W, Prakriya M, Feske S, Rao A. Dual functions for the endoplasmic reticulum calcium sensors STIM1 and STIM2 in T cell activation and tolerance. Nat Immunol. 2008;9:432–443. doi: 10.1038/ni1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pampliega O, Orhon I, Patel B, Sridhar S, Diaz-Carretero A, Beau I, Codogno P, Satir BH, Satir P, Cuervo AM. Functional interaction between autophagy and ciliogenesis. Nature. 2013;502:194–200. doi: 10.1038/nature12639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HW, Lee JH. Calcium channel blockers as potential therapeutics for obesity-associated autophagy defects and fatty liver pathologies. Autophagy. 2014;10:2385–2386. doi: 10.4161/15548627.2014.984268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen KF, Befroy D, Dufour S, Dziura J, Ariyan C, Rothman DL, DiPietro L, Cline GW, Shulman GI. Mitochondrial dysfunction in the elderly: possible role in insulin resistance. Science. 2003;300:1140–1142. doi: 10.1126/science.1082889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakriya M, Feske S, Gwack Y, Srikanth S, Rao A, Hogan PG. Orai1 is an essential pore subunit of the CRAC channel. Nature. 2006;443:230–233. doi: 10.1038/nature05122. [DOI] [PubMed] [Google Scholar]
- Rakhshandehroo M, Knoch B, Muller M, Kersten S. Peroxisome proliferator-activated receptor alpha target genes. PPAR Res. 2010 doi: 10.1155/2010/612089. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakhshandehroo M, Sanderson LM, Matilainen M, Stienstra R, Carlberg C, de Groot PJ, Muller M, Kersten S. Comprehensive analysis of PPARalpha-dependent regulation of hepatic lipid metabolism by expression profiling. PPAR Res. 2007;2007:26839. doi: 10.1155/2007/26839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee J, Inoue Y, Yoon JC, Puigserver P, Fan M, Gonzalez FJ, Spiegelman BM. Regulation of hepatic fasting response by PPARγamma coactivator-1alpha (PGC-1): requirement for hepatocyte nuclear factor 4alpha in gluconeogenesis. Proc Natl Acad Sci U S A. 2003;100:4012–4017. doi: 10.1073/pnas.0730870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer PJ, Wende AR, Magee CJ, Neilson JR, Leone TC, Chen F, Kelly DP. Calcineurin and calcium/calmodulin-dependent protein kinase activate distinct metabolic gene regulatory programs in cardiac muscle. J Biol Chem. 2004;279:39593–39603. doi: 10.1074/jbc.M403649200. [DOI] [PubMed] [Google Scholar]
- Shungin D, Winkler TW, Croteau-Chonka DC, Ferreira T, Locke AE, Magi R, Strawbridge RJ, Pers TH, Fischer K, Justice AE, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soboloff J, Rothberg BS, Madesh M, Gill DL. STIM proteins: dynamic calcium signal transducers. Nat Rev Mol Cell Biol. 2012;13:549–565. doi: 10.1038/nrm3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian M, Metya SK, Sadaf S, Kumar S, Schwudke D, Hasan G. Altered lipid homeostasis in Drosophila InsP3 receptor mutants leads to obesity and hyperphagia. Dis Model Mech. 2013;6:734–744. doi: 10.1242/dmm.010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadi A, Lebel L, Hashim Y, Mehta Z, Ashcroft SJ, Turner R. Sequence variants of the sarco(endo)plasmic reticulum Ca(2+)-transport ATPase 3 gene (SERCA3) in Caucasian type II diabetic patients (UK Prospective Diabetes Study 48) Diabetologia. 1999;42:1240–1243. doi: 10.1007/s001250051298. [DOI] [PubMed] [Google Scholar]
- Vega RB, Huss JM, Kelly DP. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor alpha in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol Cell Biol. 2000;20:1868–1876. doi: 10.1128/mcb.20.5.1868-1876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vig M, Peinelt C, Beck A, Koomoa DL, Rabah D, Koblan-Huberson M, Kraft S, Turner H, Fleig A, Penner R, et al. CRACM1 is a plasma membrane protein essential for store-operated Ca2+ entry. Science. 2006;312:1220–1223. doi: 10.1126/science.1127883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willoughby D, Cooper DM. Ca2+ stimulation of adenylyl cyclase generates dynamic oscillations in cyclic AMP. J Cell Sci. 2006;119:828–836. doi: 10.1242/jcs.02812. [DOI] [PubMed] [Google Scholar]
- Willoughby D, Cooper DM. Organization and Ca2+ regulation of adenylyl cyclases in cAMP microdomains. Physiol Rev. 2007;87:965–1010. doi: 10.1152/physrev.00049.2006. [DOI] [PubMed] [Google Scholar]
- Willoughby D, Everett KL, Halls ML, Pacheco J, Skroblin P, Vaca L, Klussmann E, Cooper DM. Direct binding between Orai1 and AC8 mediates dynamic interplay between Ca2+ and cAMP signaling. Sci Signal. 2012;5:ra29. doi: 10.1126/scisignal.2002299. [DOI] [PubMed] [Google Scholar]
- Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- Ye L, Kleiner S, Wu J, Sah R, Gupta RK, Banks AS, Cohen P, Khandekar MJ, Bostrom P, Mepani RJ, et al. TRPV4 is a regulator of adipose oxidative metabolism, inflammation, and energy homeostasis. Cell. 2012;151:96–110. doi: 10.1016/j.cell.2012.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechner R, Kienesberger PC, Haemmerle G, Zimmermann R, Lass A. Adipose triglyceride lipase and the lipolytic catabolism of cellular fat stores. J Lipid Res. 2009;50:3–21. doi: 10.1194/jlr.R800031-JLR200. [DOI] [PubMed] [Google Scholar]
- Zhang SL, Yeromin AV, Zhang XH, Yu Y, Safrina O, Penna A, Roos J, Stauderman KA, Cahalan MD. Genome-wide RNAi screen of Ca(2+) influx identifies genes that regulate Ca(2+) release-activated Ca(2+) channel activity. Proc Natl Acad Sci U S A. 2006;103:9357–9362. doi: 10.1073/pnas.0603161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SL, Yu Y, Roos J, Kozak JA, Deerinck TJ, Ellisman MH, Stauderman KA, Cahalan MD. STIM1 is a Ca2+ sensor that activates CRAC channels and migrates from the Ca2+ store to the plasma membrane. Nature. 2005;437:902–905. doi: 10.1038/nature04147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.