Abstract
Purpose
We aimed to detect, map and quantify a novel nuclear Overhauser enhancement (NOE)-mediated magnetization transfer (MT) with water at around −1.6 ppm (NOE(−1.6)) in rat brain using MRI.
Methods
Continuous wave MT sequences with a variety of RF irradiation powers were optimized to achieve the maximum contrast of this NOE(−1.6) effect at 9.4 T. The distribution of effect magnitudes, resonance frequency offsets, and line widths in healthy rat brains and the differences of the effect between tumors and contralateral normal brains were imaged and quantified using a multi-Lorentzian fitting method. MR measurements on reconstituted model phospholipids as well as two cell lines (HEK293 and 9L) were also performed to investigate the possible molecular origin of this NOE.
Results
Our results demonstrate that the NOE(−1.6) effect can be reliably detected in rat brain. Pixel-wise fittings demonstrate the regional variations of the effect. Measurements on rodent tumor model show that the amplitude of NOE(−1.6) in brain tumor is significantly diminished compared with that in normal brain tissue. Measurements on reconstituted phospholipids suggest that this effect may originate from choline phospholipids.
Conclusion
The NOE(−1.6) could be used as a new biomarker for the detection of brain tumor.
Keywords: MRI, magnetization transfer (MT), chemical exchange saturation transfer (CEST), nuclear Overhauser enhancement (NOE), brain tumor
INTRODUCTION
Magnetization transfer (MT) between tissue water and various metabolites and macromolecules has been widely exploited to produce useful contrast in MR images (1-8). Conventionally, an off-resonance RF irradiation is applied to saturate targeted protons with specific chemical shifts, and the bulk water signal is subsequently affected by ‘magnetic coupling’ including chemical exchange and/or dipolar coupling with these saturated nuclei. By measuring the attenuated water signal as a function of the irradiation frequency, an MT Z-spectrum is obtained, and peaks or dips in the spectrum can be quantified to obtain information on the irradiated species. MT techniques, including chemical exchange saturation transfer (CEST) and nuclear Overhauser enhancement (NOE)-mediated MT, act as ‘chemical amplifiers’, enabling the measurement of millimolar concentrations of solute molecules that would otherwise be undetectable, and thus provide molecular MR imaging with relatively high sensitivity (9).
The ‘magnetic coupling’ between water protons and biological tissue components has been investigated previously using water-exchange filter spectroscopy (WEX) or two-dimensional NOE spectroscopy (2D NOESY). For example, Amide Proton transfer (APT), a specific form of CEST, has been applied to detect amides in tissues at 3.5 ppm and has been applied to study of several pathological conditions including solid tumors (1,10-13), ischemic stroke (14-17), and multiple sclerosis (18). In addition, some chemical sites with faster exchanging protons also show significant MT effects. For example, amine-proton water exchange effects at offsets ranging from 2 to 3 ppm can be detected by CEST (19), and have been applied to detect creatine (4,20) and glutamate (2). Hydroxyl-proton water exchange effects can also be detected and have been applied to detect myo-inositol (5), glycogen (21), glycosaminoglycans (GAG) (22,23), and administered glucose analogues (3,24-29).
In addition to chemical exchange with downfield protons, NOE effects in the upfield MR spectral range have been reported. For example, NOE-mediated MT effects at offsets ranging from −3 to −5 ppm have been reported in animal and human brains and have been applied to image tumors (8,30-32). NOE effects ranging from −3 to −5 ppm between water protons and mobile proteins or methylene protons of membrane lipids, as well as an NOE effect at around −1.6 ppm between water protons and methyl protons on the choline head group of membrane phospholipids, have been reported from tissue extracts, intact cell lines, and reconstituted lipid samples (33). By using magic angle spinning, significant changes of water signals due to cross-relaxation and/or chemical exchange between water and various constituent molecules could be detected. However, an NOE-mediated MT effect at around −1.6 ppm has not been reported previously in vivo. Here, we aimed to detect and quantify such an NOE-mediated effect on water at around −1.6 ppm upfield in rat brain and to show that it varies in brain tumors. We also investigated the possible molecular origin of this signal.
METHODS
Animal Preparation
Adult male F344 rats were purchased from Harlan Laboratories (Indianapolis, IN). Animals were group housed in large colony rooms under a 12-h light/dark cycle with food and water available ad libitum. All procedures were approved by the Vanderbilt University Institutional Animal Care and Use Committee. For in vivo imaging, all rats were immobilized and anesthetized before MR imaging. Respiration and rectal temperature were continuously measured. Respiration was monitored to be stable, and a rectal temperature of 37°C was maintained throughout the experiments using a warm-air feedback system (SA Instruments, Stony Brook, NY). Animals were anesthetized with 2-3% isoflurane (ISO) for induction and 2% for maintenance during the experiments.
Three sets of experiments were completed:
[a] Three rats were scanned to optimize the RF irradiation power.
[b] Nine healthy rats were scanned to record the distributions of the amplitude, resonance frequency offset, and line width of the NOE-mediated MT effect at −1.6 ppm.
[c] Eight rats were prepared with 9L tumors. Each rat was injected with 1 × 105 9L glioblastoma cells in the right brain hemisphere. Rats were then imaged after 14-18 days of tumor inoculation.
Phantom Preparation
Reconstituted phospholipids were prepared by adding egg phosphatidylcholine (PC) with cholesterol (1: 0.5 Egg PC: cholesterol by weight) to a chloroform solution. Solutions were then evaporated/dried under vacuum, and re-suspended to 1: 3 Egg PC: water by weight. The lipids were completely disposed with a glass rod by mechanic force, and then went through sonication at 55°C (30 mins) and freeze cycle (10-20 mins) for 3 times. All chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Cell Culture
A cancerous cell line (9L) and a non-cancerous embryonic kidney cell line (HEK293) were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). Both cells were cultured in Dulbecco’s Modified Eagles Medium (DMEM) (ATCC) supplemented with 10% heat inactivated fetal bovine serum (Sigma–Aldrich) in a 5% CO2 humidified incubator. After the cells were collected, the cells were transferred into 0.65 ml eppendorf tubes and were centrifuged at 3,000 rpm for 2 minutes at 4°C. Then the supernatant were removed and the cell pellets were ready for MR measurement.
MRI
All phantom and animal measurements were performed on a Varian DirectDrive™ horizontal 9.4 T magnet with a 38-mm Litz RF coil (Doty Scientific Inc. Columbia, SC). A continuous wave (CW)-MT sequence with a 5 s hard irradiation pulse followed by single-shot spin-echo echo planar imaging (SE-EPI) acquisition was used to study all animals. A CW-MT sequence followed by a 2 ms 90° excitation pulse and free induction decay (FID) readout was used to study the phospholipid samples and cells. The maximum data points of the free induction decay (FID) were used for data analysis. Time of echo (TE) in this FID acquisition was defined to be the duration between the center of the 90° excitation pulse and beginning of the FID readout. Z-spectra, which are the measured signals (S) normalized by a reference signal (S0), were acquired with RF offsets at ±4000, ±3500, ±3000, ±2500, and from −2000 to 2000 Hz with a step of 50 Hz (−10 to 10 ppm on 9.4 T). S0 was obtained by setting the RF offset to 100000 Hz (250 ppm on 9.4 T). Observed water longitudinal relaxation rate (R1obs) and semi-solid MT pool size ratio (PSR) were obtained using a selective inversion recovery (SIR) method (34). Specifically, a 1 ms 180° hard pulse was applied to invert the free water pool and the subsequent longitudinal recovery times were set to 4, 5, 6, 8, 10, 12, 15, 20, 50, 200, 500, 800, 1000, 2000, 4000, and 6000 ms. SE-EPI was used for the readout scheme followed by a saturation pulse train to shorten total acquisition time as described previously (35). A constant delay time of 3.5 s was set between the saturation pulse train and the next inversion pulse. Note that in this sequence, TR varies depending on the choice of inversion recovery time and delay time (35). For the parameters used in this study, TR varied from ~3.6 to ~10s. All images were acquired with matrix size 64 × 64, field of view 30 mm × 30 mm, and one acquisition.
For optimization of the RF irradiation powers (B1) on animal experiments, B1 was varied between 0.5, 1.0, and 1.5 μT with RF irradiation duration of 5 s. For experiments on reconstituted phospholipids and cultured cell lines, B1 was set to be 0.25 μT and 0.5 μT respectively, and data were acquired at 37 °C.
Data analysis
We performed multi-Lorentzian fitting of Z-spectrum using a non-linear optimization algorithm. Eq. (1) and (2) gives the model function of the Lorentzian fit method.
| (1) |
| (4) |
Here, Li represents a Lorentzian line with central frequency offset from water (Δi), peak full width at half maximum (Wi), peak amplitude (Ai). N is the number of fitted proton pools. Δω is the irradiation offset from water. We first performed a multi-Loentzian fit to remove the background direct water saturation and MT effects. In this method, a two-pool (semi-solid MT and water pools) model Lorentzian fit of Eq. (1) was performed to process the Z-spectra with frequency offsets of ±4000, ±3500, ±3000, ±2500 ±200, ±150, ±100, ±50, and 0 Hz (−10 to −6.25 ppm, −0.5 to 0.5 ppm, and 6.25 to 10 ppm on 9.4 T). The fitted spectra were used as reference signals which represent the background direct water saturation and MT effects. Previously, CEST residual spectra (defined in Eq. (3)) have been obtained by subtracting the mesured Z-spectra (Smea) with the fitted reference spectra (Sfit) to remove the background signal. However, recent studies show that all CEST, NOE-mediated MT, and the background signals have non-linear mutual interactions which violates the assumption of the multi-Lorentzian fit that all these signal can linearly add up (36). As a result, the direct subtraction of reference and label signals can only remove the 0-order of background signal. However, the 1st-order of background signal is still significant compared with the weak CEST or NOE-mediated MT effects in vivo. This will be very serious when the solute is close to water and the direct water saturation effect is significant. An inverse subtraction of label and reference signals thus has been provided to solve this problem. In addition, it has been proved that water longitudinal relaxation scales the CEST and NOE-mediated MT signals (36-40). Here, we obtain CEST exchange-dependent relaxation (AREX) residual spectra (defined in Eq. (4)) by inversely subtracting Smea from Sfit and correcting R1obs (41),
| (3) |
| (4) |
where fc is the semi-solid pool concentration which is also called PSR in this paper. 1+ fc was added in Eq. (4) to make the inverse method more specific, as shown in the previous publication (41).
After this first round of fitting, a secondary fitting was performed to resolve the two NOE-mediated MT peaks at around −1.6 and −3.5 ppm, respectively. In the secondary fitting, another two-pool (NOEs at −1.6 and −3.5 ppm) model Lorentzian fit of Eq. (2) was performed to process the AREX residual spectra ranging from −1 to −5 ppm. The NOE amplitude, offset, and line width are quantified by the fitted parameters of the Lorentzian lines. A nonlinear optimization algorithm with the maximum iteration of 2 × 105 was applied for the fit. The fitting was performed to achieve the lowest root mean square (RMS) of residuals between the data and model in the selected segment. Z-spectrum from each voxel was interpolated and B0 correction was then done by assuming the actual water resonance to be at the frequency with the lowest signal intensity. All fits for in vivo experiments were performed voxel by voxel with images smoothed by a 3 × 3 median filter before fitting. Table 1 lists the starting points and boundaries of the fit. The goodness of fit was observed by the sum of squared errors.
Table 1.
Starting points and boundaries of the amplitude, width, and offset of the exchanging pools in the Lorentzian fit. The unit of peak width and offset is ppm.
| Start | Lower | Upper | |
|---|---|---|---|
| Awater | 0.9 | 0.02 | 1 |
| Wwater | 1.4 | 0.3 | 10 |
| Δ water | 0 | −1 | 1 |
| Amt | 0.1 | 0 | 1 |
| Wmt | 25 | 10 | 100 |
| Δ MT | 0 | −4 | 4 |
| ANOE(−1.6) | 0.001 | 0 | 0.2 |
| WNOE(−1.6) | 1 | 0 | 1.5 |
| ANOE(−1.6) | −1.5 | −2 | −1 |
| ANOE(−3.5) | 0.02 | 0 | 1 |
| WNOE(−3.5) | 3 | 1 | 5 |
| ANOE(−3.5) | −3.5 | −4.5 | 2 |
Statistics
One-way analysis of variance (ANOVA) was employed to evaluate whether the distributions of fitted parameters (i.e. amplitude, offset, and line width) are different in different brain regions (i.e. cortex (C), singular cortex (SC), corpus callosum (CC), and caudate putamen (CP)). To evaluate the difference between tumor and normal tissue, a Wilcoxon rank sum test was performed. P < 0.05 was considered to be statistically significant.
RESULTS
Parameter optimization for detecting the NOE(−1.6) signal at 9.4 T
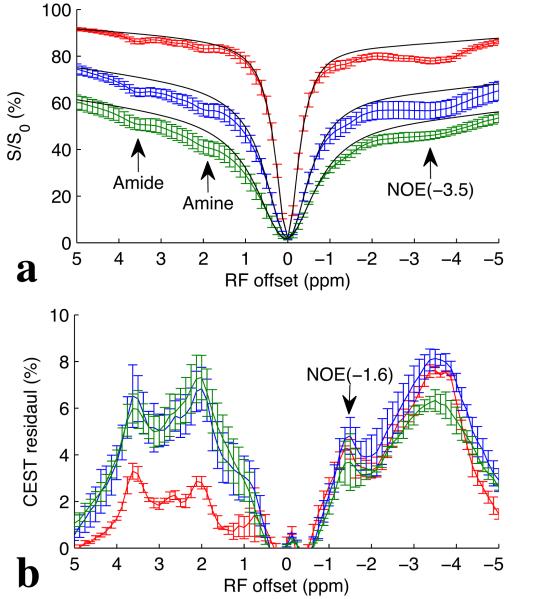
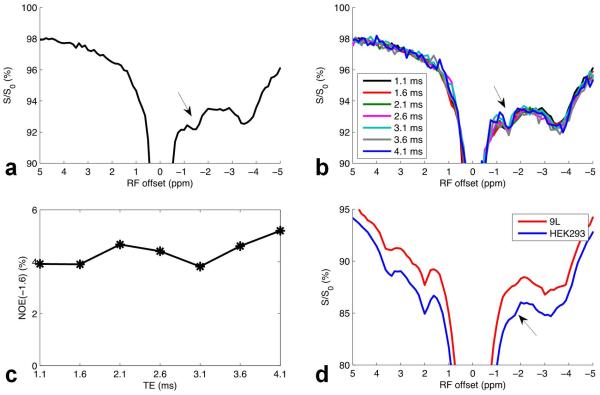
Fig. 1a and 1b show the CEST Z-spectra and CEST residual spectra, respectively, from three healthy rat brains obtained with B1 of 0.5 μT (red), 1.0 μT (blue), and 1.5 μT (green). It was found that the most distinct CEST and NOE signals in brain in the range from −5 to 5 ppm on CEST Z-spectra arise from amide protons at around 3.5 ppm, amine protons at around 2 ppm, and aliphatic protons at around −3.5 ppm (NOE(−3.5)). However, after removal of the background signal, a new NOE-mediated MT signal at around −1.6 ppm (NOE(−1.6)) can be identified on the CEST residual spectra, although it is not clear on the CEST Z-spectra. Also note in Fig. 1b that the NOE(−1.6) signal is optimized at B1 of 1.0 μT. In addition, it can be found from the CEST residuals that the NOE(−1.6) peaks overlap with the nearby NOE(−3.5) peaks, indicating the necessary to further resolve and quantify this signal.
Fig. 1.
CEST Z-spectra (a) and CEST residual spectra (b) with B1 of 0.5 μT (red), 1.0 μT (blue), and 1.5 μT (green) from three healthy rat brains. Black lines in (a) are the reference signals obtained with the Lorentzian fit of background direct water saturation and MT effects. Note that the NOE(−1.6) is not easily identified from the Z-spectra in (a), but can be identified from the CEST residual spectra in (b). Also note in (b) that the NOE(−1.6) signal (arrow) is optimized at B1 of 1.0 μT. Regions of interest (ROIs) were drawn from the whole brain. Error bars are the standard deviations across subjects.
Quantification of the NOE(−1.6) signal in rat brain
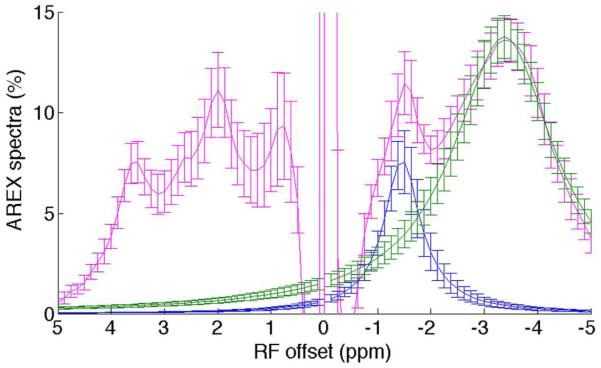
To quantify the NOE(−1.6) signal, AREX residual spectra were created using Eq. (4), and another two-pool Lorentzian fit was then performed to process the AREX residual spectra. Fig. 2 shows the AREX residual spectra (magenta) and the fitted NOE(−1.6) peak (blue) and NOE(−3.5) peak (green) from nine healthy rat brains. It was found that the NOE(−1.6) signal becomes more significant than amide signal on the AREX residual spectra, but not on the CEST residual spectra. This suggests that the NOE(−1.6) is actually a significant MT effect which, however, is attenuated by the greater direct water saturation effect at frequency offset close to water line. The separation of the NOE(−1.6) and NOE(−3.5) peaks in Fig. 2 indicates the successful fitting of this method.
Fig. 2.
AREX residual spectra (magenta), fitted NOE(−1.6) peak (blue), and fitted NOE(−3.5) peak (green) from nine healthy rat brains. Note that the NOE(−1.6) peak can be isolated from the NOE(−3.5) peak by using Lorentzian fit. Regions of interest (ROIs) were drawn from the whole brain. Error bars are the standard deviations across subjects.
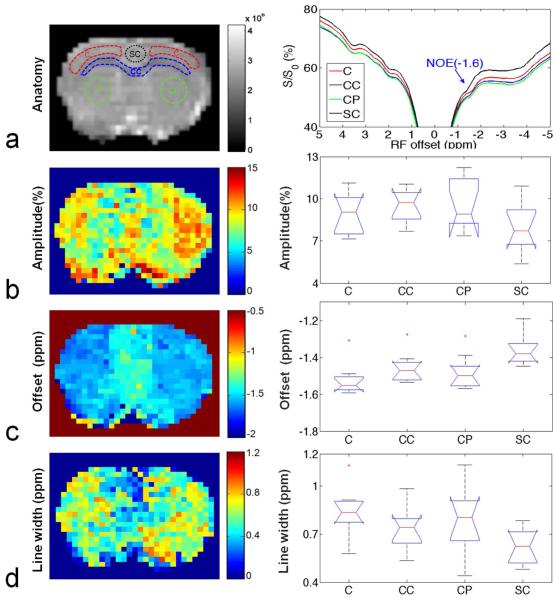
Using the optimized irradiation power and the Lorentzian fit method, we investigated different regions to evaluate the distribution of the NOE(−1.6) in rat brain. Fig. 3a shows four regions of interest (ROIs): the cortex (C), corpus callosum (CC), caudate putamen (CP), and singular cortex (SC) (left), and the averaged Z-spectra from these four ROIs (right) on nine healthy rat brains. Fig. 3b shows that the amplitudes of the NOE(−1.6) are 8.96 ± 1.45 (%), 9.49 ± 1.22 (%), 9.72± 1.82 (%), and 8.06 ± 1.76 (%) in C, CC, CP, and SC, respectively. Fig. 3c shows that the resonance frequency offsets of NOE(−1.6) are −1.52 ± 0.09 ppm, −1.46 ± 0.08 ppm, −1.48 ± 0.09 ppm and −1.36 ± 0.08 ppm in C, CC, CP, and SC, respectively. Fig. 3d shows that the line widths of NOE(−1.6) are 0.84 ± 0.15 ppm, 0.73 ± 0.15 ppm, 0.79 ± 0.22 ppm, and 0.63 ± 0.11 ppm in C, CC, CP, and SC, respectively. The one-way ANOVA provided P=0.3529 for the amplitude in the four brain regions, indicating there are no significant differences of amplitude in different rat brain regions. However, there are slight differences of the line width (P=0.0509) and significant differences of the resonance frequency offset (P=0.0062) in different rat brain regions.
Fig. 3.
Left column: anatomic image to show the four ROI (C: Cortex; CC: Corpus Callosum; CP: Caudate Putamen; SC: Singular Cortex) (a) and pixel-by-pixel fitted maps for amplitude (b), resonance frequency offset (c), and line width (d) of the NOE(−1.6) signal from a healthy rat brain. Right column: averaged Z-spectra from the four ROIs (a) and the statistical analysis of amplitude (b), resonance frequency offset (c), and line width (d) of the NOE(−1.6) signal from nine healthy rat brains. (n=9)
The NOE(−1.6) is reduced in tumor
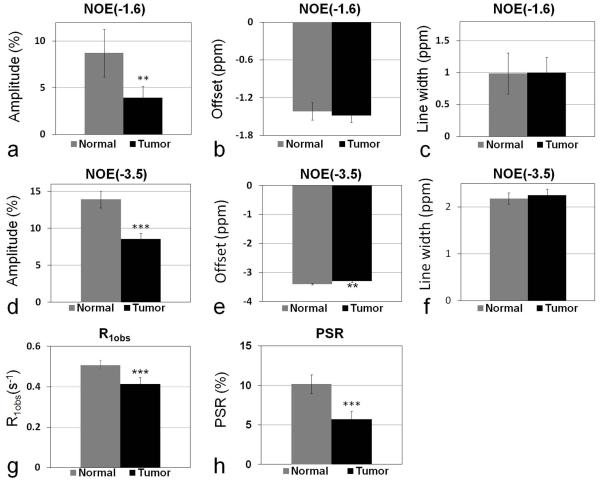
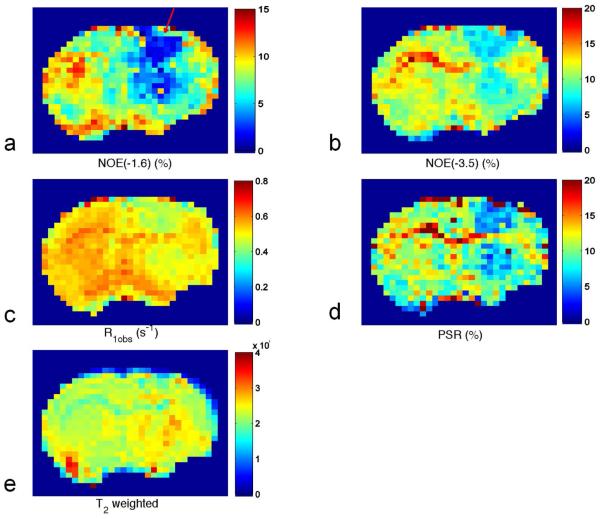
Fig. 4 shows a quantitative comparison between tumors and the contralateral normal tissues, for eight rat brains bearing 9L tumors, of the amplitude of NOE(−1.6), offset of NOE(−1.6), line width of NOE(−1.6), amplitude of NOE(−3.5), offset of NOE(−3.5), line width of NOE(−3.5), R1obs, and PSR. The amplitude of NOE(−1.6) is significantly reduced in tumor compared with that in the contralateral normal tissue (3.93 ± 1.23 (%) vs. 8.73 ± 2.55 (%), P=0.0011). However, there are no significant differences between tumor and contralateral normal tissue for the resonance frequency offset and the line widths of NOE(−1.6). The amplitude of NOE(−3.5) is also significantly reduced in tumor compared with that in contralateral normal tissue (8.54 ± 0.79 (%) vs. 13.92 ± 1.13 (%), P<0.001). The resonance frequency offset of NOE(−3.5) in tumor is slightly higher than that in contralateral normal tissue (−3.3 ± 0.04 (ppm) vs. −3.4 ± 0.02 (ppm), P=0.0032). However, there is no significant difference between tumor and contralateral normal tissue for the line width of NOE(−3.5). The R1obs in tumor (0.41 ± 0.03 (s−1)) is significantly lower than that in the contralateral normal tissue (0.51 ± 0.02 (s−1)) (P<0.001). The PSR in tumor (5.7 ± 1.0 (%)) is also significantly lower than that in the contralateral normal tissue (10.2 ± 1.2 (%)) (P<0.001). Fig. 5 shows representative maps of amplitude of NOE(−1.6), amplitude of NOE(−3.5), R1obs, and PSR, as well as T2 weighted image from a rat brain bearing a 9L tumor. Note that the tumor is clearly shown (red arrow) in the NOE(−1.6) image.
Fig. 4.
Statistics of amplitude of NOE(−1.6) (a), offset of NOE(−1.6) (b), line width of NOE(−1.6) (c), amplitude of NOE(−3.5) (d), offset of NOE(−3.5) (e), line width of NOE(−3.5) (f), R1obs (g), and PSR (h) from tumors and contralateral normal tissue from eight rat brains bearing 9L tumors. (n=8, **P<0.01, *P<0.001 )
Fig. 5.
Images of amplitude of NOE(−1.6) (a), amplitude of NOE(−3.5) (b), R1obs (c), and PSR (d), as well as T2 weighted image (e) from a representative rat brain bearing a 9L tumor. Note the tumor indicated by the red arrow in (a).
Molecular origin of the NOE(−1.6) effect
As illustrated in Fig. 6a, both NOE-mediated MT signals at around −1.6 ppm and −3.5 ppm were isolated in Egg PC. This sample shows the same resonance as identified as the NOE(−1.6) in rat brain, suggesting that the origin of the NOE(−1.6) in brain might be closely related to the interaction of water with choline phospholipids in cell membranes. The direct measurement of signals from lipid protons can also cause dips on MT Z-spectra, which is called pseudo NOE (42). To study the possible contributions from the direct measurement on the NOE(−1.6) signal, we further performed MT experiments on Egg PC with a variety of TEs. In this FID acquisition, the measured signal should be the vector sum of signals from both the lipid protons and water protons, and an oscillation with a period of ~ 1.6 ms would indicate the presence of the pseudo NOE from protons around −1.6 ppm (640 Hz at 9.4 T). Fig. 6b and 6c shows the MT Z-spectra and the fitted amplitude of the NOE(−1.6), respectively, with a variety of TEs. However, although an oscillation with a period of ~ 2 ms is found in Fig. 6c, it is very small compared with the amplitude of NOE(−1.6) signal, indicating that the NOE-mediated MT signal dominates the pseudo NOE. In animal experiments with SE readout, the MT signal at 0 ppm, which should has contributions from the direct measurement of lipids (42), is very small compared with the CEST residual signal at −1.6 ppm (see Fig.1), which indicates that the in vivo NOE(−1.6) is not a pseudo NOE, but a real MT effect. Fig. 6d shows the Z-spectra from cultured HEK293 and 9L gliosarcoma cells, respectively. The NOE(−1.6) signal can be observed in both cell lines, and the NOE(−1.6) signal in 9L cells is less significant than that in HEK293 cells.
Fig. 6.
Z-spectra from reconstituted Egg PC with TE of 1.1 ms (a), a variety of TEs (b), fitted amplitude of NOE(−1.6) as a function of TE (c), and Z-spectra from cultured HEK293 and 9L cell lines (d).
DISCUSSION
Previously, an NOE-mediated MT signal only at around −3.5 ppm offset from water has been studied in brain tissue in vivo. In the present work, we report a new NOE-mediated MT signal at around −1.6 ppm which can be reliably detected in rat brains. This NOE-mediated MT signal is close to water and thus is significantly influenced by direct water saturation effect, resulting in a dip that is not easily identified from the Z-spectrum. However, this NOE-mediated MT signal becomes apparent after removal of the direct water saturation effect. A similar fitting result showing an MT signal at around −1.6 ppm was also observed in a previous publication on a 7 T human scanner (Fig. 8a in Ref (8)), but it has not been discussed. Note that in both the previous human study and in our animal study (see Fig.1b), the NOE(−1.6) signal is comparable to other MT signals from amide, amine, and aliphatic protons at −3.5 ppm, indicating that it is a significant MT effect.
As an indirect method to detect solute molecules through measurements of water signals, MT effects depend on several tissue parameters including direct water saturation, background semi-solid MT effects, and other overlapping MT effects. To improve the quantification of the NOE(−1.6) effect, we fitted the Z-spectra to a Lorentzian function. Lorentzian functions were recently used to fit Z-spectra as a combination of several components: using three pools (solute, water, and background semi-solid pool) (43) and four pools (amide, amine, water, and NOE pool at −3.5 ppm) (44). However, the NOE pool at −1.6 ppm was not considered as a component in previous studies. In this work, we used a two-step Lorentzian fitting which first removes the background direct water saturation and MT effects and then separates the NOE(−1.6) signal from the overlapping NOE(−3.5) signal. In our fitting method, an inverse analysis of the reference and label signals was applied and the T1 normalization was considered so that the fitted NOE(−1.6) signal is more specific to the ‘coupling’ effect of the target molecule. Different from other MT effects which have constant resonance frequency offsets and line widths, the NOE(−1.6) appears to have a variable resonance frequency offset and line width.
In the present work, a dense saturation frequency sampling of Z-spectra (sampling interval is 0.125 ppm in the range from −5 to 5 ppm) was used for the Lorentzian fit. However, a sparsely sampling is usually required for fast acquisitions. We fitted the NOE(−1.6) with two other sampling intervals (0.25 and 0.5 ppm) (Sup. Fig. S1). It was found that when the sampling point around −1.6 ppm was obtained, the fitted amplitudes, offsets, and line widths of NOE(−1.6) do not show significant changes with the sampling interval of 0.25 and 0.5 ppm compared with those with sampling interval of 0.125 ppm, although there are slight changes in tumors with sampling interval of 0.5 ppm. This indicates that the sampling points or intervals can be further adjusted, and the acquisition time can be further shortened. Signal to noise ratio (SNR) also affects the fitting results. We have investigated the influences of SNR on the fittings of NOE(−1.6). Specifically, high-SNR (roughly 310 for tumor and 250 for contralateral normal tissue) Z-spectra were first created from the average of the Z-spectra acquired from 8 rat brains. Random noises at different SNR levels (20-100) were then added to the Z-spectra according to the procedure described in Ref (45). Results show that the contrast between the tumor and the contralateral normal tissue depends on SNR. When SNR is larger than 40, the tumor and the contralateral normal tissue can be reliably differentiated by the NOE(−1.6) (Sup. Fig. S2).
The NOE(−1.6) effect should be considered in the quantification of other MT effects by using conventional MT asymmetry analysis (MTRasym). MTRasym removes effects caused by direct water saturation and other symmetric MT contributions, and works well for analyzing pure samples with exchangeable protons. However, in vivo MTRasym experiments are still confounded by asymmetric contributions, including semi-solid MT asymmetry (46) and NOE-mediated MT effects. For example, asymmetry analyses of APT at 3.5 ppm can be affected by the NOE(−3.5) signal (8). For the same reason, asymmetry analyses of hydroxyl (1.2 ppm) signals (27,28) downfield may also be confounded by the new NOE(−1.6) signal, which has a spread ranging from −1 to −2 ppm.
On an MT Z-spectrum, dips at different frequency offsets represent MT effects from different molecules. Specifically, previous reports have shown that a negative NOE-mediated MT effect could be originated from restricted metabolites (47,48) and macromolecules (40), but not from mobile small molecules. In biological tissues, the main metabolites that have MRS signals with frequency offsets from water at around −1.6 ppm include free choline and choline-containing metabolites (−1.6 ppm), taurine (−1.4 ppm), as well as creatine and phosphocreatine (−1.8 ppm). However, these water-soluble metabolites are mobile and thus cannot contribute to the negative NOE-mediated MT effect. Choline phospholipids, which contain choline head groups, are the major phospholipid component of eukaryotic cells accounting for approximate 60% of total membrane lipids (49). Therefore, it is plausible that the observed in vivo NOE(−1.6) signal stems from choline phospholipids. Reconstituted choline phospholipids have been previously used to model conventional MT effect (50-52). In this paper, we used this sample as a model to study the possible origin of the NOE(−1.6) effect. Our results on reconstituted Egg PC in Fig. 6a confirm the presence of an NOE-mediated MT signal at −1.6 ppm from choline phospholipids.
As a through-space dipolar coupling effect, the NOE depends on the correlation time of the coupled protons. It has been reported that the restriction of molecules plays a key role in causing negative NOE effects (53,54). Choline methyl proton has a resonance frequency at around −1.6 ppm from water. In eukaryotic cells, choline forms soluble choline-containing metabolites or appears in the head group of phospholipids. Although the soluble free choline and choline-containing metabolites are in a mobile state, the choline head groups are in a restricted state through strong binding to a rigid membrane, which may enhance a negative NOE between water protons and choline methyl protons. Actually, this NOE effect has been previously reported for tissue extracts and intact cell lines by using 2D NOESY and magic angle spinning (33). Therefore, the MT effect relayed by this NOE should be observable by selectively saturating the choline phospholipids and detecting the reduction of water signal.
Fig. 6c indicates the presence of direct measurement of signals from phospholipid choline head groups. This directly measured signal could be greater in lipids that are more mobile and thus have longer T2. However, for lipids with rigid structures, the correlation time is longer and T2 is shorter, which would decrease the direct measurement, but facilitate NOE effects between the protons in these molecules and water. Hence, the NOE-mediated MT signal may provide complementary information to the direct measurements of lipids. In our lipid sample, an appropriate amount of cholesterol was added to the Egg PC to stiffen the lipid structures that cause NOE effects. According to the power dependence of this NOE-mediated MT signal at −1.6 ppm in Fig. 1, its coupling rate should be slow. The exact mechanism of this NOE effect at −1.6 ppm remains unclear. There might be two possible pathways: (1) the NOE at −1.6 ppm is a direct dipole-dipole interaction between the metabolite carbon bound protons and water protons; (2) the NOE at −1.6 ppm is due to a multi-step process involving both dipole-dipole and chemical exchange contributions.
Fig. 6d shows that NOE(−1.6) signals from 9L and HEK293 cells are different suggesting that this signal may be a new potential biomarker for detecting tumor cells. The reduction of NOE(−1.6) in tumors may correspond to variations of membrane lipid components in tumors (55). Here, we named the new NOE-mediated MT signal as NOE(−1.6) because we postulated that it is related to choline which has a resonance frequency at −1.6 ppm from water (3.2 ppm from Tetramethylsilane (TMS)), although Fig. 3c shows that this signal is more close to −1.5 ppm and varies in different regions. The shift of the resonance frequency of this NOE signal suggests its complicated signal mechanisms. Further investigations of the molecular origins and the signal mechanisms of the NOE(−1.6) effect in vivo are underway.
CONCLUSION
A novel NOE(−1.6) effect upfield from water, which has not been noticed from in vivo imaging studies, can be reliably detectable in normal rat brain tissues and may reflect an NOE effect between membrane choline phospholipids and water protons. The amplitude of this effect is dramatically reduced in brain tumors. Further studies of this NOE-mediated MT signal are warranted as it may represent a new biomarker of pathological or other physiological variations of interest.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to acknowledge funding from the National Institutes of Health (R21EB17873, R01CA109106, R01CA184693, R01EB017767, and K25CA168936)
Grant Sponsor: R21EB17873, R01CA109106, R01CA184693, R01EB017767, and K25CA168936
Footnotes
SUPPORTING INFORMATION
Additional supporting information can be found in the online version of this article.
REFERENCES
- 1.Zhou JY, Tryggestad E, Wen ZB, Lal B, Zhou TT, Grossman R, Wang SL, Yan K, Fu DX, Ford E, Tyler B, Blakeley J, Laterra J, van Zijl PCM. Differentiation between glioma and radiation necrosis using molecular magnetic resonance imaging of endogenous proteins and peptides. Nature Medicine. 2011;17(1):130–U308. doi: 10.1038/nm.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cai KJ, Haris M, Singh A, Kogan F, Greenberg JH, Hariharan H, Detre JA, Reddy R. Magnetic resonance imaging of glutamate. Nature Medicine. 2012;18(2):302–306. doi: 10.1038/nm.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walker-Samuel S, Ramasawmy R, Torrealdea F, Rega M, Rajkumar V, Johnson SP, Richardson S, Goncalves M, Parkes HG, Arstad E, Thomas DL, Pedley RB, Lythgoe MF, Golay X. In vivo imaging of glucose uptake and metabolism in tumors. Nature Medicine. 2013;19(8):1067. doi: 10.1038/nm.3252. -+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haris M, Singh A, Cai KJ, Kogan F, McGarvey J, DeBrosse C, Zsido GA, Witschey WRT, Koomalsingh K, Pilla JJ, Chirinos JA, Ferrari VA, Gorman JH, Hariharan H, Gorman RC, Reddy R. A technique for in vivo mapping of myocardial creatine kinase metabolism. Nature Medicine. 2014;20(2):209–214. doi: 10.1038/nm.3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haris M, Cai KJ, Singh A, Hariharan H, Reddy R. In vivo mapping of brain myo-inositol. Neuroimage. 2011;54(3):2079–2085. doi: 10.1016/j.neuroimage.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ling W, Regatte RR, Navon G, Jerschow A. Assessment of glycosaminoglycan concentration in vivo by chemical exchange-dependent saturation transfer (gagCEST) Proceedings of the National Academy of Sciences of the United States of America. 2008;105(7):2266–2270. doi: 10.1073/pnas.0707666105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou JY, Payen JF, Wilson DA, Traystman RJ, van Zijl PCM. Using the amide proton signals of intracellular proteins and peptides to detect pH effects in MRI. Nature Medicine. 2003;9(8):1085–1090. doi: 10.1038/nm907. [DOI] [PubMed] [Google Scholar]
- 8.Jones CK, Huang A, Xu JD, Edden RAE, Schar M, Hua J, Oskolkov N, Zaca D, Zhou JY, McMahon MT, Pillai JJ, van Zijl PCM. Nuclear Overhauser enhancement (NOE) imaging in the human brain at 7 T. Neuroimage. 2013;77:114–124. doi: 10.1016/j.neuroimage.2013.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou JY, van Zijl PCM. Chemical exchange saturation transfer imaging and spectroscopy. Progress in Nuclear Magnetic Resonance Spectroscopy. 2006;48(2-3):109–136. [Google Scholar]
- 10.Salhotra A, Lal B, Laterra J, Sun PZ, van Zijl PCM, Zhou JY. Amide proton transfer imaging of 9L gliosarcoma and human glioblastoma xenografts. Nmr in Biomedicine. 2008;21(5):489–497. doi: 10.1002/nbm.1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones CK, Schlosser MJ, van Zijl PCM, Pomper MG, Golay X, Zhou JY. Amide proton transfer imaging of human brain tumors at 3T. Magnetic Resonance in Medicine. 2006;56(3):585–592. doi: 10.1002/mrm.20989. [DOI] [PubMed] [Google Scholar]
- 12.Jia GA, Abaza R, Williams JD, Zynger DL, Zhou JY, Shah ZK, Patel M, Sammet S, Wei L, Bahnson RR, Knopp MV. Amide Proton Transfer MR Imaging of Prostate Cancer: A Preliminary Study. Journal of Magnetic Resonance Imaging. 2011;33(3):647–654. doi: 10.1002/jmri.22480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou JY, Lal B, Wilson DA, Laterra J, van Zijl PCM. Amide proton transfer (APT) contrast for imaging of brain tumors. Magnetic Resonance in Medicine. 2003;50(6):1120–1126. doi: 10.1002/mrm.10651. [DOI] [PubMed] [Google Scholar]
- 14.Sun PZ, Wang EF, Cheung JS. Imaging acute ischemic tissue acidosis with pH-sensitive endogenous amide proton transfer (APT) MRI-Correction of tissue relaxation and concomitant RF irradiation effects toward mapping quantitative cerebral tissue pH. Neuroimage. 2012;60(1):1–6. doi: 10.1016/j.neuroimage.2011.11.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun PZ, Zhou JY, Sun WY, Huang J, van Zijl PCM. Detection of the ischemic penumbra using pH-weighted MRI. Journal of Cerebral Blood Flow and Metabolism. 2007;27(6):1129–1136. doi: 10.1038/sj.jcbfm.9600424. [DOI] [PubMed] [Google Scholar]
- 16.Sun PZ, Benner T, Copen WA, Sorensen AG. Early Experience of Translating pH-Weighted MRI to Image Human Subjects at 3 Tesla. Stroke. 2010;41(10):S147–S151. doi: 10.1161/STROKEAHA.110.595777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Zu ZL, Zaiss M, Khan IS, Singer RJ, Gochberg DF, Bachert P, Gore JC, Xu J. Imaging of amide proton transfer and nuclear Overhauser enhancement in ischemic stroke with corrections for competing effects. Nmr in Biomedicine. 2015;28(2):200–209. doi: 10.1002/nbm.3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dula AN, Asche EM, Landman BA, Welch EB, Pawate S, Sriram S, Gore JC, Smith SA. Development of Chemical Exchange Saturation Transfer at 7T. Magnetic Resonance in Medicine. 2011;66(3):831–838. doi: 10.1002/mrm.22862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin T, Wang P, Zong XP, Kim SG. Magnetic resonance imaging of the Amine-Proton EXchange (APEX) dependent contrast. Neuroimage. 2012;59(2):1218–1227. doi: 10.1016/j.neuroimage.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haris M, Nanga RPR, Singh A, Cai K, Kogan F, Hariharan H, Reddy R. Exchange rates of creatine kinase metabolites: feasibility of imaging creatine by chemical exchange saturation transfer MRI. Nmr in Biomedicine. 2012;25(11):1305–1309. doi: 10.1002/nbm.2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Zijl PCM, Jones CK, Ren J, Malloy CR, Sherry AD. MR1 detection of glycogen in vivo by using chemical exchange saturation transfer imaging (glycoCEST) Proceedings of the National Academy of Sciences of the United States of America. 2007;104(11):4359–4364. doi: 10.1073/pnas.0700281104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim M, Chan Q, Anthony MP, Cheung KMC, Samartzis D, Khong PL. Assessment of glycosaminoglycan distribution in human lumbar intervertebral discs using chemical exchange saturation transfer at 3 T: feasibility and initial experience. Nmr in Biomedicine. 2011;24(9):1137–1144. doi: 10.1002/nbm.1671. [DOI] [PubMed] [Google Scholar]
- 23.Liu Q, Tawackoli W, Pelled G, Fan ZY, Jin N, Natsuaki Y, Bi XM, Gart A, Bae H, Gazit D, Li DB. Detection of Low Back Pain Using pH Level-Dependent Imaging of the Intervertebral Disc Using the Ratio of R-1 rho Dispersion and -OH Chemical Exchange Saturation Transfer (RROC) Magnetic Resonance in Medicine. 2015;73(3):1196–1205. doi: 10.1002/mrm.25186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rivlin M, Horev J, Tsarfaty I, Navon G. Molecular imaging of tumors and metastases using chemical exchange saturation transfer (CEST) MRI. Scientific Reports. 2013;3 doi: 10.1038/srep03045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zu ZL, Spear J, Li H, Xu JZ, Gore JC. Measurement of regional cerebral glucose uptake by magnetic resonance spin-lock imaging. Magnetic Resonance Imaging. 2014;32(9):1078–1084. doi: 10.1016/j.mri.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rivlin M, Tsarfaty I, Navon G. Functional Molecular Imaging of Tumors by Chemical Exchange Saturation Transfer MRI of 3-O-Methyl-D-Glucose. Magnetic Resonance in Medicine. 2014;72(5):1375–1380. doi: 10.1002/mrm.25467. [DOI] [PubMed] [Google Scholar]
- 27.Nasrallah FA, Pages G, Kuchel PW, Golay X, Chuang KH. Imaging brain deoxyglucose uptake and metabolism by glucoCEST MRI. Journal of Cerebral Blood Flow and Metabolism. 2013;33(8):1270–1278. doi: 10.1038/jcbfm.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan KWY, McMahon MT, Kato Y, Liu GS, Bulte JWM, Bhujwalla ZM, Artemov D, van Zijl PCM. Natural D-glucose as a biodegradable MRI contrast agent for detecting cancer. Magnetic Resonance in Medicine. 2012;68(6):1764–1773. doi: 10.1002/mrm.24520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin T, Mehrens H, Hendrich KS, Kim SG. Mapping brain glucose uptake with chemical exchange-sensitive spin-lock magnetic resonance imaging. Journal of Cerebral Blood Flow and Metabolism. 2014;34(8):1402–1410. doi: 10.1038/jcbfm.2014.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Zu ZL, Zaiss M, Khan IS, Singer RJ, Gochberg DF, Bachert P, Gore JC, Xu JZ. Imaging of amide proton transfer and nuclear Overhauser enhancement in ischemic stroke with corrections for competing effects. Nmr in Biomedicine. 2015;28(2):200–209. doi: 10.1002/nbm.3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mougin O, Clemence M, Peters A, Pitiot A, Gowland P. High-resolution imaging of magnetisation transfer and nuclear Overhauser effect in the human visual cortex at 7 T. Nmr in Biomedicine. 2013;26(11):1508–1517. doi: 10.1002/nbm.2984. [DOI] [PubMed] [Google Scholar]
- 32.Paech D, Zaiss M, Meissner JE, Windschuh J, Wiestler B, Bachert P, Neumann JO, Kickingereder P, Schlemmer HP, Wick W, Nagel AM, Heiland S, Ladd ME, Bendszus M, Radbruch A. Nuclear Overhauser Enhancement Mediated Chemical Exchange Saturation Transfer Imaging at 7 Tesla in Glioblastoma Patients. Plos One. 2014;9(8) doi: 10.1371/journal.pone.0104181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen JH, Sambol EB, DeCarolis P, O'Connor R, Geha RC, Wu YV, Singer S. High-resolution MAS NMR spectroscopy detection of the spin magnetization exchange by cross-relaxation and chemical exchange in intact cell lines and human tissue specimens. Magnetic Resonance in Medicine. 2006;55(6):1246–1256. doi: 10.1002/mrm.20889. [DOI] [PubMed] [Google Scholar]
- 34.Gochberg DF, Gore JC. Quantitative magnetization transfer imaging via selective inversion recovery with short repetition times. Magnetic Resonance in Medicine. 2007;57(2):437–441. doi: 10.1002/mrm.21143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu JZ, Li K, Zu ZL, Li X, Gochberg DF, Gore JC. Quantitative magnetization transfer imaging of rodent glioma using selective inversion recovery. Nmr in Biomedicine. 2014;27(3):253–260. doi: 10.1002/nbm.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zaiss M, Bachert P. Exchange-dependent relaxation in the rotating frame for slow and intermediate exchange - modeling off-resonant spin-lock and chemical exchange saturation transfer. Nmr in Biomedicine. 2013;26(5):507–518. doi: 10.1002/nbm.2887. [DOI] [PubMed] [Google Scholar]
- 37.Aime S, Barge A, Delli Castelli D, Fedeli F, Mortillaro A, Nielsen FU, Terreno E. Paramagnetic lanthanide(III) complexes as pH-sensitive chemical exchange saturation transfer (CEST) contrast agents for MRI applications. Magnetic Resonance in Medicine. 2002;47(4):639–648. doi: 10.1002/mrm.10106. [DOI] [PubMed] [Google Scholar]
- 38.Ward KM, Balaban RS. Determination of pH using water protons and chemical exchange dependent saturation transfer (CEST) Magnetic Resonance in Medicine. 2000;44(5):799–802. doi: 10.1002/1522-2594(200011)44:5<799::aid-mrm18>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 39.Zhou JY, Wilson DA, Sun PZ, Klaus JA, van Zijl PCM. Quantitative description of proton exchange processes between water and endogenous and exogenous agents for WEX, CEST, and APT experiments. Magnetic Resonance in Medicine. 2004;51(5):945–952. doi: 10.1002/mrm.20048. [DOI] [PubMed] [Google Scholar]
- 40.van Zijl PCM, Zhou J, Mori N, Payen JF, Wilson D, Mori S. Mechanism of magnetization transfer during on-resonance water saturation. A new approach to detect mobile proteins, peptides, and lipids. Magnetic Resonance in Medicine. 2003;49(3):440–449. doi: 10.1002/mrm.10398. [DOI] [PubMed] [Google Scholar]
- 41.Zaiss M, Zu ZL, Xu JZ, Schuenke P, Gochberg DF, Gore JC, Ladd ME, Bachert P. A combined analytical solution for chemical exchange saturation transfer and semi-solid magnetization transfer. Nmr in Biomedicine. 2015;28(2):217–230. doi: 10.1002/nbm.3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu JH, Zhou JY, Cai CB, Cai SH, Chen Z. Observation of True and Pseudo NOE Signals Using CEST-MRI and CEST-MRS Sequences with and without Lipid Suppression. Magnetic Resonance in Medicine. 2015;73(4):1615–1622. doi: 10.1002/mrm.25277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zaiss M, Schmitt B, Bachert P. Quantitative separation of CEST effect from magnetization transfer and spillover effects by Lorentzian-line-fit analysis of z-spectra. Journal of Magnetic Resonance. 2011;211(2):149–155. doi: 10.1016/j.jmr.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 44.Desmond KL, Moosvi F, Stanisz GJ. Mapping of Amide, Amine, and Aliphatic Peaks in the CEST Spectra of Murine Xenografts at 7 T. Magnetic Resonance in Medicine. 2014;71(5):1841–1853. doi: 10.1002/mrm.24822. [DOI] [PubMed] [Google Scholar]
- 45.Pierpaoli C, Basser PJ. Toward a quantitative assessment of diffusion anisotropy. Magnetic Resonance in Medicine. 1996;36(6):893–906. doi: 10.1002/mrm.1910360612. [DOI] [PubMed] [Google Scholar]
- 46.Hua J, Jones CK, Blakeley J, Smith SA, van Zijl PCM, Zhou JY. Quantitative description of the asymmetry in magnetization transfer effects around the water resonance in the human brain. Magnetic Resonance in Medicine. 2007;58(4):786–793. doi: 10.1002/mrm.21387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leibfritz D, Dreher W. Magnetization transfer MRS. Nmr in Biomedicine. 2001;14(2):65–76. doi: 10.1002/nbm.681. [DOI] [PubMed] [Google Scholar]
- 48.de Graaf RA, van Kranenburg A, Nicolay K. Off-resonance metabolite magnetization transfer measurements on rat brain in situ. Magnetic Resonance in Medicine. 1999;41(6):1136–1144. doi: 10.1002/(sici)1522-2594(199906)41:6<1136::aid-mrm9>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 49.Jao CY, Roth M, Welti R, Salic A. Metabolic labeling and direct imaging of choline phospholipids in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(36):15332–15337. doi: 10.1073/pnas.0907864106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fralix TA, Ceckler TL, Wolff SD, Simon SA, Balaban RS. Lipid Bilayer and Water Proton Magnetization Transfer - Effect of Cholesterol. Magnetic Resonance in Medicine. 1991;18(1):214–223. doi: 10.1002/mrm.1910180122. [DOI] [PubMed] [Google Scholar]
- 51.Kucharczyk W, Macdonald PM, Stanisz GJ, Henkelman RM. Relaxivity and Magnetization-Transfer of White-Matter Lipids at Mr-Imaging - Importance of Cerebrosides and Ph. Radiology. 1994;192(2):521–529. doi: 10.1148/radiology.192.2.8029426. [DOI] [PubMed] [Google Scholar]
- 52.Ceckler TL, Wolff SD, Yip V, Simon SA, Balaban RS. Dynamic and Chemical Factors Affecting Water Proton Relaxation by Macromolecules. Journal of Magnetic Resonance. 1992;98(3):637–645. [Google Scholar]
- 53.Swanson SD. Protein mediated magnetic coupling between lactate and water protons. Journal of Magnetic Resonance. 1998;135(1):248–255. doi: 10.1006/jmre.1998.1535. [DOI] [PubMed] [Google Scholar]
- 54.Estilaei MR, Matson GB, Meyerhoff DJ. Indirect imaging of ethanol via magnetization transfer at high and low magnetic fields. Magnetic Resonance in Medicine. 2003;49(4):755–759. doi: 10.1002/mrm.10413. [DOI] [PubMed] [Google Scholar]
- 55.Yates AJ, Thompson DK, Boesel CP, Albrightson C, Hart RW. Lipid-Composition of Human Neural Tumors. Journal of Lipid Research. 1979;20(4):428–436. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.