Abstract
Obesity, a chronic state of energy overload, is characterized by adipose tissue dysfunction that is considered to be the major driver for obesity associated metabolic complications. The reasons for adipose tissue dysfunction are incompletely understood, but one potential contributing factor is adipose tissue mitochondrial dysfunction. Derangements of adipose tissue mitochondrial biogenesis and pathways associate with obesity and metabolic diseases. Mitochondria are central organelles in energy metabolism through their role in energy derivation through catabolic oxidative reactions. The mitochondrial processes are dependent on the proper NAD+/NADH redox balance and NAD+ is essential for reactions catalyzed by the key regulators of mitochondrial metabolism, sirtuins (SIRTs) and poly(ADP-ribose) polymerases (PARPs). Notably, obesity is associated with disturbed adipose tissue NAD+ homeostasis and the balance of SIRT and PARP activities. In this review we aim to summarize existing literature on the maintenance of intracellular NAD+ pools and the function of SIRTs and PARPs in adipose tissue during normal and obese conditions, with the purpose of comprehending their potential role in mitochondrial derangements and obesity associated metabolic complications. Understanding the molecular mechanisms that are the root cause of the adipose tissue mitochondrial derangements is crucial for developing new effective strategies to reverse obesity associated metabolic complications.
Keywords: Obesity, Mitochondria, Adipose tissue, Sirtuins, Poly(ADP-ribose) polymerases, NAD+
Graphical abstract
Highlights
-
•
NAD+ homeostasis is critical for mitochondrial and adipose tissue function.
-
•
Obesity is linked with disturbed adipose tissue mitochondrial and NAD+ homeostasis.
-
•
Imbalance of NAD+ consumers SIRTs and PARPs impairs mitochondrial function.
-
•
SIRT activation via NAD+ boosting could be used to prevent metabolic complications.
1. Introduction
Adipose tissue is the most plastic organ in the body, with an ability to enlarge and contract several-fold in response to alterations in energy balance. This process requires fine-tuned regulation of the number of adipocytes through adipogenesis of new and removal of nonviable cells, and the size of the existing adipocytes through lipogenesis and lipolysis. In addition, the other cell types present in adipose tissue namely the preadipocytes, fibroblasts, endothelial cells and immune cells must work in concert with the adipocytes to support proper adipose tissue function and homeostasis. Adipose tissue can be categorized into white and brown adipose tissue (WAT and BAT, respectively) based on cell morphology and tissue function. While brown adipocytes have multiple lipid droplets and are rich in mitochondria, enough to contribute to their brown color, white adipocytes have a unilocular lipid droplet with relative rarefaction of mitochondria and other cytosolic organelles.
The primary function of WAT is considered to be to buffer whole body energy balance through lipid handling, whereas BAT is specialized to produce heat by an energy dissipating process of non-shivering thermogenesis – both relevant for systemic energy balance homeostasis. Moreover, adipose tissue secretes hormones and adipocytokines that affect metabolic health through systemic effects on the metabolism of other tissues. Adipose tissue needs to continuously respond to physiological stimuli, such as alterations in energy yield, fasting and meals (WAT), or cold exposure (BAT). In these responses, adipose tissue mitochondria are relevant organelles for both the energy handling and endocrine function [1], [2]. Thus it is perhaps not surprising that activity of adipose tissue mitochondria seems to be especially important for the maintenance of whole-body metabolic health, characterized by high insulin sensitivity, low liver fat, and absence of low-grade inflammation [3], [4].
According to the adipose tissue expandability hypothesis [5], [6], the root cause for ectopic lipid accumulation is the inability of the adipose tissue to enlarge and handle excess nutrients leading to lipid spill over to ectopic sites (such as to the liver and muscle) and to the systemic lipotoxic insults [6]. Interestingly, subsequent studies have demonstrated that downregulation of mitochondrial pathways in adipose tissue correlates with hepatic steatosis and insulin resistance [7]. Thus, mitochondrial dysfunction may be the underlying reason for the adipose tissue expandability problem. In line with this notion, adipose tissue derived stem cells from obese individuals have reduced proliferative capacity and loss of viability [8]. As a source of building blocks for biosynthetic processes and ATP, mitochondrial metabolism is essential for active adipocyte proliferation and differentiation for tissue maintenance and expansion [9]. Moreover, earlier global transcriptomics results showed that pathways regulating adipocyte differentiation, together with mitochondrial oxidative metabolism, were significantly downregulated in obese adipose tissue [7], especially in the metabolically unhealthy obese subjects [10]. Thus, loss of mitochondrial function may result in reduction of adipogenic capacity and maintenance of mature adipocytes, which can contribute to cell death, inflammation and global adipose tissue dysfunction. This mitochondrial downregulation may also explain the paradox why adipose cell differentiation and storage of triglycerides has been proposed to be hampered in obesity [7], [11], [12].
Mitochondria are thus key organelles in several fundamental aspects of adipose tissue physiology. Mitochondrial metabolic processes are largely dependent on the NAD+/NADH redox couple, and the disruption of NAD+ homeostasis as well as mitochondrial dysfunction have been suggested to be major causes for obesity and metabolic complications. Mitochondrial metabolism is regulated via two NAD+-dependent enzyme families, sirtuins (SIRTs) and poly(ADP-ribose) polymerases (PARPs), which have opposing effects on metabolism [13], [14]. Until now, most studies on SIRTs and PARPs have been performed in nonadipose tissue and mouse models. Less is known about their functions in adipose tissue or in obesity, especially in humans. The purpose of this review is to discuss the NAD+/NADH redox couple dependent mitochondrial pathways in obesity and the maintenance of adequate NAD+ homeostasis in adipose tissue. Moreover, we will present the current knowledge of SIRTs and PARPs in adipose tissue and elucidate what is known about their role in mitochondrial metabolism, obesity and metabolic health.
2. Mitochondrial metabolism and obesity
2.1. Mitochondrion as an organelle
Mitochondria are essential organelles of eukaryotic organisms and are present in all nucleated mammalian cells. They are double membrane bound subcellular structures where the membranes line two distinct compartments: the matrix within the inner membrane and the intermembrane space between the two membranes. Mitochondria possess their own genome, mitochondrial DNA (mtDNA), which encodes for 13 proteins essential for the function of the organelle and the RNA molecules for their translation. However, as over a 1000 proteins have been found in mitochondria [15], the majority of them are encoded in the nucleus and the proteins are imported into mitochondria after synthesis in the cytosol, thus, necessitating a uniquely coordinated effort of the nuclear mitochondrial genes and mtDNA encoded genes for proper mitochondrial function.
Mitochondria are highly dynamic structures and their morphology, which is relevant for function, can vary from fragmented separate mitochondria to elongated interconnected reticulum depending on the cell type or tissue and physiological signals [16]. The morphology is determined by the balance of fusion and fission events mediated by a set of dedicated GTPase proteins, namely optic atrophy 1, mitofusin 1 and mitofusin 2 for fusion and dynamin-1-like for fission. Moreover, mitochondria form functionally relevant and stable contacts with other subcellular organelles, especially the endoplasmic reticulum. These contacts with the endoplasmic reticulum have recently been shown to participate in determination of mitochondrial morphology through mitochondrial fission [17].
In addition to morphology, mitochondrial mass and activity, determined by the balance of mitochondrial biogenesis and turnover, vary greatly between different tissues and in response to physiological signals. The biogenesis of mitochondria is a tightly regulated process. Key regulators in coordinating mitochondrial biogenesis and metabolism in response to physiological demand are two co-transcriptional regulators, peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) and nuclear receptor co-repressor 1, which have opposing effects on mitochondrial biogenesis and oxidative metabolism. While PGC-1α increases mitochondrial biogenesis and oxidative metabolism during conditions of increased cellular energy demands, such as exercise and cold exposure, [18], [19], [20], nuclear receptor co-repressor 1 represses mitochondrial biogenesis during energy excess [21], [22], [23]. These regulators affect several transcription factors, the peroxisome proliferator-activated receptor (PPAR) and estrogen related receptor families for example, to regulate gene expression of mitochondrial genes [24].
Less is known about mitochondrial turnover, which is governed by several processes. Mitochondrial proteins can be degraded through proteases within the mitochondria and by the proteasome system on the mitochondrial outer membrane [25], [26]. If the proteolytic system is overwhelmed or mitochondrial proteostasis is otherwise disrupted, a protective stress response called the mitochondrial unfolded protein response is triggered. The mitochondrial unfolded protein response leads to upregulation of mitochondrial chaperones and proteases to alleviate the stress [27]. Mitochondria can also bud off vesicles containing proteins destined for degradation in the lysosomes [28]. Furthermore, degradation of entire organelles through macroautophagy (referred to as mitophagy) has been described. During mitophagy damaged mitochondria are segregated from the mitochondrial network and targeted for degradation through a mechanism dependent on PTEN-induced putative kinase 1 and E3 ubiquitin ligase parkin proteins [29]. However, the relative contribution of these various mechanisms to turnover of mitochondria in vivo in different tissues is at present unclear.
2.2. Mitochondrial metabolism and NAD+/NADH redox reactions
Mitochondria have many important functions in the cell, but given the scope of this review, we will here focus on their vital role in cellular energy and metabolic homeostasis. One of the most prominent mitochondrial functions is oxidative metabolism, in which carbon fuels from food (carbohydrates, proteins and fats) are catabolized and converted to ATP, the major cellular energy currency, through mitochondrial respiration. ATP is also produced in the cytosol through glycolysis, but mitochondrial oxidative energy production is more efficient than anaerobic energy metabolism. Both anaerobic and aerobic metabolism rely on the NAD+/NADH redox couple for the production of energy. In these reactions NAD+ accepts a hydride ion (H-) from the coupled reactant, reducing NAD+ consequently to NADH and oxidizing the reactant. The role of NAD+ in transferring hydrogen in biochemical reactions was first discovered almost 100 years ago by Otto Warburg and colleagues [30].
In the cytoplasm, aerobic glycolysis requires two NAD+ molecules to convert one glucose molecule to pyruvate (Fig. 1) [31]. In this process glucose is first converted to two glyceraldehyde-3-phosphate molecules and successively oxidized into 1–3-biphosphoglycerate by glyceraldehyde-3-phosphate dehydrogenase. In parallel, glyceraldehyde-3-phosphate dehydrogenase reduces NAD+ to NADH. Through a few intermediate steps 1–3-biphosphoglycerate is converted to pyruvate, which is then directed to mitochondria for aerobic respiration, i.e. glucose oxidation. In mitochondria, pyruvate undergoes irreversible oxidative decarboxylation, catalyzed by pyruvate dehydrogenase complex, resulting in the formation of acetyl-CoA and the reduction of NAD+ to NADH. As the mitochondrial inner membrane is impermeable to both NAD+ and NADH, NADH is transferred into the mitochondria via either the malate-aspartate or the glycerol-3-phosphate shuttles [32].
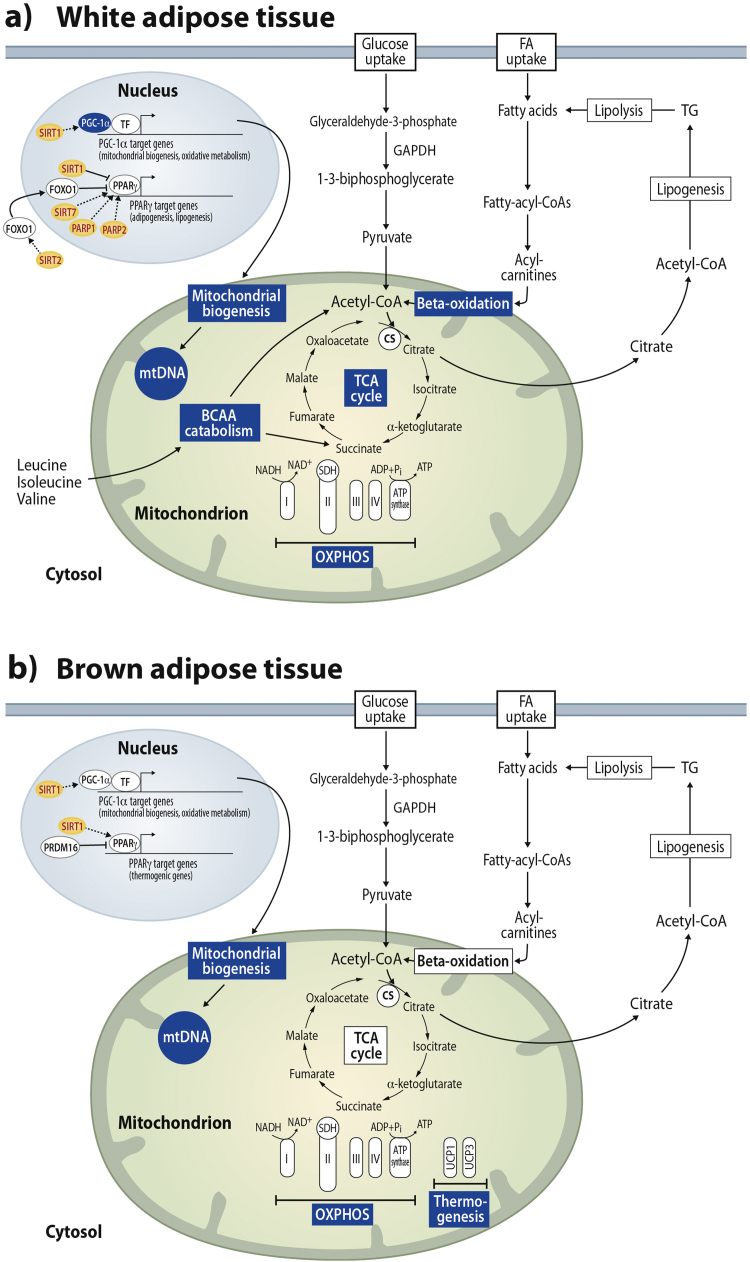
Fig. 1.
Adipose tissue metabolic and mitochondrial pathways in white (A) and brown (B) adipose tissue. Pyruvate and fatty-acyl-carnitines derived from glycolysis and break-down of fatty acids, respectively, enter the mitochondria where they are further catabolized to acetyl-CoA by the pyruvate dehydrogenase complex and beta-oxidation. The acetyl-CoA enters the TCA cycle and the high-energy electrons derived from the TCA cycle are used to power ATP production through oxidative phosphorylation (OXPHOS). Citrate derived from the TCA cycle is used a precursor for lipogenesis. In white adipose tissue, acetyl-CoA and succinyl-CoA derived from branched chain amino acid (BCAA) catabolism also enter the TCA cycle (A). In brown adipose tissue, the uncoupling proteins (UCPs) induce thermogenesis by uncoupling mitochondrial respiration from ATP production (B). Pathways downregulated by obesity are highlighted in blue. CS; citrate synthase, FA; fatty acid, FOXO1; forkhead box O1, GAPDH; glyceraldehyde-3-phosphate dehydrogenase, mtDNA;mitochondrial DNA, PARP; poly(ADP-ribose) polymerases, PGC-1α; peroxisome proliferator-activated receptor gamma coactivator 1-alpha, PPARγ; peroxisome proliferator-activated receptor gamma, PRDM16; PR domain containing 16, SDH; succinate dehydrogenase, SIRT; sirtuin, TCA; tricarboxylic acid, TF; transcription factor and TG; triglycerides.
In addition to glucose oxidation, acetyl-CoA and NADH can be generated from fatty acids through β-oxidation (Fig. 1) [33]. Fatty acids are conjugated with a CoA group in the cytosol, converted to long-chain acyl carnitine to be transported across the mitochondrial membranes and converted back to long-chain acyl-CoA, which enters the fatty acid β-oxidation pathway, i.e. degradation of acyl-CoA to acetyl-CoA. One cycle of β-oxidation is a four-step process during which acetyl-CoA, NADH, FADH2 and new two carbons shorter acyl-CoA are produced.
The acetyl-CoA produced either through glucose oxidation or fatty acid β-oxidation is directed to the mitochondrial tricarboxylic acid (TCA) cycle, the final common pathway for oxidation of fuel molecules (Fig. 1). In the TCA cycle, in concert with other enzymatic reactions isocitrate dehydrogenase, α-ketoglutarate dehydrogenase and malate dehydrogenase oxidize their substrates and at the same time reduce NAD+ to NADH [34].
Finally, the high-energy electrons derived from the TCA cycle carried by NADH and FADH2 are sequentially transferred through the respiratory chain complexes (Fig. 1) [31] and their energy is used to generate a proton motive force across the inner mitochondrial membrane by coupling the electron transport to pumping of protons across the inner mitochondrial membrane by complexes I, III and IV. This electrochemical gradient is utilized by the ATP synthase to generate ATP in oxidative phosphorylation (OXPHOS). In the respiratory chain, the oxidation of NADH by complex I yields NAD+.
Mitochondria are also central for other metabolic processes. One of them is catabolism of branched chain amino acids (BCAAs), valine, leucine and isoleucine. BCAAs are first deaminated to α-ketoacids and subsequently converted into TCA cycle intermediates acetyl-CoA or succinyl-CoA through oxidative reactions that yield NADH (Fig. 1A) [35]. In addition to its role in oxidative metabolism, the TCA cycle is an essential anabolic pathway providing precursor carbon metabolites for biosynthetic processes, such as citrate for fatty acid and sterol synthesis, and oxaloacetate and α-ketoglutarate for non-essential amino acid synthesis [31].
Overall, many of the metabolic reactions in mitochondria are dependent on the NAD+/NADH redox couple. Thus, disruption of the cellular NAD+/NADH homeostasis has the potential to cause pervasive disturbances in mitochondrial function and metabolism.
3. Obesity-associated adipose tissue mitochondrial derangements
3.1. WAT derangements
The relatively low abundance of mitochondria in white adipocytes has led to the underestimation of their role in WAT. However, quite recently, the WAT mitochondrial function has gained more interest, especially after the notion that mitochondrial biogenesis and oxidative functions are downregulated in human subcutaneous WAT in obesity [11]. Importantly, downregulation of mitochondrial oxidative pathways in obesity was recapitulated in isolated mature adipocytes [36], showing that despite their assumed primary function as lipid storage, white adipocytes do have an important oxidative role in regulating the energy balance. As mentioned before, mitochondrial transcriptional downregulation in WAT has been closely linked to metabolically unhealthy obese phenotypes [10] and may thus have an effect on the development of metabolic disturbances in obesity.
The majority of the data available from mouse and human studies show that mitochondrial mass is reduced in obese WAT. Mitochondrial mass in tissues is assessed through analysis of several parameters including relative mtDNA copy number, protein amount of mitochondrial proteins and transmission electron microscopy based evaluation of mitochondrial area. The copy number of mtDNA is reduced in genetic mouse models of obesity, human obese diabetic patients, the heavier co-twin of monozygotic twins discordant for BMI, and correlates inversely with BMI Fig. 1 [1], [7], [11], [37], [38], [39]. A reduction in the WAT OXPHOS subunit protein amount is also seen in high-fat diet (HFD) and genetically induced obesity in mice, in the heavier monozygotic twins compared to their leaner co-twins and in isolated adipocytes from obese individuals [1], [11], [36], [38], [40]. Moreover, a reduction in mitochondrial size or total area within the adipocytes was observed by transmission electron microscopy analysis in obese and obese diabetic mice, respectively [38].
To link changes in mitochondrial mass to biogenesis and turnover, the gene expression and protein levels of key regulators in these pathways have been analyzed. In mice, the expression of PGC-1α is reduced in WAT in both HFD and genetically induced obesity (Fig. 1) [40], [41]. Moreover, PGC-1α expression is diminished in both WAT and isolated adipocytes of the heavier co-twin of BMI discordant monozygotic twins [11], [36] and in obese diabetic patients [37], indicating a reduced activity of the mitochondrial biogenesis program. Data on mitochondrial turnover in obese WAT are less clear. In HFD induced obese mouse WAT, the protein levels of mitophagy markers including PTEN-induced putative kinase 1 and E3 ubiquitin ligase parkin are increased, indicating increased mitochondrial turnover [40]. In contrast, WAT mRNA levels of the same markers correlate inversely with body weight and positively with insulin sensitivity in mice [42]. Current data on the mitochondrial unfolded protein response pathway are obese WAT is also limited, but expression level of genes in this pathway were decreased in the heavier co-twin of BMI discordant monozygotic twins [43], and increased by treatment with anti-diabetic medication in WAT of obese diabetic mice [41].
Transcriptomics analyses of WAT in obesity have consistently shown a global downregulation of gene expression in pathways related mitochondrial metabolism and function, such as OXPHOS, fatty acid beta-oxidation and BCAA catabolism (Fig. 1A), in both mouse models and humans. In mice, HFD and genetically induced obesity led to reduction in gene expression in mitochondrial pathways including the TCA cycle, OXPHOS, fatty acid beta-oxidation and BCAA catabolism in WAT [41], [44], [45]. In diabetic obese patients, genes in OXPHOS pathway were downregulated in both visceral and subcutaneous WAT [46]. In healthy monozygotic BMI discordant twins, many mitochondrial pathways including the TCA cycle, OXPHOS, fatty acid beta-oxidation and BCAA catabolism are downregulated in the heavier co-twin [7], [11], and the downregulation of OXPHOS and BCAA catabolism is also seen in isolated adipocytes of the heavier co-twin [36]. Global proteomics analysis supports the transcriptomics findings of downregulation of mitochondrial pathways [47], [48]. Hence, it seems clear that obesity leads to a systemic downregulation of mRNA and protein level expression in mitochondrial pathways in both mice and humans.
The key enzyme of the mitochondrial TCA cycle, citrate synthase (Fig. 1A), catalyzes the formation of citrate that is used as a precursor for fatty acid synthesis. The activity of citrate synthase is often used as a marker for mitochondrial mass, but given the role of this enzyme in fatty acid metabolism, it is likely that its abundance and activity is regulated by HFD/obesity and should thus be considered separately. The activity of citrate synthase is reduced in HFD and genetically induced obesity mouse models, and it should be noted that mtDNA copy number was not significantly altered in these models [40], [41]. Reduced citrate synthase activity has also been demonstrated in mitochondria of both subcutaneous and visceral adipocytes of obese humans [49], [50], [51], but no difference in citrate synthase activity in mitochondria isolated from subcutaneous WAT between lean controls, obese or obese diabetic patients has also been reported [52]. Despite this discrepancy the majority of the existing data support a reduction of citrate synthase activity in obese WAT in mice and humans.
The mitochondrial oxidative capacity has also been directly assessed in WAT. A recent study used high-resolution respirometry to show reduced respiration capacity in mitochondria isolated from both subcutaneous and visceral adipocytes of several genetically and HFD induced obesity mouse models, providing strong evidence for impaired WAT mitochondrial oxidative capacity in obesity (Fig. 1A) [53]. These results are in agreement with previous studies that showed reduced oxygen consumption and fatty acid oxidation rate when normalized to cell number in white adipocytes isolated from obese mice [38], [54]. However, no alterations in basal oxygen consumption levels has been reported when measured from WAT explants and normalized to tissue mass [40]. Most human studies show that respiration or enzymatic activities of the respiratory chain complexes are reduced in isolated mitochondria from obese subcutaneous adipocytes and adipose tissue [50], [51] or visceral adipocytes [51]. Recent findings examining preadipocytes isolated from mouse and human WAT in cell culture setting indicate that the obesity-associated defects in mitochondrial respiration persist in cell culture [8].
Overall, the association between mitochondrial abnormalities in WAT and obesity is strong, but the causal pathogenic relationship and whether mitochondria are per se defective or if mitochondrial dysfunction is a reflection of decreased mitochondrial content is uncertain. Based on the limited number of studies showing impaired mitochondrial respiration in isolated mitochondria, it appears probable that functional defects intrinsic to mitochondria are present in obese WAT in addition to decreased content. Interestingly, acquired obesity associates with impaired mitochondrial biogenesis and low mitochondria number in young metabolically healthy monozygotic obesity-discordant twin pairs [11]. Thus, we suspect that mitochondrial abnormalities precede the development of metabolic syndrome in obese subjects.
3.2. BAT derangements
BAT is abundant and active in small rodents and newborn humans but also in adult humans upon cold exposure [55], [56]. Brown adipocytes regulate body temperature via thermogenesis by uncoupling ATP synthesis from OXPHOS (Fig. 1B), and thus, dissipating energy as heat. An essential gene for the induction of thermogenic response and characteristic of brown adipocytes is uncoupling protein 1 (UCP1) (Fig. 1B). Its transcription is activated by the co-operation of the transcriptional regulators PGC-1α, PPARγ and PR domain containing 16 (Prdm16), which also promote the expression of the fatty acid oxidation and respiratory complex subunit genes in BAT [57], [58], [59].
Obesity is characterized by impaired function, enlargement and whitening of BAT, as evidenced by several animal and human studies [60], [61], [62], [63]. Impaired function of BAT in obesity has been proposed to be caused by the rarefaction of vasculature and development of hypoxia [62]. The impaired BAT function is described by diminished β-adrenergic signaling, reduction of thermogenesis and the UCP1 protein amount, decreased mitochondrial content, and impaired respiratory complex activities (Fig. 1B) [62], [64]. This results in the formation of a large, unilocular lipid droplet in brown adipocytes, and hence, increased brown adipocyte size and expansion of the overall tissue size giving obese BAT a whitened appearance.
4. The maintenance of NAD+ homeostasis in adipose tissue
4.1. Introduction
As discussed in the previous section, obesity is characterized with derangements of mitochondrial metabolism. As many of the mitochondrial enzymatic pathways rely on the redox couple NAD+/NADH, it is not surprising that obesity was recently noticed to associate with deteriorated NAD+ metabolism in adipose tissue in mice [65], [66], [67] and humans [68], [69]. Thus, the preservation of adequate NAD+ homeostasis is most likely essential for the function of adipose tissue mitochondrial metabolism and metabolic health. In the next section, we will thus discuss intracellular NAD+ pools, NAD+ biosynthesis and consumption pathways, and potential NAD+ boosting strategies in obesity.
4.2. Intracellular NAD+ levels and compartmentalization
Total intracellular NAD+ levels are normally maintained around 50 pmol/mg tissue in mouse WAT whereas NAD+ levels are markedly higher (approximately 100–500 pmol/mg tissue) in mouse BAT and other peripheral tissues [70], [71]. There are four different subcellular NAD+ compartments inside the mammalian cell: nuclear/cytosolic, mitochondrial, peroxisomal and endoplasmic reticulum/golgi [72]. Nuclear and cytosolic pools of NAD+ are considered to form one combined pool as NAD+ has been suggested to be freely exchangeable between these two cell compartments. As mitochondrial membrane is impermeable to NAD+, the mitochondrial and nuclear/cytoplasmic NAD+ pools are connected via the malate/aspartate and the glycerol-3-phosphate shuttles [73]. Adequate NAD+ homeostasis is maintained in nuclear/cytosolic and mitochondrial pools by autonomous NAD+ biosynthesis [72], [73]. The peroxisomal NAD+ pool is possibly preserved by the import of the NAD+ from the cytosol through the carrier protein. Currently, the role of NAD+ in endoplasmic reticulum and golgi is not well understood. Of the different subcellular compartments, NAD+ concentration is highest in the mitochondrial compartment which contains almost 70% of total cellular NAD+ [73]. However, this notion needs to be confirmed when future technical improvements will allow us to measure NAD+ levels in different cell compartments.
4.3. NAD+ biosynthesis
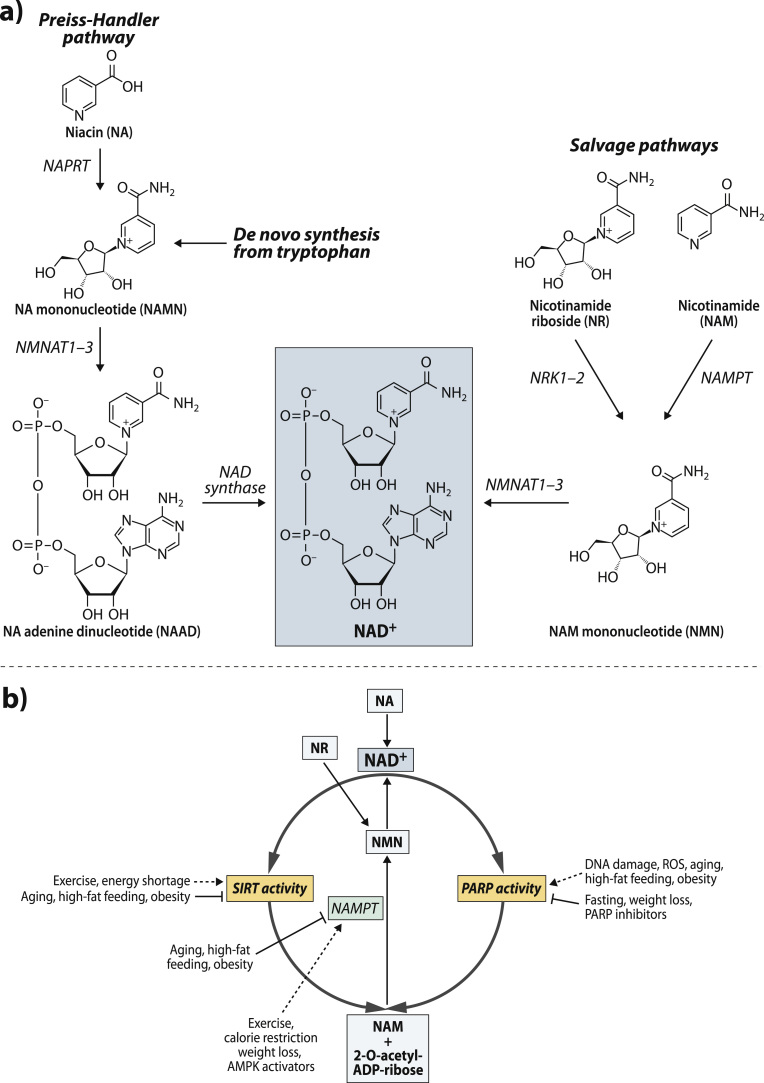
Intracellular NAD+ availability is controlled by biosynthesis of NAD+, which can occur via de novo synthesis from the dietary derived amino acid tryptophan or salvage pathways (Fig. 2A). As enzymes involved in the de novo synthesis pathway are not well expressed in adipose tissue [74], this pathway most likely has a negligible role in NAD+ biosynthesis in adipose tissue in mammals. Thus, the majority of the NAD+ in adipose tissue is synthesized via salvage pathways, which require the uptake of NAD+ precursors from the diet [74]. Forms of the dietary vitamin B3, niacin (NA), nicotinamide (NAM) and nicotinamide riboside (NR), can serve as the NAD+ precursor in the salvage pathways (Fig. 2A). Unfortunately, the preference for the NAD+ precursor use in WAT and BAT has not yet been properly investigated. As BAT is more metabolically active than WAT, BAT is likely to possess higher capacity for NAD+ biosynthesis than WAT. This notion is supported by the finding that expression of the key enzymes involved in the salvage pathways is higher in BAT versus WAT [75].
Fig. 2.
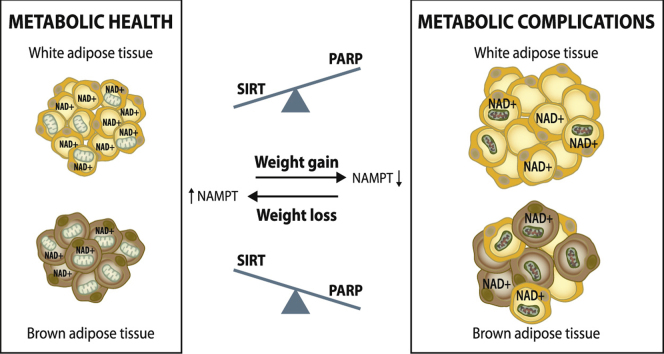
Cellular NAD+ biosynthesis and consumption processes. (A) NAD+ can be synthesized de novo from the amino acid tryptophan and through the salvage pathway from nicotinamide (NAM) or nicotinamide riboside (NR) or niacin (NA). (B) The competition of SIRTs and PARPs for the same intracellular NAD+ pool in the cell. In addition, effect of physiological stimuli on cellular NAD+ biosynthesis via nicotinamide phosphoribosyltransferase (NAMPT) and the activities of sirtuins (SIRTs) and poly(ADP-ribose) polymerases (PARPs). AMPK; AMP-activated protein kinase, NRK; nicotinamide riboside kinase, NMNAT; nicotinamide mononucleotide adenylyltransferase, NAPRT; niacin phosphoribosyltransferase and ROS; reactive oxygen species.
The NAD+ precursor NAM can be derived from the diet but it is also the end product of several intracellular NAD+ degradation reactions (see next Section 4.4). WAT and BAT most likely recycle and use intracellularly produced NAM readily for NAD+ synthesis. In the cytosol, NAM is first converted to NAM mononucleotide (NMN) by the rate-limiting enzyme, NAM phosphoribosyltransferase (NAMPT) (also known as visfatin) (Fig. 2A). Interestingly, in addition to an intracellular form of NAMPT, an extracellular circulating form [76] secreted from both WAT and BAT exists, but the function of this extracellular form is still unclear. However, it has been suggested to maintain normal NAD+ levels in hypothalamus [70] and regulate insulin secretion in pancreas [76]. After the rate-limiting NAMPT step, NMN is converted to NAD+ by NMN adenylyltransferases (NMNATs) (Fig. 2A). There are three NMNAT isoforms (NMNAT1-3) with different subcellular distributions in mammalian adipose tissue. NMNAT1 is mainly a nuclear enzyme, while NMNAT2 is localized to the golgi complex and cytosol, and NMNAT3 is found in cytosol and mitochondria. Thus, these different NMNAT isoforms are most likely responsible for modulating cell compartment-specific NAD+ pools.
Adipose tissue seems to also utilize NR as a precursor for NAD+ biosynthesis [71]. After entering the cell via equilibrative nucleoside transporters, NR is converted to NMN in the cytosol by the rate-limiting enzyme, NR kinase 1 (NRK1) (Fig. 2A) [75]. The subsequent conversion of NMN to NAD+ is catalyzed by NMNATs (Fig. 2A). NRK1 gene and protein expression is higher in BAT by comparison to WAT suggesting that NR is more readily metabolized in BAT [75]. This notion is supported by the finding that NR supplementation increased intracellular NAD+ more efficiently in BAT than WAT in wild-type mice [71]. The other isoform of NRK enzyme, NRK2, is not expressed in WAT or BAT [75].
The third NAD+ precursor, NA, can be converted to NAD+ inside the cell via the Preiss-Handler pathway in adipose tissue [74] (Fig. 2A). NA is initially metabolized to form NA mononucleotide (NAMN) in the reaction catalyzed by NA phosphoribosyltransferase (NAPRT) (Fig. 2A). This is followed by the conversion of NAMN to NA adenine dinucleotide (NAAD) by NMNATs. In the final step in this pathway NAAD is amidated to form NAD+ by the NAD synthase in an ATP-dependent manner (Fig. 2A).
4.4. NAD+ consumers SIRTs and PARPS
The intracellular NAD+ levels are also influenced via the action of several NAD+ consumer families such as SIRTs, PARPs and the NAD+ glycohydrolases CD38 and CD157 [77]. All these enzymes cleave NAD+ to produce different ADP-ribosyl products and NAM. However, in this review we focus on two most studied enzyme families: SIRTs and PARPs, which are critical regulators of several distinct cellular processes, such as DNA repair, inflammation, and differentiation but also have an important role in mitochondrial metabolism with opposing functions.
SIRTs are regulators that affect their target proteins activities through protein post-translational modifications such as deacetylation [14]. The most of the SIRTs transfer an acetyl group from their target proteins to ADP-ribose moiety of NAD+ to form 2-O-acetyl-ADP-ribose and NAM (Fig. 2B) [14]. The family comprises of seven proteins (SIRT1–SIRT7), which vary in tissue specificity, subcellular localization, enzymatic activities and targets. Importantly, SIRTs and especially SIRT1, the most-studied SIRT family member, play an important role in the regulation of metabolic health by activating mitochondrial biogenesis and function.
PARPs, the enzyme family of up to 17 members, constitute the major NAD+ consuming activity in the cell [78]. PARP1 is considered to be the isoform responsible for the main PARP activity (85–90%) in the cell while PARP2 accounts for the remaining PARP activity. PARP1 and PARP2 are ubiquitously expressed in mammalian tissues and mainly localized in nucleus. PARPs catalyze the reaction in which ADP-ribose moiety from NAD+ is transferred to form branched poly(ADP-ribose) polymers on their target proteins and as a side-product NAM is generated (Fig. 2B) [79]. PARPs have an opposing effect of mitochondrial function than SIRT1. Deletion of Parp1 or pharmacological PARP inhibition improve mitochondrial function in mice [80], [81].
It has been suggested that SIRT1 and PARP1 can interact and subsequently influence each other's enzyme activities because they compete for the same intracellular NAD+ pool (Fig. 2B) [78]. However, as the Km of PARP1 for NAD+ is many fold lower than intracellular NAD+ levels, SIRT1 activity is unlikely to reduce NAD+ content to the level limiting for PARP1 activity. Instead, SIRT1 can reduce PARP1 activity via deacetylation of PARP1 [82]. In contrast to PARP1, the Km of SIRT1 for NAD+ is within the physiological range of intracellular NAD+ concentrations. Therefore, SIRT1 activity is critically affected by changes in NAD+ levels and PARP1 may be able to dampen SIRT1 activity as its activation can deplete intracellular NAD+ pools even by 80–90% [74]. In support of this notion, genetic deletion of Parp1 or pharmacological PARP inhibition increase intracellular NAD+ levels and activates SIRT1 in muscle and BAT in mice [80], [81]. It has been also suggested that PARP1 might inhibit SIRT1 via PARylation but this type of cross-modification has not been observed at least in myotubes [80]. Overall, the SIRT1-PARP1 interaction seems to exist in muscle and BAT but needs to be studied in more detail in adipose tissue.
4.5. The effect of physiological conditions and obesity on NAD+ biosynthesis and consumption in adipose tissue
Intracellular NAD+ levels fluctuate through the day in response to various physiological stimuli but normally not more than 2-fold [74]. The regulation of the rate-liming enzyme for conversion of NAM, NAMPT, is the best studied and understood. Gene and protein expression of NAMPT shows diurnal oscillation regulated by a core clock machinery in WAT in mice [83]. In addition, gene and protein expression of NAMPT can be increased by physiological conditions that activate AMP-activated protein kinase (AMPK) (Fig. 2B). For example, exercise, caloric restriction and weight-loss have been shown to increase expression of NAMPT in WAT (Fig. 2B) [69], [84], [85], [86]. In contrast, obesity, high-fat feeding and aging decrease NAMPT in WAT (Fig. 2B) [43], [66], [67], [68], [87]. The regulation of BAT NAMPT is currently poorly understood.
The NAD+ consumers, SIRTs and PARPs, respond to similar type of physiological and stress conditions. Energy shortage, such as caloric restriction, starvation and exercise, activate SIRTs due to an increase in NAD+ availability (Fig. 2B). In contrast, aging and high-fat feeding blunt SIRT activity by decreasing NAD+ biosynthesis (Fig. 2B). High-fat feeding, obesity, aging, oxidative stress and DNA damage stimulate PARPs while fasting suppresses PARP enzymes in peripheral tissues (Fig. 2B) [80], [88]. Moreover, long-term calorie restriction was demonstrated to lower PARP activity in subcutaneous WAT in obese subjects (Fig. 2B) [69]. The observation that obesity is characterized with low adipose tissue NAD+ levels [43], [66], [67], [68], [87] has raised an important question: what are the factors diminishing NAD+ levels in adipose tissue in obesity? One likely factor is that energy or fat excess, such as HFD-feeding, disturbs WAT NAMPT expression (Fig. 2B) [43], [66], [67], [68], [87] and stimulates PARP activity (Fig. 2B) [80]. Inflammation and oxidative stress may also reduce Nampt expression [67] and activate PARPs [88], but whether this occurs in adipose tissue has not been investigated. Sedentary life-style may also impair NAD+ biosynthesis as evidence supports the role of exercise in the maintenance of adequate NAD+ homeostasis in adipose tissue [84]. Collectively, deteriorated NAD+ homeostasis in obesity is likely to reflect poor life-style and dietary habits as well as adipose tissue stress condition.
4.6. Strategies to boost adipose tissue NAD+ levels and SIRT1 in obesity
Recent animal studies have demonstrated that NAD+ restoration therapy is a promising approach to activate SIRT1 [67], [71]. As SIRT1 activation has been shown to protect against diet-induced weight and fat gain during HFD [71], [89], [90], NAD+ restoration therapy has recently emerged as a potential treatment option for obesity [71], [89], [90]. Cellular NAD+ levels can be restored by stimulating NAD+ biosynthesis or by inhibiting the activity of NAD+ consuming enzymes. Currently, the most commonly tested approach has been the stimulation of NAD+ biosynthesis, which can be achieved either via treatment with pharmacological compounds that induce NAD+ biosynthesis enzymes or dietary supplementation of NAD+ precursors, vitamin B3 forms, 5′ AMP-activated protein kinase activators such as resveratrol, AICAR and metformin have been established to increase mRNA and protein expression of NAMPT in several tissues (Fig. 2B) [18]. However, metformin is the only 5′ AMP-activated protein kinase activator demonstrated to stimulate Nampt gene expression in adipose tissue so far [87]. PPARα, PPARγ and PPARδ agonists may also provide a tool to stimulate WAT Nampt expression [91], [92]. Recently, the therapeutic possibilities of NAD+ precursors, have been widely investigated in different disease models. NR and the NAD+ intermediate generated from NR or NAM, NMN (Fig. 2B), have been observed to efficiently boost intracellular NAD+ levels in BAT [71], [75] and WAT [67], [93], respectively, in mice (Fig. 2B). NA and NAM are known to robustly increase intracellular NAD+ levels in liver [14], but unfortunately their effect on adipose tissue NAD+ levels have not been examined. NAD+ precursors are well-tolerated and not associated with severe side-effects. However, NA is known to cause cutaneous flushing via GPR109A receptor-activation mediated vasodilatation [94]. As NR and NMN do not activate GPR109A receptor [71], their use in the tissue NAD+ restoration therapy is nowadays preferred over NA. Both of these vitamin B3 metabolites, as water-soluble compounds are probably safe and cost-effective potential therapy options. However, long-term data in humans are still mostly lacking.
An attractive NAD+ restoration therapy approach is also the inhibition of main NAD+ consuming enzymes PARPs (Fig. 2B). Deficiency of Parp1 and Parp2 can cause an enormous rise in tissue NAD+ levels in BAT and muscle [13], [80], [95]. Development of pharmacological pan-PARP [88] inhibitors have opened new avenues for targeting NAD+ consuming enzymes. As expected, PARP inhibitors have been shown to elevate NAD+ levels in multiple tissues such as muscle and liver [81], [96], [97], but adipose tissue has not been investigated in these studies. Furthermore, there are no human studies in which the effect of PARP inhibitor treatment on tissue NAD+ levels has been investigated. Thus, further work is required to evaluate the efficacy of PARP inhibitors on adipose tissue NAD+ levels in mice and humans. It is also of interest to develop specific PARP1 inhibitors since Parp2 deletion leads to glucose intolerance due to failure of pancreatic function in mice [13]. As PARP1 is involved in DNA repair, further studies are also needed to ensure the safety of chronic inhibition of this enzyme. However, long-term treatment with pan-PARP inhibitor was not observed to cause DNA damage in treated mice [81].
In spite of above mentioned pharmacological approaches, the most cost-effective therapy options to replenish the adipose tissue NAD+ levels in obesity are diet-induced weight-loss and exercise, as they are known to activate NAMPT in WAT [69], [84] and attenuate PARP activation in subcutaneous WAT in obese subjects (Fig. 2B) [69]. Notably, lifestyle modifications combined with the vitamin B3 nutritional therapies could be a powerful approach to improve NAD+ metabolism. Several ongoing human clinical trials with NAD+ precursors will reveal their efficacy in humans in the near future.
5. SIRTs and PARPs in adipose tissue function in basal and obese conditions
5.1. Introduction
The role of NAD+-dependent SIRTs and PARPs in the regulation of metabolic and mitochondrial functions has been mostly investigated in tissues considered to be metabolically active, such as muscle and liver, but less is known about their function in adipose tissue and obesity. In the following section we aim to review what is known about these two families in regard to the central functions of WAT and BAT. We provide an update on the physiological actions of SIRTs and PARPs in adipose tissue based on the lessons learned from mouse and human studies (Table 1).
Table 1.
Animal models and human studies of SIRTs and PARPs in WAT and BAT.
| Study model | Intervention (length) | Overall metabolic phenotype | WAT phenotype | BAT phenotype | Reference |
|---|---|---|---|---|---|
| Rodents | |||||
| Adipose tissue-specific | Regular diet | Body weight ↑ | SAT and VAT mass ↑ | Mass ↑ due to increased adiposity | [101] |
| SIRT1 KO | Insulin sensitivity and glucose tolerance ↓ | Large adipocytes with enlarged lipid droplets | |||
| FABP4 promoter | Plasma leptin ↑, adiponectin - | ||||
| C57BL/6 | |||||
| HFD (8–12 weeks) | Insulin sensitivity and glucose tolerance ↓ | Plasma leptin ↑, adiponectin ↓ | Mass ↑ | ||
| SIRT1 KO | Insulin sensitivity and glucose tolerance ↓ | [107], [115] | |||
| aP2 promoter | |||||
| C57BL/6 | HFD (5 weeks) | Insulin sensitivity and glucose tolerance ↓ | Large adipocytes | Mass ↑ | |
| Inflammation ↑ | |||||
| HFD (15 weeks) | Insulin sensitivity and glucose tolerance ↑ | Small adipocytes, hyperplasia | Mass ↑ | ||
| Inflammation ↓ | |||||
| Thermogenic genes ↑ | |||||
| PPARg signaling ↑ | |||||
| SIRT1 KO | Regular diet | Postnatal lethal, small in size | Thermogenic genes ↓ | Thermogenic genes not changed | [148] |
| Homozygote | Only small fraction of mice survive to adulthood | ||||
| 129/J×C57BL/6J | |||||
| SIRT1 KO | Regular diet | Overall metabolism normal | Phenotype not changed | Phenotype not changed | [134], [138] |
| Heterozygote | |||||
| C57BL/6 | Medium/high-fat diet | Body weight ↑ | SAT mass ↑ | Enlarged lipid droplets | |
| (12–16 weeks) | Energy expenditure ↓ | Inflammation ↑ | Thermogenic response ↓ | ||
| Insulin sensitivity and glucose tolerance ↓ | Expression of leptin ↓/- and adiponectin - | Mitochondrial content ↓ | |||
| Liver steatosis ↑ | Insulin signaling ↓ | FA oxidation ↓ | |||
| Fat/liver | Regular diet | Body weight ↓ | WAT mass ↓ | Phenotype not reported | [107], [156] |
| SIRT1 deficiency rat | Food intake ↓ | Plasma leptin ↓, adiponectin ↓ | |||
| Antisense oligonucleotide (ASO) | Plasma cholesterol ↓ and FA ↓ | ||||
| Sprague-Dawley rats | |||||
| HFD | Food intake ↓ | VAT mass ↓ | |||
| Inflammation↑ | |||||
| Dbc1 KO | Regular diet | Thermogenic response ↑ upon cold exposure | Phenotype not changed | [148] | |
| SIRT1 inhibitor | |||||
| C57BL/6 × 129/J | |||||
| Adipose tissue-specific | Regular diet | Body weight ↓ | Fat mass not changed | Phenotype not reported | [126] |
| human SIRT1 overexpression | Energy expenditure ↑ | Lipolysis ↑ | |||
| aP2 promoter | Insulin sensitivity and glucose tolerance ↑ | Expression of and plasma adiponectin ↑ | |||
| C57BL/6J | Liver and muscle lipid content ↓ | Lipogenesis ↓ | |||
| SIRT1 overexperssion | Regular diet/HFD | Body weight not changed/comparable weight gain | Phenotype not changed | Lipid droplet size ↓ | [127] |
| Homozygote | Energy expenditure ↑ | Thermogenic response ↑ | |||
| C57BL/6N | Insulin sensitivity and glucose tolerance ↑ | Mitochondrial content not changed | |||
| FA oxidation ↑ | |||||
| SIRT1 overexperssion | Regular diet | Body weight ↓ | VAT mass ↓ | Phenotype not reported | [140] |
| beta-actin promoter | Plasma FA and cholesterol ↓ | Plasma leptin ↓, adiponectin ↓ | |||
| Heterozygotes | Energy expenditure ↑ | ||||
| C57BL/6×129/Sv | Glucose tolerance ↑ | ||||
| SIRT1 overexperssion | Regular diet | Overall metabolism normal | Phenotype not changed | Phenotype not changed | [154] |
| Heterozygote | |||||
| C57BL/6 | HFD (19 weeks) | Energy expenditure ↑ | Fat mass not changed | Thermogenic response ↓ | |
| Glucose tolerance ↑ | |||||
| Protected from liver steatosis | |||||
| SIRT1 overexpression | Regular diet | Energy expenditure ↓ | Fat mass not changed | Phenotype not changed | [107], [139], [148] |
| Heterozygote | Glucose/insulin metabolism normal | Plasma adiponectin ↑ | |||
| C57BL/6J | Thermogenic response ↑ upon cold exposure | ||||
| Crossbred with db/db mice | Comparable weight gain | Comparable fat gain | |||
| Glucose tolerance ↑ | |||||
| HFD (6–16 weeks) | Comparable weight gain | Comparable fat gain | |||
| Insulin sensitivity and glucose tolerance ↑ | Inflammation ↓ | ||||
| SIRT3 KO | Fasting | Impaired glucose metabolism upon cold exposure | Phenotype not changed | FA oxidation ↓ | [142], [157] |
| 129/Sv | Cold tolerance ↓ | ||||
| SIRT4 KO | Regular diet | Body weight not changed | Lipogenesis ↑ | [104] | |
| 129/J | Overall metabolism normal | ||||
| HFD (16 weeks) | Weight gain ↓ | WAT gain ↓ | Mass not changed | ||
| Energy expenditure ↑ | Lipogenesis ↑ | ||||
| Comparable insulin and glucose tolerance | |||||
| SIRT6 KO | Regular diet | Lethal hypoglycemia | SAT mass ↓ | Glucose uptake ↑ | [155], [158] |
| 129/SvJ | Small body size | ||||
| SIRT6 overexpression | Regular diet | Overall metabolism normal | Fat mass not changed | Not reported | [131] |
| HFD (16 weeks) | Comparable weight gain | VAT gain ↓ | |||
| Glucose tolerance ↑ | Triglyceride synthesis ↓ | ||||
| Plasma FA, TG and cholesterol ↓ | |||||
| SIRT7 KO | HFD (22 weeks) | Liver steatosis ↓ | WAT mass ↓ | BAT mass ↓ | [135] |
| C57BL/6J | Glucose tolerance ↑ | Plasma leptin ↓ and adiponectin - | Thermogenic response ↑ | ||
| Inflammation ↓ | |||||
| PARP1 KO | Regular diet | Body weight ↓ | WAT mass ↓ | Thermogenic response ↑ | [13] |
| C57BL/6 | Energy expenditure ↑ | Mitochondrial content ↑ | |||
| Plasma FA and TG ↓ | FA oxidation ↑ | ||||
| Glucose tolerance ↑ | |||||
| HFD (8 weeks) | Weight gain ↓ | WAT gain ↓ | |||
| Insulin sensitivity and glucose tolerance ↑ | |||||
| Plasma FA ↓ | |||||
| PARP1 KO | Regular diet | Overall metabolism normal | Phenotype unaffected | Phenotype not reported | [122] |
| C57BL/6 | |||||
| HFD (14 weeks) | Weight gain ↓ | Adipocyte size ↓ | |||
| Liver steatosis ↑ | Adipogenesis ↓ | ||||
| Plasma FA, TG and cholesterol ↑ | Inflammation ↓ | ||||
| Glucose tolerance ↓ | |||||
| PARP1 KO | Regular diet | Overall metabolism normal | Plasma leptin - | Phenotype not reported | [159] |
| 129/SvImJ | |||||
| HFD (19 weeks) | Weight gain ↑ | WAT mass ↑ | Lipid content ↑ | ||
| Energy expenditure ↓ | Adipocyte size ↑ | Thermogenic response ↓ | |||
| Insulin sensitivity and glucose tolerance ↓ | Plasma leptin ↑ | ||||
| PARP2 KO | Regular diet | Body weight ↓ | WAT mass ↓ | Phenotype not changed | [13], [125] |
| C57BL/6J×SV129 | Energy expenditure ↑ | Adipocyte size ↓ | |||
| Adipogenesis ↓ | |||||
| Inflammation ↑ | |||||
| HFD (8 weeks) | Weight gain ↓ | WAT mass ↓ | |||
| Energy expenditure ↑ | Inflammation ↑ | ||||
| Insulin sensitivity ↑ | |||||
| beta cell dysf->glucose intolerant | |||||
| SIRT1 activator | Regular diet | Body weight unaffected | [90] | ||
| Resveratrol | |||||
| C57BL/6 | HFD (15 weeks) | Weight gain ↓ | WAT gain ↓ | Lipid droplet size ↓ | |
| Energy expenditure ↑ | Adipocyte size ↓ | Thermogenic response ↑ | |||
| Insulin sensitivity and glucose tolerance ↑ | Mitochondrial content ↑ | ||||
| SIRT1 activator SRT1720 | Body weight unaffected | [89] | |||
| C57BL/6 | Glucose tolerance ↑ | ||||
| Plasma TG ↓ | |||||
| HFD (13–20 weeks) | Weight gain ↓ | WAT gain ↓ | Lipid droplet size ↓ | ||
| Energy expenditure ↑ | Adipocyte size ↓ | Expression of UCP1 unaffected, UCP3 ↑ | |||
| Insulin sensitivity and glucose tolerance ↑ | FA oxidation ↑ | ||||
| Plasma TG and cholesterol ↓ | |||||
| Liver steatosis ↓ | |||||
| NAD+ precursor | Regular diet | Body weight unaffected | SIRT1 and SIRT3 not activated | SIRT1 and SIRT3 activated | [71] |
| nicotinamide riboside | Insulin sensitivity ↑ | Cold tolerance ↑ | |||
| C57BL/6 | |||||
| HFD (8–16 weeks) | Weight gain ↓ | VAT mass ↓ | Cold tolerance ↑ | ||
| Energy expenditure ↑ | Mitochondrial content ↑ | ||||
| Pan-PARP inhibitor | Regular diet | Body weight unaffected | WAT mass unaffected | Mass unaffected | [81], [124] |
| (MRLB-45696) | |||||
| C57BL/6 | HFD (18 weeks) | Weight gain ↓ | SAT gain ↓ | Mass unaffected | |
| Energy expenditure ↑ | Adipocyte size ↓ | ||||
| Expression of adiponectin ↓ | |||||
| Inflammation ↓ | |||||
| Humans | |||||
| Obese | Mixed group of subjects with/without metabolic syndrome | SIRT1 expression ↓ | [108] | ||
| SIRT2-SIRT6 expression unaffected | |||||
| SIRT7 expression ↑ | |||||
| Obese | Clinically healthy | SIRT1/SIRT3/SIRT7 expression ↓ | [69] | ||
| PARP activity ↑ | |||||
| Correlation (+): PARP activity and WAT mass | |||||
| Correlation (-): SIRT1 expression and inflammation | |||||
| Correlation (+): SIRT1 and insulin sensitivity | |||||
| Caloric restriction diet | SIRT1/SIRT7 expression ↑ | ||||
| SIRT3 expression ↓ | |||||
| PARP activity ↓ | |||||
| Obese | Metabolic syndrome not reported | SIRT1 expression ↓ | [107] | ||
| Correlation (-): SIRT1 expression and inflammation | |||||
| BMI discordant | Clinically healthy | SIRT1/SIRT3/SIRT5 expression ↓ | [43] | ||
| Monozygotic twins | SIRT7 expression unaffected | ||||
| Correlation (-): SIRT1/SIRT5 expression and inflammation | |||||
| Correlation (+): SIRT1/SIRT5 and insulin sensitivity | |||||
| Correlation (-): SIRT1/SIRT5 and insulin resistance | |||||
| Obese | Non-diabetic | SIRT1 expression ↓ | [110] | ||
| Correlation (+): SIRT1 and adiponectin expression | |||||
| Obese | Mixed group of subjects with (10%)/without diabetes | Correlation (+): SIRT1/SIRT3/SIRT6 and adiponectin expression | [111] | ||
| Gastric band surgery | SIRT1/SIRT3/SIRT6 expression ↑ | ||||
| Obese | Not reported | SIRT1 expression ↓ | [106] | ||
| Obese | Clinically healthy | SIRT1 expression ↓ | [109] | ||
| Obese | Clinically healthy | SIRT2/SIRT3/SIRT5 expression ↓ | [102] | ||
5.2. Expression of SIRTs and PARPs in WAT in basal and obese conditions
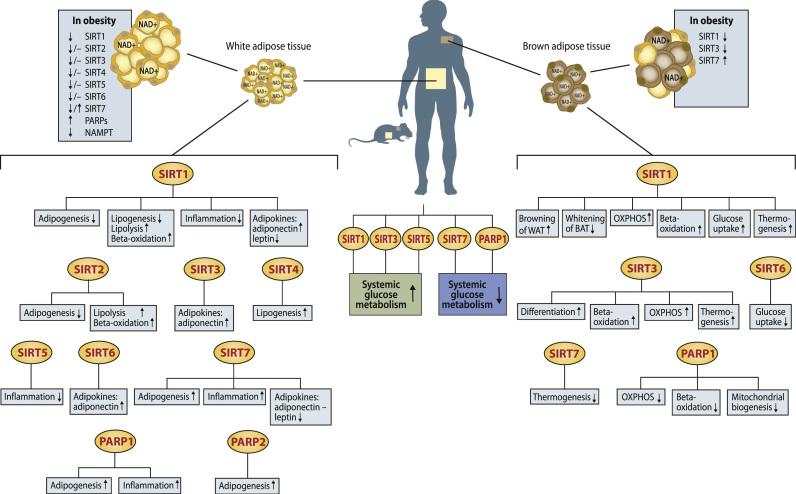
Studies in both rodent models and humans have shown that all SIRT genes are expressed in WAT and regulated by obesity (Fig. 3). In rodents, HFD induced obesity decreases the WAT expression level of Sirt1-4 and Sirt6, whereas acute HFD increases the WAT expression of Sirt5 [40], [98], [99], [100], [101], [102]. In contrast, caloric restriction increases Sirt1 and Sirt2 but reduces Sirt4 mRNA and protein level in WAT [85], [103], [104], [105]. Human studies have similarly shown that SIRT1 is expressed in both subcutaneous and visceral WAT and reduced by obesity [43], [69], [106], [107], [108], [109], [110]. The expressions of SIRT2, SIRT3 and SIRT5 were reported to be either reduced [43], [69], [102], or unaffected in obese subcutaneous WAT [108]. For SIRT7 results between different studies have also been varied: Rappou and colleagues [69] found decreased expression in obese subcutaneous WAT, whereas no expression difference [43] or increased expression [108] has also been reported. Caloric restriction and weight loss induced by gastric band operation increased the expression of SIRT1, SIRT3 and SIRT6 in subcutaneous WAT in obese subjects [111], whereas long-term weight loss based on conventional caloric restriction diet regiment increased the mRNA levels of SIRT1 and SIRT7, but in contrast to gastric banding, SIRT3 expression was further reduced [69].
Fig. 3.
Adipose tissue pathways regulated by sirtuins (SIRTs) and poly(ADP-ribose) polymerases (PARPs) based on current literature in mouse models, adipocyte cell lines and human studies. The effect of obesity on white and brown adipose tissue NAD+ levels and expression levels of SIRTs, PARPs and NAD+ biosynthesis gene nicotinamide phosphoribosyltransferase (NAMPT) are also shown in separate boxes next to both tissues. Arrows indicate increased (↑), decreased (↓) or unchanged (−) pathway activity/gene expression level. OXPHOS; oxidative phosphorylation.
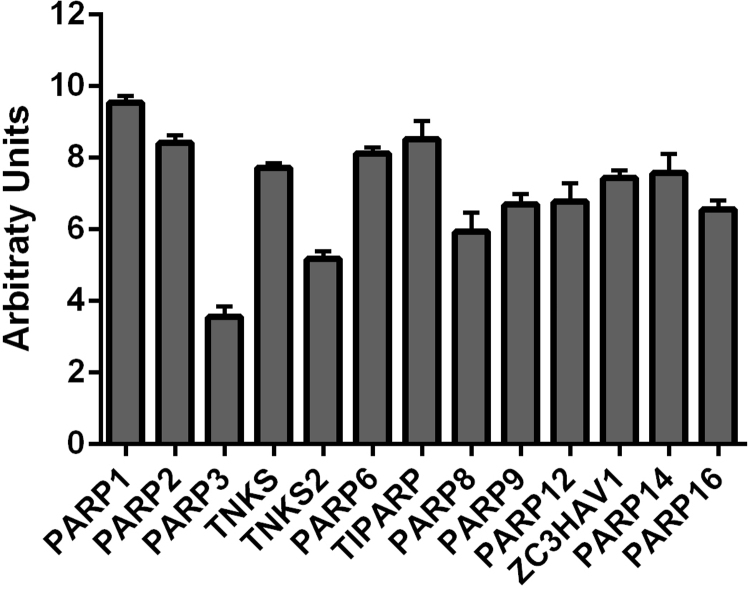
PARP expression has been less studied in WAT. Thus, we examined the mRNA expression of all 17 PARP family members from microarray data of the lean, metabolically healthy twin of BMI discordant monozygotic twins from our earlier publication [43] to elucidate WAT expression of PARP family genes. Expression of 13 out of 17 PARP gene family members was detected in human WAT, with the expression level of PARP1 being the highest (Supplemental Fig. 1). Recent human studies showed that total PARP activity is elevated in obesity and is reduced by weight loss (Fig. 3) [43], [69].
5.3. The function of SIRTs and PARPs in WAT
5.3.1. Adipogenesis
Adipose tissue is renewed by recruitment of mesenchymal stem cells to differentiation into adipocytes, adipogenesis. Adipogenesis is orchestrated by transcriptional program where the master regulator is Pparγ that works in concert with other transcriptional regulators, most notably the CAAT enhancer binding proteins transcription factor family to promote adipocyte differentiation (Fig. 1A) [112]. Interestingly, activation of mitochondrial biogenesis program and an increase of mitochondrial mass and oxidative capacity characterize adipocyte differentiation, highlighting the importance of mitochondrial function for adipocytes [113].
Studies with the mouse 3T3-L1 cell line capable of differentiating into adipocytes have shown that Sirt1 inhibits adipogenesis (Fig. 3). Overexpression of Sirt1 reduces gene expression of the adipogenic transcriptional program and markers of terminal differentiation, whereas Sirt1 knock-down has the opposite effect [114]. Mechanistically Sirt1 interacts with Pparγ and its co-repressors, suggesting that Sirt1 impairs adipogenesis through inhibiting Pparγ signaling by docking with these co-repressors [114]. In vivo data confirm Sirt1 as a negative regulator of adipogenesis. Adipocyte-specific Sirt1 deletion increased the adipogenic capacity of WAT in mice in response to chronic HFD as demonstrated by white adipocyte hyperplasia and hyperacetylation mediated activation of Pparγ [115], and increased WAT mass [101], [115] even under normal conditions. In agreement, mouse embryonic fibroblasts derived from mice homo- or heterozygous for Sirt1 deletion, were shown to possess higher adipogenic capacity in cell culture [116], [117]. By contrast, treatment with pharmacological SIRT1 activators decreases WAT mass during HFD [71], [89], [90]. Bone marrow derived mesenchymal stem cells with Sirt1 deletion have a serious defect in self-renewal and osteoblast differentiation, while adipocyte differentiation was only modestly affected but adipocytes were smaller containing less lipids [118]. Taken together these results demonstrate that while Sirt1 inhibits adipocyte differentiation, it is required for maintenance of the mesenchymal stem cell pool and sustained renewal of WAT in vivo.
Based on studies in cultured adipocytes Sirt2 has a similar inhibitory effect on adipogenesis as Sirt1; overexpression of Sirt2 inhibits adipogenesis whereas Sirt2 inhibition promotes it (Fig. 3). The underlying mechanism is suggested to be Sirt2 mediated deacetylation of forkhead box protein O1, which binds and represses Pparγ after nuclear localization (Fig. 3) [105], [119]. However, the effect of Sirt2 on adipogenesis has not yet been investigated in vivo.
In contrast to Sirt1 and Sirt2, Sirt7 appears to be a positive regulator of adipogenesis (Fig. 3) [120]. Gene expression level of the adipogenic transcriptional program in WAT from mice homozygous for Sirt7 deletion was reduced, and freshly isolated early adipogenic precursor cells were diminished in number and had impaired adipogenic capacity in culture. Furthermore SIRT7 knockdown in primary human preadipocytes impaired lipid accumulation and decreased the number of cells that expressed the terminal adipocyte differentiation marker fatty acid-binding protein 4 [120]. It has been proposed that the opposite effects of Sirt1 and Sirt7 on adipogenesis could be mediated by direct inhibition of Sirt1 by Sirt7 [121]. However, SIRT7 has been reported to either behave similarly to SIRT1 and be downregulated [69], or in an opposite manner and be upregulated [108] in obese by comparison to lean subjects in humans. Thus, further research is required to clarify whether SIRT7 truly has opposing effects on adipogenesis as SIRT1.
Parp1 and Parp2 have been shown to promote adipogenesis in cell lines and in vivo (Fig. 3). Parp activity increases during adipogenesis and lentiviral Parp1 knockdown or Parp inhibition in 3T3-L1 cells impairs adipogenic differentiation through reduced gene expression of Pparγ and Pparγ responsive adipogenic genes [122]. Parp1 deletion in mice leads to morphologically abnormal WAT with small adipocytes upon HFD and preadipocytes isolated from these mice exhibit impaired capacity for adipogenic differentiation in culture [123]. In agreement, Parp inhibition during HFD leads to small adipocyte morphology similar to Parp1 deletion in mice and is associated with reduced gene expression level of Pparγ target genes in WAT [124]. Mechanistically Parp1 was shown to associate with Pparγ responsive promoters and enhance Pparγ ligand binding, which mediates co-activator and co-repressor exchange in a poly(ADP-ribosyl)ation dependent manner (Fig. 1A) [124]. Similarly to Parp1, Parp2 deletion in mice resulted in small and irregular adipocyte morphology and reduced gene expression Pparγ target genes, even under unchallenged conditions [125]. In agreement, differentiation of mouse embryonic fibroblasts with Parp2 deletion into adipocytes was impaired in culture [125]. Mechanistically Parp2 seems to directly interact with Pparγ on the chromatin and increase Pparγ-driven gene expression. In humans, the effect of PARPs on adipogenesis has not yet been studied. However, recent study demonstrated that WAT PARP activity correlates positively with fat mass in humans [69], which may in part reflect the capability of PARPs to promote adipogenic capacity.
5.3.2. Lipid homeostasis
WAT lipid homeostasis is determined by lipid synthesis, storage and utilization through lipolysis and mitochondrial fatty acid β-oxidation. Based on current evidence SIRTs regulate lipid homeostasis in WAT, but to our knowledge the role of PARPs has not been investigated.
The majority of in vitro and vivo data indicate that Sirt1 regulates adipocyte lipolysis (Fig. 3). Pharmacological Sirt1 activation increased lipolysis and fatty acid release from rodent adipocytes in culture under basal and β-adrenergic stimulated conditions, whereas Sirt1 inhibition had the opposite effect [114]. In agreement, heterozygous loss of Sirt1 decreases plasma free fatty acid concentrations in mice through a reduction of β-adrenergic mediated fatty acid release from adipocytes [114]. Adipocyte specific overexpression of human SIRT1 in mice increases basal lipolytic rate and the expression level of adipose triglyceride lipase, a key enzyme in the lipolytic pathway [126]. However, in another Sirt1 overexpression model, WAT lipolysis rate was unaffected [127]. Mechanistic studies in cultured adipocytes have indicated that Sirt1 promotes lipolysis via deacetylation and activation of forkhead box protein O1, which in turn stimulates the expression of adipose triglyceride lipase [128]. SIRT1 may also have an effect on adipocyte fatty acid β-oxidation as a trend for increased WAT fatty acid β-oxidation was observed in SIRT1 overexpressing mice (Fig. 3), but these results did not reach statistical significance [126]. Similar to Sirt1, Sirt2 overexpression also promoted basal and insulin stimulated lipolysis [105] and increased fatty acid oxidation in 3T3-L1 differentiated adipocytes, while Sirt2 knock down impaired fatty acid oxidation (Fig. 3) [102]. Further research is required to investigate the role of Sirt2 in WAT lipolysis and fat oxidation in vivo.
SIRTs are also involved in the regulation of lipogenesis in mouse WAT. Genetic or pharmacological Sirt1 activation reduces the rate-limiting enzyme in de novo lipogenesis, acetyl-CoA carboxylase, and expression of genes in the lipogenic pathway in mice or a human preadipocyte line, respectively [126], [129]. Therefore, based on these studies, Sirt1 appears to inhibit lipogenesis in WAT (Fig. 3). In contrast, deletion of the mitochondrial Sirt4 decreased lipogenesis in freshly isolated adipocytes and WAT (Fig. 3) [104]. This finding can be explained by the dual role of Sirt4 in the regulation of fat catabolism and anabolism. In fed state Sirt4 promotes lipid synthesis by deacetylating and inactivating the malonyl-CoA decarboxylase, which increases the amount of malonyl-CoA, the metabolite that represses lipid oxidation [130]. During fasting WAT Sirt4 expression reduces leading to an activation of malonyl-CoA carboxylase and lipid oxidation. Sirt6 has also been implicated in the control of lipogenesis. Sirt6 may act as a negative regulator of lipid triglyceride synthesis based on transcriptional changes in WAT of Sirt6 overexpressing mice during HFD (Fig. 3). Notably, the expression levels of Pparγ responsive lipid metabolism genes and diacylglycerol acyltransferases, key enzymes in triglyceride synthesis, were significantly downregulated in the Sirt6 overexpressing mice in comparison with controls [131].
5.3.3. OXPHOS
SIRTs are described as regulators of mitochondrial respiration and oxidative metabolism in non-adipose tissues [14], but their role in WAT respiration during basal conditions or obesity is poorly investigated. Fasting has been shown to induce both Sirt1 expression and increase in mitochondrial function assessed as mtDNA copy number, expression of OXPHOS genes and mitochondrial respiration capacity in WAT [100]. Moreover, pharmacological activation of Sirt1 protects from HFD induced obesity [71], [89], [90], which may in part be explained by upregulation of genes involved in oxidative metabolism (such as PGC-1α) in WAT along with non-adipose tissues and inhibition of adipogenesis [89]. In contrast, Sirt1 overexpression in mice did not affect mitochondrial respiration capacity determined by high-resolution respirometry or OXPHOS complex subunit protein amount in mouse WAT during unchallenged conditions or caloric restriction [127], [132]. However, it is worth noting that the caloric restriction employed in this study, every other day feeding, did not induce an increase in Sirt1 expression in WAT in wild type mice. Therefore, further research is needed to clarify the role of SIRTs in regulation of WAT mitochondrial respiration under basal conditions and during metabolic challenges such as obesity or fasting. Studies examining the role of PARPs on WAT respiration are at present lacking.
5.3.4. WAT inflammation and cytokine secretion
WAT inflammation is a key characteristic of obesity and is considered to contribute to obesity related pathologies. Current in vitro and in vivo data support an anti-inflammatory role for Sirt1 in WAT (Fig. 3). In 3T3-L1 adipocytes genetic or pharmacological Sirt1 inhibition increased the expression of inflammatory cytokines upon tumor necrosis factor 1 alpha stimulation, whereas Sirt1 activation had the opposite effect [133]. In agreement, genetic or antisense oligonucleotide mediated Sirt1 deficiency in rodents leads to elevated inflammation characterized by increased macrophage infiltration and mRNA expression of inflammatory cytokines in WAT under normal [107] and HFD conditions [101], [115], [117], [134]. However, during long-term HFD adipocyte specific Sirt1 deletion reduced WAT inflammation in contrast to the other findings [115]. Consistent with a role for Sirt1 in repressing inflammation, overexpression of Sirt1 reduced WAT macrophage infiltration and nuclear factor kappa B and tumor necrosis factor 1 alpha signaling during HFD [107]. In humans, WAT SIRT1 expression levels correlate inversely to markers of inflammation in obese subjects [43], [69], [107]. Notably, as mentioned earlier, obesity reduces expression of SIRT1 in WAT both in mouse and humans. This can be likely explained by the finding that caspase-1, activated by the inflammasome, promotes Sirt1 degradation [101]. Therefore, inflammation and reduced Sirt1 amount appeared to be associated with obesity and could contribute to WAT dysfunction.
The role of other SIRTs and Parps in WAT inflammation has been poorly investigated. In humans, WAT SIRT5 expression levels correlate inversely to markers of inflammation (Fig. 3) [43]. Deletion of Sirt7 in mice reduced WAT inflammatory gene expression during HFD (Fig. 3) [135], providing another piece of evidence for the opposite functions of Sirt1 and Sirt7 in WAT as discussed in relation to adipogenesis. Similarly, Parps may play a role in promoting WAT inflammation as Parp1 deletion or Parp inhibitor treatment reduced interleukin-6 and other inflammatory gene expression levels (Fig. 3) [123], [124].
5.3.5. Adipokine secretion
Adipose tissue affects other tissues involved in systemic energy homeostasis through secretion of adipokines, most notably the satiety hormone leptin and the insulin sensitizer adiponectin. Mitochondrial function has been tied to adipokine secretion through studies in cultured adipocytes showing that inhibition of mitochondrial function reduces while activation increases the production and secretion of adiponectin [1], [2]. Obesity is characterized by increased leptin and decreased adiponectin levels.
Initial studies with 3T3-L1 adipocytes were inconclusive in regard to the role of Sirt1 in the regulation of adiponectin. Qiao and colleagues showed that Sirt1 promotes adiponectin expression through deacetylation of forkhead boxO1, which facilitates the transcription factor CAAT enhancer binding protein alpha binding to the adiponectin promoter [136]. In contrast, Qiang and colleagues showed that Sirt1 inhibits adiponectin secretion [137]. However, based on in vivo studies Sirt1 is a positive regulator of adiponectin production and secretion (Fig. 3). Genetic Sirt1 deficiency leads to decreased circulating adiponectin level and gene expression in WAT during HFD whereas circulating leptin levels were either increased [101] or unaffected [138] by Sirt1 depletion. SIRT1 activation has the opposite effect increasing mRNA and circulating levels of adiponectin upon aging or HFD [138], [139], but in contrast SIRT1 activation decreased circulating adiponectin level in unchallenged conditions [140]. Sirt7 deletion reduced plasma leptin during HFD, whereas adiponectin level was not affected (Fig. 3) [135]. In obese patients expression levels of WAT SIRT1 [110], [111], SIRT3 and SIRT6 [111] correlate positively to the expression of adiponectin and its receptor (Fig. 3).
5.3.6. Systemic glucose homeostasis
Systemic glucose homeostasis is determined by the intricate interplay of different tissues participating in glucose metabolism. Loss of WAT Sirt1 during aging or short to intermediate HFD in mice leads exacerbated insulin resistance and glucose intolerance (Fig. 3) [101], [115]. Overexpression of human SIRT1 in adipose tissue during aging has the opposite effect and delays the development of insulin resistance and glucose intolerance [126]. In humans, WAT SIRT1 and SIRT5 correlate positively with insulin sensitivity (Fig. 3) [43], [69]. Overall, WAT Sirt1 appears to be a modifier of glucose homeostasis especially in challenged conditions such as HFD or aging. The whole-body Parp1 deletion as well as pharmacological inhibition of Parps have been shown to improve glucose homeostasis [13], [80], [81] but the role of WAT PARP activity and other SIRTs in glucose homeostasis remains to be investigated by future studies.
5.4. SIRT and PARP expression in BAT
All seven members of mammalian SIRT family are expressed in BAT. The expression levels of Sirt2, Sirt4 and Sirt6 are comparable to that seen in WAT whereas levels of Sirt1 and Sirt7 are lower and levels of Sirt3 and Sirt5 higher in BAT by comparison to WAT [141]. Calorie restriction and short-term food deprivation as well as cold exposure increase the expression of Sirt1, Sirt2 and Sirt3 in BAT [105], [141], [142], [143], [144]. The knowledge of factors affecting the expression of other SIRTs in BAT is currently very restricted as well as SIRT expression in human BAT. The effect of obesity on BAT Sirt expression has remained obscure.
Out of the 17 members of PARP family, Parp1-12 and Parp16 have been shown to be expressed in BAT in mice, the expression level of Parp5b/Tnks2 (tankyrase 2) being the highest [80]. However, it is not known how physiological stimuli related changes in cellular energy demand or metabolic state affects the expression level of Parps in BAT and how PARPs are expressed in human BAT.
5.5. The function of SIRTs and PARPs in BAT
5.5.1. SIRTs and brown adipocyte differentiation
The acquisition of brown adipocyte phenotype depends on the coordinated action of transcriptional co-regulators PGC-1α PPARγ and Prdm16. See chapter 3 for details. A limited number of studies have been conducted to investigate the role of SIRTs in brown adipocyte differentiation. Boutant and colleagues [127], showed that the differentiation capacity of immortalized brown preadipocytes isolated from homozygous Sirt1 overexpressing mice is similar to that of wild-type cells. In contrast, Sirt3 has been shown to participate in PGC-1α mediated activation of BAT-specific thermogenic response in differentiating primary brown adipocytes [141], [145]. In agreement, PGC-1α was not able to fully induce the expression of thermogenic genes in response to β3-adrenergic stimuli in Sirt3 deficient brown adipocytes [146]. Thus, of the SIRTs, Sirt3 seems to be required for the differentiation of brown adipocytes (Fig. 3). The role of Parps in the brown adipocyte differentiation is not yet understood.
5.5.2. Browning of WAT
Thermogenic adipocytes are not exclusive to BAT but also adipocytes in WAT can gain thermogenic capacity through browning in response to various stimuli, at least in rodents [147]. These cells, called beige or brite adipocytes, show comparable amounts of UCP-1 to brown adipocytes when stimulated. Activation of PGC-1α, PPARγ and Prdm16 in white adipocytes also drives them towards brown adipocyte phenotype by enhancing brown adipocyte characteristics.
Sirt1 has been shown to be an important factor in facilitating browning of WAT by mediating Prdm16 driven activation of thermogenic program in white adipocytes (Fig. 1, Fig. 3). SIRT1 deacetylates PPARγ which leads to recruitment of coactivator Prdm16 and concomitant clearance of the nuclear co-repressor 1 from the PPARγ complex, and thus, induction of thermogenic genes [148]. Qiang and colleagues [148] showed that Sirt1 promotes browning of white adipocytes in vivo in response to cold exposure but not under basal conditions in different models of increased Sirt1 activity. Deficiency of Sirt1 has the opposite effect and results in lower levels of BAT markers in subcutaneous WAT in mice [138], [148]. Currently, there are no studies available of the role of other SIRTs or PARPs in WAT browning. These processes in humans are also unclear.
5.5.3. Whitening of BAT in obesity
Whitening of BAT results from mitochondrial dysfunction. Therefore, as an important regulator of cellular energy homeostasis and mitochondrial biogenesis, SIRT1 has the potential to regulate BAT whitening. Indeed, while Sirt1 gain-of-function models lead to browning of white adipocytes (see above), Sirt1 deficiency in BAT is related to whitening of the tissue (Fig. 3). Sirt1 deficiency in mice results in accumulation of lipid droplets as well as in reduction of mitochondrial content and respiratory chain complex subunit abundance in brown adipocytes upon HFD [138]. Moreover, adipose tissue specific Sirt1 deletion increases the weight of BAT by increasing its adiposity [101]. Mechanistically SIRT1 has also been shown to control the angiogenic activity of endothelial cells [149], and thus, it remains to be clarified whether Sirt1 deficiency mediates BAT whitening through affecting vasculature of BAT, mitochondrial metabolism of brown adipocytes, or both. As a whole, it seems clear that SIRT1 is needed for the proper function of BAT. The role of other SIRTs and Parps in whitening of BAT has been poorly investigated and their role remains to be elucidated by future research.
5.5.4. Mitochondrial biogenesis and OXPHOS
Among the key factors activating BAT mitochondrial biogenesis and OXPHOS are cofactors PGC-1α and PGC-1β [59]. Genetic and pharmacological activation of Sirt1 have been shown to increase expression and/or activity of PGC-1α and OXPHOS genes in BAT in mice but unexpectedly this has not been translated into higher mitochondrial content in most of the studies [71], [89], [90], [127]. Of the other SIRTs, SIRT3 has been suggested to play a role in the stimulation of OXPHOS in BAT as it deacetylates and activates subunits of the respiratory chain complex IV subunits as well as activates complex II (SDH) [141], [150]. The most potent activator of BAT mitochondrial biogenesis and OXPHOS has been shown to be Parp1. Deletion of Parp1 activates both PGC-1α activity and mitochondrial biogenesis, but also OXPHOS in BAT [80]. Surprisingly, deletion of Parp2 does not influence PGC-1α expression and mitochondrial biogenesis in BAT despite of increased SIRT1 content [13]. Thus, it can be speculated that maybe additional mediators, such as PGC-1β, are required in addition to PGC-1α and SIRT1 for the activation of mitochondrial biogenesis in some situations. In summary, SIRT1 activation likely has beneficial impacts on BAT OXPHOS while Parp1 may dampen both mitochondrial biogenesis and OXPHOS in BAT (Fig. 3).
5.5.5. Fatty acid oxidation
SIRTs have been shown to be involved in the control of BAT fatty acid oxidation, which is the main energy production pathway in this tissue. Pharmacological activation of SIRT1 enhances expression of the fatty acid oxidation promoting gene, PPARα, in BAT [90]. Moreover, overexpression of Sirt1 leads to a strong induction of the genes involved in fatty acid β-oxidation in BAT via activation of UCP-1, PPARα, forkhead box O1 and forkhead box O3 [127]. It has been also suggested that SIRT1 activation may mediate its stimulatory effect on fatty acid oxidation in brown adipocytes by enhancing the transcriptional response to β3-adrenergic stimuli. Of the other SIRTs, SIRT3 may enhance fatty acid oxidation by deacetylating and activating the long-chain acyl coenzyme A dehydrogenase [142] in BAT [151]. Indeed, fatty-acid oxidation is decreased upon fasting in BAT as well as in other peripheral tissues of Sirt3 deficient mice [142]. Parp1 may also play a role in regulating BAT fatty acid oxidation as whole-body Parp1 deletion increases the expression of medium-chain acyl-CoA dehydrogenase gene probably via activation of SIRT1 (Fig. 3) [80]. Overall, in the light of current data Sirt1 and Sirt3 enhance whereas Parp1 possibly inhibits fatty acid oxidation of brown adipocytes (Fig. 3), while the potential role of other SIRTs or Parps has not been investigated.
5.5.6. Thermogenesis and systemic glucose homeostasis
Cold-activation increases BAT thermogenesis with concomitant increase in glucose uptake and utilization, thus, subsequently improving overall glucose metabolism of an organism [152], [153]. Genetic and pharmacological activation of SIRT1 have been consistently shown to increase BAT thermogenesis and improve systemic glucose tolerance in mice [71], [89], [90], [127], [154]. In agreement, Sirt1 deficiency in mice has the opposite effect and results in diminished cold tolerance and exacerbated glucose intolerance and insulin resistance in response to HFD [138]. Sirt3 has also been suggested to play a role in adaptive thermogenesis. Sirt3 deficient mice exhibit cold intolerance and impaired glucose metabolism during simultaneous fasting and cold exposure [142]. Furthermore, overexpression of Sirt3 in HIB1B brown adipocytes enhances the expression of thermogenic genes. However, due to the lack of Sirt3 overexpression mouse model it remains to be clarified whether activation of Sirt3 can lead to beneficial effects on systemic glucose metabolism. The role of SIRT6 in thermogenesis is not well understood but Sirt6 deficient mice exhibit drastic hypoglycemia due to increased BAT uptake of glucose [155]. In contrast, Sirt7 deficient mice show increased body temperature and increased expression of thermogenic genes in BAT along with enhanced glucose tolerance [135]. At present the role of other SIRTs in BAT in systemic glucose homeostasis is not known. Nevertheless, based on current data, BAT SIRT1, SIRT3 and SIRT7 potentially participate in the control of overall glucose homeostasis through thermogenesis (Fig. 3).
Parp1 deficiency leads to similar beneficial effects on cold tolerance and glucose metabolism as SIRT1 activation models (Fig. 3) [80], whereas Parp2 deletion does not affect adaptive thermogenesis of BAT [13] despite increased SIRT1 activity in BAT in both models. The role of other Parps on activation of BAT thermogenesis and glucose metabolism has not been investigated. Like mentioned earlier whole body deletion of Parp1 and PARP inhibition improve whole body glucose homeostasis [13], [80], [81] but as there are no BAT specific models for Parps available, it is difficult to estimate their role in the regulation of BAT and whole body glucose metabolism conclusively.
6. Conclusions
Based on the current knowledge reviewed here we are for the very first time closer to understanding what is the complex mechanism behind obesity-associated adipose tissue dysfunction; the key driver for metabolic complications. It has recently become evident that adipose tissue dysfunction is tightly associated with mitochondrial derangements and disturbed NAD+ homeostasis. This implies that the maintenance of NAD+ intracellular levels and mitochondrial function is required for adipose tissue health. Mitochondrial metabolism is regulated by two NAD+-consuming enzyme families, SIRTs and PARPs, which sense energy and stress status of the cell and translate this information to mitochondria with opposing effects on adipose tissue metabolism. These two enzyme families are relevant for myriad aspects of adipose tissue function and metabolic health. Obesity interferes with the balance of SIRT and PARP activities in adipose tissue, which may be one of the underlying causes for the adipose tissue mitochondrial dysfunction and adverse metabolic health complications associated with obesity.
In WAT SIRT1 is by far the most studied member of the SIRT family. It appears to be involved in the regulation of several adipose tissue processes such as adipogenesis, lipid homeostasis, inflammation, adipokine secretion and the influence on systemic glucose homeostasis. In general, SIRT1 promotes healthy adipose tissue function and preserves systemic metabolic health. In the coming years, further research is needed to elucidate and confirm the role of other SIRTs in WAT function. The impact of PARPs on adipose tissue health has been less investigated and so far, the information is mainly limited to Parp1 and Parp2. Collectively, Parp1 and Parp2 seem to promote adipogenesis and inflammation in WAT. However, we need to investigate whether other PARP family members also have important functions in adipose tissue. Currently, another caveat in the research field is the lack of understanding how SIRTs and PARPs affect WAT mitochondrial function. Therefore, further research should focus on elucidating their potential crosstalk in WAT in the regulation of mitochondrial metabolism during normal and obese conditions. This could solve the still open question whether the opposing effects of SIRTs and PARPs on metabolic health are mediated by alterations in WAT mitochondrial function.
In BAT, an antagonistic crosstalk of SIRTs and PARPs in the regulation of mitochondrial metabolism has been observed in different mouse models. Sirt1 and Sirt3 seem to increase mitochondrial thermogenesis with concomitant activation of OXPHOS and fatty acid beta-oxidation, and improvement of systemic glucose metabolism while Parp1 has the opposite effect on BAT mitochondrial pathways. What is lacking in the research field, however, are studies using tissue-specific loss-of-function and gain-of-function approaches in mice to clarify the role of SIRTs and PARPs in the BAT metabolic health. Despite recent improvements in the understanding of SIRTs and PARPs mitochondrial functions in BAT in mice, their biology and the effect on mitochondrial metabolism in humans has remained obscure. Thus, new methods and techniques to study human BAT metabolism are welcomed.
Increasing tissue level SIRT activity through boosting intracellular NAD+ levels either by stimulating NAD+ synthesis with vitamin B3 derived NAD+ precursors or inhibiting PARPs is currently a focus of intensive research as a potential treatment for obesity and associated metabolic complications. However, improving adipose tissue metabolic health through NAD+ restoration therapy will require further understanding of efficacies of different NAD+ boosters in increasing the intracellular NAD+ levels, SIRT activity and mitochondrial function in WAT. Importantly, future studies with NAD+ boosters should strive to target WAT and/or BAT in mouse models and humans, as systemic metabolic health is likely to be dependent on the concerted effort of both adipose tissues. As NAD+ precursors, vitamin B3 forms are water-soluble vitamins, they are considered to be safe and cost-effective potential therapy options. In the coming years, new vitamin B3 based nutritional therapies to activate SIRTs may be developed, not only for treatment of obesity and associated metabolic complications, but also for disease prevention and health maintenance.
Acknowledgements
The authors apologize to colleagues whose original work could not be cited due to the vast amount of literature. The study was supported by Academy of Finland (285963 to E.P.), Biocentrum Helsinki (E.P.), Sigrid Jusélius Foundation (E.P.), Academy of Finland Centre of Excellence in Research on Mitochondria, Metabolism, and Disease (FinMIT) (272376 to K.H.P.), Academy of Finland project grant (266286 to K.H.P.), the Novo Nordisk Foundation (K.H.P.), Finnish Foundation for Cardiovascular Research (K.H.P.), University of Helsinki (K.H.P.), Finnish Diabetes Research Foundation (K.H.P.) and Helsinki University Hospital research funds (K.H.P).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2017.02.011.
Appendix A. Supplementary material
Fig. S1.
Supplementary Fig 1. Relative gene expression level poly(ADP-ribose) polymerases in human subcutaneous WAT. Gene expression values are presented as means with standard deviation from normalized and log2 transformed values from Affymetrix U133 Plus 2.0 chips (arbitrary units) from the lean twins of BMI discordant monozygotic twins ([43], see original publication for technical details). Expression of PARP4, PARP10, PARP11 or PARP15 was not detected in this study material.
.
References
- 1.Koh E.H., Park J.-Y., Park H.-S. Essential role of mitochondrial function in adiponectin synthesis in adipocytes. Diabetes. 2007;56:2973–2981. doi: 10.2337/db07-0510. [DOI] [PubMed] [Google Scholar]
- 2.Wang C.-H., Wang C.-C., Huang H.-C., Wei Y.-H. Mitochondrial dysfunction leads to impairment of insulin sensitivity and adiponectin secretion in adipocytes. FEBS J. 2013;280:1039–1050. doi: 10.1111/febs.12096. [DOI] [PubMed] [Google Scholar]
- 3.Björntorp P., Bengtsson C., Blohmé G. Adipose tissue fat cell size and number in relation to metabolism in randomly selected middle-aged men and women. Metabolism. 1971;20:927–935. doi: 10.1016/0026-0495(71)90013-8. [DOI] [PubMed] [Google Scholar]
- 4.Heinonen S., Saarinen L., Naukkarinen J. Adipocyte morphology and implications for metabolic derangements in acquired obesity. Int. J. Obes. 2014;38:1423–1431. doi: 10.1038/ijo.2014.31. [DOI] [PubMed] [Google Scholar]
- 5.Heilbronn L., Smith S.R., Ravussin E. Failure of fat cell proliferation, mitochondrial function and fat oxidation results in ectopic fat storage, insulin resistance and type II diabetes mellitus. Int. J. Obes. Relat. Metab. Disord. 2004;28:S12–S21. doi: 10.1038/sj.ijo.0802853. [DOI] [PubMed] [Google Scholar]
- 6.Virtue S., Vidal-Puig A. Adipose tissue expandability, lipotoxicity and the metabolic syndrome – an allostatic perspective. Biochim. Et. Biophys. Acta (BBA) - Mol. Cell Biol. Lipids. 2010;1801:338–349. doi: 10.1016/j.bbalip.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 7.Pietiläinen K.H., Naukkarinen J., Rissanen A. Global transcript profiles of fat in monozygotic twins discordant for BMI: pathways behind acquired obesity. PLoS Med. 2008;5:e51. doi: 10.1371/journal.pmed.0050051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pérez L.M., Bernal A., de Lucas B. Altered metabolic and stemness capacity of adipose tissue-derived stem cells from obese mouse and human. PloS One. 2015;10:e0123397. doi: 10.1371/journal.pone.0123397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.V. Tormos Kathryn, E. Anso, B.R.Hamanaka, et al. Mitochondrial complex III ROS regulate adipocyte Differentiation, Cell Metab. 14: pp. 537–544. [DOI] [PMC free article] [PubMed]
- 10.Naukkarinen J., Heinonen S., Hakkarainen A. Characterising metabolically healthy obesity in weight-discordant monozygotic twins. Diabetologia. 2014;57:167–176. doi: 10.1007/s00125-013-3066-y. [DOI] [PubMed] [Google Scholar]
- 11.Heinonen S., Buzkova J., Muniandy M. Impaired mitochondrial biogenesis in adipose tissue in acquired obesity. Diabetes. 2015;64:3135. doi: 10.2337/db14-1937. [DOI] [PubMed] [Google Scholar]
- 12.Christodoulides C., Lagathu C., Sethi J.K., Vidal-Puig A. Adipogenesis and WNT signalling. Trends Endocrinol. Metab.: TEM. 2009;20:16–24. doi: 10.1016/j.tem.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bai P., Canto C., Brunyanszki A. PARP-2 regulates SIRT1 expression and whole-body energy expenditure. Cell Metab. 2011;13:450–460. doi: 10.1016/j.cmet.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Houtkooper R.H., Pirinen E., Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat. Rev. Mol. Cell Biol. 2012;13:225–238. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pagliarini D.J., Calvo S.E., Chang B. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wai T., Langer T. Mitochondrial dynamics and metabolic regulation. Trends Endocrinol. Metab. 2016;27:105–117. doi: 10.1016/j.tem.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 17.Phillips M.J., Voeltz G.K. Structure and function of ER membrane contact sites with other organelles. Nat. Rev. Mol. Cell Biol. 2016;17:69–82. doi: 10.1038/nrm.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cantó C., Gerhart-Hines Z., Feige J.N. AMPK regulates energy expenditure by modulating NAD(+) metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jäger S., Handschin C., St.-Pierre J., Spiegelman B.M. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1α. Proc. Natl. Acad. Sci. 2007;104:12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Puigserver P., Wu Z., Park C.W., Graves R., Wright M., Spiegelman B.M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto H., Williams E.G., Mouchiroud L. NCoR1 is a conserved physiological modulator of muscle mass and oxidative function. Cell. 2011;147:827–839. doi: 10.1016/j.cell.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pérez-Schindler J., Summermatter S., Salatino S. The corepressor NCoR1 antagonizes PGC-1α and estrogen-related receptor α in the regulation of skeletal muscle function and oxidative metabolism. Mol. Cell. Biol. 2012;32:4913–4924. doi: 10.1128/MCB.00877-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li P., Fan W., Xu J. Adipocyte NCoR knockout decreases PPARγ phosphorylation and enhances PPARγ activity and insulin sensitivity. Cell. 2011;147:815–826. doi: 10.1016/j.cell.2011.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dominy J.E., Puigserver P. Mitochondrial biogenesis through activation of nuclear signaling proteins. Cold Spring Harb. Perspect. Biol. 2013;5:a015008. doi: 10.1101/cshperspect.a015008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quiros P.M., Langer T., Lopez-Otin C. New roles for mitochondrial proteases in health, ageing and disease. Nat. Rev. Mol. Cell Biol. 2015;16:345–359. doi: 10.1038/nrm3984. [DOI] [PubMed] [Google Scholar]
- 26.Karbowski M., Youle R.J. Regulating mitochondrial outer membrane proteins by ubiquitination and proteasomal degradation. Curr. Opin. Cell Biol. 2011;23:476–482. doi: 10.1016/j.ceb.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jovaisaite V., Mouchiroud L., Auwerx J. The mitochondrial unfolded protein response, a conserved stress response pathway with implications in health and disease. J. Exp. Biol. 2014;217:137–143. doi: 10.1242/jeb.090738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sugiura A., McLelland G.-L., Fon E.A., McBride H.M. A new pathway for mitochondrial quality control: mitochondrial-derived vesicles. Embo J. 2014;33:2142–2156. doi: 10.15252/embj.201488104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Youle R.J., Narendra D.P. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Canto C., Menzies K.J., Auwerx J. NAD(+) metabolism and the control of energy homeostasis: a balancing act between mitochondria and the nucleus. Cell Metab. 2015;22:31–53. doi: 10.1016/j.cmet.2015.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dashty M. A quick look at biochemistry: carbohydrate metabolism. Clin. Biochem. 2013;46:1339–1352. doi: 10.1016/j.clinbiochem.2013.04.027. [DOI] [PubMed] [Google Scholar]
- 32.McKenna M.C., Waagepetersen H.S., Schousboe A., Sonnewald U. Neuronal and astrocytic shuttle mechanisms for cytosolic-mitochondrial transfer of reducing equivalents: current evidence and pharmacological tools. Biochem. Pharmacol. 2006;71:399–407. doi: 10.1016/j.bcp.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 33.Eaton S. Control of mitochondrial beta-oxidation flux. Prog. Lipid Res. 2002;41:197–239. doi: 10.1016/s0163-7827(01)00024-8. [DOI] [PubMed] [Google Scholar]
- 34.Ussher J.R., Jaswal J.S., Lopaschuk G.D. Pyridine nucleotide regulation of cardiac intermediary metabolism. Circ. Res. 2012;111:628–641. doi: 10.1161/CIRCRESAHA.111.246371. [DOI] [PubMed] [Google Scholar]
- 35.Lynch C.J., Adams S.H. Branched-chain amino acids in metabolic signalling and insulin resistance. Nat. Rev. Endocrinol. 2014;10:723–736. doi: 10.1038/nrendo.2014.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heinonen S., Muniandy M., Buzkova J. Mitochondria-related transcriptional signature is downregulated in adipocytes in obesity: a study of young healthy MZ twins. Diabetologia. 2016:1–13. doi: 10.1007/s00125-016-4121-2. [DOI] [PubMed] [Google Scholar]
- 37.Bogacka I., Xie H., Bray G.A., Smith S.R. Pioglitazone induces mitochondrial biogenesis in human subcutaneous adipose tissue in vivo. Diabetes. 2005;54:1392–1399. doi: 10.2337/diabetes.54.5.1392. [DOI] [PubMed] [Google Scholar]
- 38.Choo H.-J., Kim J.-H., Kwon O.-B. Mitochondria are impaired in the adipocytes of type 2 diabetic mice. Diabetologia. 2006;49:784–791. doi: 10.1007/s00125-006-0170-2. [DOI] [PubMed] [Google Scholar]
- 39.Kaaman M., Sparks L.M., van Harmelen V. Strong association between mitochondrial DNA copy number and lipogenesis in human white adipose tissue. Diabetologia. 2007;50:2526–2533. doi: 10.1007/s00125-007-0818-6. [DOI] [PubMed] [Google Scholar]
- 40.Cummins T.D., Holden C.R., Sansbury B.E. Metabolic remodeling of white adipose tissue in obesity. Am. J. Physiol. - Endocrinol. Metab. 2014;307:E262–E277. doi: 10.1152/ajpendo.00271.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rong J.X., Qiu Y., Hansen M.K. Adipose mitochondrial biogenesis is suppressed in db/db and high-fat diet–fed mice and improved by rosiglitazone. Diabetes. 2007;56:1751–1760. doi: 10.2337/db06-1135. [DOI] [PubMed] [Google Scholar]
- 42.Tol M.J., Ottenhoff R., van Eijk M. A PPARγ-Bnip3 axis couples adipose mitochondrial fusion-fission balance to systemic insulin sensitivity. Diabetes. 2016;65:2591–2605. doi: 10.2337/db16-0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jukarainen S., Heinonen S., Rämö J.T. Obesity is associated with low NAD+/SIRT pathway expression in adipose tissue of BMI-discordant monozygotic twins. J. Clin. Endocrinol. Metab. 2016;101:275–283. doi: 10.1210/jc.2015-3095. [DOI] [PubMed] [Google Scholar]
- 44.Moreno-Viedma V., Amor M., Sarabi A. Common dysregulated pathways in obese adipose tissue and atherosclerosis. Cardiovasc. Diabetol. 2016;15:120. doi: 10.1186/s12933-016-0441-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.She P., Van Horn C., Reid T., Hutson S.M., Cooney R.N., Lynch C.J. Obesity-related elevations in plasma leucine are associated with alterations in enzymes involved in branched-chain amino acid metabolism. Am. J. Physiol. Endocrinol. Metab. 2007;293:E1552–E1563. doi: 10.1152/ajpendo.00134.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dahlman I., Forsgren M., Sjögren A. Downregulation of electron transport chain genes in visceral adipose tissue in type 2 diabetes independent of obesity and possibly involving tumor necrosis factor-α. Diabetes. 2006;55:1792–1799. doi: 10.2337/db05-1421. [DOI] [PubMed] [Google Scholar]
- 47.Lindinger P.W., Christe M., Eberle A.N. Important mitochondrial proteins in human omental adipose tissue show reduced expression in obesity. J. Proteom. 2015;124:79–87. doi: 10.1016/j.jprot.2015.03.037. [DOI] [PubMed] [Google Scholar]
- 48.Xie X., Yi Z., Bowen B. Characterization of the human adipocyte proteome and reproducibility of protein abundance by one-dimensional gel electrophoresis and HPLC-ESI-MS/MS. J. Proteome Res. 2010;9:4521–4534. doi: 10.1021/pr100268f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Christe M., Hirzel E., Lindinger A. Obesity affects mitochondrial citrate synthase in human omental adipose tissue. ISRN Obes. 2013;2013:826027. doi: 10.1155/2013/826027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fischer B., Schöttl T., Schempp C. Inverse relationship between body mass index and mitochondrial oxidative phosphorylation capacity in human subcutaneous adipocytes. Am. J. Physiol. - Endocrinol. Metab. 2015;309:E380–E387. doi: 10.1152/ajpendo.00524.2014. [DOI] [PubMed] [Google Scholar]
- 51.Yin X., Lanza I.R., Swain J.M., Sarr M.G., Nair K.S., Jensen M.D. Adipocyte mitochondrial function is reduced in human obesity independent of fat cell size. J. Clin. Endocrinol. Metab. 2014;99:E209–E216. doi: 10.1210/jc.2013-3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chattopadhyay M., GuhaThakurta I., Behera P. Mitochondrial bioenergetics is not impaired in nonobese subjects with type 2 diabetes mellitus. Metab. - Clin. Exp. 2011;60:1702–1710. doi: 10.1016/j.metabol.2011.04.015. [DOI] [PubMed] [Google Scholar]
- 53.Schöttl T., Kappler L., Fromme T., Klingenspor M. Limited OXPHOS capacity in white adipocytes is a hallmark of obesity in laboratory mice irrespective of the glucose tolerance status. Mol. Metab. 2015;4:631–642. doi: 10.1016/j.molmet.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilson-Fritch L., Nicoloro S., Chouinard M. Mitochondrial remodeling in adipose tissue associated with obesity and treatment with rosiglitazone. J. Clin. Investig. 2004;114:1281–1289. doi: 10.1172/JCI21752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Calderon-Dominguez M., Mir J.F., Fucho R., Weber M., Serra D., Herrero L. Fatty acid metabolism and the basis of brown adipose tissue function. Adipocyte. 2016;5:98–118. doi: 10.1080/21623945.2015.1122857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Virtanen K.A., Lidell M.E., Orava J. Functional brown adipose tissue in healthy adults. New Engl. J. Med. 2009;360:1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- 57.Hondares E., Rosell M., Diaz-Delfin J. Peroxisome proliferator-activated receptor alpha (PPARalpha) induces PPARgamma coactivator 1alpha (PGC-1alpha) gene expression and contributes to thermogenic activation of brown fat: involvement of PRDM16. J. Biol. Chem. 2011;286:43112–43122. doi: 10.1074/jbc.M111.252775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seale P., Kajimura S., Yang W. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007;6:38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Uldry M., Yang W., St-Pierre J., Lin J., Seale P., Spiegelman B.M. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 2006;3:333–341. doi: 10.1016/j.cmet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 60.Goodbody A.E., Trayhurn P. GDP binding to brown-adipose-tissue mitochondria of diabetic--obese (db/db) mice. Decreased binding in both the obese and pre-obese states. Biochem. J. 1981;194:1019–1022. doi: 10.1042/bj1941019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Himms-Hagen J., Desautels M. A mitochondrial defect in brown adipose tissue of the obese (ob/ob) mouse: reduced binding of purine nucleotides and a failure to respond to cold by an increase in binding. Biochem. Biophys. Res. Commun. 1978;83:628–634. doi: 10.1016/0006-291x(78)91036-7. [DOI] [PubMed] [Google Scholar]
- 62.Shimizu I., Aprahamian T., Kikuchi R. Vascular rarefaction mediates whitening of brown fat in obesity. J. Clin. Invest. 2014;124:2099–2112. doi: 10.1172/JCI71643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vijgen G.H., Bouvy N.D., Teule G.J., Brans B., Schrauwen P., van Marken Lichtenbelt W.D. Brown adipose tissue in morbidly obese subjects. PloS One. 2011;6:e17247. doi: 10.1371/journal.pone.0017247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vernochet C., Damilano F., Mourier A. Adipose tissue mitochondrial dysfunction triggers a lipodystrophic syndrome with insulin resistance, hepatosteatosis, and cardiovascular complications. FASEB J. : Off. Publ. Fed. Am. Soc. Exp. Biol. 2014;28:4408–4419. doi: 10.1096/fj.14-253971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Caton P.W., Kieswich J., Yaqoob M.M., Holness M.J., Sugden M.C. Nicotinamide mononucleotide protects against pro-inflammatory cytokine-mediated impairment of mouse islet function. Diabetologia. 2011;54:3083–3092. doi: 10.1007/s00125-011-2288-0. [DOI] [PubMed] [Google Scholar]
- 66.Mercader J., Granados N., Caimari A., Oliver P., Bonet M.L., Palou A. Retinol-binding protein 4 and nicotinamide phosphoribosyltransferase/visfatin in rat obesity models. Horm. Metab. Res. 2008;40:467–472. doi: 10.1055/s-2008-1065324. [DOI] [PubMed] [Google Scholar]
- 67.Yoshino J., Mills K.F., Yoon M.J., Imai S. Nicotinamide mononucleotide, a key NAD(+) intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metab. 2011;14:528–536. doi: 10.1016/j.cmet.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pagano C., Pilon C., Olivieri M. Reduced plasma visfatin/pre-B cell colony-enhancing factor in obesity is not related to insulin resistance in humans. J. Clin. Endocrinol. Metab. 2006;91:3165–3170. doi: 10.1210/jc.2006-0361. [DOI] [PubMed] [Google Scholar]
- 69.Rappou E., Jukarainen S., Rinnankoski-Tuikka R. Weight loss is associated with increased NAD+/SIRT1 expression but reduced PARP activity in white adipose tissue. J. Clin. Endocrinol. Metab. 2016;101:1263–1273. doi: 10.1210/jc.2015-3054. [DOI] [PubMed] [Google Scholar]
- 70.Yoon M.J., Yoshida M., Johnson S. SIRT1-mediated eNAMPT secretion from adipose tissue regulates hypothalamic NAD+ and function in mice. Cell Metab. 2015;21:706–717. doi: 10.1016/j.cmet.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Canto C., Houtkooper R.H., Pirinen E. The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab. 2012;15:838–847. doi: 10.1016/j.cmet.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nikiforov A., Kulikova V., Ziegler M. The human NAD metabolome: functions, metabolism and compartmentalization. Crit. Rev. Biochem. Mol. Biol. 2015;50:284–297. doi: 10.3109/10409238.2015.1028612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stein L.R., Imai S. The dynamic regulation of NAD metabolism in mitochondria. Trends Endocrinol. Metab. 2012;23:420–428. doi: 10.1016/j.tem.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Houtkooper R.H., Canto C., Wanders R.J., Auwerx J. The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr. Rev. 2010;31:194–223. doi: 10.1210/er.2009-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ratajczak J., Joffraud M., Trammell S.A. NRK1 controls nicotinamide mononucleotide and nicotinamide riboside metabolism in mammalian cells. Nat. Commun. 2016;7:13103. doi: 10.1038/ncomms13103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Revollo J.R., Korner A., Mills K.F. Nampt/PBEF/Visfatin regulates insulin secretion in beta cells as a systemic NAD biosynthetic enzyme. Cell Metab. 2007;6:363–375. doi: 10.1016/j.cmet.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mouchiroud L., Houtkooper R.H., Auwerx J. NAD(+) metabolism: a therapeutic target for age-related metabolic disease. Crit. Rev. Biochem. Mol. Biol. 2013;48:397–408. doi: 10.3109/10409238.2013.789479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bai P., Cantó C. The role of PARP-1 and PARP-2 enzymes in metabolic regulation and disease. Cell Metab. 2012;16:290–295. doi: 10.1016/j.cmet.2012.06.016. [DOI] [PubMed] [Google Scholar]
- 79.Bai P., Nagy L., Fodor T., Liaudet L., Pacher P. Poly(ADP-ribose) polymerases as modulators of mitochondrial activity. Trends Endocrinol. Metab. 2015;26:75–83. doi: 10.1016/j.tem.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 80.Bai P., Canto C., Oudart H. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab. 2011;13:461–468. doi: 10.1016/j.cmet.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pirinen E., Canto C., Jo Y.S. Pharmacological inhibition of poly(ADP-ribose) polymerases improves fitness and mitochondrial function in skeletal muscle. Cell Metab. 2014;19:1034–1041. doi: 10.1016/j.cmet.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rajamohan S.B., Pillai V.B., Gupta M. SIRT1 promotes cell survival under stress by deacetylation-dependent deactivation of poly(ADP-ribose) polymerase 1. Mol. Cell. Biol. 2009;29:4116–4129. doi: 10.1128/MCB.00121-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ramsey K.M., Yoshino J., Brace C.S. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Frydelund-Larsen L., Akerstrom T., Nielsen S., Keller P., Keller C., Pedersen B.K. Visfatin mRNA expression in human subcutaneous adipose tissue is regulated by exercise. Am. J. Physiol. Endocrinol. Metab. 2007;292:E24–E31. doi: 10.1152/ajpendo.00113.2006. [DOI] [PubMed] [Google Scholar]
- 85.Chen D., Bruno J., Easlon E. Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev. 2008;22:1753–1757. doi: 10.1101/gad.1650608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Song J., Ke S.F., Zhou C.C. Nicotinamide phosphoribosyltransferase is required for the calorie restriction-mediated improvements in oxidative stress, mitochondrial biogenesis, and metabolic adaptation. J. Gerontol. A Biol. Sci. Med. Sci. 2014;69:44–57. doi: 10.1093/gerona/glt122. [DOI] [PubMed] [Google Scholar]
- 87.Caton P.W., Kieswich J., Yaqoob M.M., Holness M.J., Sugden M.C. Metformin opposes impaired AMPK and SIRT1 function and deleterious changes in core clock protein expression in white adipose tissue of genetically-obese db/db mice. Diabetes Obes. Metab. 2011;13:1097–1104. doi: 10.1111/j.1463-1326.2011.01466.x. [DOI] [PubMed] [Google Scholar]
- 88.Curtin N.J., Szabo C. Therapeutic applications of PARP inhibitors: anticancer therapy and beyond. Mol. Asp. Med. 2013;34:1217–1256. doi: 10.1016/j.mam.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Feige J.N., Lagouge M., Canto C. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 2008;8:347–358. doi: 10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 90.Lagouge M., Argmann C., Gerhart-Hines Z. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 91.Choi K.C., Ryu O.H., Lee K.W. Effect of PPAR-alpha and -gamma agonist on the expression of visfatin, adiponectin, and TNF-alpha in visceral fat of OLETF rats. Biochem. Biophys. Res. Commun. 2005;336:747–753. doi: 10.1016/j.bbrc.2005.08.203. [DOI] [PubMed] [Google Scholar]
- 92.Choi K.C., Lee S.Y., Yoo H.J. Effect of PPAR-delta agonist on the expression of visfatin, adiponectin, and resistin in rat adipose tissue and 3T3-L1 adipocytes. Biochem. Biophys. Res. Commun. 2007;357:62–67. doi: 10.1016/j.bbrc.2007.03.114. [DOI] [PubMed] [Google Scholar]
- 93.Stromsdorfer K.L., Yamaguchi S., Yoon M.J. NAMPT-mediated NAD(+) biosynthesis in adipocytes regulates adipose tissue function and multi-organ insulin sensitivity in mice. Cell Rep. 2016;16:1851–1860. doi: 10.1016/j.celrep.2016.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Birjmohun R.S., Hutten B.A., Kastelein J.J., Stroes E.S. Efficacy and safety of high-density lipoprotein cholesterol-increasing compounds: a meta-analysis of randomized controlled trials. J. Am. Coll. Cardiol. 2005;45:185–197. doi: 10.1016/j.jacc.2004.10.031. [DOI] [PubMed] [Google Scholar]
- 95.Barbosa M.T., Soares S.M., Novak C.M. The enzyme CD38 (a NAD glycohydrolase, EC 3.2.2.5) is necessary for the development of diet-induced obesity. FASEB J. : Off. Publ. Fed. Am. Soc. Exp. Biol. 2007;21:3629–3639. doi: 10.1096/fj.07-8290com. [DOI] [PubMed] [Google Scholar]
- 96.Gariani K., Ryu D., Menzies K.J. Inhibiting poly ADP-ribosylation increases fatty acid oxidation and protects against fatty liver disease. J. Hepatol. 2016;66:132–141. doi: 10.1016/j.jhep.2016.08.024. [DOI] [PubMed] [Google Scholar]
- 97.Haffner C.D., Becherer J.D., Boros E.E. Discovery, synthesis, and biological evaluation of thiazoloquin(az)olin(on)es as potent CD38 inhibitors. J. Med. Chem. 2015;58:3548–3571. doi: 10.1021/jm502009h. [DOI] [PubMed] [Google Scholar]
- 98.Drew J.E., Farquharson A.J., Horgan G.W., Williams L.M. Tissue-specific regulation of sirtuin and nicotinamide adenine dinucleotide biosynthetic pathways identified in C57Bl/6 mice in response to high-fat feeding. J. Nutr. Biochem. 2016;37:20–29. doi: 10.1016/j.jnutbio.2016.07.013. [DOI] [PubMed] [Google Scholar]
- 99.Liu Z., Gan L., Liu G. Sirt1 decreased adipose inflammation by interacting with Akt2 and inhibiting mTOR/S6K1 pathway in mice. J. Lipid Res. 2016;57:1373–1381. doi: 10.1194/jlr.M063537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nisoli E., Tonello C., Cardile A. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science. 2005;310:314–317. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- 101.Chalkiadaki A., Guarente L. High-fat diet triggers inflammation-induced cleavage of SIRT1 in adipose tissue to promote metabolic dysfunction. Cell Metab. 2012;16:180–188. doi: 10.1016/j.cmet.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Krishnan J., Danzer C., Simka T. Dietary obesity-associated Hif1α activation in adipocytes restricts fatty acid oxidation and energy expenditure via suppression of the Sirt2-NAD(+) system. Genes Dev. 2012;26:259–270. doi: 10.1101/gad.180406.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cohen H.Y., Miller C., Bitterman K.J. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 104.Laurent G., German N.J., Saha A.K. SIRT4 coordinates the balance between lipid synthesis and catabolism by repressing malonyl CoA decarboxylase. Mol. Cell. 2013;50:686–698. doi: 10.1016/j.molcel.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang F., Tong Q. SIRT2 suppresses adipocyte differentiation by deacetylating FOXO1 and enhancing FOXO1's repressive interaction with PPARγ. Mol. Biol. Cell. 2009;20:801–808. doi: 10.1091/mbc.E08-06-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Clark S.J., Falchi M., Olsson B. Association of sirtuin 1 (SIRT1) gene SNPs and transcript expression levels with severe obesity. Obesity. 2012;20:178–185. doi: 10.1038/oby.2011.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gillum M.P., Kotas M.E., Erion D.M. SirT1 regulates adipose tissue inflammation. Diabetes. 2011;60:3235–3245. doi: 10.2337/db11-0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kurylowicz A., Owczarz M., Polosak J. SIRT1 and SIRT7 expression in adipose tissues of obese and normal-weight individuals is regulated by microRNAs but not by methylation status. Int. J. Obes. 2016;40:1635–1642. doi: 10.1038/ijo.2016.131. [DOI] [PubMed] [Google Scholar]
- 109.Pedersen S.B., Olholm J., Paulsen S.K., Bennetzen M.F., Richelsen B. Low Sirt1 expression, which is upregulated by fasting, in human adipose tissue from obese women. Int. J. Obes. 2008;32:1250–1255. doi: 10.1038/ijo.2008.78. [DOI] [PubMed] [Google Scholar]
- 110.Song Y.S., Lee S.K., Jang Y.J. Association between low SIRT1 expression in visceral and subcutaneous adipose tissues and metabolic abnormalities in women with obesity and type 2 diabetes. Diabetes Res. Clin. Pract. 2013;101:341–348. doi: 10.1016/j.diabres.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 111.Moschen A.R., Wieser V., Gerner R.R. Adipose tissue and liver expression of SIRT1, 3, and 6 increase after extensive weight loss in morbid obesity. J. Hepatol. 2013;59:1315–1322. doi: 10.1016/j.jhep.2013.07.027. [DOI] [PubMed] [Google Scholar]
- 112.Rosen E.D., Walkey C.J., Puigserver P., Spiegelman B.M. Transcriptional regulation of adipogenesis. Genes Dev. 2000;14:1293–1307. [PubMed] [Google Scholar]
- 113.Wilson-Fritch L., Burkart A., Bell G. Mitochondrial biogenesis and remodeling during adipogenesis and in response to the insulin sensitizer rosiglitazone. Mol. Cell. Biol. 2003;23:1085–1094. doi: 10.1128/MCB.23.3.1085-1094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Picard F., Kurtev M., Chung N. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-[gamma] Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mayoral R., Osborn O., McNelis J. Adipocyte SIRT1 knockout promotes PPARγ activity, adipogenesis and insulin sensitivity in chronic-HFD and obesity. Mol. Metab. 2015;4:378–391. doi: 10.1016/j.molmet.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Xu F., Burk D., Gao Z. Angiogenic deficiency and adipose tissue dysfunction are associated with macrophage malfunction in SIRT1(−/−) mice. Endocrinology. 2012;153:1706–1716. doi: 10.1210/en.2011-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhou Y., Song T., Peng J. SIRT1 Suppresses Adipogenesis by Activating Wnt/β-Catenin Signaling In Vivo and In Vitro. 2016;7:77707–77720. doi: 10.18632/oncotarget.12774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Simic P., Zainabadi K., Bell E. SIRT1 regulates differentiation of mesenchymal stem cells by deacetylating β-catenin. EMBO Mol. Med. 2013;5:430–440. doi: 10.1002/emmm.201201606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jing E., Gesta S., Kahn C.R. SIRT2 regulates adipocyte differentiation through FoxO1 acetylation/deacetylation. Cell Metab. 2007;6:105–114. doi: 10.1016/j.cmet.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cioffi M., Vallespinos-Serrano M., Trabulo Sara M. MiR-93 controls adiposity via inhibition of Sirt7 and Tbx3. Cell Rep. 2015;12:1594–1605. doi: 10.1016/j.celrep.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 121.Bober E., Fang J., Smolka C. Sirt7 promotes adipogenesis by binding to and inhibiting Sirt1. BMC Proc. 2012;6:P57. [Google Scholar]
- 122.Erener S., Hesse M., Kostadinova R., Hottiger M.O. Poly(ADP-Ribose)polymerase-1 (PARP1) controls adipogenic gene expression and adipocyte function. Mol. Endocrinol. 2012;26:79–86. doi: 10.1210/me.2011-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Erener S., Mirsaidi A., Hesse M. ARTD1 deletion causes increased hepatic lipid accumulation in mice fed a high-fat diet and impairs adipocyte function and differentiation. FASEB J. 2012;26:2631–2638. doi: 10.1096/fj.11-200212. [DOI] [PubMed] [Google Scholar]
- 124.Lehmann M., Pirinen E., Mirsaidi A. ARTD1-induced poly-ADP-ribose formation enhances PPARγ ligand binding and co-factor exchange. Nucleic Acids Res. 2015;43:129–142. doi: 10.1093/nar/gku1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bai P., Houten S.M., Huber A. Peroxisome proliferator-activated receptor (PPAR)-2 controls adipocyte differentiation and adipose tissue function through the regulation of the activity of the retinoid X receptor/PPARγ heterodimer. J. Biol. Chem. 2007;282:37738–37746. doi: 10.1074/jbc.M701021200. [DOI] [PubMed] [Google Scholar]
- 126.Xu C., Bai B., Fan P. Selective overexpression of human SIRT1 in adipose tissue enhances energy homeostasis and prevents the deterioration of insulin sensitivity with ageing in mice. Am. J. Transl. Res. 2013;5:412–426. [PMC free article] [PubMed] [Google Scholar]
- 127.Boutant M., Joffraud M., Kulkarni S.S. SIRT1 enhances glucose tolerance by potentiating brown adipose tissue function. Mol. Metab. 2015;4:118–131. doi: 10.1016/j.molmet.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chakrabarti P., English T., Karki S. SIRT1 controls lipolysis in adipocytes via FOXO1-mediated expression of ATGL. J. Lipid Res. 2011;52:1693–1701. doi: 10.1194/jlr.M014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Fischer-Posovszky P., Kukulus V., Tews D. Resveratrol regulates human adipocyte number and function in a Sirt1-dependent manner. Am. J. Clin. Nutr. 2010;92:5–15. doi: 10.3945/ajcn.2009.28435. [DOI] [PubMed] [Google Scholar]
- 130.Foster D.W. Malonyl-CoA: the regulator of fatty acid synthesis and oxidation. J. Clin. Invest. 2012;122:1958–1959. doi: 10.1172/JCI63967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kanfi Y., Peshti V., Gil R. SIRT6 protects against pathological damage caused by diet-induced obesity. Aging Cell. 2010;9:162–173. doi: 10.1111/j.1474-9726.2009.00544.x. [DOI] [PubMed] [Google Scholar]
- 132.Boutant M., Kulkarni Sameer S., Joffraud M. SIRT1 gain of function does not mimic or enhance the adaptations to intermittent fasting. Cell Rep. 2016;14:2068–2075. doi: 10.1016/j.celrep.2016.02.007. [DOI] [PubMed] [Google Scholar]
- 133.Lin Q.-Q., Yan C.-F., Lin R. SIRT1 regulates TNF-α-induced expression of CD40 in 3T3-L1 adipocytes via NF-κB pathway. Cytokine. 2012;60:447–455. doi: 10.1016/j.cyto.2012.05.025. [DOI] [PubMed] [Google Scholar]
- 134.Xu F., Gao Z., Zhang J. Lack of SIRT1 (mammalian sirtuin 1) activity leads to liver steatosis in the SIRT1+/− mice: a role of lipid mobilization and inflammation. Endocrinology. 2010;151:2504–2514. doi: 10.1210/en.2009-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yoshizawa T., Karim M.F., Sato Y. SIRT7 controls hepatic lipid metabolism by regulating the ubiquitin-proteasome pathway. Cell Metab. 2014;19:712–721. doi: 10.1016/j.cmet.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 136.Qiao L., Shao J. SIRT1 regulates adiponectin gene expression through Foxo1-C/enhancer-binding protein α transcriptional complex. J. Biol. Chem. 2006;281:39915–39924. doi: 10.1074/jbc.M607215200. [DOI] [PubMed] [Google Scholar]
- 137.Qiang L., Wang H., Farmer S.R. Adiponectin secretion is regulated by SIRT1 and the endoplasmic reticulum oxidoreductase Ero1-Lα. Mol. Cell. Biol. 2007;27:4698–4707. doi: 10.1128/MCB.02279-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Xu F., Zheng X., Lin B. Diet-induced obesity and insulin resistance are associated with brown fat degeneration in SIRT1-deficient mice. Obesity. 2016;24:634–642. doi: 10.1002/oby.21393. [DOI] [PubMed] [Google Scholar]
- 139.Banks A.S., Kon N., Knight C. SirT1 gain-of-function increases energy efficiency and prevents diabetes in mice. Cell Metab. 2008;8:333–341. doi: 10.1016/j.cmet.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bordone L., Cohen D., Robinson A. SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell. 2007;6:759–767. doi: 10.1111/j.1474-9726.2007.00335.x. [DOI] [PubMed] [Google Scholar]
- 141.Shi T., Wang F., Stieren E., Tong Q. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J. Biol. Chem. 2005;280:13560–13567. doi: 10.1074/jbc.M414670200. [DOI] [PubMed] [Google Scholar]
- 142.Hirschey M.D., Shimazu T., Goetzman E. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Noriega L.G., Feige J.N., Canto C. CREB and ChREBP oppositely regulate SIRT1 expression in response to energy availability. EMBO Rep. 2011;12:1069–1076. doi: 10.1038/embor.2011.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gerhart-Hines Z., Dominy J.E., Jr., Blattler S.M. The cAMP/PKA pathway rapidly activates SIRT1 to promote fatty acid oxidation independently of changes in NAD(+) Mol. Cell. 2011;44:851–863. doi: 10.1016/j.molcel.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Giralt A., Hondares E., Villena J.A. Peroxisome proliferator-activated receptor-gamma coactivator-1alpha controls transcription of the Sirt3 gene, an essential component of the thermogenic brown adipocyte phenotype. J. Biol. Chem. 2011;286:16958–16966. doi: 10.1074/jbc.M110.202390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Giralt A., Hondares E., Villena J.A. Peroxisome proliferator-activated receptor-γ coactivator-1α controls transcription of the Sirt3 gene, an essential component of the thermogenic brown adipocyte phenotype. J. Biol. Chem. 2011;286:16958–16966. doi: 10.1074/jbc.M110.202390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Tiraby C., Tavernier G., Lefort C. Acquirement of brown fat cell features by human white adipocytes. J. Biol. Chem. 2003;278:33370–33376. doi: 10.1074/jbc.M305235200. [DOI] [PubMed] [Google Scholar]
- 148.Qiang L., Wang L., Kon N. Brown remodeling of white adipose tissue by SirT1-dependent deacetylation of Pparγ. Cell. 2012;150:620–632. doi: 10.1016/j.cell.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Potente M., Ghaeni L., Baldessari D. SIRT1 controls endothelial angiogenic functions during vascular growth. Genes Dev. 2007;21:2644–2658. doi: 10.1101/gad.435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Finley L.W., Haas W., Desquiret-Dumas V. Succinate dehydrogenase is a direct target of sirtuin 3 deacetylase activity. PloS One. 2011;6:e23295. doi: 10.1371/journal.pone.0023295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Giralt A., Villarroya F. SIRT3, a pivotal actor in mitochondrial functions: metabolism, cell death and aging. Biochem. J. 2012;444:1–10. doi: 10.1042/BJ20120030. [DOI] [PubMed] [Google Scholar]
- 152.Bartelt A., Bruns O.T., Reimer R. Brown adipose tissue activity controls triglyceride clearance. Nat. Med. 2011;17:200–205. doi: 10.1038/nm.2297. [DOI] [PubMed] [Google Scholar]
- 153.Cannon B., Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol. Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 154.Pfluger P.T., Herranz D., Velasco-Miguel S., Serrano M., Tschop M.H. Sirt1 protects against high-fat diet-induced metabolic damage. Proc. Natl. Acad. Sci. USA. 2008;105:9793–9798. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhong L., D'Urso A., Toiber D. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1α. Cell. 2010;140:280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Erion D.M., Yonemitsu S., Nie Y. SirT1 knockdown in liver decreases basal hepatic glucose production and increases hepatic insulin responsiveness in diabetic rats. Proc. Natl. Acad. Sci. USA. 2009;27:11288–11293. doi: 10.1073/pnas.0812931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lombard D.B., Alt F.W., Cheng H.L. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol. Cell. Biol. 2007;24:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mostoslavsky R., Chua K.F., Lombard D.B. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 159.Devalaraja-Narashimha K., Padanilam B.J. PARP1 deficiency exacerbates diet-induced obesity in mice. J. Endocrinol. 2010;205:243–252. doi: 10.1677/JOE-09-0402. [DOI] [PubMed] [Google Scholar]