Abstract
Lipases are important industrial enzymes. Most of the lipases operate at lipid–water interfaces enabled by a mobile lid domain located over the active site. Lid protects the active site and hence responsible for catalytic activity. In pure aqueous media, the lid is predominantly closed, whereas in the presence of a hydrophobic layer, it is partially opened. Hence, the lid controls the enzyme activity. In the present review, we have classified lipases into different groups based on the structure of lid domains. It has been observed that thermostable lipases contain larger lid domains with two or more helices, whereas mesophilic lipases tend to have smaller lids in the form of a loop or a helix. Recent developments in lipase engineering addressing the lid regions are critically reviewed here. After on, the dramatic changes in substrate selectivity, activity, and thermostability have been reported. Furthermore, improved computational models can now rationalize these observations by relating it to the mobility of the lid domain. In this contribution, we summarized and critically evaluated the most recent developments in experimental and computational research on lipase lids.
Keywords: lipase, lid domain, thermostability, interfacial activation, protein engineering
Introduction
Lipases (triacylglycerol ester hydrolases EC 3.1.1.3) are among the most important industrial enzymes due to their specificity in hydrolysis, interesterification, alcoholysis, acidolysis, esterification, and aminolysis. These enzymes are generally used in different chemical sectors such as detergents, food, bioenergy, flavors, pharmaceuticals, and enantiopure esters and amino acid derivatives used in fine chemicals and agrochemicals (Hasan et al., 2006).
Lipases operate at the interface between lipid and water (Reis et al., 2009). The important feature of many lipases is the presence of a mobile subdomain lid or flap located over the active site (Brocca et al., 2003). If the lid is closed, the active site is protected from the environment and inaccessible to the substrates, hence the lipase is inactive. In an open conformation, substrates can enter the lipases’ active sites and be converted. In other words, only “open” lipases display catalytic activity (Barbe et al., 2009). For example, the structure of lipase from Thermomyces lanuginosus has been resolved both in its closed conformation (PDB code: 1DT3) as well as in its open conformation (PDB code: 1EIN). A comparison of both conformations is shown in Figure 1. In accordance with the generally very low catalytic activity of lipases in mainly aqueous media, it may be assumed that the “closed” conformation prevails under these conditions. Contrarily, in more hydrophobic (organic) media or in the presence of an organic–aqueous interphase, the “open” form may be assumed to be the predominant structure, also in accordance with the generally higher activity of lipases. The increased activity of lipases in the presence of apolar–aqueous interphases is known as “interfacial activation” (Reis et al., 2009).
Figure 1.
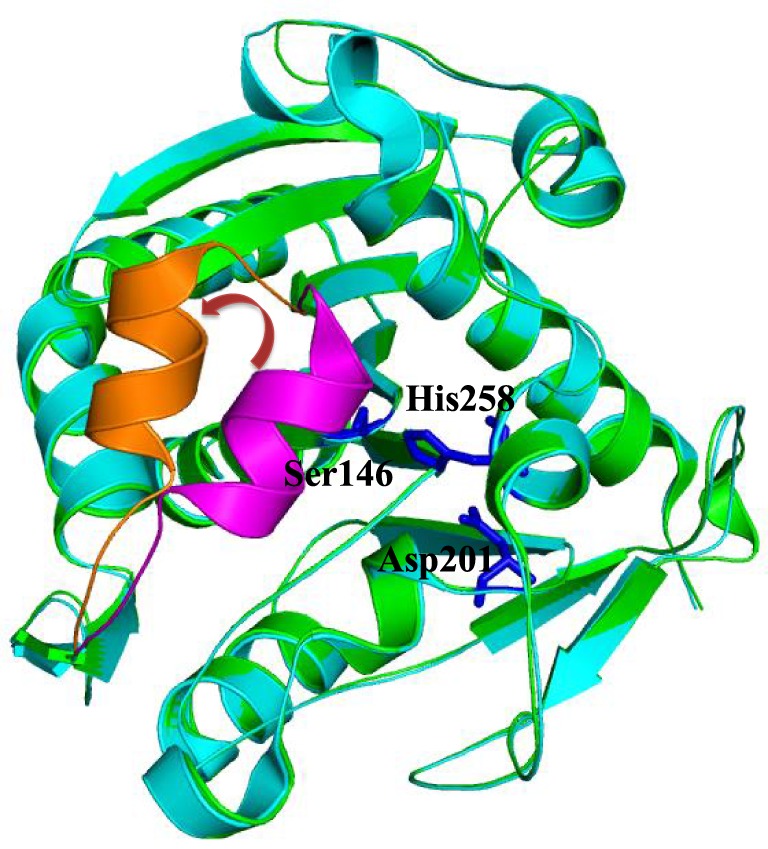
Superimposition of close (green, PDB code: 1DT3) and open (cyan, PDB code: 1EIN) conformations of Thermomyces lanuginosus lipase. The close lid, open lid, and catalytic triads were highlighted by magenta, orange, and blue colors, respectively.
Lids of lipases are amphipathic structures; in the closed conformation, their hydrophilic side faces the solvent, while the hydrophobic side is directed toward the catalytic pocket (Brocca et al., 2003). As the enzyme shifts to the open conformation, the hydrophobic face becomes exposed and contributes to the substrate-binding region (Yang and Lowe, 2000). Therefore, not only the amphipathic nature of the lid but also its specific amino acid sequence is important for activity and specificity of lipases (Holmquist et al., 1995).
It is known that lipid hydrolysis by lipase is activated by an oil–water interface (Maruyama et al., 2000). X-ray crystallographic analysis has showed that opening of the lid might occur during oil–water interfacial activation, hence allowing substrates access to the active site (Brzozowski et al., 1991).
Most lipases have a lid domain that covers its catalytic triad and the movement of their α-helical lid by rotating around two hinge regions at the lipid–water interface creates a large hydrophobic patch around the catalytic triad, resulting in activation of the lipase (Derewenda et al., 1992; Berg et al., 1998; Cajal et al., 2000).
Nowadays, there are many microorganism genomes that have been sequenced and millions of released raw sequences data have been deposited in database, such as Genbank, DDBJ, and EMBL. Genomic mining by combining bioinformatics analysis and functional screening provides opportunities to find out novel biocatalysts with desired properties for industry application, such as lipases (Masuch et al., 2015; Ufarte et al., 2015). Further modification of the catalytic behaviors of lipase can be achieved by engineering its lid domain. Therefore, the current review article focuses on the classification, mechanism, function, protein engineering, and computational analysis of lid domain of lipases. The following sections will explore the detailed study of lid domain of lipases.
Classification of Lids
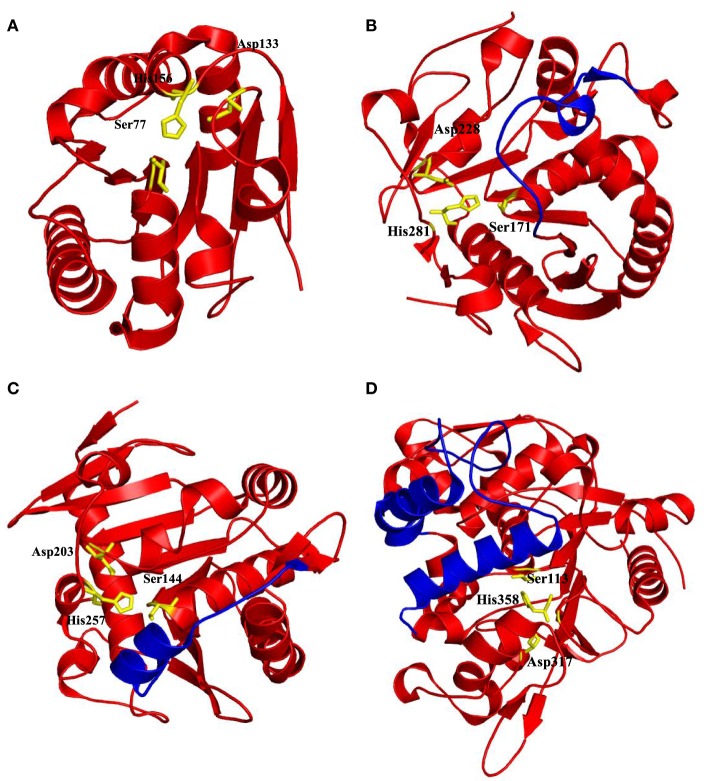
Lipases can be classified into different groups based on the similarity of sequence, structure, and function. Arpigny and Jaeger (1999) suggested that bacterial lipases can be classified into eight classes according to their conserved amino acid sequences and biochemical properties. Fischer and Pleiss (2003) have generated a lipase engineered database for analyzing sequence–structure–function relationship of α/β-hydrolase fold enzymes, and they proposed a classification of the lipolytic enzymes into GX- and GGGX-hydrolases groups based on the composition of their oxyanion hole. Kourist et al. (2010b) employed 3DM, a commercial structure-based sequence alignment and analysis tool, to analyze 1,172 structurally relative α/β-hydrolase fold enzymes, suggesting that the α/β-hydrolase fold enzyme superfamily can be divided into six families in term of their composition of the catalytic elbow. In this review, we have collected 149 structures of 44 different lipases from the Protein Data Bank (http://www.rcsb.org/). Of which, 25 lipases belong to eukaryotes, and 19 lipases to prokaryotes. Based on the type of lid domain, we have classified these lipases into three groups such as lipases without lids (Table 1), lipases with one loop or one helix lids (Table 2), and lipases with two or more helixes lids (Table 3). A structural comparison of lipases devoid of lids and with lids composed of one or two helices is shown in Figure 2. The position of the lid domain in the structure and the optimum reaction temperature were also summarized. It has been observed that high temperature lipases contain larger lid domains with two or more helices, and all mono- and diacylglycerol lipases have a small lid in the form of a loop or a helix (Table 4).
Table 1.
The structure of lipases without lid present in the Protein Data Bank.
| Organisms | PDB code | Reference | Optimum temperature (°C) | Optimum pH |
|---|---|---|---|---|
| Bacillus subtilis | 1I6W | Pouderoyen et al. (2001) | 35 (Lesuisse et al., 1993) | 10 (Lesuisse et al., 1993) |
| 1ISP | Kawasaki et al. (2002) | |||
| 1R4Z, 1R50 | Droge et al. (2006) | |||
| 1T2N, 1T4M | Acharya et al. (2004) | |||
| 2QXT, 2QXU | Rajakumara et al. (2008) | |||
| 3D2A, 3D2B, 3D2C | Ahmad et al. (2008) | |||
| 3QMM | Kamal et al. (2011) | |||
| 3QZU | Augustyniak et al. (2012) | |||
| 5CRI, 5CT4, 5CT5, 5CT6, 5CT8, 5CT9, 5CTA, 5CUR | Nordwald et al. (2015) | |||
| Streptomyces exfoliatus | 1JFR | Wei et al. (1998) | – | – |
| Pseudomonas mendocina | 2FX5 | – | – | – |
| Candida Antarctica | 1LBS, 1LBT | Uppenberg et al. (1996) | 45 (Eom et al., 2013) | 7 (Eom et al., 2013) |
| 1TCA, 1TCB, 1TCC | Uppenberg et al. (1994) | |||
| 3ICV, 3IVW | Qian et al. (2009) | |||
| 3W9B | – | |||
| 4K5Q, 4K6G, 4K6H, 4K6K | Xie et al. (2014) | |||
| 4ZV7 | Strzelczyk et al. (2016) | |||
| 5A6V5A71 | Benjamin et al. (2015) | |||
| Cavia porcellus | 1GPL | Withersmartinez et al. (1996) | – | – |
Table 2.
The structures of lipases with a loop or helical lid present in the Protein Data Bank.
| Organism | PDB code | Reference | Lid | Optimum temperature (°C) | Optimum pH |
|---|---|---|---|---|---|
| Bacillus sp. (strain H-257) | 3RLI, 3RM3 | Rengachari et al. (2012) | 119I-164T | 75 (Imamura and Kitaura, 2000) | 6–8 (Imamura and Kitaura, 2000) |
| 4KE6, 4KE7, 4KE8, 4KE9, 4KEA | Rengachari et al. (2013) | ||||
| Malassezia globosa | 3UUE, 3UUF | Xu et al. (2012) | 99E-116W | 25 (Zisis et al., 2015) | 6 (Zisis et al., 2015) |
| 4ZRD, 4ZRE | Guo et al. (2015) | ||||
| Bos taurus | 1AKN, 1AQL | Wang et al. (1997) | 116G-129E | – | – |
| 2BCE | Chen et al. (1998) | ||||
| Homo sapiens (bile salt-activated lipase) | 1F6W | Terzyan et al. (2000) | 115H-125Y | – | – |
| 1JMY | Moore et al. (2001) | ||||
| Burkholderia cepacia | 1HQD | Nardini et al. (2000) | 130D-158Q | 45 (Rathi et al., 2001) | 6 (Rathi et al., 2001) |
| 1OIL | Luic et al. (2001) | ||||
| 1YS1, 1YS2 | Kim et al. (1997) | ||||
| 2LIP, 3LIP | Mezzetti et al. (2005) | ||||
| 2NW6 | Schomburg et al. (1997) | ||||
| 4LIP, 5LIP | Luic´ et al. (2008) | ||||
| Acinetobacter baumannii | 4OPM | – | 178T-195K | – | – |
| Photobacterium sp. M37 | 2ORY | Jung et al. (2008) | 91G-104D | – | – |
| Thermomyces lanuginosus | 1DT3, 1DT5, 1DTE, 1DU4, 1EIN | Brzozowski et al. (2000) | 81R-96D | 35 (Fernandes et al., 2004) | 8 (Fernandes et al., 2004) |
| 1GT6 | Yapoudjian et al. (2002) | ||||
| 1TIB, 1TIC | Derewenda et al. (1994b) | ||||
| 4DYH, 4EA6, 4FLF, 4GBG, 4GHW, 4GI1, 4GLB, 4GWL, 4KJX, 4N8S, 4S0X, 4ZGB | – | ||||
| Gibberella zeae | 3NGM | Derewenda et al. (1994b) | 80R-90D | 35 (Long et al., 2010) | 7 (Long et al., 2010) |
| Rhizomucor miehei | 1TGL | Brady et al. (1990) | 80R-95V | 45 (Huang et al., 2014) | 8 (Huang et al., 2014) |
| 3TGL | Brzozowski et al. (1992) | ||||
| 4TGL | Derewenda et al. (1992) | ||||
| 5TGL | Brzozowski et al. (1991) | ||||
| Rhizopus niveus | 1LGY | Kohno et al. (1996) | 81R-95F | 35–40 (Kohno et al., 1994) | 6–6.5 (Kohno et al., 1994) |
| Candida cylindracea | 1LLF | Pletnev et al. (2003) | 66E-92P | ||
| Yarrowia lipolytica | 3O0D | Bordes et al. (2010) | 88T-105L | 37 (Corzo and Revah, 1999) | 6 (Corzo and Revah, 1999) |
| Penicillium camemberti | 1TIA | Derewenda et al. (1994a) | 82G-96V | 40 (Isobe et al., 1992) | 6 (Isobe et al., 1992) |
| Arabidopsis thaliana | 2YIJ | – | 154R-169G | 30 (Kim et al., 2011) | 6.5 (Kim et al., 2011) |
| Burkholderia glumae | 1CVL | Lang et al. (1996) | 130D-156T | – | – |
| 1TAH | Noble et al. (1993) | ||||
| 2ES4 | Pauwels et al. (2006) | ||||
| 1QGE | – | ||||
| Serratia marcescens | 2QUA, 2QUB | Meier et al. (2007) | 141R-169K | – | – |
| H. sapiens (human monoglyceride lipase) | 3HJU | Labar et al. (2010) | 156A-172P | – | – |
| 3JW8, 3JWE | Bertrand et al. (2009) | ||||
| 3PE6 | Schalk-Hihi et al. (2011) | ||||
| 4UUQ | Griebel et al. (2015) | ||||
Table 3.
The structures of lipases with multiple helical lid present in the Protein Data Bank.
| Organism | PDB code | Reference | Lid | Optimum temperature (°C) | Optimum pH |
|---|---|---|---|---|---|
| Proteus mirabilis | 4GW3, 4GXN | Korman and Ju (2012) | 121K-160L | 35 (Gao et al., 2009) | 9 (Gao et al., 2009) |
| Pseudomonas aeruginosa | 1EX9 | Nardini et al. (2000) | 122P-163N | 50 (Gilbert et al., 1991) | 8.5–8 (Gilbert et al., 1991) |
| Pseudomonas sp. MIS38 | 2Z8X, 2Z8Z | Angkawidjaja et al. (2007) | 45F-74P | – | – |
| 2ZJ6, 2ZJ7 | Kuwahara et al. (2008) | 146P-167G | |||
| 2ZVD, 3A6Z, 3A70 | Angkawidjaja et al. (2010) | ||||
| Geobacillus stearothermophilus | 1KU0 | Jeong et al. (2002) | 173M-238D | 60–65 (Kim et al., 1998) | 9 (Kim et al., 1998) |
| G. stearothermophilus | 1JI3 | Safra et al. (2002) | 173M-238D | 65 (Kim et al., 2000) | 9 (Kim et al., 2000) |
| Geobacillus zalihae | 2DSN | Matsumura et al. (2008) | 173M-238D | 70 (Schmidt-Dannert et al., 1997) | 9 (Schmidt-Dannert et al., 1997) |
| Geobacillus thermocatenulatus | 2W22 | Carrascolópez et al. (2009) | 174M-239D | 50 (Schmidt-Dannert et al., 1996) | 9 (Schmidt-Dannert et al., 1996) |
| Staphylococcus hyicus | 2HIH | Tiesinga et al. (2007) | 185D-240D | 37 (Schmidt-Dannert et al., 1996) | 8.5 (Schmidt-Dannert et al., 1996) |
| Uncultured bacterium | 3FAK | Nam et al. (2009b) | 1M-36V | 35 (Nam et al., 2009b) | 5 (Nam et al., 2009b) |
| Uncultured Bacterium | 3DNM | Nam et al. (2009a) | 16M-49C | 40 (Nam et al., 2009a) | 5 (Nam et al., 2009a) |
| 3K6K | 193S-223E | ||||
| Archaeoglobus fulgidus | 2ZYH, 2ZYI, 2ZYR, 2ZYS | Chen et al. (2009) | 62T-101K | 70–90 (Chen et al., 2009) | 10–11 (Chen et al., 2009) |
| Geotrichum candidum | 1THG | Schrag and Cygler (1993) | 61C-105C | ||
| Candida rugosa | 1CRL | Grochulski et al. (1993) | 60C-97C | 30 (Korbekandi et al., 2008) | 7 (Korbekandi et al., 2008) |
| 1LPM, 1LPS | Cygler et al. (1994) | ||||
| 1LPN, 1LPO, 1LPP | Grochulski et al. (1994) | ||||
| 1TRH | Pawel et al. (2008) | ||||
| 3RAR | Colton et al. (2011) | ||||
| Candida antarctica | 2VEO | Ericsson et al. (2008) | 217S-308E | 50–70 (Pfeffer et al., 2006) | 7 (Pfeffer et al., 2006) |
| 3GUU | |||||
| Homo sapiens | 1HLG | Roussel et al. (1999) | 209D-251F | – | – |
| Canis lupus | 1K8Q | Roussel et al. (2002) | 208G-251L | – | – |
| Sus scrofa | 1ETH | Hermoso et al. (1996) | 238C-262C | – | – |
| C. lupus | 1RP1 | Roussel et al. (1998a) | 237C-261C | – | – |
| Rattus norvegicus | 1BU8 | Roussel et al. (1998b) | 237C-261C | – | – |
| H. sapiens | 1N8S | Tilbeurgh et al. (1992) | 237C-261C | – | – |
| 1LPA, 1LPB | Van et al. (1993) | ||||
| Equus caballus | 1HPL | Bourne et al. (1994) | 237C-261C | – | – |
Figure 2.
Some of the lipases shown with the position of catalytic triads residues and lid domains. (A) Bacillus subtilis Lipase A with no lid domain (PDB code: 1I6W) (van Pouderoyen et al., 2001), (B) Malassezia globosa lipase “SMG1” with a lid domain Thr101-Asp119 contains loop form of lid (PDB code: 3UUE) (Xu et al., 2012), (C) Rhizomucor miehei lipase with a helix lid (PDB code: 3TGL) (Brzozowski et al., 1992), and (D) Geobacillus zalihae lipase with two helices in its lid domain (PDB code: 2DSN) (Matsumura et al., 2008). Catalytic triad residues were highlighted by yellow sticks and lid domains were highlighted by blue color, respectively.
Table 4.
The list of lipases with some special substrate selectivity.
| Organism | PDB code | Substrate specificity | Lid type |
|---|---|---|---|
| Bacillus sp. (strain H-257) | 3RM3 | Monoacylglycerol lipase | Loop |
| Malassezia globosa | 3UUF | Mono- and diacylglycerol lipase | Loop |
| Penicillium camemberti | 1TIA | Mono- and diacylglycerol lipase | One helix |
| Arabidopsis thaliana | 2YIJ | Mono- and diacylglycerol lipase | One helix |
| Homo sapiens | 3HJU | Monoacylglycerol lipase | One helix |
Interfacial Activation
The lipolytic activity of some lipases significantly increases beyond the critical micellar concentration of substrate. This “interfacial activation” phenomenon generally ascribes to the presence of an amphiphilic lid structure, which undergoes conformational changes in contact with the micellar substrates (Cambillau et al., 1996). The movement of the lid in lipases structure with substrate analogs has been also found (Brzozowski et al., 1991), which provides structural evidence for this phenomenon. Further, Cheng et al. (2012) reported that lipase from Pseudomonas sp. MIS38 (PML) does not undergo interfacial activation after deletion of its lid2, and they proposed that lid2 is important for interfacial activation of PML. However, lipases with mini- or without lid domains such as guinea pig and lipase B from Candida antarctica are found not to show any interfacial activation (Hjorth et al., 1993; Martinelle et al., 1995). It is surprising that coypu lipase containing a 23-amino acid lid domain did not exhibit interfacial activation (Thirstrup et al., 1994). On the basis of these observations, Verger (1997) suggested that interfacial activation and the existence of lid domain are not suitable criteria to determine a lipolytic enzyme as a lipase.
Effects on Activity and Substrate Specificity
The vital role of the lipase lid domain in substrate selectivity and activity has been confirmed by several approaches such as lid swapping and site-directed mutagenesis. Dugi et al. (1995) constructed chimeras of hepatic lipase (HL) with lipoprotein lipase (LPL) lid, and LPL with HL lid to analyze their activity with triacylglycrerols and phospholipid as substrate. Chimeric LPL that contains the lid of HL had reduced triacylglycrerol hydrolyzing activity, but increased phospholipase activity. In contrast, chimeric HL that contains the LPL lid was found to be more active against triacylglycrerols and less active against phospholipid substrate. This study clearly showed that the triglyceride and phospholipid hydrolysis activity of lipase can be alerted by swapping the lids of LPL and HL.
Accordingly, Brocca et al. (2003) generated a Candida rugosa lipase (CRL) 1 mutant with lid domain of CRL3. This CRL1 mutant displayed 200-fold higher activity toward cholesterol esters, showing that the lid was involved in determining the cholesterol esterase activity of CRL. Santarossa et al. (2005) performed site-directed substitution of residual T137 and T138 in lid domain of Pseudomonas fragi lipase with the valine and asparagines, respectively. The mutants showed a different chain length preference profile as compared to the wild-type lipase. The lipase activity can be modulated by mutagenesis in the lid domain. Substitution of serine 154 and glycine 152 in lid of Pseudomonas sp. CR611 Lip I.3 lipase with threonine and leucine resulting in fivefold and twofold increase in activity on 4-methylumbelliferyl-heptanoate, respectively (Panizza et al., 2015). Similarly, Bacillus thermocatenulatus lipase activity was increased up to 2.6-fold by substitution of F181 with alanine due to decrease in steric hindrance in the lid domain (Karkhane et al., 2009). A detailed analysis of shape and physicochemical properties of lipase-binding sites were analyzed by Pleiss et al. (1998) in order to understand the molecular basis of substrate specificity. Overall, these studies confirm the pivotal role of lids in selectivity and activity of lipases.
Effects on Thermostability
Next to selectivity and activity, thermostability is one of the most desirable traits of lipases (Dizge et al., 2009; Avila-Cisneros et al., 2014; Khan et al., 2016). Several factors define this property such as the number of hydrogen bonds, salt bridges, stabilization of secondary structures, occurrence of disulfide bonds, higher number of proline residues, higher polar surface area, shortening of loops, and stabilization of the lid domain (Pack and Yoo, 2004; Santarossa et al., 2005; Zhou et al., 2008; Khan et al., 2015a,b).
It has been found that the activity and thermostability of lipases can be altered by modifications in their lid domains. Timucin and Sezerman (2013) found that the conserved tryptophan of the lid region potentiates the thermostability and thermoactivity in bacterial thermoalkalophilic lipases from B. thermocatenulatus that stabilizes the aggregates by forming new intermolecular interactions. Yang et al. (2015) characterized the thermostable lipase from Pseudomonas sp. R0-14 and found that, when the lid is in the open conformation, the proportion of α-helices increased. An increase in the number of α-helices may make the lipase more thermostable in open conformation. Dror et al. (2014) employed protein engineering to enhance the stability of Geobacillus stearothermophilus Lipase T6 in methanol. They found that Gln185 situated on the lipase α-helix lid has an important role in the lipase interfacial activation. The substitution of Gln185 to Leu resulted in an improved stability in organic solvents due to the replacement of the polar glutamine by the more hydrophobic leucine. This hydrophobization also improved the structural stability of the enzyme by facilitating the interaction between the solvent molecules and the lid surface.
The substitutions of amino acids in the lid region of R. chinensis lipase affect not only its substrate specificity but also its thermostability. Probably, this is due to destabilization of lid structure by disrupting the H-bond interaction in the lid region (Zhu et al., 2013). Yu et al. (2012) demonstrated that introducing a disulfide bond in the lid hinge region of R. chinensis lipase increases thermostability and alters the acyl chain length specificity due to stabilization of the geometric structure of the lid region. Wu et al. (2010) suggested that the conserved residue Tyr224 of Geobacillus sp. RD-2 lipase is very close to the lid domain and is the key amino acid residue, which determines the thermostability of lipase. Santarossa et al. (2005) found that the mutations in the lid region of P. fragi lipase effect the chain length specificity and thermostability. The above studies concluded that the lid region not only plays an important role in the function of the lipase but also stabilizes the helix.
It has been also found that substitutions such as Val72Gly and Val72Ala causes higher activity and enantioselectivity of Penicillium expansum lipase, but decreases the thermostability (Tang et al., 2013). The substitution of Asp189 residue in the lid domain of Geobacillus sp. NTU 03 lipase also leads to a loss in its thermostability but exhibited higher activity (Shih and Pan, 2011). Sheng et al. (2014) employed the circular permutation protein engineering technique to acquire active mutants of Yarrowia lipolytica lipase. They also found that most of the functional mutations are seen in the surface-exposed loop region in close proximity to the lid domain, which implies the steric effect of the lid on lipase activity and substrate specificity, but there were no change in thermostability. So, the change in amino acid residues of lid region may lead to increase as well as decrease in stability depending upon nature of amino acid substitution.
Engineering the Lid Domains of Lipases
Most lipases bear a flexible lid close to the active site. This dynamic domain is very likely to affect both stability as well as catalytic properties of the biocatalyst and is, therefore, an attractive target for protein design (Kourist et al., 2010a). For instance, the modification of the lid region by site-directed mutagenesis of lid domain or hinge region or by lid swapping (Table 5), resulted in changes in the substrate specificity (Yu et al., 2014), enantioselectivity (Secundo et al., 2004; Gao et al., 2011), and stability (Yu et al., 2012) toward detergents (Brocca et al., 2003) and organic solvents (Secundo et al., 2004), which could turn into lack of oil–water interfacial activation (Shu et al., 2011; Tang et al., 2015).
Table 5.
Properties of lipase variants generated by lid and hinge region modification.
| Enzyme | Mutants | Mutants description | Mutants property | Reference |
|---|---|---|---|---|
| Proteus sp. LipK107 | E130L + K131I | The hydrophobicity of the lid domain increases | The eep (%) and E on the resolution of racemic 1-phenylethanol increased by 1.36 and 137.6%, respectively | Gao et al. (2011) |
| T138V | The eep (%) and E on the resolution of racemic 1-phenylethanol increased by 0.52 and 30.6%, respectively | |||
| Rhizopus chinensis lipase S4-3 | S4-3M | The lid of S4-3 was swapped with ferulic acid esterase from Aspergillus niger | Specific activity toward short-chain substrates increased by 7.2-fold (C3) and 38.0-fold (C2), respectively | Yu et al. (2014) |
| S4-3N | The lid of S4-3 was swapped with Rhizomucor miehei lipase | Specific activity toward substrates (C2, C6, C8, C12, and C16) increased by 1.5- to 3.3-fold and reduced 40% toward tristearin (C18) | ||
| Candida rugosa lipase (CRL) Trx-LIP4 | CRL4LID1 | The lid of CRL4 was swapped with CRL1 | Hydrolytic activity decreased by 85%, changed CLP, and reduced enantioselectivity | Akoh et al. (2004) |
| CRL LIP1 | CRL1LID3 | The lid of CRL1 was swapped with CRL3 | Specific activity toward cholesterol esters increased by 200-fold, enantioselectivity and activity reduced in organic solvent | Akoh et al. (2004) |
| Candida antarctica lipase B (CALB) | CALB-N. crassa | The lid of CALB was swapped with CALB homolog from Neurospora crassa lipase | Hydrolytic activity increased on simple esters, specifically, substrates with Ca branching on the carboxylic side, and increased enantioselectivity in hydrolysis of racemic ethyl 2-phenylpropanoate (E > 50) | Skjot et al. (2009) |
| CALB-G. zeae | The lid of CALB was swapped with CALB homologs from Gibberella zeae lipase | |||
| Penicillium expansum lipase (PEL) | T66L + D70N | The mutant residues are located at the lid (D70N) and the lid hinge region (T66L, E83K) of PEL | Specific activity toward p-nitrophenyl palmitate increased by 136.4-fold | Tang et al. (2015) |
| E83K | Specific activity toward p-nitrophenyl butyrate increased by 136.4-fold, but lack interfacial activation | |||
| A. niger lipase (ANL) | S84G | The mutant residues are located at the lid hinge region of ANL | Specific activity toward p-nitrophenyl esters decreased and displayed a pronounced interfacial activation | Shu et al. (2011) |
| D99P | Specific activity toward p-nitrophenyl palmitate increased by 2.2-fold and displayed no interfacial activation | |||
| R. chinensis lipase | F95C + F214C | A disulfide bridge was introduced into the lipase from R. chinensis in the hinge region of the lid | The half-life t1/2 value increased by 11-fold at 60°C and the Tm increase by 7°C, but the catalytic efficiency toward pNPP decreased by 1.5-fold | Yu et al. (2012) |
Site-Directed Mutagenesis of the Lid Domains
Site-directed mutagenesis of the lid domain of Proteus sp. LipK107 lipase improved the conversion of 1-phenylethanol with a slight increase in enantiodiscrimination (Gao et al., 2011). Increase in the hydrophobicity of the lid in case of mutants Glu130Leu + Lys131Ile and Thr138Val resulted in higher conversions of 1-phenylethanol than LipK107. On the contrary, the mutant Ile128Glu + Val129Asp has lower conversions than that of LipK107, and the E value (enantiomeric ratio) of the resolution changed in accordance with the conversions. Several studies were carried out to understand the mechanism of lid-opening and closing. It has been shown that site-directed mutagenesis of the lid region in T. lanuginosus lipase could possibly generate lipase variants with attractive features such as high lipase activity, fast activation at the lipid interface, ability to act on water soluble substrates, and enhanced calcium independence (Skjold-Jorgensen et al., 2014). Skjold-Jorgensen et al. (2017) studied controlled lid-opening in T. lanuginosus lipase by introducing disulfide bond between C86 and C255 residues that causes strained closure of the lid-domain. The formation of disulfide bind leads to locking of lid in a closed conformation. Upon unlocking, enzymatic activity was fully restored. They showed that this intrinsic bond enables control of both lipase activity and interfacial binding. They also suggested the key role that the lid plays in determining the polarity-dependent activation of lipases using a combination of methods measuring enzymatic activity, detecting structural changes using the tryptophan-induced quenching method, and calculating the lid opening energies using an MD simulations, and suggested that mutagenesis of the lid can lower the energy barrier associated with lid opening (Skjold-Jorgensen et al., 2016). Tryptophan-induced quenching fluorescence method has been applied to successfully measure the lid movements in T. lanuginosus lipase and its variants in solvents with different dielectric constants (Skjold-Jorgensen et al., 2015). The results indicated that lid movement is highly dependent on the particular lid residue composition as well as solvent polarity. In other words, lipases are more active in low polarity solvents because the lid adopts an open conformation, and relatively small conformational changes in the lid region play a key role in the activation mechanism.
Lid Swapping
An interesting approach for the protein engineering of lipases is the exchange of lids of homologous enzymes, also referred to as “lid swapping.” The amphipathic nature of the lid is very important for the substrate specificity, and it provides new insight into the structural basis of lipase substrate specificity and a way to tune the substrate preference of lipases.
The substrate specificity of R. chinensis lipase S4-3 was successfully modified by replacing the hydrophobic lid (85.7% polar residues) with a hydrophilic lid (57.1% polar residues) of ferulic acid esterase from Aspergillus niger (AnFaeA) or a hydrophobic lid of Rhizomucor miehei lipase (RML) (Yu et al., 2014). The most apparent changes by lid swapping were that the replacement of the S4-3 lid with that of AnFaeA shifted the specificity toward short-chain substrates (C2–C6) compared with that of the parent (C12), increased by 7.2-fold (C3) and 38.0-fold (C2), respectively. While the replacement of the S4-3 lid with that of RML caused a 1.5- to 3.3-fold increase in the specific activity toward those substrates (C2, C6, C8, C12, and C16) and a 40% reduction toward tristearin (C18) compared with the corresponding activity of the parent.
In one of the study, novel Candida antarctica lipase B (CALB) mutants in which the entire CALB lid region is substituted with that of homologs (Neurospora crassa and Gibberella zeae) were characterized (Skjot et al., 2009). It revealed several interesting properties such as increased hydrolytic activity on simple esters and much increased enantioselectivity in hydrolysis of racemic ethyl 2-phenylpropanoate (E > 50). C. rugosa LIP4 lipase was also studied by exchanging the lid regions from the other four C. rugosa isoforms (LIP1, LIP2, LIP3, and LIP5; and corresponding lids 1, 2, 3, and 5) with that of LIP4, respectively (Akoh et al., 2004). Lid swapping resulted in increased hydrolytic activities toward tributyrin of the chimeric LIP4/lid2 and LIP4/lid3, whereas chimeric LIP4/lid1 and LIP4/lid5 activities decreased, compared with the native LIP4. Furthermore, Brocca et al. (2003) substituted the lid sequences from isoenzymes C. rugosa LIP3, which has high activity toward cholesterol esters, to the LIP1, which had little cholesterol esterase activity in its native form. It revealed that the chimeric LIP1/lid3 specific activity toward cholesterol esters increased 200-fold. Secundo et al. (2004) found that the chimeric C. rugosa LIP1/lid3 was less active and enantioselective than the wild type for reactions of alcoholysis of chloroethyl-2-hydroxy hexanoate with methanol and of vinyl acetate with 6-methyl-5-hepten-2-ol in organic solvent. They postulated that the decrease in activity may be due to the chimera enzyme having a lower proportion of enzyme molecule in the open form, thereby hindering access to the enzyme active site.
Site-Directed Mutagenesis of Hinge Region
It is believed that interfacial activation of lipases involves conformational changes of the mobile lid domains. Derewenda et al. (1992) reported that the specific dihedral angles (φ and ψ) conformations of the hinge region amino acids experienced dramatic changes during the process of R. miehei interfacial activation. At the N-terminal end of the lid, Ser83 and Ser84 undergo conformational changes. Ser83, in spite of a change in the φ angle of 60° remains within the γR region of the Ramachandran plot, while Ser84 changes its conformation from δ to β region with a change in the ψ angle of 90°.
Shu et al. (2011) found that Asp99Pro and Ly108Glu mutants of A. niger lipase (ANL) become oil–water interface independent lipase, probably because of change in the β-sheet configuration of the second hinge region at the side of the lid domain. Three ANL mutants such as ANL-Ser84Gly, ANL-Asp99Pro, and ANL-Lys108Glu were constructed base on the fact that Ser84, Asp99, and Lys108 might be in the hinge region of the lid domain of ANL. ANL-Ser84Gly displayed interfacial activation, while ANL-Asp99Pro and ANL-Lys108Glu displayed no interfacial activation. The specific activity of ANL-Ser84Gly toward p-nitrophenyl esters decreases as compared to the wild-type enzyme, while the specific activity of ANL-Asp99Pro increases toward p-nitrophenyl palmitate by 2.2-fold.
Computational Approaches
The behavior of the lid domain and the dynamics of lipase at different temperatures and solvent conditions can be understood by in silico methods and computational approaches. The computational methods can help to predict the impact of mutations in the lid of lipase in order to understand the importance of a particular residue. Molecular dynamics (MD) simulations studies can predict the behavior of the lid domain at different temperature, pH and in a particular solvent. Recently, Haque and Prabhu (2016) performed MD simulation of double mutant porcine pancreatic lipase in open and closed conformations using ethanol, toluene, and octanol as solvent to explain the dynamics of lid opening. They found that the Asp250Val and Glu254Leu mutants showed lid opening at higher temperature suggesting the important role of these residues in holding the lid in closed conformation. Also, the dynamics of lid opening was faster in octanol than in water, due to the fact that non-polar solvents favor open conformation of the lid.
Likewise, Jiang et al. (2014) performed a MD simulation study on Y. lipolytica lipase in methanol and hexane and proposed a lid closure mechanism. They suggested that the lipase undergoes a greater conformational change in methanol, where several regions such as Ser274-Asn288 and Thr106-His126 were found to interact with the lid region. They proposed that the closure mechanism of the Y. lipolytica lipase is due to a double-lid movement in methanol.
Candida antarctica lipase B is one of the lipase that displays an enhanced catalytic rate for bulky substrates when adsorbed to a hydrophobic interface. It was proposed that the increased activity of this lipase is due to conformational changes leading to a more open active site. This hypothesis is supported by MD simulations and docking studies suggesting the presence of a highly mobile lid. Molecular docking study confirmed that a highly open conformation is required for binding large, bulky substrates (Zisis et al., 2015). Ganjalikhany et al. (2012) demonstrated the flexibility of the lid region of CALB using comparative MD simulation and essential dynamics analysis carried out at different temperatures, showing that the opening of the lid is temperature dependent. A similar approach was used by Rahman et al. (2012) T1 lipase confirming this temperature-dependency. They found that the lid movement was only observed in the presence of an interface and that the activation process is temperature-dependent. The structural rearrangement of the lid domain was caused by the interaction between the hydrophobic residues of the lid with octane. So, there may be several factors responsible for the mobility of the lid.
Disulfide bonds near the lid region play an important role in stabilization of its helical structure. Recently, Singh et al. (2016) predicted that the disruption of disulfide bonds lower the activation energy and improved catalytic efficiency of Trichosporon asahii MSR54 lipase. Using MD simulation methods, they predicted a mutant of this lipase with fourfold increased specific activity with a lower temperature optimum. In silico analysis suggested that there are two lids in this lipase and both of them are opened at 40°C through clockwise and anticlockwise rotations, respectively.
The computational analyses were also helpful in understanding the mechanism of thermoactivity and thermostability and the role of conserved tryptophan residue in bacterial thermoalkalophilic lipases (Timucin and Sezerman, 2013). It has been found that residue Trp211 in the lid region stabilized the intermolecular interactions in the dimeric lipase and that it is critical to the stability of the monomeric lipase. Dror et al. (2014) applied in silico modeling technique and concluded that the amino acid substitution Gln185Leu facilitates a closed lid conformation, and the enhanced stability of His86Tyr and Ala269Thr mutants was due to formation of new hydrogen bonds in case of G. stearothermophilus lipase.
Computational methods are also helpful to change the position of the lid to generate an open conformation of lipase in the absence of crystal structure. Nasr et al. (2013) generated open conformation of monoacylglycerol lipase and performed MD simulations. They suggested that lid region was found to interact with the nanodisc phospholipid bilayer and penetrated into the phospholipid bilayer.
The lipase from Pseudomonas sp. MIS38 has two lids, which greatly change its conformation upon substrate binding. Cheng et al. (2012) employed computational approaches to compare the tertiary structures in closed and open conformations. They proposed that a hydrophobic surface is formed by these lids, which is necessary to hold the substrates firmly in the active site. Barbe et al. (2011) applied an advanced computational molecular modeling robotics approach with fully atomistic description to investigate the geometrically feasible transition pathways between Burkholderia cepacia lipase lid conformations and classical molecular mechanics to evaluate pathway energetic under the influence of solvent. They proposed a descriptive analysis of intermediate conformations of B. cepacia lid. Rehm et al. (2010) performed MD simulations study on different lipases from C. rugosa, R. miehei, and Thermomyces lanuginosa. The results from MD analysis suggested that in all the three lipases, opening and closing of lids were driven by the solvent and independent of a bound substrate molecule.
Conclusion and Future Perspectives
The role of the lid on enzyme activity is very complex because it involves specific interactions with substrate molecules and controls the equilibria between active and inactive enzyme conformations. The lid is important for substrate binding as it undergoes dramatic shift that changes the secondary structure of lipase-binding site from closed lid structure to an open structure. We have classified lipases based on the different types of lid domain. Some common and novel characteristics of lipases can be deduced from the nature of lid domain. Lipases that have similar sequence or length of lid could have similar mechanism of action. Different characteristics of lipases including substrate preference, themostablility, and interfacial properties can also be predicted by comparing the lid domain. The lid domain has a close relationship with the substrate specificity of lipases. This makes it a “hot spot” for protein engineering to modulate the lipases catalytic properties that might fulfill the demand of industrial application. Various efforts such as modifications of lid domain using site-directed mutagenesis, lid swapping, introduction of extra bonds, and computational approaches have been employed to modify the activity and thermostability of lipases. Further advancement in the bioinformatics tools will help to predict the accurate function of amino acids present near the lid region of lipases. Protein engineering of lid may provide an opportunity for better understanding of the structural basis of the lipases property. There is a possibility of using these protein engineered thermostable lipases as industrial enzymes at high temperatures.
Author Contributions
FK and DL wrote the manuscript; FK has drawn the figures, RD, ZZ, and WH prepared the table; YW revised the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer SS and handling editor declared their shared affiliation, and the handling editor states that the process nevertheless met the standards of a fair and objective review.
Funding
This work was supported by Science and Technology Planning Project of Guangdong Province (2014B020204003) and National Natural Science Foundation of China (21406076, 31471690, and 31501412).
References
- Acharya P., Rajakumara E., Sankaranarayanan R., Rao N. M. (2004). Structural basis of selection and thermostability of laboratory evolved Bacillus subtilis lipase. J. Mol. Biol. 341, 1271–1281. 10.1016/j.jmb.2004.06.059 [DOI] [PubMed] [Google Scholar]
- Ahmad S., Kamal M. Z., Sankaranarayanan R., Rao N. M. (2008). Thermostable Bacillus subtilis lipases: in vitro evolution and structural insight. J. Mol. Biol. 381, 324–340. 10.1016/j.jmb.2008.05.063 [DOI] [PubMed] [Google Scholar]
- Akoh C. C., Lee G. C., Shaw J. F. (2004). Protein engineering and applications of Candida rugosa lipase isoforms. Lipids 39, 513–526. 10.1007/s11745-004-1258-7 [DOI] [PubMed] [Google Scholar]
- Angkawidjaja C., Matsumura H., Koga Y., Takano K., Kanaya S. (2010). X-ray crystallographic and MD simulation studies on the mechanism of interfacial activation of a family I.3 lipase with two lids. J. Mol. Biol. 400, 82–95. 10.1016/j.jmb.2010.04.051 [DOI] [PubMed] [Google Scholar]
- Angkawidjaja C., You D. J., Matsumura H., Kuwahara K., Koga Y., Takano K., et al. (2007). Crystal structure of a family I.3 lipase from Pseudomonas sp. MIS38 in a closed conformation. FEBS Lett. 581, 5060–5064. 10.1016/j.febslet.2007.09.048 [DOI] [PubMed] [Google Scholar]
- Arpigny J. L., Jaeger K. E. (1999). Bacterial lipolytic enzymes: classification and properties. Biochem. J. 343(Pt 1), 177. 10.1042/0264-6021:3430177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustyniak W., Brzezinska A. A., Pijning T., Wienk H., Boelens R., Dijkstra B. W., et al. (2012). Biophysical characterization of mutants of Bacillus subtilis lipase evolved for thermostability: factors contributing to increased activity retention. Protein Sci. 21, 487–497. 10.1002/pro.2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila-Cisneros N., Velasco-Lozano S., Huerta-Ochoa S., Cordova-Lopez J., Gimeno M., Favela-Torres E. (2014). Production of thermostable lipase by Thermomyces lanuginosus on solid-state fermentation: selective hydrolysis of sardine oil. Appl. Biochem. Biotechnol. 174, 1859–1872. 10.1007/s12010-014-1159-9 [DOI] [PubMed] [Google Scholar]
- Barbe S., Cortes J., Simeon T., Monsan P., Remaud-Simeon M., Andre I. (2011). A mixed molecular modeling-robotics approach to investigate lipase large molecular motions. Proteins 79, 2517–2529. 10.1002/prot.23075 [DOI] [PubMed] [Google Scholar]
- Barbe S., Lafaquiere V., Guieysse D., Monsan P., Remaud-Siméon M., Andre I. (2009). Insights into lid movements of Burkholderia cepacia lipase inferred from molecular dynamics simulations. Proteins 77, 509–523. 10.1002/prot.22462 [DOI] [PubMed] [Google Scholar]
- Benjamin S., Fisher S. J., Michele C. (2015). Open and closed states of Candida antarctica lipase B: protonation and the mechanism of interfacial activation. J. Lipid Res. 56, 2348–2358. 10.1194/jlr.M063388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg O. G., Cajal Y., Butterfoss G. L., Grey R. L., Alsina M. A., Yu B.-Z., et al. (1998). Interfacial activation of triglyceride lipase from Thermomyces (Humicola) lanuginosa: kinetic parameters and a basis for control of the lid. Biochemistry 37, 6615–6627. 10.1021/bi972998p [DOI] [PubMed] [Google Scholar]
- Bertrand T., Augé F., Houtmann J., Rak A., Vallée F., Mikol V., et al. (2009). Structural basis for human monoglyceride lipase inhibition. J. Mol. Biol. 396, 663–673. 10.1016/j.jmb.2009.11.060 [DOI] [PubMed] [Google Scholar]
- Bordes F., Barbe S., Escalier P., Mourey L., André I., Marty A., et al. (2010). Exploring the conformational states and rearrangements of Yarrowia lipolytica lipase. Biophys. J. 99, 2225–2234. 10.1016/j.bpj.2010.07.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne Y., Martinez C., Kerfelec B., Lombardo D., Chapus C., Cambillau C. (1994). Horse pancreatic lipase. The crystal structure refined at 2.3 A resolution. J. Mol. Biol. 238, 709–732. 10.1006/jmbi.1994.1331 [DOI] [PubMed] [Google Scholar]
- Brady L., Brzozowski A. M., Derewenda Z. S., Dodson E., Dodson G., Tolley S., et al. (1990). A serine protease triad forms the catalytic centre of a triacylglycerol lipase. Nature 343, 767–770. 10.1038/343767a0 [DOI] [PubMed] [Google Scholar]
- Brocca S., Secundo F., Ossola M., Alberghina L., Carrea G., Lotti M. (2003). Sequence of the lid affects activity and specificity of Candida rugosa lipase isoenzymes. Protein Sci. 12, 2312–2319. 10.1110/ps.0304003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzozowski A. M., Derewenda U., Derewenda Z. S., Dodson G. G., Lawson D. M., Turkenburg J. P., et al. (1991). A model for interfacial activation in lipases from the structure of a fungal lipase-inhibitor complex. Nature 351, 491–494. 10.1038/351491a0 [DOI] [PubMed] [Google Scholar]
- Brzozowski A. M., Derewenda Z. S., Dodson E. J., Dodson G. G., Turkenburg J. P. (1992). Structure and molecular model refinement if Rhizomucor miehei triacylglyceride lipase: a case study of the use of simulated annealing in partial model refinement. Acta Crystallogr. B 48, 307–319. 10.1107/S0108768191014647 [DOI] [Google Scholar]
- Brzozowski A. M., Savage H., Verma C. S., Turkenburg J. P., Lawson D. M., Svendsen A., et al. (2000). Structural origins of the interfacial activation in Thermomyces (Humicola) lanuginosa lipase. Biochemistry 39, 15071–15082. 10.1021/bi0013905 [DOI] [PubMed] [Google Scholar]
- Cajal Y., Svendsen A., Girona V., Patkar S. A., Alsina M. A. (2000). Interfacial control of lid opening in Thermomyces lanuginosa Lipase. Biochemistry 39, 413–423. 10.1021/bi991927i [DOI] [PubMed] [Google Scholar]
- Cambillau C., Longhi S., Nicolas A., Martinez C. (1996). Acyl glycerol hydrolases: inhibitors, interface and catalysis. Curr. Opin. Struct. Biol. 6, 449–455. 10.1016/s0959-440x(96)80108-4 [DOI] [PubMed] [Google Scholar]
- Carrascolópez C., Godoy C., De L. R. B., Fernándezlorente G., Palomo J. M., Guisán J. M., et al. (2009). Activation of bacterial thermoalkalophilic lipases is spurred by dramatic structural rearrangements. J. Biol. Chem. 284, 4365–4372. 10.1074/jbc.M808268200 [DOI] [PubMed] [Google Scholar]
- Chen J. C., Miercke L. J., Krucinski J., Starr J. R., Saenz G., Wang X., et al. (1998). Structure of bovine pancreatic cholesterol esterase at 1.6 A: novel structural features involved in lipase activation. Biochemistry 37, 5107–5117. 10.1021/bi972989g [DOI] [PubMed] [Google Scholar]
- Chen K. M., Lee G. C., Ko T. P., Guo R. T., Huang L. M., Liu H. J., et al. (2009). Structure of the alkalohyperthermophilic Archaeoglobus fulgidus lipase contains a unique c-terminal domain essential for long-chain substrate binding. J. Mol. Biol. 390, 672–685. 10.1016/j.jmb.2009.05.017 [DOI] [PubMed] [Google Scholar]
- Cheng M., Angkawidjaja C., Koga Y., Kanaya S. (2012). Requirement of lid2 for interfacial activation of a family I.3 lipase with unique two lid structures. FEBS J. 279, 3727–3737. 10.1111/j.1742-4658.2012.08734.x [DOI] [PubMed] [Google Scholar]
- Colton I. J., Yin D. L., Grochulski P., Kazlauskas R. J. (2011). Molecular basis of chiral acid recognition by Candida rugosa lipase: X-ray structure of transition state analog and modeling of the hydrolysis of methyl 2-methoxy-2-phenylacetate. Adv. Synth. Catal. 353, 2529–2544. 10.1002/adsc.201100459 [DOI] [Google Scholar]
- Corzo G., Revah S. (1999). Production and characteristics of lipase from Yarrowia lipolytica 681. Bioresour. Technol. 70, 173–180. 10.1016/S0960-8524(99)00024-3 [DOI] [Google Scholar]
- Cygler M., Grochulski P., Kazlauskas R. J., Schrag J. D., Bouthillier F., Rubin B., et al. (1994). A structural basis for the chiral preferences of lipases. J. Am. Chem. Soc. 116, 3180–3186. 10.1021/ja00087a002 [DOI] [Google Scholar]
- Derewenda U., Brzozowski A. M., Lawson D. M., Derewenda Z. S. (1992). Catalysis at the interface: the anatomy of a conformational change in a triglyceride lipase. Biochemistry 31, 1532–1541. 10.1021/bi00120a034 [DOI] [PubMed] [Google Scholar]
- Derewenda U., Swenson L., Green R., Wei Y., Dodson G. G., Yamaguchi S., et al. (1994a). An unusual buried polar cluster in a family of fungal lipases. Nat. Struct. Biol. 1, 36–47. 10.1038/nsb0194-36 [DOI] [PubMed] [Google Scholar]
- Derewenda U., Swenson L., Wei Y., Green R., Kobos P. M., Joerger R., et al. (1994b). Conformational lability of lipases observed in the absence of an oil-water interface: crystallographic studies of enzymes from the fungi Humicola lanuginosa and Rhizopus delemar. J. Lipid Res. 35, 524–534. [PubMed] [Google Scholar]
- Dizge N., Aydiner C., Imer D. Y., Bayramoglu M., Tanriseven A., Keskinler B. (2009). Biodiesel production from sunflower, soybean, and waste cooking oils by transesterification using lipase immobilized onto a novel microporous polymer. Bioresour. Technol. 100, 1983–1991. 10.1016/j.biortech.2008.10.008 [DOI] [PubMed] [Google Scholar]
- Droge M. J., Boersma Y. L., Van Pouderoyen G., Vrenken T. E., Ruggeberg C. J., Reetz M. T., et al. (2006). Directed evolution of Bacillus subtilis lipase A by use of enantiomeric phosphonate inhibitors: crystal structures and phage display selection. Chembiochem 7, 149–157. 10.1002/cbic.200500308 [DOI] [PubMed] [Google Scholar]
- Dror A., Shemesh E., Dayan N., Fishman A. (2014). Protein engineering by random mutagenesis and structure-guided consensus of Geobacillus stearothermophilus lipase T6 for enhanced stability in methanol. Appl. Environ. Microbiol. 80, 1515–1527. 10.1128/AEM.03371-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugi K. A., Dichek H. L., Santamarina-Fojo S. (1995). Human hepatic and lipoprotein lipase: the loop covering the catalytic site mediates lipase substrate specificity. J. Biol. Chem. 270, 25396–25401. 10.1074/jbc.270.43.25396 [DOI] [PubMed] [Google Scholar]
- Eom G. T., Lee S. H., Song B. K., Chung K. W., Kim Y. W., Song J. K. (2013). High-level extracellular production and characterization of Candida antarctica lipase B in Pichia pastoris. J. Biosci. Bioeng. 116, 165–170. 10.1016/j.jbiosc.2013.02.016 [DOI] [PubMed] [Google Scholar]
- Ericsson D. J., Kasrayan A., Johansson P., Bergfors T., Sandström A. G., Bäckvall J. E., et al. (2008). X-ray structure of Candida antarctica lipase A shows a novel lid structure and a likely mode of interfacial activation. J. Mol. Biol. 376, 109–119. 10.1016/j.jmb.2007.10.079 [DOI] [PubMed] [Google Scholar]
- Fernandes M. L. M., Krieger N., Baron A. M., Zamora P. P., Ramos L. P., Mitchell D. A. (2004). Hydrolysis and synthesis reactions catalysed by Thermomyces lanuginosa lipase in the AOT/Isooctane reversed micellar system. J. Mol. Catal. B Enzym. 30, 43–49. 10.1016/j.molcatb.2004.03.004 [DOI] [Google Scholar]
- Fischer M., Pleiss J. (2003). The lipase engineering database: a navigation and analysis tool for protein families. Nucleic Acids Res. 31, 319–321. 10.1093/nar/gkg015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganjalikhany M. R., Ranjbar B., Taghavi A. H., Tohidi Moghadam T. (2012). Functional motions of Candida antarctica lipase B: a survey through open-close conformations. PLoS ONE 7:e40327. 10.1371/journal.pone.0040327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B., Su E., Lin J., Jiang Z., Ma Y., Wei D. (2009). Development of recombinant Escherichia coli whole-cell biocatalyst expressing a novel alkaline lipase-coding gene from Proteus sp for biodiesel production. J. Biotechnol. 139, 169–175. 10.1016/j.jbiotec.2008.10.004 [DOI] [PubMed] [Google Scholar]
- Gao B., Xu T., Lin J. P., Zhang L. J., Su E. Z., Jiang Z. B., et al. (2011). Improving the catalytic activity of lipase LipK107 from Proteus sp. by site-directed mutagenesis in the lid domain based on computer simulation. J. Mol. Catal. B Enzym. 68, 286–291. 10.1016/j.molcatb.2010.12.001 [DOI] [Google Scholar]
- Gilbert E. J., Cornish A., Jones C. W. (1991). Purification and properties of extracellular lipase from Pseudomonas aeruginosa EF2. J. Gen. Microbiol. 137, 2223–2229. 10.1099/00221287-137-9-2223 [DOI] [PubMed] [Google Scholar]
- Griebel G., Pichat P., Beeské S., Leroy T., Redon N., Jacquet A., et al. (2015). Selective blockade of the hydrolysis of the endocannabinoid 2-arachidonoylglycerol impairs learning and memory performance while producing antinociceptive activity in rodents. Sci. Rep. 5, 5859–5868. 10.1038/srep07642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grochulski P., Bouthillier F., Kazlauskas R. J., Serreqi A. N., Schrag J. D., Ziomek E., et al. (1994). Analogs of reaction intermediates identify a unique substrate binding site in Candida rugosa lipase. Biochemistry 33, 3494–3500. 10.2210/pdb1lpp/pdb [DOI] [PubMed] [Google Scholar]
- Grochulski P., Li Y., Schrag J. D., Bouthillier F., Smith P., Harrison D., et al. (1993). Insights into interfacial activation from an open structure of Candida rugosa lipase. J. Biol. Chem. 268, 12843–12847. [PubMed] [Google Scholar]
- Guo S., Xu J., Pavlidis I. V., Lan D., Bornscheuer U. T., Liu J., et al. (2015). Structure of product-bound SMG1 lipase: active site gating implications. FEBS J. 282, 4538–4547. 10.1111/febs.13513 [DOI] [PubMed] [Google Scholar]
- Haque N., Prabhu N. P. (2016). Lid dynamics of porcine pancreatic lipase in non-aqueous solvents. Biochim. Biophys. Acta 1860, 2326–2334. 10.1016/j.bbagen.2016.05.006 [DOI] [PubMed] [Google Scholar]
- Hasan F., Shah A. A., Hameed A. (2006). Industrial applications of microbial lipases. Enzyme Microb. Technol. 39, 235–251. 10.1016/j.enzmictec.2005.10.016 [DOI] [Google Scholar]
- Hermoso J., Pignol D., Kerfelec B., Crenon I., Chapus C., Fontecillacamps J. C. (1996). Lipase activation by nonionic detergents. The crystal structure of the porcine lipase-colipase-tetraethylene glycol monooctyl ether complex. J. Biol. Chem. 271, 18007–18016. 10.1074/jbc.271.30.18007 [DOI] [PubMed] [Google Scholar]
- Hjorth A., Carriere F., Cudrey C., Woldike H., Boel E., Lawson D. M., et al. (1993). A structural domain (the lid) found in pancreatic lipases is absent in the guinea pig (phospho)lipase. Biochemistry 32, 4702–4707. 10.1021/bi00069a003 [DOI] [PubMed] [Google Scholar]
- Holmquist M., Clausen I. G., Patkar S., Svendsen A., Hult K. (1995). Probing a functional role of Glu87 and Trp89 in the lid of Humicola lanuginosa lipase through transesterification reactions in organic solvent. J. Protein Chem. 14, 217–224. 10.1007/BF01886762 [DOI] [PubMed] [Google Scholar]
- Huang J., Ji X., Zhen Y., Guan F., Di C., Guan G., et al. (2014). Improved production of a recombinant Rhizomucor miehei lipase expressed in Pichia pastoris and its application for conversion of microalgae oil to biodiesel. Biotechnol. Biofuels 7, 1–11. 10.1186/1754-6834-7-111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura S., Kitaura S. (2000). Purification and characterization of a monoacylglycerol lipase from the moderately thermophilic Bacillus sp. H-257. J. Biochem. 127, 419–425. 10.1093/oxfordjournals.jbchem.a022623 [DOI] [PubMed] [Google Scholar]
- Isobe K., Nokihara K., Yamaguchi S., Mase T., Schmid R. D. (1992). Crystallization and characterization of monoacylglycerol and diacylglycerol lipase from Penicillium camemberti. Eur. J. Biochem. 203, 233–237. 10.1111/j.1432-1033.1992.tb19851.x [DOI] [PubMed] [Google Scholar]
- Jeong S. T., Kim H. K., Kim S. J., Chi S. W., Pan J. G., Oh T. K., et al. (2002). Novel zinc-binding center and a temperature switch in the Bacillus stearothermophilus L1 lipase. J. Biol. Chem. 277, 17041–17047. 10.1074/jbc.M200640200 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Li L., Zhang H., Feng W., Tan T. (2014). Lid closure mechanism of Yarrowia lipolytica lipase in methanol investigated by molecular dynamics simulation. J. Chem. Inf. Model 54, 2033–2041. 10.1021/ci500163y [DOI] [PubMed] [Google Scholar]
- Jung S. K., Jeong D. G., Mi S. L., Lee J. K., Kim H. K., Ryu S. E., et al. (2008). Structural basis for the cold adaptation of psychrophilic M37 lipase from Photobacterium lipolyticum. Proteins 71, 476–484. 10.1002/prot.21884 [DOI] [PubMed] [Google Scholar]
- Kamal M. Z., Ahmad S., Molugu T. R., Vijayalakshmi A., Deshmukh M. V., Sankaranarayanan R., et al. (2011). In vitro evolved non-aggregating and thermostable lipase: structural and thermodynamic investigation. J. Mol. Biol. 413, 726–741. 10.1016/j.jmb.2011.09.002 [DOI] [PubMed] [Google Scholar]
- Karkhane A. A., Yakhchali B., Jazii F. R., Bambai B. (2009). The effect of substitution of Phe181 and Phe182 with Ala on activity, substrate specificity and stabilization of substrate at the active site of Bacillus thermocatenulatus lipase. J. Mol. Catal. B Enzym. 61, 162–167. 10.1016/j.molcatb.2009.06.006 [DOI] [Google Scholar]
- Kawasaki K., Kondo H., Suzuki M., Ohgiyai S., Tsuda S. (2002). Alternate conformations observed in catalytic serine of Bacillus subtilis lipase determined at 1.3 A resolution. Acta Crystallogr. 58, 1168–1174. 10.1107/S090744490200714X [DOI] [PubMed] [Google Scholar]
- Khan F. I., Bisetty K., Singh S., Permaul K., Hassan M. I. (2015a). Chitinase from Thermomyces lanuginosus SSBP and its biotechnological applications. Extremophiles 19, 1055–1066. 10.1007/s00792-015-0792-8 [DOI] [PubMed] [Google Scholar]
- Khan F. I., Govender A., Permaul K., Singh S., Bisetty K. (2015b). Thermostable chitinase II from Thermomyces lanuginosus SSBP: cloning, structure prediction and molecular dynamics simulations. J. Theor. Biol. 374, 107–114. 10.1016/j.jtbi.2015.03.035 [DOI] [PubMed] [Google Scholar]
- Khan F. I., Nizami B., Anwer R., Gu K. R., Bisetty K., Hassan M. I., et al. (2016). Structure prediction and functional analyses of a thermostable lipase obtained from Shewanella putrefaciens. J. Biomol. Struct. Dyn. 1–13. 10.1080/07391102.2016.1206837 [DOI] [PubMed] [Google Scholar]
- Kim E. Y., Seo Y. S., Kim W. T. (2011). AtDSEL, an Arabidopsis cytosolic DAD1-like acylhydrolase, is involved in negative regulation of storage oil mobilization during seedling establishment. J. Plant Physiol. 168, 1705–1709. 10.1016/j.jplph.2011.03.004 [DOI] [PubMed] [Google Scholar]
- Kim H. K., Park S. Y., Lee J. K., Oh T. K. (1998). Gene cloning and characterization of thermostable lipase from Bacillus stearothermophilus L1. Biosci. Biotechnol. Biochem. 62, 66–71. 10.1271/bbb.62.66 [DOI] [PubMed] [Google Scholar]
- Kim K. K., Song H. K., Dong H. S., Hwang K. Y., Suh S. W. (1997). The crystal structure of a triacylglycerol lipase from Pseudomonas cepacia reveals a highly open conformation in the absence of a bound inhibitor. Structure 5, 173–185. 10.1016/S0969-2126(97)00271-2 [DOI] [PubMed] [Google Scholar]
- Kim M. H., Kim H. K., Lee J. K., Park S. Y., Oh T. K. (2000). Thermostable lipase of Bacillus Stearothermophilus: high-level production, purification, and calcium-dependent thermostability. Biosci. Biotechnol. Biochem. 64, 280–286. 10.1271/bbb.64.280 [DOI] [PubMed] [Google Scholar]
- Kohno M., Funatsu J., Mikami B., Kugimiya W., Matsuo T., Morita Y. (1996). The crystal structure of lipase II from Rhizopus niveus at 2.2 A resolution. J. Biochem. 120, 505–510. 10.1093/oxfordjournals.jbchem.a021442 [DOI] [PubMed] [Google Scholar]
- Kohno M., Kugimiya W., Hashimoto Y., Morita Y. (1994). Purification, characterization, and crystallization of two types of lipase from Rhizopus niveus. Biosci. Biotechnol. Biochem. 58, 1007–1012. 10.1271/bbb.58.1007 [DOI] [PubMed] [Google Scholar]
- Korbekandi H., Abedi D., Pourhossein M., Motovali-Bashi M., Hejazi M., Narimousaei M., et al. (2008). Optimisation of Candida rugosa lipase (CRL) esterase activity. J. Biotechnol. 131, S121. 10.1155/2015/594238 [DOI] [Google Scholar]
- Korman T. P., Ju B. (2012). Crystal structure of Proteus mirabilis lipase, a novel lipase from the Proteus/Psychrophilic subfamily of lipase family I.1. PLoS ONE 7:5806–5819. 10.1371/journal.pone.0052890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourist R., Brundiek H., Bornscheuer U. T. (2010a). Protein engineering and discovery of lipases. Eur. J. Lipid Sci. Technol. 112, 64–74. 10.1002/ejlt.200900143 [DOI] [Google Scholar]
- Kourist R., Jochens H., Bartsch S., Kuipers R., Padhi S. K., Gall M., et al. (2010b). The α/β-hydrolase fold 3DM database (ABHDB) as a tool for protein engineering. Chembiochem 11, 1635–1643. 10.1002/cbic.201000213 [DOI] [PubMed] [Google Scholar]
- Kuwahara K., Angkawidjaja C., Matsumura H., Koga Y., Takano K., Kanaya S. (2008). Importance of the Ca2+-binding sites in the N-catalytic domain of a family I.3 lipase for activity and stability. Protein Eng. Des. Sel. 21, 737–744. 10.1093/protein/gzn057 [DOI] [PubMed] [Google Scholar]
- Labar G., Bauvois C., Borel F., Ferrer J. L., Wouters J., Lambert D. M. (2010). Crystal structure of the human monoacylglycerol lipase, a key actor in endocannabinoid signaling. Chembiochem 11, 218–227. 10.1002/cbic.200900621 [DOI] [PubMed] [Google Scholar]
- Lang D., Hofmann B., Haalck L., Hecht H. J., Spener F., Schmid R. D., et al. (1996). Crystal structure of a bacterial lipase from Chromobacterium viscosum ATCC 6918 refined at 1.6 Å resolution. J. Mol. Biol. 259, 704–717. 10.1006/jmbi.1996.0352 [DOI] [PubMed] [Google Scholar]
- Lesuisse E., Schanck K., Colson C. (1993). Purification and preliminary characterization of the extracellular lipase of Bacillus subtilis 168, an extremely basic pH-tolerant enzyme. Eur. J. Biochem. 216, 155–160. 10.1111/j.1432-1033.1993.tb18127.x [DOI] [PubMed] [Google Scholar]
- Long N. N., Dao T. T., Živković T., Fehrholz M., Schäfer W., Salomon S. (2010). Enzymatic properties and expression patterns of five extracellular lipases of Fusarium graminearum in vitro. Enzyme Microb. Technol. 46, 479–486. 10.1016/j.enzmictec.2010.02.005 [DOI] [PubMed] [Google Scholar]
- Luić M., Stefanić Z., Ceilinger I., Hodoscek M., Janezic D., Lenac T., et al. (2008). Combined X-ray diffraction and QM/MM study of the Burkholderia cepacia lipase-catalyzed secondary alcohol esterification. J. Phys. Chem. B 112, 4876–4883. 10.1021/jp077717u [DOI] [PubMed] [Google Scholar]
- Luic M., Tomic S., Lescic I., Ljubovic E., Sepac D., Sunjic V., et al. (2001). Complex of Burkholderia cepacia lipase with transition state analogue of 1-phenoxy-2-acetoxybutane – biocatalytic, structural and modelling study. Eur. J. Biochem. 268, 3964–3973. 10.1046/j.1432-1327.2001.02303.x [DOI] [PubMed] [Google Scholar]
- Martinelle M., Holmquist M., Hult K. (1995). On the interfacial activation of Candida antarctica lipase A and B as compared with Humicola lanuginosa lipase. Biochim. Biophys. Acta 1258, 272–276. 10.1016/0005-2760(95)00131-u [DOI] [PubMed] [Google Scholar]
- Maruyama T., Nakajima M., Uchikawa S., Nabetani H., Furusaki S., Seki M. (2000). Oil-water interfacial activation of lipase for interesterification of triglyceride and fatty acid. J. Am. Oil Chem. Soc. 77, 1121–1127. 10.1007/s11746-000-0176-4 [DOI] [Google Scholar]
- Masuch T., Kusnezowa A., Nilewski S., Bautista J. T., Kourist R., Leichert L. I. (2015). A combined bioinformatics and functional metagenomics approach to discovering lipolytic biocatalysts. Front. Microbiol. 6:1110. 10.3389/fmicb.2015.01110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura H., Yamamoto T., Leow T. C., Mori T., Salleh A. B., Basri M., et al. (2008). Novel cation-pi interaction revealed by crystal structure of thermoalkalophilic lipase. Proteins 70, 592–598. 10.1002/prot.21799 [DOI] [PubMed] [Google Scholar]
- Meier R., Drepper T., Svensson V., Jaeger K. E., Baumann U. (2007). A calcium-gated lid and a large beta-roll sandwich are revealed by the crystal structure of extracellular lipase from Serratia marcescens. J. Biol. Chem. 282, 31477–31483. 10.1074/jbc.M704942200 [DOI] [PubMed] [Google Scholar]
- Mezzetti A., Schrag J. D., Chan S. C., Kazlauskas R. J. (2005). Mirror-image packing in enantiomer discrimination: molecular basis for the enantioselectivity of B.cepacia lipase toward 2-methyl-3-phenyl-1-propanol. Chem. Biol. 12, 427–437. 10.1016/j.chembiol.2005.01.016 [DOI] [PubMed] [Google Scholar]
- Moore S. A., Kingston R. L., Loomes K. M., Hernell O., Bläckberg L., Baker H. M., et al. (2001). The structure of truncated recombinant human bile salt-stimulated lipase reveals bile salt-independent conformational flexibility at the active-site loop and provides insights into heparin binding. J. Mol. Biol. 312, 511–523. 10.1006/jmbi.2001.4979 [DOI] [PubMed] [Google Scholar]
- Nam K. H., Kim M. Y., Kim S. J., Priyadarshi A., Kwon S. T., Koo B. S., et al. (2009a). Structural and functional analysis of a novel hormone-sensitive lipase from a metagenome library. Proteins 74, 1036–1040. 10.1002/prot.22313 [DOI] [PubMed] [Google Scholar]
- Nam K. H., Kim M. Y., Kim S. J., Priyadarshi A., Lee W. H., Hwang K. Y. (2009b). Structural and functional analysis of a novel EstE5 belonging to the subfamily of hormone-sensitive lipase. Biochem. Biophys. Res. Commun. 379, 553–556. 10.1016/j.bbrc.2008.12.085 [DOI] [PubMed] [Google Scholar]
- Nardini M., Lang D. A., Liebeton K., Jaeger K. E., Dijkstra B. W. (2000). Crystal structure of Pseudomonas aeruginosa lipase in the open conformation. J. Biol. Chem. 275, 31219–31225. 10.1074/jbc.M003903200 [DOI] [PubMed] [Google Scholar]
- Nasr M. L., Shi X., Bowman A. L., Johnson M., Zvonok N., Janero D. R., et al. (2013). Membrane phospholipid bilayer as a determinant of monoacylglycerol lipase kinetic profile and conformational repertoire. Protein Sci. 22, 774–787. 10.1002/pro.2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble M. E., Cleasby A., Johnson L. N., Egmond M. R., Frenken L. G. (1993). The crystal structure of triacylglycerol lipase from Pseudomonas glumae reveals a partially redundant catalytic aspartate. FEBS Lett. 331, 123–128. 10.1016/0014-5793(93)80310-q [DOI] [PubMed] [Google Scholar]
- Nordwald E. M., Plaks J. G., Snell J. R., Sousa M. C., Kaar J. L. (2015). Crystallographic Investigation of imidazolium ionic liquid effects on enzyme structure. Chembiochem 16, 2456–2459. 10.1002/cbic.201500398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pack S. P., Yoo Y. J. (2004). Protein thermostability: structure-based difference of amino acid between thermophilic and mesophilic proteins. J. Biotechnol. 111, 269–277. 10.1016/j.jbiotec.2004.01.018 [DOI] [PubMed] [Google Scholar]
- Panizza P., Cesarini S., Diaz P., Rodriguez Giordano S. (2015). Saturation mutagenesis in selected amino acids to shift Pseudomonas sp. acidic lipase Lip I.3 substrate specificity and activity. Chem. Commun. (Camb) 51, 1330–1333. 10.1039/c4cc08477b [DOI] [PubMed] [Google Scholar]
- Pauwels K., Lustig A., Wyns L., Tommassen J., Savvides S. N., Van G. P. (2006). Structure of a membrane-based steric chaperone in complex with its lipase substrate. Nat. Struct. Mol. Biol. 13, 374–375. 10.1038/nsmb1065 [DOI] [PubMed] [Google Scholar]
- Pawel G., Li Y., Schrag J. D., Miroslaw C. (2008). Two conformational states of Candida rugosa lipase. Protein Sci. 3, 82–91. 10.1002/pro.5560030111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer J., Richter S., Nieveler J., Hansen C. E., Rhlid R. B., Schmid R. D., et al. (2006). High yield expression of lipase A from Candida antarctica in the methylotrophic yeast Pichia pastoris and its purification and characterisation. Appl. Microbiol. Biotechnol. 72, 931–938. 10.1007/s00253-006-0400-z [DOI] [PubMed] [Google Scholar]
- Pleiss J., Fischer M., Schmid R. D. (1998). Anatomy of lipase binding sites: the scissile fatty acid binding site. Chem. Phys. Lipids 93, 67–80. 10.1016/S0009-3084(98)00030-9 [DOI] [PubMed] [Google Scholar]
- Pletnev V., Addlagatta A., Wawrzak Z., Duax W. (2003). Three-dimensional structure of homodimeric cholesterol esterase–ligand complex at 1.4 Å resolution. Acta Crystallogr. 59, 50–56. 10.1107/S0907444902018851 [DOI] [PubMed] [Google Scholar]
- Pouderoyen G. V., Eggert T., Jaeger K. E., Dijkstra B. W. (2001). The crystal structure of Bacillus subtili lipase: a minimal α/β hydrolase fold enzyme 1. J. Mol. Biol. 309, 215–226. 10.1006/jmbi.2001.4659 [DOI] [PubMed] [Google Scholar]
- Qian Z., Horton J. R., Cheng X., Lutz S. (2009). Structural redesign of lipase B from Candida antarctica by circular permutation and incremental truncation. J. Mol. Biol. 393, 191–201. 10.1016/j.jmb.2009.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M. Z. A., Salleh A., Rahman R., Rahman M. B. A., Basri M., Leow T. C. (2012). Unlocking the mystery behind the activation phenomenon of T1 lipase: a molecular dynamics simulations approach. Protein Sci. 21, 1210–1221. 10.1002/pro.2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumara E., Acharya P., Ahmad S., Sankaranaryanan R., Rao N. M. (2008). Structural basis for the remarkable stability of Bacillus subtilis lipase (Lip A) at low pH. Biochim. Biophys. Acta 1784, 302–311. 10.1016/j.bbapap.2007.10.012 [DOI] [PubMed] [Google Scholar]
- Rathi P., Saxena R. K., Gupta R. (2001). A novel alkaline lipase from Burkholderia cepacia for detergent formulation. Process Biochem. 37, 187–192. 10.1016/S0032-9592(01)00200-X [DOI] [Google Scholar]
- Rehm S., Trodler P., Pleiss J. (2010). Solvent-induced lid opening in lipases: a molecular dynamics study. Protein Sci. 19, 2122–2130. 10.1002/pro.493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis P., Holmberg K., Watzke H., Leser M., Miller R. (2009). Lipases at interfaces: a review. Adv. Colloid Interface Sci. 147, 237–250. 10.1016/j.cis.2008.06.001 [DOI] [PubMed] [Google Scholar]
- Rengachari S., Aschauer P., Schittmayer M., Mayer N., Gruber K., Breinbauer R., et al. (2013). Conformational plasticity and ligand binding of bacterial monoacylglycerol lipase. J. Biol. Chem. 288, 31093–31104. 10.1074/jbc.M113.491415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengachari S., Bezerra G. A., Riegler-Berket L., Gruber C. C., Sturm C., Taschler U., et al. (2012). The structure of monoacylglycerol lipase from Bacillus sp. H257 reveals unexpected conservation of the cap architecture between bacterial and human enzymes. Biochim. Biophys. Acta 1821, 1012–1021. 10.1016/j.bbalip.2012.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel A., Canaan S., Egloff M. P., Riviere M., Dupuis L., Verger R., et al. (1999). Crystal structure of human gastric lipase and model of lysosomal acid lipase, two lipolytic enzymes of medical interest. J. Biol. Chem. 274, 16995–17002. 10.1074/jbc.274.24.16995 [DOI] [PubMed] [Google Scholar]
- Roussel A., Caro J. D., Bezzine S., Gastinel L., Caro A. D., Carrière F., et al. (1998a). Reactivation of the totally inactive pancreatic lipase RP1 by structure-predicted point mutations. Proteins 32, 523–531. [DOI] [PubMed] [Google Scholar]
- Roussel A., Yang Y., Ferrato F., Verger R., Cambillau C., Lowe M. (1998b). Structure and activity of rat pancreatic lipase-related protein 2. J. Biol. Chem. 273, 32121–32128. 10.1074/jbc.273.48.32121 [DOI] [PubMed] [Google Scholar]
- Roussel A., Miled N., Berti-Dupuis L., Riviere M., Spinelli S., Berna P., et al. (2002). Crystal structure of the open form of dog gastric lipase in complex with a phosphonate inhibitor. J. Biol. Chem. 277, 2266–2274. 10.1074/jbc.M109484200 [DOI] [PubMed] [Google Scholar]
- Safra T., Shamai S., Greenberg J., Veizman A., Shpigel S., Matcejevsky D., et al. (2002). Crystal structure of a thermostable lipase from Bacillus stearothermophilus P1. J. Mol. Biol. 323, 859–869. 10.1016/S0022-2836(02)01004-5 [DOI] [PubMed] [Google Scholar]
- Santarossa G., Lafranconi P. G., Alquati C., Degioia L., Alberghina L., Fantucci P., et al. (2005). Mutations in the “lid” region affect chain length specificity and thermostability of a Pseudomonas fragi lipase. FEBS Lett. 579, 2383–2386. 10.1016/j.febslet.2005.03.037 [DOI] [PubMed] [Google Scholar]
- Schalk-Hihi C., Schubert C., Alexander R., Bayoumy S., Clemente J. C., Deckman I., et al. (2011). Crystal structure of a soluble form of human monoglyceride lipase in complex with an inhibitor at 1.35 angstrom resolution. Protein Sci. 20, 670–683. 10.1002/pro.596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Dannert C., Rúa M. L., Atomi H., Schmid R. D. (1996). Thermoalkalophilic lipase of Bacillus thermocatenulatus. I. Molecular cloning, nucleotide sequence, purification and some properties. Biochim. Biophys. Acta 1301, 105–114. 10.1016/0005-2760(96)00027-6 [DOI] [PubMed] [Google Scholar]
- Schmidt-Dannert C., Rúa M. L., Schmid R. D. (1997). Two novel lipases from thermophile Bacillus thermocatenulatus: screening, purification, cloning, overexpression, and properties. Meth. Enzymol. 284, 194–220. 10.1016/S0076-6879(97)84013-X [DOI] [PubMed] [Google Scholar]
- Schomburg D., Rydel T. J., Oliver J. D. (1997). The open conformation of a Pseudomonas lipase: structure. Structure 5, 187–202. 10.1016/S0969-2126(97)00178-0 [DOI] [PubMed] [Google Scholar]
- Schrag J. D., Cygler M. (1993). 1·8 Å refined structure of the lipase from Geotrichum candidum. J. Mol. Biol. 230, 575–591. 10.1006/jmbi.1993.1171 [DOI] [PubMed] [Google Scholar]
- Secundo F., Carrea G., Tarabiono C., Brocca S., Lotti M. (2004). Activity and enantioselectivity of wildtype and lid mutated Candida rugosa lipase isoform 1 in organic solvents. Biotechnol. Bioeng. 86, 236–240. 10.1002/bit.20034 [DOI] [PubMed] [Google Scholar]
- Sheng J., Ji X. F., Wang F., Sun M. (2014). Engineering of Yarrowia lipolytica lipase Lip8p by circular permutation to alter substrate and temperature characteristics. J. Ind. Microbiol. Biotechnol. 41, 757–762. 10.1007/s10295-014-1428-1 [DOI] [PubMed] [Google Scholar]
- Shih T. W., Pan T. M. (2011). Substitution of Asp189 residue alters the activity and thermostability of Geobacillus sp. NTU 03 lipase. Biotechnol. Lett. 33, 1841–1846. 10.1007/s10529-011-0635-3 [DOI] [PubMed] [Google Scholar]
- Shu Z. Y., Wu J. G., Xue L. Y., Lin R. F., Jiang Y. M., Tang L. H., et al. (2011). Construction of Aspergillus niger lipase mutants with oil-water interface independence. Enzyme Microb. Technol. 48, 129–133. 10.1016/j.enzmictec.2010.10.011 [DOI] [PubMed] [Google Scholar]
- Singh Y., Gupta N., Verma V. V., Goel M., Gupta R. (2016). Selective disruption of disulphide bonds lowered activation energy and improved catalytic efficiency in TALipB from Trichosporon asahii MSR54: MD simulations revealed flexible lid and extended substrate binding area in the mutant. Biochem. Biophys. Res. Commun. 472, 223–230. 10.1016/j.bbrc.2016.01.189 [DOI] [PubMed] [Google Scholar]
- Skjold-Jorgensen J., Bhatia V. K., Vind J., Svendsen A., Bjerrum M. J., Farrens D. (2015). The enzymatic activity of lipases correlates with polarity-induced conformational changes: a trp-induced quenching fluorescence study. Biochemistry 54, 4186–4196. 10.1021/acs.biochem.5b00328 [DOI] [PubMed] [Google Scholar]
- Skjold-Jorgensen J., Vind J., Moroz O. V., Blagova E., Bhatia V. K., Svendsen A., et al. (2017). Controlled lid-opening in Thermomyces lanuginosus lipase – an engineered switch for studying lipase function. Biochim. Biophys. Acta 1865, 20–27. 10.1016/j.bbapap.2016.09.016 [DOI] [PubMed] [Google Scholar]
- Skjold-Jorgensen J., Vind J., Svendsen A., Bjerrum M. J. (2014). Altering the activation mechanism in Thermomyces lanuginosus lipase. Biochemistry 53, 4152–4160. 10.1021/bi500233h [DOI] [PubMed] [Google Scholar]
- Skjold-Jorgensen J., Vind J., Svendsen A., Bjerrum M. J. (2016). Lipases that activate at high solvent polarities. Biochemistry 55, 146–156. 10.1021/acs.biochem.5b01114 [DOI] [PubMed] [Google Scholar]
- Skjot M., De Maria L., Chatterjee R., Svendsen A., Patkar S. A., Ostergaard P. R., et al. (2009). Understanding the plasticity of the alpha/beta hydrolase fold: lid swapping on the Candida antarctica lipase B results in chimeras with interesting biocatalytic properties. Chembiochem 10, 520–527. 10.1002/cbic.200800668 [DOI] [PubMed] [Google Scholar]
- Strzelczyk P., Bujacz G. D., Kiełbasiński P., Błaszczyk J. (2016). Crystal and molecular structure of hexagonal form of lipase B from Candida antarctica. Acta Biochim. Pol. 63, 103–109. 10.18388/abp.2015_1065 [DOI] [PubMed] [Google Scholar]
- Tang L., Su M., Zhu L., Chi L., Zhang J., Zhou Q. (2013). Substitution of Val72 residue alters the enantioselectivity and activity of Penicillium expansum lipase. World J. Microbiol. Biotechnol. 29, 145–151. 10.1007/s11274-012-1167-2 [DOI] [PubMed] [Google Scholar]
- Tang L. H., Su M., Yan J. Z., Xie S., Zhang W. H. (2015). Lid hinge region of Penicillium expansum lipase affects enzyme activity and interfacial activation. Process Biochem. 50, 1218–1223. 10.1016/j.procbio.2015.04.022 [DOI] [Google Scholar]
- Terzyan S., Wang C. S., Downs D., Hunter B., Zhang X. C. (2000). Crystal structure of the catalytic domain of human bile salt activated lipase. Protein Sci. 9, 1783–1790. 10.1110/ps.9.9.1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirstrup K., Verger R., Carriere F. (1994). Evidence for a pancreatic lipase subfamily with new kinetic properties. Biochemistry 33, 2748–2756. 10.1021/bi00176a002 [DOI] [PubMed] [Google Scholar]
- Tiesinga J. J. W., Pouderoyen G. V., Nardini M., Ransac S., Dijkstra B. W. (2007). Structural basis of phospholipase activity of Staphylococcus hyicus lipase. J. Mol. Biol. 371, 447–456. 10.1016/j.jmb.2007.05.041 [DOI] [PubMed] [Google Scholar]
- Tilbeurgh H. V., Sarda L., Verger R., Cambillau C. (1992). Structure of the pancreatic-procolipase complex. Nature 359, 159–162. 10.1038/359159a0 [DOI] [PubMed] [Google Scholar]
- Timucin E., Sezerman O. U. (2013). The conserved lid tryptophan, W211, potentiates thermostability and thermoactivity in bacterial thermoalkalophilic lipases. PLoS ONE 8:e85186. 10.1371/journal.pone.0085186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ufarte L., Potocki-Veronese G., Laville E. (2015). Discovery of new protein families and functions: new challenges in functional metagenomics for biotechnologies and microbial ecology. Front. Microbiol. 6:563. 10.3389/fmicb.2015.00563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppenberg J., Hansen M. T., Patkar S., Jones T. A. (1994). The sequence, crystal structure determination and refinement of two crystal forms of lipase B from Candida antarctica. Structure 2, 293–308. 10.1016/S0969-2126(00)00031-9 [DOI] [PubMed] [Google Scholar]
- Uppenberg J., Ohrner N., Norin M., Hult K., Kleywegt G. J., Patkar S., et al. (1996). Crystallographic and molecular-modeling studies of lipase B from Candida antarctica reveal a stereospecific pocket for secondary alcohols. Biochemistry 34, 16838–16851. 10.1021/bi00051a035 [DOI] [PubMed] [Google Scholar]
- Van T. H., Egloff M. P., Martinez C., Rugani N., Verger R., Cambillau C. (1993). Interfacial activation of the lipase-procolipase complex by mixed micelles revealed by X-ray crystallography. Nature 362, 814–820. 10.1038/362814a0 [DOI] [PubMed] [Google Scholar]
- van Pouderoyen G., Eggert T., Jaeger K. E., Dijkstra B. W. (2001). The crystal structure of Bacillus subtilis lipase: a minimal alpha/beta hydrolase fold enzyme. J. Mol. Biol. 309, 215–226. 10.1006/jmbi.2001.4659 [DOI] [PubMed] [Google Scholar]
- Verger R. (1997). ‘Interfacial activation’ of lipases: facts and artifacts. Trends Biotechnol. 15, 32–38. 10.1016/S0167-7799(96)10064-0 [DOI] [Google Scholar]
- Wang X., Wang C. S., Tang J., Dyda F., Zhang X. C. (1997). The crystal structure of bovine bile salt activated lipase: insights into the bile salt activation mechanism. Structure 5, 1209–1218. 10.1016/S0969-2126(97)00271-2 [DOI] [PubMed] [Google Scholar]
- Wei Y., Swenson L., Castro C., Derewenda U., Minor W., Arai H., et al. (1998). Structure of a microbial homologue of mammalian platelet-activating factor acetylhydrolases: Streptomyces exfoliatus lipase at 1.9 å resolution. Structure 6, 511–519. 10.1016/S0969-2126(98)00052-5 [DOI] [PubMed] [Google Scholar]
- Withersmartinez C., Carriere F., Verger R., Bourgeois D., Cambillau C. (1996). A pancreatic lipase with a phospholipase A1 activity: crystal structure of a chimeric pancreatic lipase-related protein 2 from guinea pig. Structure 4, 1363–1374. 10.1016/S0969-2126(96)00143-8 [DOI] [PubMed] [Google Scholar]
- Wu L., Liu B., Hong Y., Sheng D., Shen Y., Ni J. (2010). Residue Tyr224 is critical for the thermostability of Geobacillus sp. RD-2 lipase. Biotechnol. Lett. 32, 107–112. 10.1007/s10529-009-0121-3 [DOI] [PubMed] [Google Scholar]
- Xie Y., An J., Yang G., Wu G., Zhang Y., Cui L., et al. (2014). Enhanced enzyme kinetic stability by increasing rigidity within the active site. J. Biol. Chem. 289, 7994–8006. 10.1074/jbc.M113.536045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Liu L., Hou S., Xu J., Yang B., Wang Y., et al. (2012). Crystal structure of a mono- and diacylglycerol lipase from Malassezia globosa reveals a novel lid conformation and insights into the substrate specificity. J. Struct. Biol. 178, 363–369. 10.1016/j.jsb.2012.03.006 [DOI] [PubMed] [Google Scholar]
- Yang W., Xu L., Zhang H., Yan Y. (2015). Characterizing LipR from Pseudomonas sp. R0-14 and applying in enrichment of polyunsaturated fatty acids from algal oil. J. Microbiol. Biotechnol. 25, 1880–1893. 10.4014/jmb.1506.06011 [DOI] [PubMed] [Google Scholar]
- Yang Y., Lowe M. E. (2000). The open lid mediates pancreatic lipase function. J. Lipid Res. 41, 48–57. [PubMed] [Google Scholar]
- Yapoudjian S., Ivanova M. G., Brzozowski A. M., Patkar S. A., Vind J., Svendsen A., et al. (2002). Binding of Thermomyces (Humicola) lanuginosa lipase to the mixed micelles of cis-parinaric acid/NaTDC. Eur. J. Biochem. 269, 1613–1621. 10.1046/j.1432-1327.2002.02786.x [DOI] [PubMed] [Google Scholar]
- Yu X. W., Tan N. J., Xiao R., Xu Y. (2012). Engineering a disulfide bond in the lid hinge region of Rhizopus chinensis lipase: increased thermostability and altered acyl chain length specificity. PLoS ONE 7:e46388. 10.1371/journal.pone.0046388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X. W., Zhu S. S., Xiao R., Xu Y. (2014). Conversion of a Rhizopus chinensis lipase into an esterase by lid swapping. J. Lipid Res. 55, 1044–1051. 10.1194/jlr.M043950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X. X., Wang Y. B., Pan Y. J., Li W. F. (2008). Differences in amino acids composition and coupling patterns between mesophilic and thermophilic proteins. Amino Acids 34, 25–33. 10.1007/s00726-007-0589-x [DOI] [PubMed] [Google Scholar]
- Zhu S. S., Li M., Yu X., Xu Y. (2013). Role of Met93 and Thr96 in the lid hinge region of Rhizopus chinensis lipase. Appl. Biochem. Biotechnol. 170, 436–447. 10.1007/s12010-013-0209-z [DOI] [PubMed] [Google Scholar]
- Zisis T., Freddolino P. L., Turunen P., Van Teeseling M. C., Rowan A. E., Blank K. G. (2015). Interfacial activation of Candida antarctica lipase B: combined evidence from experiment and simulation. Biochemistry 54, 5969–5979. 10.1021/acs.biochem.5b00586 [DOI] [PMC free article] [PubMed] [Google Scholar]