Abstract
Vision is a critical sense for organismal survival with visual sensitivities strongly shaped by the environment. Some freshwater fishes with a Gondwanan origin are distributed in both South American rivers including the Amazon, as well as African rivers and lakes. These different habitats likely required adaptations to murky and clear environments. In this study, we compare the molecular basis of Amazonian and African cichlid fishes’ visual systems. We used next generation sequencing of genomes and retinal transcriptomes to examine three Amazonian cichlid species. Genome assemblies revealed six cone opsin classes (SWS1, SWS2B, SWS2A, RH2B, RH2A, LWS) and rod opsin (RH1). However, the functionality of these genes varies across species with different pseudogenes found in different species. Our results support evidence of an RH2A gene duplication event that is shared across both cichlid groups, but which was probably followed by gene conversion. Transcriptome analyses show that Amazonian species mainly express three opsin classes (SWS2A, RH2A and LWS) which likely are a good match to the long wavelength oriented light environment of the Amazon basin. Furthermore, analysis of amino acid sequences suggest that the short wavelength sensitive genes (SWS2B, SWS2A) may be under selective pressures in order to shift their spectral properties to a longer wavelength visual palette. Our results agree with the ‘sensitivity hypothesis’ where the light environment causes visual adaptation. Amazonian cichlid visual systems are likely adapting through gene expression, gene loss, and possibly spectral tuning of opsin sequences. Such mechanisms may be shared across the Amazonian fish fauna.
Keywords: fish vision, opsins, cichlids, Amazon
Introduction
Sensory capabilities play a major role in an individual’s fitness because they specify a channel through which information from the environment is transmitted to the organism (Endler 1993). Vision plays a role in mediating behaviors like foraging, mating, social interactions with conspecifics and predator avoidance. In vertebrates, visual perception begins when light reaches the retina and is detected by the photoreceptive rods and cones. These photoreceptors contain visual pigments composed of a transmembrane protein, opsin, bound to a light sensitive chromophore. Visual pigment spectral sensitivities can vary due to the type of chromophore (derived from either vitamin A1 or A2), and the chromophore’s interaction with opsin amino acid residues facing the retinal binding pocket (Parry & Bowmaker 2000; Yokoyama 2008). Therefore, opsins are the main components controlling the spectral response of the first step in the visual transduction cascade.
Visual systems in fishes are particularly interesting because of the remarkable diversity of visual pigments and the proximate mechanisms that underlie their evolution. Genetic mechanisms that contribute to visual pigment diversity are sequence evolution, differential gene expression, gene duplication, and gene conversion (Hofmann & Carleton 2009; Rennison et al. 2012; Cortesi et al. 2015). Selective pressures from the environment greatly influence fish spectral sensitivities, providing ultimate causation for visual sensitivity shifts to better match environmental light.
Cichlids, a diverse group of percomorph fishes with a Gondwanan distribution, are a prime example of teleost visual pigment diversity. Vision research on cichlid flocks from the great African Lakes has identified the genetic basis of their visual sensitivities resulting from seven spectrally distinct cone opsins and a rod opsin gene (Carleton 2009). The cone opsins belong to four cone opsin classes including UV sensitive (SWS1), short-wavelength sensitive (SWS2A, SWS2B), rod opsin like (RH2Aα, RH2Aβ, RH2B) and long-wavelength sensitive (LWS) opsins (Carleton et al. 2016). Cichlids typically express three different opsin combinations termed short (SWS1, RH2A, RH2B), medium (SWS2B, RH2A, RH2B) and long (SW2A, RH2A, LWS) (Spady et al. 2006). These combinations are believed to match the available light in a particular environment. For example, the “long” combination would be suited to a long wavelength shifted environment like turbid waters (Hofmann et al. 2009).
While vision in African cichlids has been studied extensively, little is known about vision in Neotropical cichlid lineages. A previous study analyzed the visual system of the Trinidadian pike, Crenicichla frenata, and found that this species has a reduced opsin complement where SWS1 has been lost and RH2B pseudogenized (Weadick et al. 2012). Furthermore, a rod opsin study in Neotropical cichlids suggested there is divergent selection on this gene across cichlid lineages probably caused by ecologically and/or biogeographic differences (Schott et al. 2014). Indeed, Neotropical cichlids live in more highly contrasting light environments than clear African lakes species. In South America, rivers in the Amazon basin can be classified based on water color (white, black and clear). This is a product of different physico-chemical and geological properties of their catchments; as well as rainfall, soil, and vegetation which results in waterbodies with different concentrations of particulates and dissolved compounds. White water rivers exhibit high quantities of inorganic suspended particles, black water rivers have high concentrations of dissolved organic matter, and clear water rivers are low in both dissolved organic matter and inorganic suspended particles (Costa et al. 2012; Sawakuchi et al. 2016). Thus, Amazonian rivers display an adverse visual environment for fish due to large amounts of suspended particles and dissolved substances that absorb most of the incoming light creating an extremely long wavelength-shifted light environment. These light conditions may have an effect on the phenotypic adaptation of Amazonian cichlids. Muntz (1973, 1982) studied visual sensitivities of Amazonian fishes and found that several cichlids had ocular media with yellow pigments. Such pigments filter short wavelengths of the incoming light to the retina, reducing the detected level of background scattered light and serving as visual adaptations to the long wavelength light environments (Muntz 1973, 1982) (Muntz 1973). Vision research in Neotropical cichlids is important because it offers a unique opportunity to analyze the evolution of the visual system in different ecosystems within the same cichlid lineage.
In this study, we investigated the opsin complement of three Amazonian cichlids. We examined whole genomes and retinal transcriptomes of Pterophyllum scalare (Angelfish), Symphysodon discus (Discus) and Astronotus ocellatus (Oscar) in order to (1) examine the genomic opsin palette, (2) analyze their opsin gene expression, and (3) discuss the evolution of these species’ visual system under the influence of the Amazon environment. Based on next generation genomic and RNA sequencing, phylogenetic and opsin sequence analysis suggest dynamic evolution of opsin genes between lineages and a “long” opsin expression profile that is consistent with potential adaptations to Amazonian rivers.
Materials and Methods
For whole genome sequencing fish were caught from the Amazon basin (Table S3). Sampling permits were in accordance to Brazilian laws for environmental protection (wild collection permit, ICMBio 22984-1 e 32556-2), and specimens stocked at INPA (National Amazon Institute of Research, Manaus – AM, Brazil). The geographic coordinates of the collected points were 0°52’2.29"S/62°48’35.61"W for S. discus and 3°09’43.0"S/59°54’59.4"W for P. scalare and A. ocellatus. Fish were euthanized through immersion in a benzocaine (250 mg/liter) water bath for 10 minutes, according to the international guidelines of Sao Paulo State University approved by the Institutional Animal Care and Use Committee (IACUC) (Protocol no. 34/08 - CEEA/IBB/UNESP). For RNAseq fish were obtained from the aquarium trade and were around five months old. Fish were euthanized with buffered MS-222 according to University of Maryland (UM) IACUC approved protocols (R12–90).
Genome Analysis
Muscle tissue from two fish (female and male) was used for DNA extraction (Sambrook & Russel 2001). Whole genome sequencing was performed through paired-end library preparation (Truseq DNA Library Preparation Kit) and sequenced on an Illumina HiSeq1500 platform. Obtained reads (150 bp length) were trimmed based on base-pair sequencing quality (minimum 90% of read base pairs with Phred quality score greater than 30) and removal of sequencing adaptors using FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/index.html) and in house scripts. The trimmed reads were assembled with Velvet (Zerbino & Birney 2008) using a k-mer value estimated by Kmergenie (Chikhi & Medvedev 2014). Finally, the assembled contigs were evaluated using the CEGMA pipeline (Parra et al. 2007) and Assemblathon2 scripts (Bradnam et al. 2013).
RNA analysis
To ensure high quality transcriptome assemblies, one specimen of each species was used for transcriptomes. Fish eyes were enucleated and retinas including retinal pigment epithelia were dissected out and preserved in RNAlater. RNA was isolated from two whole retinas for each species. Total RNA was extracted with an RNeasy kit (Qiagen) and RNA quality was verified on an Agilent Bioanalyzer. RNAseq libraries were made using the Illumina TruSeq RNA library preparation kit (Illumina Inc, San Diego) by the UM Institute for Bioscience & Biotechnology Research sequencing core, and 100 bp paired end reads were obtained on an Illumina HiSeq1500 sequencer with samples multiplexed in one lane. The data was quality checked using FastQC version 0.10.1. Data were trimmed using Trimmomatic version 0.32 (Bolger et al. 2014) in order to remove over-represented sequences and to retain sequences with a minimum quality score of 20 and a minimum length of 80 bp. The final assembly was performed using Trinity version r20140413 (Haas et al. 2013) using only paired sequences with a minimum coverage of two to join contigs.
Opsin sequencing by PCR
We used two additional wild-caught individuals of each species for genomic DNA sequencing. If transcripts or contigs of a certain opsin were incomplete, primers were designed and the complete opsin sequence was recovered with PCR and Sanger sequencing (Table S4). DNA was extracted with a DNeasy kit (Qiagen) and DNA quality was verified in a spectrometer.
Phylogenetic Trees
Putative opsin sequences of the three South American cichlids were identified from genome and transcriptome assembled FASTA files by Tblastx querying with the cichlid opsin genes of Oreochromis niloticus (Spady et al. 2006). We confirmed that sequences for each species were assigned to a particular opsin class based on phylogenetic relationships of the opsin sequences with those from other teleost lineages obtained from Genbank (Danio rerio, Lucania goodei, Oryzias latipes, Oreochromis niloticus, Metriaclima zebra, Neolamprologus brichardi, Pundamilia nyererei) (Benson et al. 2005). We used MAFFT (Katoh et al. 2002) to align nucleotide sequences, and GARLI for building maximum likelihood trees. We ran 10 searches for the best tree and 2000 bootstrap replicates performed at the GARLI 2.0 (Zwickl 2006) web service (molecularevolution.org, (Bazinet et al. 2014)).
Gene Expression
For estimating gene expression of each opsin, reads were mapped back to the assembled transcripts using RSEM as part of the Trinity package (Haas et al. 2013). Read counts for each opsin class were extracted from the RSEM output (quantified as fragments per kilobase of transcript per million reads, FPKM). Cone opsin read counts were normalized by dividing the sum of all the cone opsin genes to get the proportion of each expressed opsin. For RH2A duplicates, we used Geneious 8.1 to map reads to the distinct first exons to estimate expression levels.
Spectral sensitivities
To determine potential tuning sites we aligned South American cichlid opsin sequences of each opsin class with bovine rhodopsin (GenBank. NP_001014890.1) and compared them to known spectral tuning sites (Hunt et al. 2001; Yokoyama 2008). We also aligned opsin sequences to O. niloticus and C. frenata for which λmax and opsin gene sequences have been characterized (Spady et al. 2006; Weadick et al. 2012). The alignments were analyzed to identify amino acid substitutions that fell in the putative opsin transmembrane regions and the retinal binding pocket facing the chromophore (Supplementary materials 2) (Carleton et al. 2005). Substitutions that involve changes in amino acid polarity could alter interactions and potentially create a shift in peak absorption, tuning the sensitivity of the visual pigment. Substitutions were examined only in functional genes to determine their variability in physico-chemical properties.
Pseudogenes divergence times
Because pseudogenes were discovered in our analysis, we wanted to estimate when they emerged. For this, we modified the method from Li et al 1981 (Supplementary materials 1). This method is based on the assumption that the rate of change in a gene’s synonymous positions is faster than for its non-synonymous positions. However, in a pseudogene, the lack of selective constraint causes the rates of change for both positions to be the same. Hence, the increase in pseudogene nucleotide substitutions can be used to determine how long ago this increased rate began (Li et al. 1981). For both the SWS1 and SWS2B opsins we aligned the nucleotide sequences of one pseudogene (gene A) and two functional genes (genes B and C). Fig. S8 shows the plausible evolutionary outline for functional opsins and pseudogenes with O. niloticus (C) as the more distant lineage. Insertions and deletions that were not shared between pseudogenes and the functional opsin genes were excluded. Proportion of synonymous and non-synonymous sites were estimated between taxa using MEGA6 (Tamura et al. 2013). These were corrected for multiple hits using a Jukes-Cantor model to determine DNA sequence differences, dABi, dACi, and dBCi for i = s or ns (Jukes & Cantor 1969). These were then used to calculate the distance difference (yi) between pseudogenes and functional genes (equation 1a). Rates of synonymous and non-synonymous substitutions per year, as and ans, were calculated based on the divergence time (T1) between C and the other sequences (A or B) (equation 1b). For T1, we used the divergence time of O. niloticus and Neotropical cichlids (77.2 Mya; timetree.org). From these equations, we calculated the time (Tn) since pseudogenization, equation 1c.
| (1a) |
| (1b) |
| (1c) |
Results
Opsin genomic sequences
Genomes were assembled from three Amazonian cichlids including P. scalare (1.088 Gb; scaffold N50 7.559 bp), S. discus (0.684 Gb; scaffold N50 2.221 bp) and A. ocellatus (1.007Gb; scaffold N50 7.659 bp) (Escobar-Camacho et al. 2016, Ramos et al. unpublished). Blasting the three genomes identified members of six cone opsin classes (SWS1, SWS2B, SWS2A, RH2B, RH2A and LWS) as well as rod opsin (RH1). SWS1, SWS2B, and SWS2A opsin genes contained gaps in some species while RH2B was incomplete in all three genomes. The missing sequences for the SWS genes were recovered by PCR and sequencing using primers based on adjacent regions of each gap, using primers for other species where the opsin genes were present, or the transcriptomes (NCBI accession numbers in Table S1).
Examination of the complete opsin sequences revealed the presence of at least one pseudogene in all three species. These occurred among different opsin classes (Table 1). SWS1 is pseudogenized in A. ocellatus exhibiting a 4 bp frame shifting insertion in the second exon (position 450) leading to numerous stop codons as well as a codon insertion in the third exon (position 634) (Fig. S1). SWS2B opsin is a pseudogene in S. discus with two deletions and a single bp insertion in the first exon (position 59 & 303 respectively) (Fig. S2). The single nucleotide insertion seems to be polymorphic as one specimen had it while others did not. SWS1 pseudogene of A. ocellatus was only found in the genome whereas SWS2B pseudogene in S. discus was present in the genome and the transcriptome. RH2B appears to be pseudogenized in P. scalare, S. discus and A. ocellatus for both genome and aquarium trade individuals. Despite significant effort, the complete sequence of RH2B was not fully recovered for most species, perhaps as a result of it being pseudogenized. In P. scalare only half of the coding region of RH2B was recovered with a 14 bp deletion in the last exon (Fig S3) whereas in S. discus, a single bp insertion (position 163) in the first exon was present that caused numerous stop codons (Fig. S4). RH2B in A. ocellatus has a 13 bp insertion and 9 bp deletion in the first exon causing a premature stop codon in the first exon (Fig. S5). Overall it seems that RH2B is a pseudogene in all three species. Further, these data suggest that RH2B mutations were acquired separately in each of the three species, although we could be overlooking a shared common pseudogenization event.
Table 1.
Opsin gene classes across species
| Species | Opsins | ||||||
|---|---|---|---|---|---|---|---|
| SWS1 | SWS2B | SWS2A | RH2B | RH2 A | LWS | RH1 | |
| Pterophylum scalare | ✓ | ✓ | ✓ | Ψ | ✓ | ✓ | ✓ |
| Symphysodon discus | ✓ | Ψ | ✓ | Ψ | ✓ | ✓ | ✓ |
| Astronotus ocellatus | Ψ | ✓ | ✓ | Ψ | ✓ | ✓ | ✓ |
Opsin genes are sorted by increasing spectral sensitivity. Ψ denotes pseudogenes.
Opsin RNAseq
Retina samples run on the Agilent Bioanalyzer had RIN (RNA integrity number) values that varied between 7.70 and 9.60. The RNAseq data obtained by multiplexing these retinal samples provided sufficient data to assemble and quantify opsin transcripts. We obtained 71,0 64.5 and 70.9 M reads for P. scalare, S. discus and A. ocellatus respectively; and after trimming used, 45.1, 38.5 and 34 M reads for the assemblies. RNAseq reads were submitted to the SRA database (SUB2057474)
Amazonian cichlid RNAseq isolated four cone opsins: SWS2A, SWS2B, RH2A, and LWS, as well as rod opsin, RH1. These were all complete transcripts except for SWS2B, which had lower transcript abundance in comparison to the other opsins. The SWS2A, RH2A, LWS and RH1 transcripts were verified with genome sequence and matched nearly exactly. There were a few SNPs since the individuals were different between RNAseq and genomic sequencing (Table S5).
Phylogenetic trees
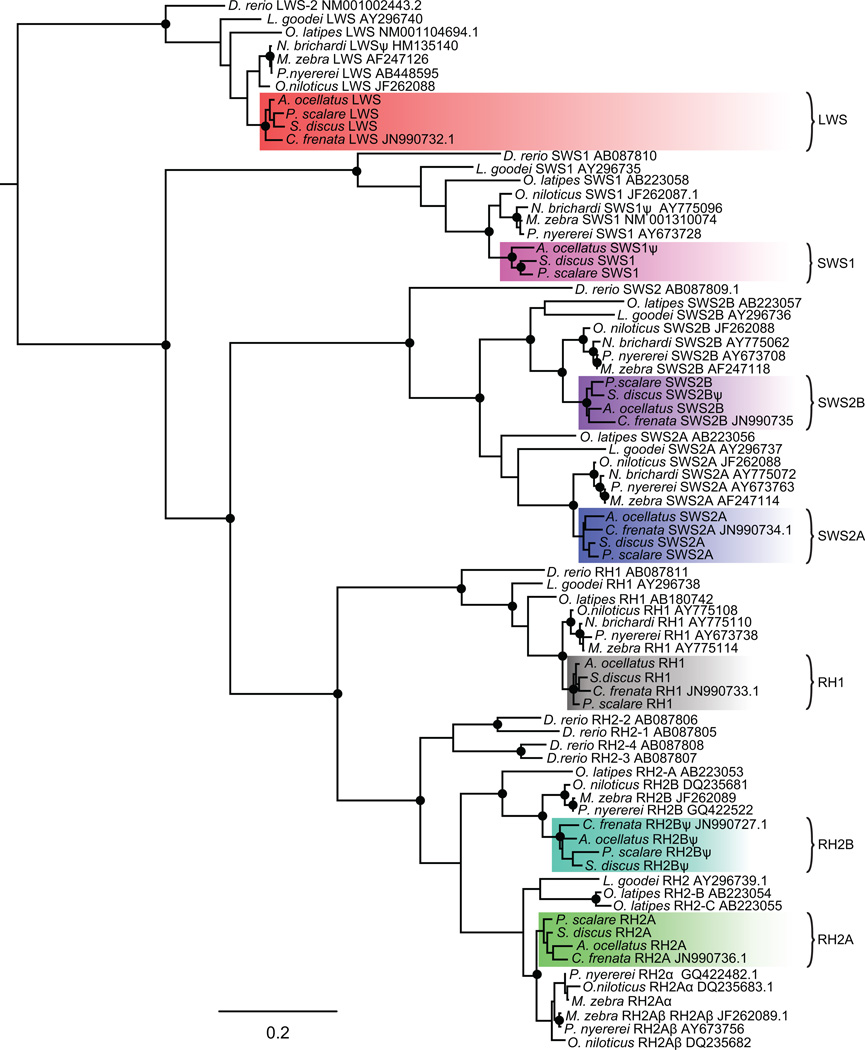
Maximum likelihood trees based on these three Amazonian species as well as from C. frenata, confirmed the identities of all South American cichlids opsins: SWS1, SWS2B, SWS2A, RH2B, RH2A, LWS and RH1. The cichlid opsin lineages are exclusively monophyletic between the New World and African lineages, with New World opsins placed as sister groups to the respective Nile Tilapia orthologs in all opsin classes. P. scalare and S. discus were placed together in the SWS1, SWS2B, SWS2A, and LWS opsin clades. Opsins of C. frenata were also nested within the South American clade (Fig. 1).
Figure 1.
Opsin maximum likelihood phylogenetic tree of Cichlids, Danio rerio, Oryzias latipes, and Lucania goodei. Black circles represent bootstrap support over 95%. Color shades indicate the Neotropical cichlid lineage. Ψ denotes pseudogenes.
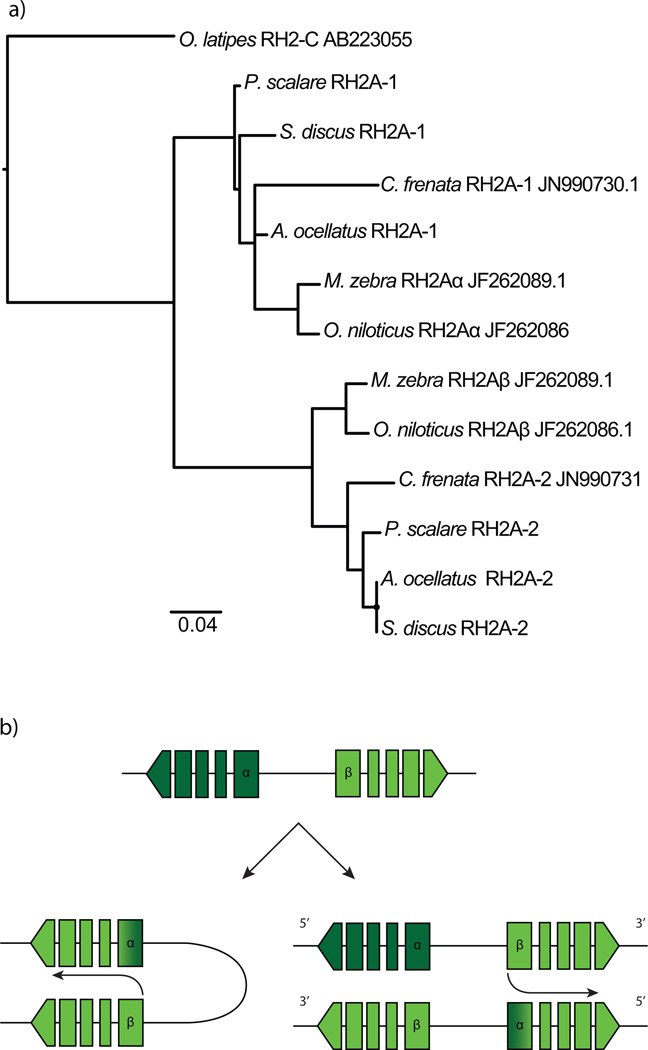
Genome and transcriptome analysis suggested that a gene duplication event generated two copies of RH2A (Fig. S6). Due to the high similarity of the genes, we were only able to obtain full sequences of one expressed RH2A in the RNAseq data. Within the genomes, we noticed that RH2A duplicates in each species were almost identical but had differences located mostly in the 5’UTR (~80bp) and in the first 150 bp of the coding sequence, indicative of gene conversion. Blasting the exons and performing phylogenetic analysis of only the first exons and 5’UTRs showed that these RH2A copies correspond, although with low support, to the African cichlid duplicates RH2Aα and RH2Aβ (Fig. 2a). In both copies, coding regions after 150 bp were highly similar to RH2Aβ (including the first introns confirmed by PCR, Fig. S7), thus, we believe gene conversion resulted in RH2Aα being replaced in large part by RH2Aβ (Fig. 2b).
Figure 2.
a) Phylogeny of upstream UTRs and first exons of the RH2A duplicates. b) Intrachromosomal and interchromosomal RH2A gene conversion scenario.
Gene Expression
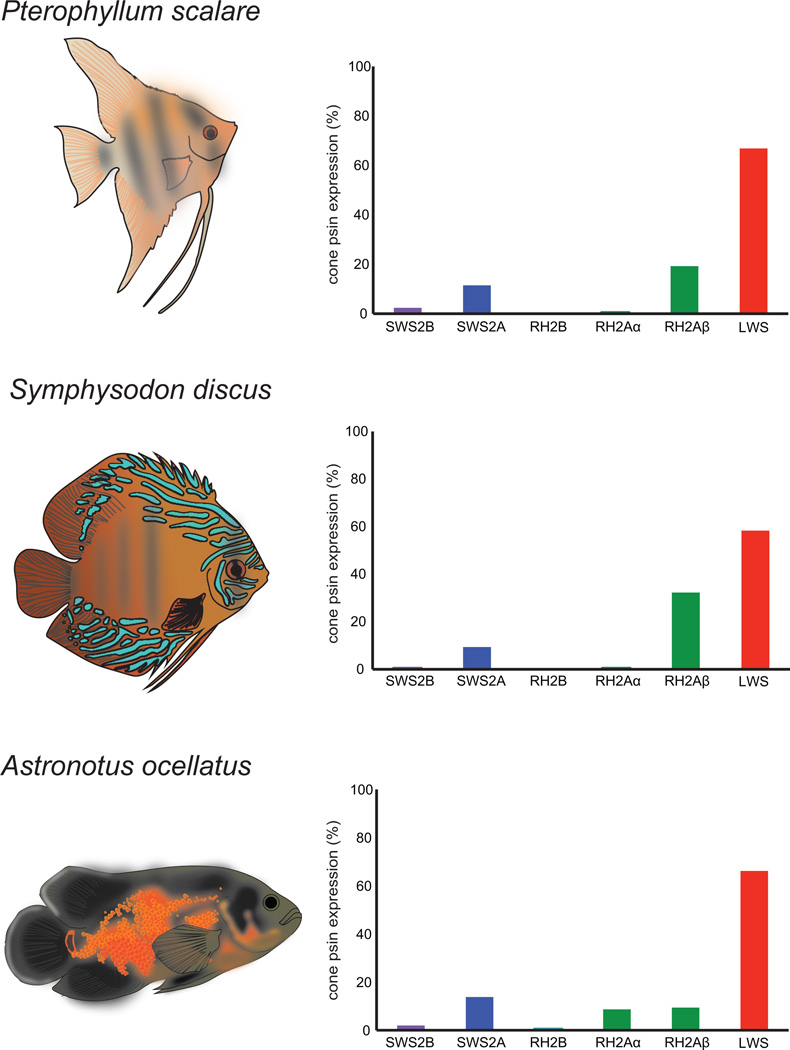
Color vision in Amazonian cichlids is based mainly on the expression of three cone opsin genes: SWS2A, RH2A, and LWS. The three species had similar expression profiles (Fig. 3). The most expressed short wavelength pigment was SWS2A whereas SWS2B accounted for less than 5% of expressed cone opsin in P. scalare, S. discus and A. ocellatus. The LWS opsin seems to be much more expressed accounting for more than 50% of expressed cone opsins. No SWS1 opsin was expressed. In A. ocellatus, only traces of RH2B were found. Interestingly, RH2Aα is lowly expressed (< 1%) in P. scalare and S. discus whereas RH2Aα and RH2Aβ are equally expressed in A. ocellatus. Pseudogenes were not expressed except for SWS2BΨ in S. discus and RH2BΨ in A. ocellatus accounting for less than 1% of all expressed opsins. RH1 was the most highly expressed visual pigment in the three species (Fig. S8).
Figure 3.
Relative cone opsin expression profiles of P. scalare, S. discus, and A. ocellatus. Drawings by D Escobar-Camacho.
Spectral tuning
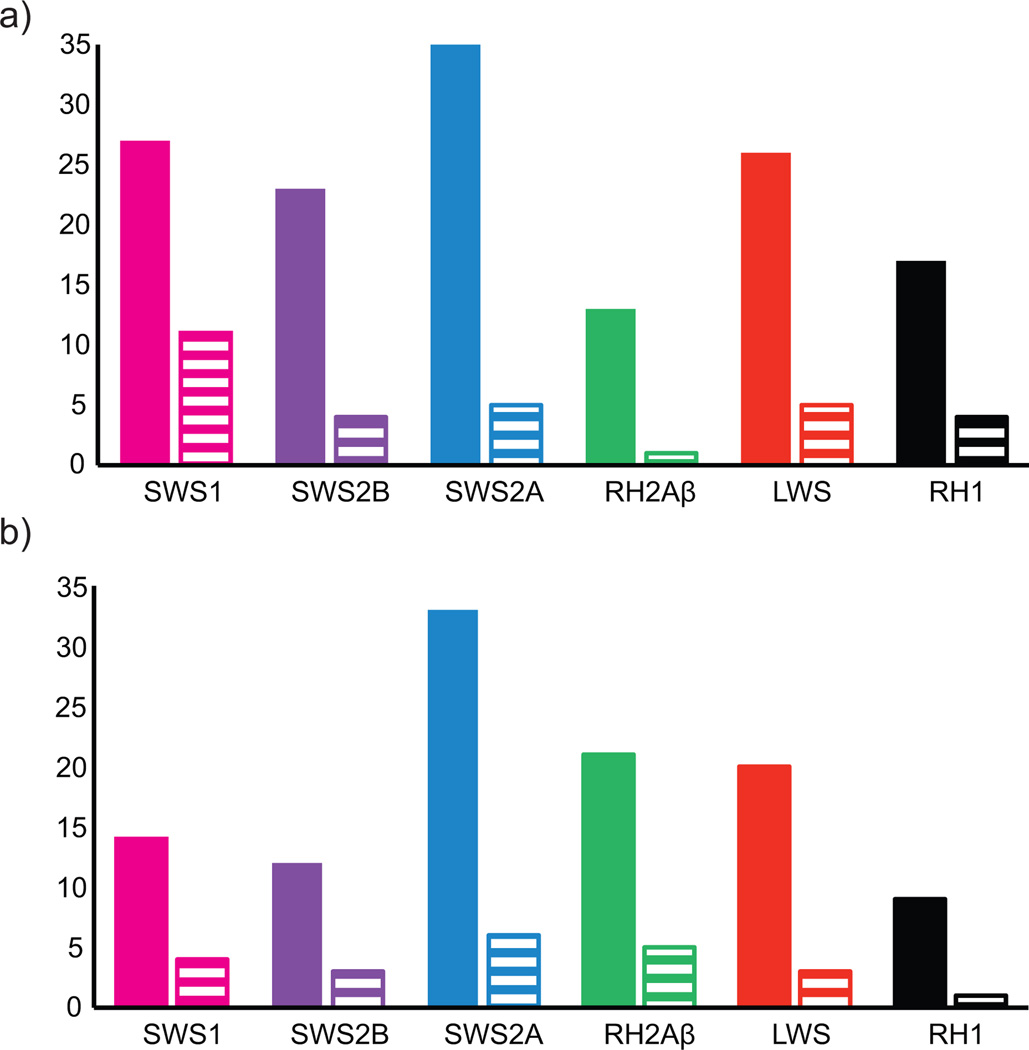
The analysis of Amazonian cichlid opsin sequences revealed evidence of changes that might impact on spectral tuning. There was great variation of amino acids substitutions between the three species in comparison to O. niloticus (Fig. 4a). The greatest diversity at transmembrane sites was in the SWS2A opsin, with 35 variable transmembrane sites with five of these in the retinal binding pocket (five polarity changes) and three (M44T, M116L, S292A) occurring in known tuning sites (Table 2) (Supplementary materials 2). SWS1 had the greatest diversity in potentially functional tuning sites with 27 variable transmembrane sites, eleven substitutions in retinal binding pocket sites (seven polarity changes) and one substitution occurring at a known tuning site, A118S. In S. discus the LWS opsin showed a λmax shifting substitution S164A, previously shown to cause a −7nm shift (Asenjo et al. 1994). SWS2B in P. scalare and A. ocellatus also exhibited three amino acid substitutions (F46V, A109G, A269T) in known tuning sites of which the A269T substitution is known to shift λmax by +6nm (Yokoyama & Tada 2003). Variable amino acid sites were also found between the three species and C. frenata with SWS2A having the most variable sites (Fig. 4b).
Figure 4.
Amino acid substitution variation for each opsin class of the three species. Filled bars represent amino acid variation in transmembrane regions of the opsin, striped bars are amino acid variation in the retinal binding pocket sites. a) Amino acid substitution variation between P. scalare, S. discus, A. ocellatus and to O. niloticus. b) Amino acid substitution variation between P. scalare, S.discus, A. ocellatus and C. frenata. SWS1 variation is based only on P. scalare and S. discus while SWS2B on P. scalare, A. ocellatus and C. frenata.
Table 2.
Summary of South American cichlids variation.
| SWS1 | SWS2B | SWS2A | RH2A | LWS | RH1 | |
|---|---|---|---|---|---|---|
| Total number of nucleotides | 1008 | 1059 | 1056 | 1059 | 1074 | 1065 |
| Total number of amino acids | 335 | 352 | 351 | 352 | 357 | 354 |
| Variable nt sites | 46 | 52 | 67 | 70 | 50 | 38 |
| Indels | 0 | 3 | 0 | 0 | 0 | 0 |
| Vs. O. niloticus | ||||||
| Variable transmembrane sites* | 27(12) | 23(4) | 35(12) | 13(3) | 26(7) | 17(4) |
| Functionally variable retinal binding pocket sites | 11(7) | 4(2) | 5(5) | 1(1) | 5(3) | 4(1) |
| Number of substitutions at known tuning sites | 1(1) | 3(1) | 3(2) | 0(0) | 1(1) | 2(0) |
| Synonymous substitution (ds) | 0.114 | 0.129 | 0.084 | 0.129 | 0.083 | 0.064 |
| Non-synonymous substitution (dn) | 0.024 | 0.024 | 0.032 | 0.022 | 0.017 | 0.012 |
| dn/ds | 0.210 | 0.186 | 0.380 | 0.170 | 0.204 | 0.187 |
| Between SA species | ||||||
| Variable transmembrane sites | 14(5) | 12(2) | 33(12) | 21(3) | 20(5) | 9(1) |
| Functionally variable retinal binding pocket sites | 4(1) | 3(2) | 6(4) | 5(1) | 3(1) | 1(0) |
| Number of substitutions at known tuning sites | 1(1) | 2(2) | 2(2) | 0(0) | 1(1) | 1(0) |
Parenthesis indicates number of amino acid substitutions that differ in polarity. Known tuning sites are based on (Hunt et al. 2001; Yokoyama 2008). Opsin genes are sorted by increasing spectral sensitivity.
Pseudogene divergence times
The time since opsins became pseudogenes (Tn) was estimated for both the SWS1 (A. ocellatus) and SWS2B (S. discus) pseudogenes. For SWS1Ψ in A. ocellatus, Tn values varied around −3.8 to 0.07 Mya when compared to functional genes of P. scalare and S. discus respectively; while for SWS2BΨ, S. discus, values were between 1.84 and 7.35 Mya compared to P. scalare and A. ocellatus (Table 3). Based on these values, it seems that SWS2BΨ appeared in the late Neogene (23-2.5 Mya) whereas SWS1Ψ may be very recent (Quaternary, 2.5 Mya to present).
Table 3.
Distances between sequences (dACi, dBCi), for nonsynonymous and synonymous sites (I = ns or s, as substitutions per site), rates of non-synonymous and synonymous substitutions (ai, in substitutions per site per year), and Time since pseudogenization (Tn given in My).
| Pseudogene Sequence A |
Sequence B | Sequence C | dACns | dACs | dBCns | dBCs | ans | as |
Tn (Mya) |
|---|---|---|---|---|---|---|---|---|---|
| SWS1 | |||||||||
| A. ocellatusΨ | P. scalare | O. niloticus | 0.042 | 0.480 | 0.034 | 0.452 | 4.50E-10 | 5.86E-9 | −3.80 |
| A. ocellatusΨ | S. discus | O. niloticus | 0.042 | 0.480 | 0.037 | 0.477 | 4.91E-10 | 6.18E-9 | 0.07 |
| SWS2B | |||||||||
| S. discusΨ | A. ocellatus | O. niloticus | 0.06 | 0.35 | 0.058 | 0.376 | 7.53E-10 | 4.87E-9 | 7.35 |
| S. discusΨ | P. scalare | O. niloticus | 0.06 | 0.35 | 0.053 | 0.348 | 6.98E-10 | 4.51E-9 | 1.84 |
Discussion
Opsin gene complements
Through genome and transcriptome sequencing analysis, we have characterized the genetic component of the visual system of three Amazonian cichlids. Genomic and PCR opsin evidence suggests that all three species have seven cone opsin genes and a rod opsin gene. This includes evidence for two separate copies of RH2A each with unique 5’UTRs that could be separately PCR amplified. This supports the idea that the common ancestor of the South American lineages had the same seven cone opsin genes found in the African cichlids, perhaps arising in the Gondwanan ancestor.
Our results show that present day Amazonian species exhibit a variable complement of functional opsins with different opsin genes inactivated in different lineages. Overall, these species exhibit a less rich palette of functional opsin genes than African cichlids (Carleton 2009). This is in concordance with previous results on the Trinidadian pike (Weadick et al. 2012) which has RH2B pseudogenized and a possible loss of SWS1. These results confirm that gene inactivation is common among percomorph fishes, thereby creating complex patterns of opsin evolution as adaptation to new spectral environments occur (Davies et al. 2012; Cortesi et al. 2015).
RH2A opsins appear to have undergone gene conversion where RH2Aβ converted RH2Aα with the recombination location in the middle of the first exon of the coding sequence. This conversion event seems to have happened early in the Neotropical cichlid lineage as it is shared across Amazonian species and C. frenata (Fig. 2). Gene conversion events can be frequent between adjacent opsin duplicates. This has been shown in primate L and M cone opsin genes where introns exhibit lower nucleotide divergence than exons (Zhao et al. 1998; Hiwatashi et al. 2011). Furthermore, gene conversion occurring between exons has been found on several teleost lineages (Owens et al. 2009; Rennison et al. 2012; Cortesi et al. 2015; Sandkam 2015). Our results suggest that the RH2A duplication was present early in the cichlid lineage prior to the breakup of Gondwana and the African – South American divergence. Furthermore, RH2 paralogs seem to have been present before the Cichlomorphae-Atherinomorphae split (Betancur-R et al. 2013) since O. latipes’ RH2 opsins (RH2-A, RH2-B, RH2-C) are grouped as sister groups in cichlids’ RH2B and RH2A clades (Fig. 1). This agrees with previous studies suggesting that opsin gene duplications promote visual pigment diversity in teleosts (Chinen et al. 2003; Spady et al. 2006; Matsumoto et al. 2006; Hofmann & Carleton 2009). Further studies on opsin genes of more divergent cichlids from India and Madagascar are needed to analyze the evolution of RH2 opsins.
In the opsin gene trees, Neotropical cichlid opsins were placed as sister taxa to the African cichlid clade as expected. There is general phylogenetic concordance of the taxonomic relationships across the opsin genes. P. scalare and S. discus are often paired among opsins clades (SWS1, SWS2B, SWS2A, RH2B, LWS), whereas A. ocellatus and C. frenata are placed as sister taxa (SWS2B, SWS2A, RH2A). This is in agreement with Neotropical cichlid phylogenetic studies where P. scalare and S. discus shared a more recent common ancestor than A. ocellatus (López-Fernández et al. 2010). This pattern is not consistent in RH2A and RH1, however, these clades exhibit low support.
Opsin gene expression
In spite of the diversity in genomic opsin complements, all species expressed the same three cone opsins: SWS2A, RH2A and LWS. A reliance on three distinct cone opsins is consistent with previous studies that found a typical cone mosaic arrangement in A. ocellatus where one single cone was surrounded by four double (twin) cones (Hibbard 1971; Braekevelt 1992). The expression profile of P. scalare, S. discus and A. ocellatus is indicative of a long-wavelength oriented visual system which is characteristic of cichlids living in murky and riverine habitats (Halstenberg et al. 2005; Carleton et al. 2008; Hofmann et al. 2009). This agrees with the light environment of Neotropical rivers, particularly in the Amazon, that have very long wavelength transmission properties. In the Amazon basin, short and medium wavelengths are scattered/absorbed by colored dissolved organic matter, suspended inorganic particles and phytoplankton resulting in a red shifted light environment. Light transmission can vary according to the type of water (white, black, and clear waters) and hydrological cycles (receding and rising) (Muntz 1982; Costa et al. 2012). Overall, Amazonian rivers exhibit a downwelling irradiance peak (λmax) beyond 650nm resulting from high attenuation for blue and green light, and high reflectance for red (Fig. S9) (Martinez et al. 2004; Rudorff et al. 2006; Costa et al. 2012). P. scalare, S. discus and A. ocellatus have been documented throughout the Amazon basin inhabiting white, black and clear waters (Farias & Hrbek 2008; Albert & Reis 2011; Froese & Pauly 2016). Furthermore, visual studies on Amazonian fishes including P. scalare and A. ocellatus, have shown that they had yellow filters in the lenses and cornea which would filter out short wavelength light (Muntz 1973, 1982). Because there is little short wavelength light transmission in Amazonian rivers and yellow pigments in cichlid eyes filter short wavelengths, we suggest that the aquatic environment in the Amazon basin has influenced the visual system adaptation of Neotropical cichlid retinas, inactivating short wavelength sensitive opsin genes (SWS1, SWS2B, RH2B) and shifting their opsin expression profile to the long wavelength palette.
The expression of SWS2A, RH2A and LWS is consistent with previous microspectrophotometry (MSP) studies of wavelength sensitivities in Neotropical cichlids (Levine & MacNichol 1979; Kröger et al. 1999; Weadick et al. 2012) (Table S2). Based on these studies, we estimate a spectral sensitivity for SWS2A between 450 to 480 nm. Similarly, for RH2A and LWS we suggest values between 530–555 nm and 560–617 nm respectively. Finally, rods sensitivity should lie around 500–525 nm. This spectral sensitivity variation may include a significant effect from the type of chromophore (A1-A2).(Levine & MacNichol 1979) The effects of changing from A1 to A2 chromophore can result in modest 15–30 nm shifts for short to medium wavelength pigments, but shifts up to 60nm in λmax for long wavelength pigments (Hárosi 1994; Parry & Bowmaker 2000; Carleton et al. 2008) Similarly, spectral sensitivities of Neotropical cichlids lie within this range suggesting different A1-A2 combinations (Table S2, Fig. S9). There are no records of the peak absorbance of the shortest wavelength sensitive pigments, SWS1 and SWS2B, yet we would expect sensitivities similar to their African counterparts between 360–378 nm for SWS1 and 415–425 nm for SWS2B (Parry et al. 2005; Spady et al. 2006). However, since Amazonian rivers have little downwelling light below 450nm, these opsins would not be sensitive to the available light and therefore need not be expressed (Fig. S9) (Costa et al. 2012).
In cichlids, opsin gene expression can change through development and species can differ in their ontogenetic profiles with shorter wavelength genes expressed earlier (SWS1, SWS2B, RH2B) followed by longer wavelength genes (SWS2A, RH2A and LWS) (Carleton et al. 2008). Specimens used for our transcriptome analyses were aquarium trade juveniles raised in fluorescent light. This could have influenced their opsin expression profile by increasing the expression of longer wavelength sensitive opsins. However, fish were at least five months old and also exhibited yellow lenses. Since changes in gene expression stabilize by around six months in African cichlids, our results suggest that either these fish were old enough to obtain the adult expression pattern, or that they do not undergo developmental shifts in expression (Carleton et al. 2008; Sabbah et al. 2012). The latter would be consistent with some species having lost the SWS1 and SWS2B opsins that are normally expressed in the larval to juvenile stages.
Opsin sequence variation
Studies in African cichlids have found that the short and the long wavelength sensitive pigments are the most variable with shifts in peak sensitivity of 30 nm in the short, and 50 nm in the long wavelength sensitive opsins (Carleton et al. 2008) (Table S2). This is concordant with the diversity of total amino acid substitutions in SWS2A and LWS gene sequences found in our results. Opsin sequence analysis showed that the greatest variation in amino acid substitution, and polarity changes, in the retinal binding pocket sites, was in SWS2A followed by SWS1 and LWS (Fig. 4a). These results differ from previous studies that have found the greatest variation in opsins sensitive at both ends of the wavelength spectrum, SWS1 and LWS (Hofmann et al. 2009, 2012; Phillips et al. 2015). Since the Amazon basin exhibit a long wavelength light environment, the SWS2A gene may be the shortest wavelength gene to be expressed. Changes in SWS2A might be the result of strong selection to enhance sensitivity where functional amino acids substitution would shift the λmax of SWS2A to longer wavelengths. In this way, the visual system may still be optimizing the shortest (SWS2A) and longest (LWS) opsins that are relevant to the long wavelength shifted environments where these fish are located. Indeed, divergent selective pressures driven by ecological and/or biogeographic differences have been suggested for Neotropical cichlids’ and anchovies visual pigments (Schott et al. 2014; Nynatten et al. 2015; Torres-Dowdall et al. 2015). In addition to amino acid substitutions adapting spectral sensitivities over the long term, shifts in A1-A2 use could also have great impact on λmax possibly enabling cichlids to adapt to shifts in the light environment over a shorter time scale. Chromophore shifts are known to occur in just a few weeks to provide seasonal adjustments (Munz & McFarland 1977). Indeed, Muntz suggested that Amazonian fishes use mixtures of both chromophores (Muntz 1973, 1982).
The pseudogenization time of SWS1Ψ and SWS2BΨ in A. ocellatus and S. discus, dates back to the late Neogene and the Quaternary. During the Neogene (~7 Mya), the modern Amazon river system, including the present-day configuration of white, clear and black water, had already come into place. Nevertheless, geological shifts, including the Andean uplift (12-4 Mya) on the western lowland and continuous marine incursions until the Pleistocene, played a role in habitat fragmentation and greatly influenced diversification of the Amazon biota (Hubert & Renno 2006; Albert & Reis 2011; Turchetto-Zolet et al. 2013). Furthermore, the glacio-eustatic oscillations in the Quaternary (<2.5 Mya) dynamically altered and reorganized river courses and watersheds resulting in both isolated and expanded fish populations (Albert & Reis 2011). Consequently, the changing Amazon conditions over the last 7 Mya could have selected for maintenance of expression of some opsins and pseudogenization of others. Although we were not able to date the pseudogenization time for RH2B, it is possible that it might have been inactivated multiple times across lineages. This is supported by the SWS1 inactivation in African cichlids (Neolamprologus brichardi and N. mondabu from Lake Tanganyka (O’Quin et al. 2010)) and Neotropical cichlids which arose independently. More species need to be analyzed in order to better understand the pseudogenization process. Yet it is interesting that in spite of opsin genes have been inactivated at different times in different South American lineages to generate the same expressed opsin set. This suggests that this expressed gene set arose convergently in Amazonian cichlids (Fig 5).
Figure 5.
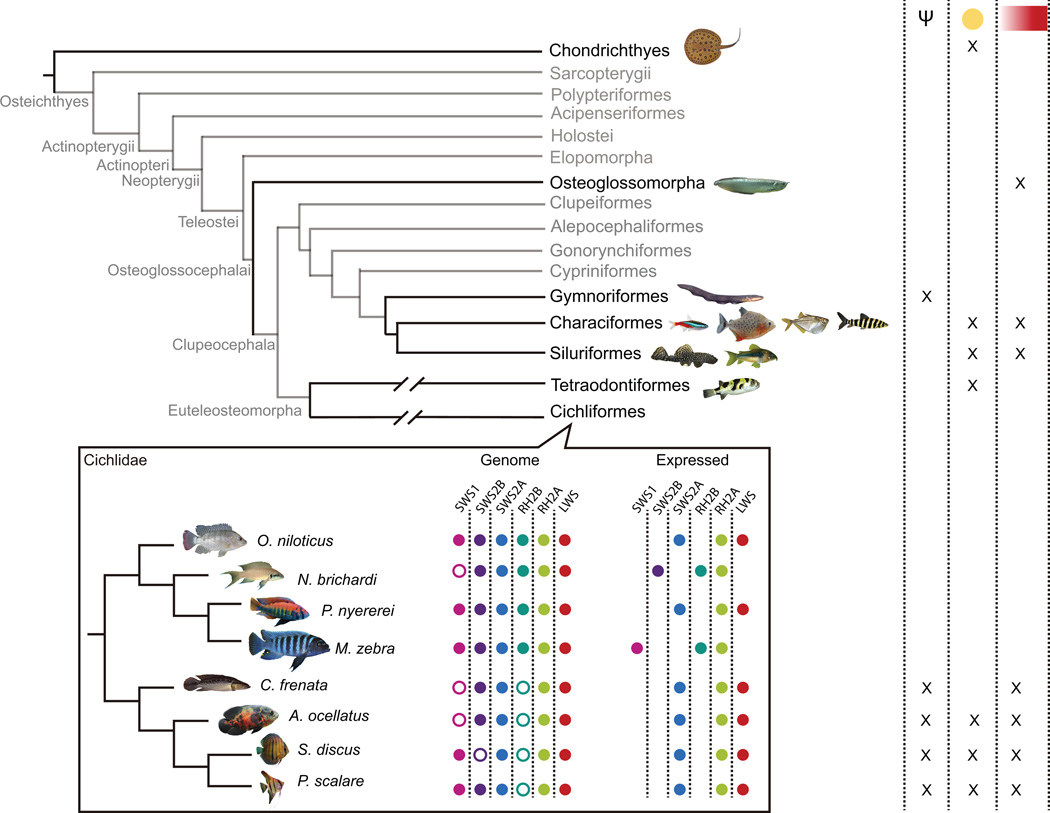
Schematic representation of visual system adaptation in a phylogenetic context. Phylogenetic tree topology is based on Betancur et al (2013). Taxonomic groups present in the Amazon where visual systems have been analyzed at some level are displayed in black. Based on previous and the present study, an “x” denotes the presence of pseudogenes or lost genes (Ψ), yellow pigments in ocular media (yellow circle), or long wavelength spectral sensitivities, either by opsin expression or MSP, (red rectangle) in a specific group. The cichlid inset denotes the opsin genes in the genome and the ones that are being expressed in the Neotropical and African lineages. Empty circles denote pseudogenes or lost genes. For simplicity, RH2A duplicates were excluded.
Ecological adaptation
Our results suggest that these Amazonian cichlids are adapting to their light environment through several different genetic mechanisms. These include changes in opsin gene expression, opsin gene sequence and the accumulation of pseudogenes relative to their African counterparts. This is further supported by yellow filters in their ocular media which filter short wavelengths, reducing the background scattered light common in these long wavelength transmitting waters. These traits are shared among other teleost lineages found in the Amazon and may be a signature of adaptation to the Amazon’s murky environment. The absence of short wavelength visual pigments seems to be common among the Amazonian ichthyofauna with a number of species having spectral sensitivities that are red shifted (Table S2). Gene loss or pseudogenization also occur in other Amazonian taxa such as the absence of the short wavelength sensitive opsin genes in the electric eel (Electrophorus electricus) and non-electric catfish (Traeger et al. 2015). Furthermore, convergent opsin inactivation is common in mammals where opsin pseudogenes have arisen independently in response to changes in their behavior and ecology (Zhao et al. 2009; Meredith et al. 2013). Additionally, adaptations such as the presence of yellow filters in ocular media have been found in Amazonian fishes besides cichlids, such as the pink-tailed chalceus (Characiformes: Chalceus macrolepidotus), the freshwater puffer fish (Tetraodontiformes: Colomessus asellus), the freshwater stingray (Myliobatiformes: Paratrygon motoro) and several other siluriforms (Fig. 5) (Muntz 1973, 1982; Muntz et al. 1973).
In conclusion, we have described the opsin complement of three Amazonian cichlids using both RNA and genomic sequences. There is evidence for visual pigment evolution in this lineage with both opsin gene pseudogenization and gene conversion taking place. This might be a consequence of the long wavelength light environment in the Amazon basin. This environment has further influenced cichlid visual system adaptation through adaptation of opsin gene expression, changes in amino acids substitution in spectral tuning sites, and yellow filters in ocular media; all traits characteristic of species living in a long wavelength environment. These traits likely arose convergently in response to environmental selection and seem to be shared among a number of Amazonian fishes. The molecular adaptive traits discussed in this study corroborates the vast body of vertebrate research where it has been shown that as animals occupy different ecological niches, their visual systems adapts through several mechanisms, enabling them to operate in these new spectral environments.
Supplementary Material
Acknowledgments
We thank the University of Maryland Institute for Bioscience & Biotechnology Research for sequencing the libraries and Ian Misner for helping with the Trinity pipeline. We also thank Carlos Schneider for his collaboration in collecting samples, Jessica Goodheart for advise in phylogenetic trees, and Fabio Cortesi for help with identifying gene duplications through read mapping. We thank three anonymous reviewers for providing helpful suggestions on this manuscript. This work was supported by the National Institute of Health (R01EY024693 to K.L.C.), the Center for Studies of Adaptation to Environmental Changes in the Amazon (INCT ADAPTA, FAPEAM/CNPq 573976/2008-2 to C.M.), Goldhaber Travel Award, International Conference Support Award (ICSSA), Biology Department Travel Award and Summer Research Fellowship through the University of Maryland Graduate School (2015–2016 to D.E-C.). D.E-C. is supported by graduate fellowship of the Secretariat of Higher Education, Science, Technology and Innovation of Ecuador (2014-AR2Q4465).
Footnotes
Data Accessibility
Genbank accessions: KX382915-57, KY082759-61. SRA database: SRR4841690-92. DRYAD: doi:10.5061/dryad.1h272.
Author Contributions
Designed research (DEC, ER, CM and KC), analyzed data including assembling the genomes (ER, CM) and transcriptomes (DEC, KC) and wrote the paper (DEC, ER, CM and KC).
References
- Albert JS, Reis RE. Historical Biogeography of Neotropical Freshwater Fishes. Berkeley, Los Angeles, London: University of California Press; 2011. [Google Scholar]
- Asenjo AB, Rim J, Oprian DD. Molecular determinants of human red/green color discrimination. Neuron. 1994;12:1131–1138. doi: 10.1016/0896-6273(94)90320-4. [DOI] [PubMed] [Google Scholar]
- Bazin AL, Zwickl DJ, Cummings MP, et al. A Gateway for Phylogenetic Analysis Powered by Grid Computing Featuring GARLI 2.0. Systematic biology. 2014;63:812–818. doi: 10.1093/sysbio/syu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Research. 2005;33:34–38. doi: 10.1093/nar/gki063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancur-R R, Broughton R, Wiley E, Carpenter K. The tree of life and a new classification of bony fishes. PLoS Currents Tree of Life. 2013 doi: 10.1371/currents.tol.53ba26640df0ccaee75bb165c8c26288. 0732988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradnam KR, Fass JN, Alexandrov A, et al. Assemblathon 2: evaluating de novo methods of genome assembly in three vertebrate species. GigaScience. 2013;2:10. doi: 10.1186/2047-217X-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braekevelt CR. Anatomy and Embryology in the velvet cichlid (Astronotus ocellatus) Anatomy and embryology. 1992;186:363–370. doi: 10.1007/BF00185986. [DOI] [PubMed] [Google Scholar]
- Carleton K. Cichlid fish visual systems: mechanisms of spectral tuning. Integrative zoology. 2009;4:75–86. doi: 10.1111/j.1749-4877.2008.00137.x. [DOI] [PubMed] [Google Scholar]
- Carleton KL, Dalton BE, Escobar-Camacho D, Nandamuri SP. Proximate and ultimate causes of variable visual sensitivities: Insights from cichlid fish radiations. Genesis. 2016;54:299–325. doi: 10.1002/dvg.22940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carleton KL, Spady TC, Cote RH. Rod and cone opsin families differ in spectral tuning domains but not signal transducing domains as judged by saturated evolutionary trace analysis. Journal of molecular evolution. 2005;61:75–89. doi: 10.1007/s00239-004-0289-z. [DOI] [PubMed] [Google Scholar]
- Carleton KL, Spady TC, Streelman JT, et al. Visual sensitivities tuned by heterochronic shifts in opsin gene expression. BMC biology. 2008;6:22. doi: 10.1186/1741-7007-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikhi R, Medvedev P. Informed and automated k-mer size selection for genome assembly. Bioinformatics. 2014;30:31–37. doi: 10.1093/bioinformatics/btt310. [DOI] [PubMed] [Google Scholar]
- Chinen A, Hamaoka T, Yamada Y, Kawamura S. Gene duplication and spectral diversification of cone visual pigments of zebrafish. Genetics. 2003;163:663–675. doi: 10.1093/genetics/163.2.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortesi F, Musilová Z, Stieb SM, et al. Ancestral duplications and highly dynamic opsin gene evolution in percomorph fishes. Proceedings of the National Academy of Sciences. 2015;112:1493–1498. doi: 10.1073/pnas.1417803112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa MPF, Novo EMLM, Telmer KH. Spatial and temporal variability of light attenuation in large rivers of the Amazon. Hydrobiologia. 2012;702:171–190. [Google Scholar]
- Dalton BE, Lu J, Leips J, Cronin TW, Carleton KL. Variable light environments induce plastic spectral tuning by regional opsin coexpression in the African cichlid fish, Metriaclima zebra. Molecular Ecology. 2015;24:4193–4204. doi: 10.1111/mec.13312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies WIL, Collin SP, Hunt DM. Molecular ecology and adaptation of visual photopigments in craniates. Molecular Ecology. 2012;21:3121–3158. doi: 10.1111/j.1365-294X.2012.05617.x. [DOI] [PubMed] [Google Scholar]
- Endler JA. Some general comments on the evolution and design of animal communication systems. Philosophical transactions of the Royal Society of London. Series B, Biological sciences. 1993;340:215–225. doi: 10.1098/rstb.1993.0060. [DOI] [PubMed] [Google Scholar]
- Escobar-Camacho D, Ramos E, Martins C, Carleton KL. Data from: The Opsin genes of Three Amazonian Cichlids. Dryad Digital Repository. 2016 [Google Scholar]
- Farias IP, Hrbek T. Patterns of diversification in the discus fishes (Symphysodon spp. Cichlidae) of the Amazon basin. Molecular phylogenetics and evolution. 2008;49:32–43. doi: 10.1016/j.ympev.2008.05.033. [DOI] [PubMed] [Google Scholar]
- Froese R, Pauly D. FishBase. World Wide Web electronic publication; 2016. [Google Scholar]
- Fuller RC, Carleton KL, Fadool JM, Spady TC, Travis J. Genetic and environmental variation in the visual properties of bluefin killifish, Lucania goodei. Journal of Evolutionary Biology. 2005;18:516–523. doi: 10.1111/j.1420-9101.2005.00886.x. [DOI] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nature protocols. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstenberg S, Lindgren KM, Samagh SPS, et al. Diurnal rhythm of cone opsin expression in the teleost fish Haplochromis burtoni . Visual neuroscience. 2005;22:135–141. doi: 10.1017/S0952523805222022. [DOI] [PubMed] [Google Scholar]
- Hárosi FI. An analysis of two spectral properties of vertebral visual pigments. Vision Research. 1994;34:1359–1367. doi: 10.1016/0042-6989(94)90134-1. [DOI] [PubMed] [Google Scholar]
- Hibbard E. Grid patterns in the retinal organization of the cichlid fish Astronotus ocellatus. Experimental Eye Research. 1971;12:175–180. doi: 10.1016/0014-4835(71)90087-x. [DOI] [PubMed] [Google Scholar]
- Hiwatashi T, Mikami A, Katsumura T, et al. Gene conversion and purifying selection shape nucleotide variation in gibbon L/M opsin genes. BMC evolutionary biology. 2011;11:312. doi: 10.1186/1471-2148-11-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann CM, Carleton KL. Gene duplication and differential gene expression play an important role in the diversification of visual pigments in fish. Integrative and Comparative Biology. 2009;49:630–643. doi: 10.1093/icb/icp079. [DOI] [PubMed] [Google Scholar]
- Hofmann CM, Marshall NJ, Abdilleh K, et al. Opsin evolution in damselfish: convergence, reversal, and parallel evolution across tuning sites. Journal of molecular evolution. 2012;75:79–91. doi: 10.1007/s00239-012-9525-0. [DOI] [PubMed] [Google Scholar]
- Hofmann CM, O’Quin KE, Justin Marshall N, et al. The eyes have it: Regulatory and structural changes both underlie cichlid visual pigment diversity. PLoS Biology. 2009;7 doi: 10.1371/journal.pbio.1000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann CM, O’Quin KE, Smith AR, Carleton KL. Plasticity of opsin gene expression in cichlids from Lake Malawi. Molecular Ecology. 2010;19:2064–2074. doi: 10.1111/j.1365-294X.2010.04621.x. [DOI] [PubMed] [Google Scholar]
- Hubert N, Renno J-F. Historical biogeography of South American freshwater fishes. Journal of Biogeography. 2006;33:1414–1436. [Google Scholar]
- Hunt DM, Dulai KS, Partridge JC, Cottrill P, Bowmaker JK. The molecular basis for spectral tuning of rod visual pigments in deep-sea fish. The Journal of experimental biology. 2001;204:3333–3344. doi: 10.1242/jeb.204.19.3333. [DOI] [PubMed] [Google Scholar]
- Jukes TH, Cantor CR. Evolution of protein molecules. In: Munro HN, editor. Mammalian Protein Metabolism, III. New York: Academic Press Inc; 1969. pp. 21–132. [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic acids research. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kröger RHH, Bowmaker JK, Wagner HJ. Morphological changes in the retina of Aequidens pulcher (Cichlidae) after rearing in monochromatic light. Vision Research. 1999;39:2441–2448. doi: 10.1016/s0042-6989(98)00256-9. [DOI] [PubMed] [Google Scholar]
- Levine JS, MacNichol EF. Visual Pigments in Teleost Fishes: Effects of Habitat, Microhabitat, and Behavior on Visual System Evolution. Sensory Processes. 1979;3:95–131. [PubMed] [Google Scholar]
- Li W-H, Gojobori T, Nei M. Pseudogenes as a paradigm of neutral evolution. Nature. 1981;292:237–239. doi: 10.1038/292237a0. [DOI] [PubMed] [Google Scholar]
- López-Fernández H, Winemiller KO, Honeycutt RL. Multilocus phylogeny and rapid radiations in Neotropical cichlid fishes (Perciformes: Cichlidae: Cichlinae) Molecular Phylogenetics and Evolution. 2010;55:1070–1086. doi: 10.1016/j.ympev.2010.02.020. [DOI] [PubMed] [Google Scholar]
- Martinez J, Seyler F, Bourgoin LM, Guyot JL. Proc. of the 2004 Envisat & ERS Symposium. 2004. Amazon Basin Water Quality Monitoring Using Meris and Modis Data; pp. 1–10. [Google Scholar]
- Matsumoto Y, Fukamachi S, Mitani H, Kawamura S. Functional characterization of visual opsin repertoire in Medaka (Oryzias latipes) Gene. 2006;371:268–278. doi: 10.1016/j.gene.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Matsumoto T, Ishibashi Y. Sequence analysis and expression patterns of opsin genes in the longtooth grouper Epinephelus bruneus. Fisheries Science. 2016;82:17–27. [Google Scholar]
- Meredith RW, Gatesy J, Emerling CA, York VM, Springer MS. Rod Monochromacy and the Coevolution of Cetacean Retinal Opsins. PLoS Genetics. 2013;9 doi: 10.1371/journal.pgen.1003432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntz WRA. Yellow filters and the absorption of light by the visual pigments of some amazonian fishes. Vision Research. 1973;13:2235–2254. doi: 10.1016/0042-6989(73)90225-3. [DOI] [PubMed] [Google Scholar]
- Muntz WRA. Visual Adaptations to Different light environments in Amazonian Fishes. Rev. Can. Biol. Experiment. 1982;41:35–46. [PubMed] [Google Scholar]
- Muntz WRA, Church E, Dartnall HJA. Visual Pigment of the Freshwater Stingray, Paratrygon motoro. Nature. 1973;246:517–517. doi: 10.1038/246517a0. [DOI] [PubMed] [Google Scholar]
- Munz FW, McFarland WN. Evolutionary Adaptations of Fishes to the photic Environment. In: Crescitelli F, editor. The visual system in vertebrates. New York: Springer-Verlag; 1977. pp. 193–274. [Google Scholar]
- Nynatten A Van, Bloom D, Chang BSW, Lovejoy NR, Lovejoy NR. Out of the blue?: adaptive visual pigment evolution accompanies Amazon invasion. Biology Letters. 2015;11:20150349. doi: 10.1098/rsbl.2015.0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Quin KE, Hofmann CM, Hofmann Ha, Carleton KL. Parallel Evolution of opsin gene expression in African cichlid fishes. Molecular Biology and Evolution. 2010;27:2839–2854. doi: 10.1093/molbev/msq171. [DOI] [PubMed] [Google Scholar]
- Owens GL, Windsor DJ, Mui J, Taylor JS. A fish eye out of water: Ten visual opsins in the four-eyed fish, Anableps anableps. PLoS ONE. 2009;4:1–7. doi: 10.1371/journal.pone.0005970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Korf I. CEGMA: A pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- Parry JWL, Bowmaker JK. Visual pigment reconstitution in intact goldfish retina using synthetic retinaldehyde isomers. Vision Research. 2000;40:9–15. doi: 10.1016/s0042-6989(00)00101-2. [DOI] [PubMed] [Google Scholar]
- Parry JWL, Carleton KL, Spady T, et al. Mix and match color vision: tuning spectral sensitivity by differential opsin gene expression in Lake Malawi cichlids. Current biology?: CB. 2005;15:1734–1739. doi: 10.1016/j.cub.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Phillips GAC, Carleton KL, Marshall NJ. Multiple Genetic Mechanisms Contribute to Visual Sensitivity Variation in the Labridae. Molecular Biology and Evolution. 2015 Oct;:1–15. doi: 10.1093/molbev/msv213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennison DJ, Owens GL, Taylor JS. Opsin gene duplication and divergence in ray-finned fish. Molecular Phylogenetics and Evolution. 2012;62:986–1008. doi: 10.1016/j.ympev.2011.11.030. [DOI] [PubMed] [Google Scholar]
- Rudorff CM, Novo EMLM, Galvão LS. Spectral mixture analysis for water quality assessment over the Amazon floodplain using Hyperion / EO-1 images. Revista Ambi-Agua. 2006;1:65–79. [Google Scholar]
- Sabbah S, Hui J, Hauser FE, Nelson Wa, Hawryshyn CW. Ontogeny in the visual system of Nile tilapia. The Journal of experimental biology. 2012;215:2684–2695. doi: 10.1242/jeb.069922. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russel DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Sandkam BA. Beauty in the Eyes of the Beholders: Colour Vision and Mate Choice in the Family poeciliidae. Simon Fraser University; 2015. [Google Scholar]
- Sandkam BA, Young CM, Margaret F, et al. Color vision varies more among populations than among species of live-bearing fish from South America. BMC Evolutionary Biology. 2015;15:1–11. doi: 10.1186/s12862-015-0501-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawakuchi HO, Bastviken D, Sawakuchi AO, et al. Oxidative mitigation of aquatic methane emissions in large Amazonian rivers. Global Change Biology. 2016;22:1075–1085. doi: 10.1111/gcb.13169. [DOI] [PubMed] [Google Scholar]
- Schott RK, Refvik SP, Hauser FE, López-Fernández H, Chang BSW. Divergent positive selection in rhodopsin from lake and riverine cichlid fishes. Molecular biology and evolution. 2014;31:1149–1165. doi: 10.1093/molbev/msu064. [DOI] [PubMed] [Google Scholar]
- Spady TC, Parry JWL, Robinson PR, et al. Evolution of the cichlid visual palette through ontogenetic subfunctionalization of the opsin gene arrays. Molecular Biology and Evolution. 2006;23:1538–1547. doi: 10.1093/molbev/msl014. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Dowdall J, Henning F, Elmer KR, Meyer A. Ecological and lineage specific factors drive the molecular evolution of rhodopsin in cichlid fishes. Molecular Biology and Evolution. 2015;32:2876–2882. doi: 10.1093/molbev/msv159. [DOI] [PubMed] [Google Scholar]
- Traeger LL, Volkening JD, Moffett H, et al. Unique patterns of transcript and miRNA expression in the South American strong voltage electric eel (Electrophorus electricus) BMC Genomics. 2015;16:243. doi: 10.1186/s12864-015-1288-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchetto-Zolet AC, Pinheiro F, Salgueiro F, Palma-Silva C. Phylogeographical patterns shed light on evolutionary process in South America. Molecular ecology. 2013;22:1193–1213. doi: 10.1111/mec.12164. [DOI] [PubMed] [Google Scholar]
- Weadick CJ, Loew ER, Rodd FH, Chang BSW. Visual pigment molecular evolution in the Trinidadian pike cichlid (Crenicichla frenata): a less colorful world for neotropical cichlids? Molecular biology and evolution. 2012;29:3045–3060. doi: 10.1093/molbev/mss115. [DOI] [PubMed] [Google Scholar]
- Yokoyama S. Evolution of dim-light and color vision pigments. Annual review of genomics and human genetics. 2008;9:259–282. doi: 10.1146/annurev.genom.9.081307.164228. [DOI] [PubMed] [Google Scholar]
- Yokoyama S, Tada T. The spectral tuning in the short wavelength-sensitive type 2 pigments. Gene. 2003;306:91–98. doi: 10.1016/s0378-1119(03)00424-4. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Research. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Hewett-Emmett D, Li WH. Frequent gene conversion between human red and green opsin genes. J Mol Evol. 1998;46:494–496. doi: 10.1007/pl00013147. [DOI] [PubMed] [Google Scholar]
- Zhao H, Rossiter SJ, Teeling EC, et al. The evolution of color vision in nocturnal mammals. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:8980–8985. doi: 10.1073/pnas.0813201106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwickl DJ. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. 2006 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.