Abstract
Chironomid larvae that inhabit in aquatic sediments play an important role as vector for bacterial pathogens. Its life cycle consists of four stages i.e. eggs, larvae, pupae and adult. In the present study we identified bacterial species associated with whole larvae of chironomids from 11 lake sediments of Bangalore region using 16s rRNA gene Sanger sequencing. We found that larvae from all lake sediments associated with bacterial species which include key pathogens. Totally we identified 65 bacterial isolates and obtained GenBank accession numbers (KX980423 - KX980487). Phylogenetic tree constructed using MEGA 7 software and tree analysis highlight the predominant bacterial community associated with larvae which include Enterobacteriaceae (43.08%; 28 isolates) and Aeromonas (24.62%; 16 isolates), Shewanella, Delftia, Bacillus (6.15%; 4 isolates each), Pseudomonas (4.62%; 3 isolates) and Exiguobacterium (3.08%; 2 isolates). Current findings state that among bacterial population Aeromonas, Enterobacter and Escherichia with serotypes are commonly associated with larvae in maximum lake points. In other hand Vibrio, Pseudomonas, Klebsiella, Shigella, Bacillus, and other bacterial species were identified moderately in all lakes. Interestingly, we identified first time Shigella Gram negative, rod shaped pathogenic organism of Enterobacteriaceae and Rheinheimera Gram negative, rod shaped organism associating chironomid larvae.
Keywords: Chironomid larvae, Bacterial species, 16s rRNA gene, Sanger sequencing, MEGA 7 software, Phylogenetic tree
1. Introduction
Chironomids are important organisms most commonly inhabited in aquatic environment and they are tolerant to extreme environmental changes [1] and are important nutrient source for both vertebrates and invertebrates [2], [3]. The complete metamorphosis of chironomids follows four stages of life cycle which include egg, larva, pupa and adult. It is well documented the association of endogenous bacterial population with chironomids and their role against toxic metal for host survival [4]. Both egg and larvae of the chironomids are associated with unique and stable bacterial community [5]. With addition of anthropogenic pollutants such as fecal excretion and domestic sewage into natural waters, the persistence of bacterial pathogens have increased in sediments with subsequent health hazards to human [6], [7], [8]. Chironomids potentially mobilize bacteria to different environments via food chain, which is a human health concern. Chironomid egg mass are natural reservoir of Vibrio cholerae [9], [10], [11], [12] and pathogenic Aeromonas species [13], [14] and associate with various bacterial genera like Acinetobacter, Brachymonas, Exiguobacterium, Klebsiella, Leucobacter, Oceanobacillus, Paracoccus, Pseudomonas, Rheinheimera, Shewanella [12], [13], [15], [16], [17], [18]. However, very few reports are available on bacterial association with chironomid larvae. Microbial communities can be extremely diverse and are underestimated by culture dependent methods. 16s rRNA gene sequencing is culture independent method used for identification of bacterial species from microbial communities. 16s rRNA is conserved gene and present in all bacteria which allow the differentiation between organisms at genus level across all major phyla of bacteria and can have greater impact on the assignment of relationship of the deeper branches [19]. In present study, we have investigated 16s rRNA gene based identification of microbial communities associated with chironomid larvae (Chironomus circumdatus) of different lake sediments by using Sanger sequencing.
2. Materials and methods
2.1. Sample collection and culturing
Chironomid larvae samples (Chironomus circumdatus) were collected from 11 lakes from Bangalore region, India (Fig. S1) with an aquatic handle net along with the sediment, species identification performed as per our previous report [20]. The samples were subjected to surface sterilization with 70% ethanol and processed for culturing. 20 larvae (4th instar) collected from different points of each lake and homogenized with the help of mortar and pestle under sterile conditions. Then homogenate was serially diluted (10− 1) and cultured on Nutrient agar plate accordingly and incubated for 48 h at 37 °C. Individual colonies were further enriched in LB broth for subsequent experiment.
Supplementary Fig. 1.
Geographical map of eleven Sample collection points from different lakes of Bengaluru city, India.
2.2. Genomic DNA preparation
Genomic DNA was extracted from 65 enriched cultures using Macherey-Nagel Nucleospin Microbial DNA kit (Germany). gDNA concentration was measured using NanoDrop8000 (Thermo Scientific, USA) and average concentration of all samples obtained was about 40–50 ng/μL for further experiment.
2.3. Polymerase chain reaction
A Polymerase Chain Reaction was performed for all the isolates with modified PCR analysis of Murugkar et al. [21]. To the MicroAmp® 96-Well reaction Plate (0.2 mL), added 3 μL buffer, 2 μL dNTPs, 0.3 μL TaqDNA polymerase (NEB, USA), 2 μL 5 M Betaine, template 2 μL, 20 pmol concentration of primer forward 2 μL, primer reverse 2 μL and HPLC water 6.7 μL and sealed accordingly with the applicator. The ± 1500 bp product of 16s rRNA gene was amplified by using primer set 27F - 5′ AGA GTT TGA TCM TGG CTC AG 3′ and 1492R- 5′ TAC GGY TAC CTT GTT ACG ACT T 3′ (Eurofins Genomics India Pvt. Ltd). The amplification by Conventional PCR process was started with an initial denaturation step (95 °C, 3 min). Each cycle consisted of three steps (denaturation, annealing, and extension). Each PCR reaction consisted of 40 cycles of amplification (initial 10 cycles was denaturation at 95 °C for1 min, annealing at 50 °C for 1 min, and DNA chain extension at 72 °C for 1 min, last 30 cycles was denaturation at 95 °C for 30s, annealing at 55 °C for 10s, and DNA chain extension at 72 °C for 30s). A final extension cycle was performed at 72 °C for 5 min (Applied Biosystems Veriti Thermal Cycler). PCR products were detected by using 2% agarose gel electrophoresis and photographed under UV illumination by using a Gel documentation system (UVITEC Cambridge).
2.4. Sanger sequencing
Amplified amplicons were purified using QIAquick PCR Purification kit (QIAGEN, Malaysia). The amplicons were sequenced in both directions using BDT v3.1 chemistry, POP7 Polymer on 3730XL Genetic Analyzer. The thermal program was made up of an initial pre-denaturation step at 95 °C for 2 min; followed by 25 cycles consisting of a denaturation step at 95 °C for 10 s, annealing step at 55 °C for 10 s and an extension step at 60 °C for 4 min. Consensus sequence of 16s rRNA genes were generated from forward and reverse sequence data using codan code aligner software. DNA sequencing data was analyzed using BLAST with NR database of NCBI GenBank. 65 species sequences were submitted to NCBI GenBank through Bankit tool. Consensus sequences were used to construct phylogenetic tree using MEGA7 software.
3. Results
The chironomid larvae were investigated for its association with endogenous bacterial communities. Successfully obtained NCBI GenBank accession numbers for 65 isolates (KX980423–KX980487) based on sequence similarity and listed Table 1. Among 65 isolates, 59 isolates had 99% similarity to the 16s rRNA data base. Furthermore, 4 isolates had 100% and 2 isolates with 96% sequence similarity to the 16s rRNA data base Table 1. The most dominant bacterial genera were associated with larvae were Aeromonas (16 isolates), Escherichia (10 isolates) and Enterobacter (10 isolates) and found in maximum lake points. Other bacterial groups presented moderately in all 11 lake points.
Table 1.
Bacterial isolates from chironomid larvae of Bengaluru Lake points.
| Isolate name_accession no. | Closest relative in GenBank database (accession no.) | Similarity (%) |
|---|---|---|
| Bacillus thuringiensis RPK1 (KX980423) | Bacillus thuringiensis (NR_043403.1) | 99% |
| Enterobacter hormaechei RPK2 (KX980424) | Enterobacter hormaechei (CP010376.2) | 99% |
| Acinetobacter bereziniae RPK3 (KX980425) | Acinetobacter bereziniae (NR_117625.1) | 99% |
| Enterobacter cancerogenus RPK4 (KX980426) | Enterobacter cancerogenus (NR_044977.1) | 99% |
| Klebsiella pneumoniae RPK5 (KX980427) | Klebsiella pneumoniae (NR_117683.1) | 99% |
| Aeromonas veronii RPK6 (KX980428) | Aeromonas veronii (NR_118947.1) | 99% |
| Enterobacter aerogenes RPK7 (KX980429) | Enterobacter aerogenes (NR_102493.1) | 99% |
| Bacillus toyonensis RPK8 (KX980430) | Bacillus toyonensis (NR_121761.1) | 99% |
| Exiguobacterium acetylicum RPK9 (KX980431) | Exiguobacterium acetylicum (NR_043479.1) | 99% |
| Pseudomonas mosselii RPK10 (KX980432) | Pseudomonas mosselii (NR_024924.1) | 99% |
| Enterobacter aerogenes RPK11 (KX980433) | Enterobacter aerogenes (NR_102493.1) | 99% |
| Pseudomonas anguilliseptica RPK12 (KX980434) | Pseudomonas anguilliseptica (NR_029319.1) | 99% |
| Pseudomonas anguilliseptica RPK13 (KX980435) | Pseudomonas anguilliseptica (NR_029319.1) | 99% |
| Aeromonas hydrophila RPK14 (KX980436) | Aeromonas hydrophila (NR_119190.1) | 99% |
| Enterobacter cloacae RPK15 (KX980437) | Enterobacter cloacae (NR_044978.1) | 99% |
| Enterobacter cloacae RPK16 (KX980438) | Enterobacter cloacae (NR_044978.1) | 99% |
| Klebsiella pneumoniae RPK17 (KX980439) | Klebsiella pneumoniae (NR_117683.1) | 99% |
| Vibrio cholera RPK18 (KX980440) | Vibrio cholera (NR_115936.1) | 99% |
| Aeromonas caviae RPK19 (KX980441) | Aeromonas caviae (NR_104824.1) | 100% |
| Bacillus cereus RPK20 (KX980442) | Bacillus cereus (NR_074540.1) | 99% |
| Aeromonas veronii RPK21 (KX980443) | Aeromonas veronii (NR_118947.1) | 100% |
| Aeromonas veronii RPK22 (KX980444) | Aeromonas veronii (NR_118947.1) | 99% |
| Enterobacter xiangfangensis RPK23 (KX980445) | Enterobacter xiangfangensis (NR_126208.1) | 99% |
| Enterobacter ludwigii RPK24 (KX980446) | Enterobacter ludwigii (NR_042349.1) | 99% |
| Delftia lacustris RPK25 (KX980447) | Delftia lacustris (NR_116495.1) | 99% |
| Aeromonas hydrophila RPK26 (KX980448) | Aeromonas hydrophila (NR_119190.1) | 99% |
| Escherichia fergusonii RPK27 (KX980449) | Escherichia fergusonii (NR_027549.1) | 99% |
| Aeromonas veronii RPK28 (KX980450) | Aeromonas veronii (NR_118947.1) | 99% |
| Escherichia fergusonii RPK29 (KX980451) | Escherichia fergusonii (NR_074902.1) | 99% |
| Aeromonas hydrophila RPK30 (KX980452) | Aeromonas hydrophila (NR_119190.1) | 99% |
| Shewanella xiamenensis RPK31 (KX980453) | Shewanella xiamenensis (NR_116732.1) | 99% |
| Shewanella xiamenensis RPK32 (KX980454) | Shewanella xiamenensis (NR_116732.1) | 99% |
| Shigella flexneri RPK33 (KX980455) | Shigella flexneri (NR_026331.1) | 99% |
| Aeromonas enteropelogenes RPK34 (KX980456) | Aeromonas enteropelogenes (NR_116026.1) | 99% |
| Enterobacter xiangfangensis RPK35 (KX980457) | Enterobacter xiangfangensis (NR_126208.1) | 99% |
| Shewanella seohaensis RPK36 (KX980458) | Shewanella seohaensis (NR_108852.1) | 99% |
| Enterobacter ludwigii RPK37 (KX980459) | Enterobacter ludwigii (NR_042349.1) | 99% |
| Aeromonas sanarellii RPK38 (KX980460) | Aeromonas sanarellii (NR_116584.1) | 99% |
| Shewanella seohaensis RPK39 (KX980461) | Shewanella seohaensis (NR_108852.1) | 100% |
| Aeromonas caviae RPK40 (KX980462) | Aeromonas caviae (NR_104824.1) | 100% |
| Rheinheimera chironomi RPK41 (KX980463) | Rheinheimera chironomi (NR_043699.1) | 99% |
| Aeromonas jandaei RPK42 (KX980464) | Aeromonas jandaei (NR_037013.2) | 99% |
| Exiguobacterium acetylicum RPK43 (KX980465) | Exiguobacterium acetylicum (NR_043479.1) | 99% |
| Aeromonas hydrophila RPK44 (KX980466) | Aeromonas hydrophila (NR_119190.1) | 99% |
| Aeromonas hydrophila RPK45 (KX980467) | Aeromonas hydrophila (NR_119190.1) | 99% |
| Delftia lacustris RPK46 (KX980468) | Delftia lacustris (NR_116495.1) | 99% |
| Aeromonas jandaei RPK47 (KX980469) | Aeromonas jandaei (NR_037013.2) | 99% |
| Delftia lacustris RPK48 (KX980470) | Delftia lacustris (NR_116495.1) | 99% |
| Delftia lacustris RPK49 (KX980471) | Delftia lacustris (NR_116495.1) | 99% |
| Aeromonas enteropelogenes RPK50 (KX980472) | Aeromonas enteropelogenes (NR_116026.1) | 99% |
| Klebsiella pneumoniae RPK51 (KX980473) | Klebsiella pneumoniae (NR_117683.1) | 99% |
| Shigella sonnei RPK52 (KX980474) | Shigella sonnei (NR_104826.1) | 99% |
| Escherichia fergusonii RPK53 (KX980475) | Escherichia fergusonii (NR_074902.1) | 99% |
| Escherichia fergusonii RPK54 (KX980476) | Escherichia fergusonii (NR_074902.1) | 99% |
| Citrobacter freundii RPK55 (KX980477) | Citrobacter freundii (NR_028894.1) | 99% |
| Staphylococcus warneri RPK56 (KX980478) | Staphylococcus warneri (NR_025922.1) | 96% |
| Escherichia fergusonii RPK57 (KX980479) | Escherichia fergusonii (NR_074902.1) | 96% |
| Escherichia fergusonii RPK58 (KX980480) | Escherichia fergusonii (NR_074902.1) | 99% |
| Shigella flexneri RPK59 (KX980481) | Shigella flexneri (NR_026331.1) | 99% |
| Bacillus safensis RPK60 (KX980482) | Bacillus safensis (NR_113945.1) | 99% |
| Escherichia fergusonii RPK61 (KX980483) | Escherichia fergusonii (NR_074902.1) | 99% |
| Escherichia fergusonii RPK62 (KX980484) | Escherichia fergusonii (NR_074902.1) | 99% |
| Klebsiella pneumoniae RPK63 (KX980485) | Klebsiella pneumoniae (NR_117683.1) | 99% |
| Escherichia fergusonii RPK64 (KX980486) | Escherichia fergusonii (NR_074902.1) | 99% |
| Escherichia fergusonii RPK65 (KX980487) | Escherichia fergusonii (NR_074902.1) | 99% |
Chironomid larvae collected from the lakes of Bangalore city and subjected for culturing. Genomic DNA was extracted from enriched cultures and performed 16s rRNA gene Sanger sequencing. The data was obtained, blasted in NCBI GenBank data base and identified pathogenic and non-pathogenic bacteria with 96–99% similarity.
3.1. Phylogenetic tree analysis
Phylogenetic tree was constructed for sixty five isolates and 10 related reference sequences from NCBI GenBank using MEGA 7 software to understand the taxonomic relationship (Fig. 1). Phylogenetic tree contain 7 clades namely Enterobacteriaceae, Aeromonas, Shewanella, Pseudomonas, Delftia, Exiguobacterium and Bacillus. Whereas Vibrio cholera, Rheinheimera chironomi, Acinetobacter bereziniae and Staphylococcus warneri form isolated branches. The first largest clade belongs to Enterobacteriaceae which is predominant one with 43.08% (28 isolates). Within this clade, obtained 5 genera of Enterobacter, Klebsiella, Shigella, Escherichia and Citrobacter having 99% sequence similarity and supported by high bootstrap values. Second largest clade in the phylogenetic tree was Aeromonas with 24.62% (16 isolates). Each clade of Shewanella, Delftia and Bacillus were moderately occurred with 6.15% (4 isolates each). Clades of Pseudomonas (4.62%; 3 isolates) and Exiguobacterium (3.08%; 2 isolates) showed 99% similarity. Interestingly, the isolates such as Vibrio cholera, Rheinheimera chironomi, Acinetobacter bereziniae and Staphylococcus warneri were also obtained from larvae in the present study (Fig. 1, Fig. 2).
Fig. 1.
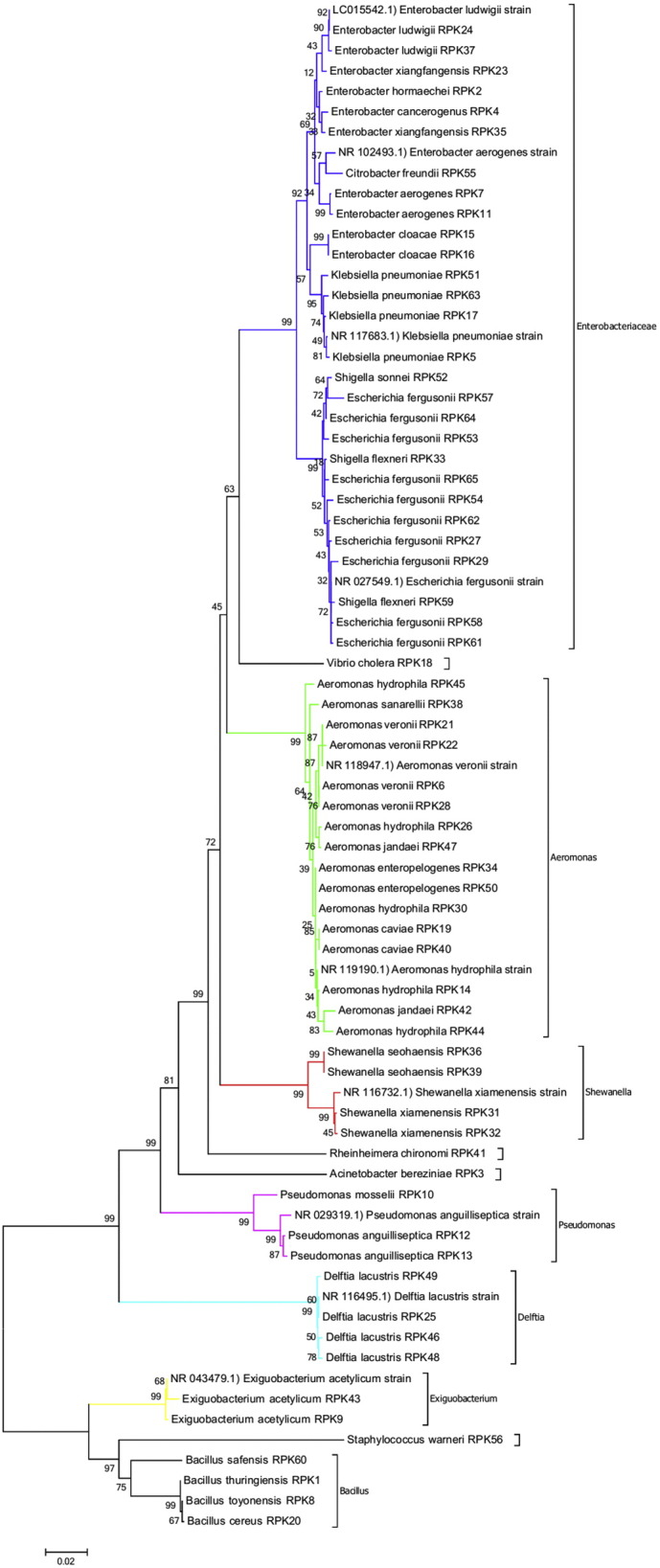
Phylogenetic relationship of partial 16s rRNA gene sequences of bacterial isolates associating with chironomid larvae samples. NJ phylogenetic tree contains 65 isolates and related reference gene sequence with accession number from NCBI GenBank. The rooted tree was constructed as Neighbor-Joining (NJ) and boot strapped with 1000 trials, using MEGA7 software.
Fig. 2.
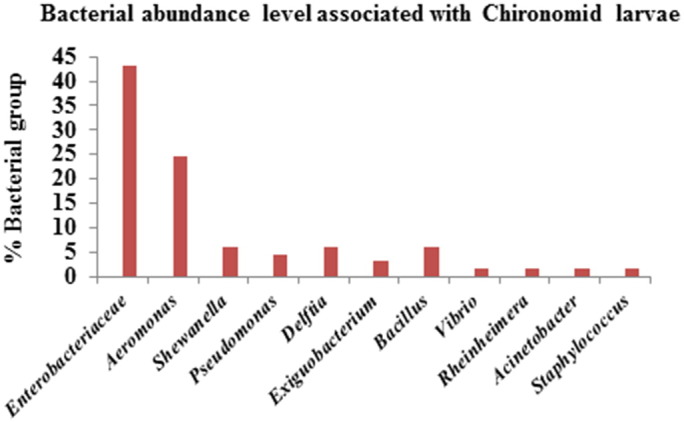
Bacterial abundance level associated with chironomid larvae.
4. Discussion
The microbial diversity revealed by investigation of microbial communities brings understanding of their characteristic features in different environmental circumstances [22], [23]. Interaction and communication of both bacterial population and host among wide range of organisms were archived previously, like with corals [24], sponges [25] and hydra [26]. Here, we studied the bacterial community associated with chironomid larvae from aquatic sediments. The chironomid larvae are surviving in aquatic sediment and make bionetwork of chironomid and endogenous bacterial communities [27], [28]. They found in almost aquatic environment and tolerable to various conditions like pH, temperature, salinity, current velocity and depth [29]. Aquatic sediments contamination with organic, inorganic pollutants [30] and chironomids as a pollutant tolerant community is well reported. Endogenous bacterial communities associating with chironomids degrade the toxicants for its host survival [5]. The bacterial population association with chironomids was identified and reported through various methods including Denaturing Gradient Gel Electrophoresis (DGGE), Clone libraries, and 454 pyrosequencing of 16s rRNA gene [4], [5]. Here, our research reveals the identification of bacterial population associated with chironomid larvae through 16s rRNA gene Sanger sequencing. The bacterial community in larvae was found to be varied significantly among different lake points. Initially, chironomid larvae samples from 11 lake points were examined individually for the bacterial identification. We obtained GenBank accession number for 65 isolates from KX980423–KX980487. Phylogenetic tree analysis explored 96–100% closed similarity on 16s rRNA sequencing for all 65 isolates based on the related reference gene sequence from NCBI GenBank accordingly.
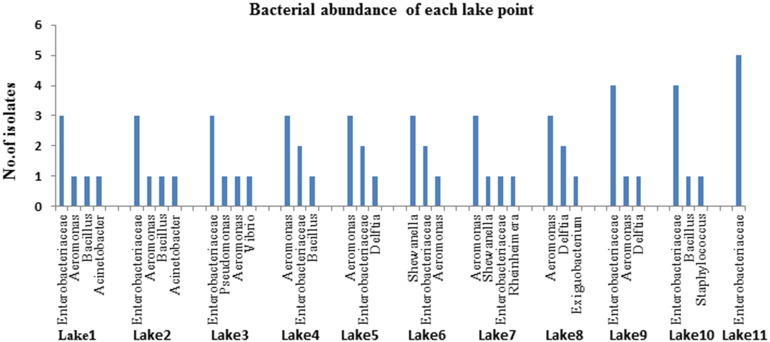
The abundance level of different bacterial population of each lake point was identified (Fig. 3) and our findings reveal that Enterobacteriaceae family (43.08%; 28 isolates) and Aeromonas (24.62%; 16 isolates) are dominating bacterial population surviving in maximum lake points. Here we found that all lakes contaminated with pathogenic organisms which include the genera of Escherichia, Klebsiella, Shigella, Enterobacter, Citrobacter, Pseudomonas, Staphylococcus, Bacillus, Vibrio, Aeromonas and also Acinetobacter, Exiguobacterium, Delftia, Rheinheimera, Shewanella. Among these identified bacterial population Aeromonas, Enterobacter and Escherichia with serotypes are commonly associated with larvae in maximum lake points. In other hand Vibrio, Pseudomonas, Klebsiella, Bacillus and other bacterial species were identified moderately in all lakes. Rouf and Rigney [31] have reported previously that chironomid larvae in lake sediment associating with following bacterial genera i.e. Achromobacter, Acinetobacter, Aeromonas, Bacillus, Citrobacter, Clostridium, Corynebacterium, Edwardsiella, Enterobacter, Escherichia, Klebsiella, Micrococcus, Pseudomonas, Serratia, Providencia, Yersinia, and Staphylococcus. Senderovich and Halpern [5] identified Exiguobacterium, Delftia and Shewanella and other metal degrading endogenous bacterial population associating with chironomid larvae and egg masses through 454-pyrosequencing. Eller et al. [32] reported that chironomid larvae and gut associating with metal detoxifying bacteria Bacteroides, Clostridium, Dysgonomonas, Hyphomicrobium, Methylobacillus, Methylobacter, Methylocaldum, and Methylomicrobium.
Fig. 3.
Bacterial abundance level of each lake point.
Very interestingly we identified the genera of Shigella and Rheinheimera from chironomid larvae with high sequence similarity with public database. Rheinheimera is Gram negative, rod shaped bacterial strain, marine isolate was previously identified from egg mass of chironomids [15]. Shigella is Gram negative, rod shaped bacteria, pathogenic organism belongs to Enterobacteriaceae family and major causative agent of dysentery and Shigella was not identified before from chironomids. In best of our knowledge, this is the first report identified Shigella and Rheinheimera from chironomids larvae of lake sediments. The hypothesis that bacterial communities could have transferred into further life stages needs to be established. Future research is required to understand the symbiotic relationship of chironomid larvae and bacterial communities.
5. Conclusion
Using 16s rRNA gene Sanger sequencing, we identified endogenous bacterial pathogens and non-pathogens from chironomid larvae of lake sediments. Our study indicates that the Bangalore city lakes were highly polluted with a diverse range of bacterial pathogens. Further study has to be taken to extend the current observation in large data set. In addition, our preliminary data may be used for the detection of antibiotic resistance gene and their relationship with other organisms found in aquatic environment.
The following are the supplementary data related to this article.
Supplementary material 1
Supplementary material 2
Conflict of interest
All authors don't have conflict of interest.
Acknowledgement
We grateful to thank Dr. Surendra K Chikara, Executive Director, Eurofins Genomics India Pvt. Ltd, India for given wonderful opportunity to perform the present study. Also we thank Eurofins Management for providing the facilities to perform the experiments. Especially we would like to thank Dr. Krishna Mohan Singh, Laboratory Manager, Eurofins Clinical Genetics India Pvt. Ltd, India for drafting this article.
References
- 1.Moor B.C., Martinez E., Gay J.M., Rice D.H. Survival of Salmonella enterica in fresh water and sediments and transmission by the aquatic midge Chironomous tetans (Chironomidae: Diptera) Appl. Environ. Microbiol. 2003;69:4556–5456. doi: 10.1128/AEM.69.8.4556-4560.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armitage P.D. Chironomidae as food. In: Armitage P.D., Cranston P.S., Pinder L.C.V., editors. The Chironomidae: Biology and Ecology of Nonbiting Midges. Chapman and Hall; London UK: 1995. pp. 423–435. [Google Scholar]
- 3.Bat L., Akbulut M. Studies on sediment toxicity bioassays using Chironomus thummi Keiffer larvae. Turk. J. Zool. 2001;25:87–93. [Google Scholar]
- 4.Senderovich Y., Halpern M. Bacterial community composition associated with chironomid egg masses. J. Insect Sci. 2012;12:149. doi: 10.1673/031.012.14901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Senderovich Y., Halpern M. The protective role of endogenous bacterial communities in chironomid egg masses and larvae. J. ISME. 2013;7:2147–2158. doi: 10.1038/ismej.2013.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hendricks C.W. Increased recovery rate of Salmonella from stream bottom sediments versus surface waters. Appl. Microbiol. 1971;21:379–380. doi: 10.1128/am.21.2.379-380.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.LaLiberte P., Grimes D.J. Survival of Escherichia coli in lake bottom sediment. Appl. Environ. Microbiol. 1982;43:623–628. doi: 10.1128/aem.43.3.623-628.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morinigo M.A., Borrego J.J., Romero P. Comparative study of different methods for detection and enumeration of Salmonella spp. in natural waters. J. Appl. Bacteriol. 1986;61:169–176. doi: 10.1111/j.1365-2672.1986.tb04272.x. [DOI] [PubMed] [Google Scholar]
- 9.Broza M., Halpern M. Chironomid egg masses and Vibrio cholerae. Nature. 2001;412:40. doi: 10.1038/35083691. [DOI] [PubMed] [Google Scholar]
- 10.Halpern M., Gancz H., Broza M., Kashi Y. Vibrio cholerae hemagglutinin/protease degrades chironomid egg masses. Appl. Environ. Microbiol. 2003;69:4200–4204. doi: 10.1128/AEM.69.7.4200-4204.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halpern M., Broza Y.B., Mittler S., Arakawa E., Broza M. Chironomid egg masses as a natural reservoir of Vibrio cholerae non-O1 and non-O139 in freshwater habitats. Microb. Ecol. 2004;47:341–349. doi: 10.1007/s00248-003-2007-6. [DOI] [PubMed] [Google Scholar]
- 12.Halpern M., Landsberg O., Raats D., Rosenberg E. Culturable and VBNC Vibrio cholerae: interactions with chironomid egg masses and their bacterial population. Microb. Ecol. 2007;53:285–293. doi: 10.1007/s00248-006-9094-0. [DOI] [PubMed] [Google Scholar]
- 13.Senderovich Y., Gershtein Y., Halewa E., Halpern M. Vibrio cholerae and Aeromonas: do they share a mutual host? J. ISME. 2008;2:276–283. doi: 10.1038/ismej.2007.114. [DOI] [PubMed] [Google Scholar]
- 14.Figueras M.J., Beaz-Hidalgo R., Senderovich Y., Laviad S., Halpern M. Re-identification of Aeromonas isolates from chironomid egg masses as the potential pathogenic bacteria Aeromonas aquariorum. Environ. Microbiol. 2011;3:239–244. doi: 10.1111/j.1758-2229.2010.00216.x. [DOI] [PubMed] [Google Scholar]
- 15.Halpern M., Senderovich Y., Snir S. Rheinheimera chironomi sp. nov., isolated from a chironomid (Diptera; Chironomidae) egg mass. Int. J. Syst. Evol. Microbiol. 2007;57:1872–1875. doi: 10.1099/ijs.0.64927-0. [DOI] [PubMed] [Google Scholar]
- 16.Halpern M., Shaked T., Pukall R., Schumann P. Leucobacter chironomi sp. nov., a chromate resistant bacterium isolated from a chironomid egg mass. Int. J. Syst. Evol. Microbiol. 2009;59:665–670. doi: 10.1099/ijs.0.004663-0. [DOI] [PubMed] [Google Scholar]
- 17.Halpern M., Shaked T., Schumann P. Brachymonas chironomi sp. nov., isolated from a chironomid egg mass, and emended description of the genus Brachymonas. Int. J. Syst. Evol. Microbiol. 2009;59:3025–3029. doi: 10.1099/ijs.0.007211-0. [DOI] [PubMed] [Google Scholar]
- 18.Raats D., Halpern M. Oceanobacillus chironomi sp. nov., a halotolerant and facultative alkaliphilic species isolated from a chironomid egg mass. Int. J. Syst. Evol. Microbiol. 2007;57:255–259. doi: 10.1099/ijs.0.64502-0. [DOI] [PubMed] [Google Scholar]
- 19.Garrity G.M., Holt J.G. Phylum BVI. Chloroflexi phy. nov. In: Boone D.R., Castenholz R.W., editors. Vol. 1: The Archaea and the Deeply Branching and Phototrophic Bacteria. In Garrity GM (ed.), Bergey's Manual of Systematic Bacteriology. second ed. Springer-Verlag; New York: 2001. pp. 427–446. [Google Scholar]
- 20.Kuncham R., Thayumanavan T., Subba Reddy G.V. Inter and intraspecific diversity of Chironomid larvae using COI and RAPD markers. J. Environ. Biol. 2016;37(6):1369–1375. [Google Scholar]
- 21.Murugkar H., Rahman H., Dulta P. Distribution of virulence genes in Salmonella serovars isolated from man and animals. Indian J. Med. Res. 2003;117:66–70. [PubMed] [Google Scholar]
- 22.Debroas D., Humbert J.F., Enault F., Bronner G., Faubladier M., Cornillot E. Metagenomic approach studying the taxonomic and functional diversity of the bacterial community in a mesotrophic lake (Lac du Bourget - France) Environ. Microbiol. 2009;11:2412–2424. doi: 10.1111/j.1462-2920.2009.01969.x. [DOI] [PubMed] [Google Scholar]
- 23.Hewson I., Paerl R.W., Tripp H.J., Zehr J.P., Karl D.M. Metagenomic potential of microbial assemblages in the surface waters of the central Pacific Ocean tracks variability in oceanic habitat. Limnol. Oceanogr. 2009;54:1981–1994. [Google Scholar]
- 24.Rohwer F., Seguritan V., Azam F., Knowlton N. Diversity and distribution of coral associated bacteria. Mar. Ecol. Prog. Ser. 2002;243:1–10. [Google Scholar]
- 25.Friedrich A.B., Fischer I., Proksch P., Hacker J., Hentschel U. Temporal variation of the microbial community associated with the mediterranean sponge Aplysina aerophoba. FEMS Microbiol. Ecol. 2001;38:105–113. [Google Scholar]
- 26.Fraune S., Bosch T.C.G. Long-term maintenance of species-specific bacterial microbiota in the basal metazoan hydra. Proc. Natl. Acad. Sci. U. S. A. 2007;104:13146–13151. doi: 10.1073/pnas.0703375104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coffman W.P., Ferrington L.C. Chironomidae. In: Merritt R.W., Cummins K.W., editors. An Introduction to the Aquatic Insects of North America. second ed. Kendall/Hunt; Dubuque: 1984. pp. 551–652. [Google Scholar]
- 28.Pinder L.C.V. Biology of freshwater chironomidae. Annu. Rev. Entomol. 1986;31:1–23. [Google Scholar]
- 29.Armitage P., Cranston P.S., Pinder L.C.V. Chapman and Hall; London, Glasgow, New York, Tokyo, Melbourne, Madras: 1995. The Chironomidae: The Biology and Ecology of Non-biting Midges; p. 572. [Google Scholar]
- 30.Salomons W., de Rooij N.M., Kerdijk H., Bril J. Sediment as a source for contaminants. Hydrobiologia. 1987;149:13–30. [Google Scholar]
- 31.Rouf M.A., Rigney M.M. Bacterial florae in larvae of the lake fly Chironomus plumosus. Appl. Environ. Microbiol. 1993;59:1236–1241. doi: 10.1128/aem.59.4.1236-1241.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eller G., Deines P., Krüger M. Possible sources of methane-derived carbon for chironomid larvae. Aquat. Microb. Ecol. 2007;46:283–293. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2