Abstract
Sphingomonas melonis TY utilizes nicotine as a sole source of carbon, nitrogen, and energy through a variant of the pyridine and pyrrolidine pathways (VPP). A 31-kb novel nicotine-degrading gene cluster, ndp, in strain TY exhibited a different genetic organization with the vpp cluster in strains Ochrobactrum rhizosphaerae SJY1 and Agrobacterium tumefaciens S33. Genes in vpp were separated by a 20-kb interval sequence, while genes in ndp were localized together. Half of the homolog genes were in different locus in ndp and vpp. Moreover, there was a gene encoding putative transporter of nicotine or other critical metabolite in ndp. Among the putative nicotine-degrading related genes, the nicotine hydroxylase, 6-hydroxy-L-nicotine oxidase, 6-hydroxypseudooxynicotine oxidase, and 6-hydroxy-3-succinyl-pyridine monooxygenase responsible for catalyzing the transformation of nicotine to 2, 5-dihydropyridine in the initial four steps of the VPP were characterized. Hydroxylation at C6 of the pyridine ring and dehydrogenation at the C2–C3 bond of the pyrrolidine ring were the key common reactions in the VPP, pyrrolidine and pyridine pathways. Besides, VPP and pyrrolidine pathway shared the same latter part of metabolic pathway. After analysis of metabolic genes in the pyridine, pyrrolidine, and VPP pathways, we found that both the evolutionary features and metabolic mechanisms of the VPP were more similar to the pyrrolidine pathway. The linked ndpHFEG genes shared by the VPP and pyrrolidine pathways indicated that these two pathways might share the same origin, but variants were observed in some bacteria. And we speculated that the pyridine pathway was distributed in Gram-positive bacteria and the VPP and pyrrolidine pathways were distributed in Gram-negative bacteria by using comprehensive homologs searching and phylogenetic tree construction.
Keywords: nicotine, Sphingomonas melonis TY, ndp gene cluster, pathway evolution, pathway distribution
Introduction
Nicotine is the most abundant alkaloid in tobacco plants, and it maintains a high content (15.680–32.536 mg/g dry weight according to the particle size of the solid powdery waste) in tobacco waste, which accumulates in large amounts (Civilini et al., 1997; Novotny and Zhao, 1999; Cosic et al., 2012). Nicotine has good hydrophilicity, can spread in the environment through water and soil, and poses a threat to human health and the environment because of its toxicity. Moreover, nicotine can be easily absorbed and traverse the blood-brain barrier (Oldendorf, 1974; Oldendorf et al., 1979; O'neill et al., 2002; Lemay et al., 2004), and it has been classified as a Toxics Release Inventory (TRI) chemical by the United States Environmental Protection Agency since 1994. Microbial bioremediation has been considered an economical and effective way to eliminate nicotine in the environment (Brandsch, 2006; Gurusamy and Natarajan, 2013).
To date, four main types of nicotine degradation pathways have been described in microorganisms. First, the pyrrolidine pathway (PRL) was found in Pseudomonas sp. No. 41, Pseudomonas putida NRRL B-8061, Pseudomonas putida S16, Pseudomonas sp. HF-1, and Pseudomonas sp. HZN6 (Wada and Yamasaki, 1953; DeTraglia and Tometsko, 1980; Wang et al., 2004; Ruan et al., 2005; Qiu et al., 2011). Second, the pyridine pathway (PD) was reported in Arthrobacter oxydans P-34, Arthrobacter nicotinovorans pAO1, Nocardioides sp. JS614, Rhodococcus opacus B4, and Arthrobacter aurescens M2012083 (Hochstein and Rittenberg, 1959a; Ganas et al., 2008; Cobzaru et al., 2011; Yao et al., 2012, 2015). Third, the methyl pathway was found only in several fungi Microsporum gypseum, Pellicularia filamentosa JTS-208, Cunninghamella echinulata IFO-4444, and Aspergillus oryzae 112822 (Sindelar et al., 1979; Uchida et al., 1983; Meng et al., 2010). Fourth, a variant of the pyridine and pyrrolidine pathway (VPP) was discovered in Agrobacterium tumefaciens S33, Sphingomonas melonis TY, Shinella sp. HZN7, and Ochrobactrum rhizosphaerae SJY1 (Wang et al., 2009, 2011; Ma et al., 2013; Yu et al., 2014). Additionally, two special pathways were recently found in Pseudomonas plecoglossicida TND35 and Pusillimonas sp. T2, the former with some new intermediates and the latter containing both the VPP and a partial PD with the formation of 2, 6-dihydroxypyridine (Raman et al., 2013; Ma et al., 2014). Interestingly, four intermediates of the nicotine metabolic pathway defined as 6-hydroxynicotine (6HN), pseudooxynicotine, 3-succinoyl-pyridine, and 6-hydroxy-3-succinoyl-pyridine (HSP) were studied in a nicotine-degrading strain Achromobacter nicotinophagum in 1958 (Hylin, 1958, 1959). It was anticipated that more new pathways would be found. Among these published pathways, the most well-established were the PD in A. nicotinovorans pAO1 (Dang et al., 1968; Brühmüller et al., 1972; Grether-Beck et al., 1994; Schenk et al., 1998; Baitsch et al., 2001; Chiribau et al., 2004, 2006; Sachelaru et al., 2005, 2006; Mihasan et al., 2007), the PRL in Pseudomonas putida S16 (Tang et al., 2008, 2009, 2011, 2012, 2013; Jiang et al., 2015), and the VPP in O. rhizosphaerae SJY1 (Yu et al., 2014, 2015). The nicotine-degrading gene clusters, for example, the nic-genes in A. nicotinovorans pAO1, nic1 and nic2 in strain S16, and the vpp cluster in strain SJY1, have been comprehensively studied.
The nomenclature and classification of microbial nicotine degradation pathway was according to the reaction position of the first step in each pathway. In the PRL, the first reaction was occurred at C2–C3 bond of the pyrrolidine ring (Tang et al., 2013); in the PD, the first reaction was occurred at C6 of the pyridine ring (Hochstein and Rittenberg, 1959b); in the methyl pathway, the first reaction was occurred at methyl group linked in the N atom of the pyrrolidine ring (Uchida et al., 1983). All these three pathways have totally different nicotine metabolism pathway from the first step of degradation (Raman et al., 2013). And VPP was designated by its feature of combing upper pathway of PD with lower pathway of PRL (Wang et al., 2012). There are two key steps shared by the PRL, PD, and VPP pathways, that were hydroxylation at C6 of the pyridine ring and dehydrogenation at the C2–C3 bond of the pyrrolidine ring, which was important for opening the ring.
Horizontal gene transfer (HGT) has been recognized as a main force in the genomes evolution for a long time (Gray, 1992). In comparison to eukaryotes, which evolve mainly through the modification of existing genetic information, bacteria obtain a considerable ratio of their genetic variants through HGT from distantly related organisms (Ochman et al., 2000). There are some commonly used criteria and methods for identifying HGT. HGT creates an unduly high degree of DNA or protein sequence similarity between the donor and the recipient strains for the character in question, and the acquired trait will be limited to the offsprings of the recipient strain and absent from the closely related taxa, thereby producing a scattered phylogenetic distribution (Ochman et al., 2000). However, the strongest evidence to identify cases of HGT derives from a molecular genetic analysis of their DNA sequences. Bacterial species display a wide range of variation in their total G+C content, but the genes in a particular species' genome are considerably similar in regard to their nucleotide compositions, patterns of codon usage and frequencies of di- and tri-nucleotides (Sueoka, 1962; Muto and Osawa, 1987; Karlin et al., 1998). Therefore, sequences that are newly transferred into the bacterial genome, namely, those introduced through HGT, retain the sequence characteristics of the original genome and have atypical nucleotide compositions, or patterns of codon usage bias with the host genome and thus can be differentiate from vertically inherited DNA (Lawrence and Ochman, 1998). Additionally, the regions contiguous to the genes that were confirmed to be horizontally transferred often contained traces of sequences promoting their integration, such as mobile element remnants, transfer origins of plasmids or attachment sites of phage integrases, further confirming their foreign origin in the genome (Ochman et al., 2000).
Although the molecular mechanism of nicotine metabolism is very clear in several representative strains, the evolutionary relationship of these nicotine degradation gene clusters and the evolutionary relationship between the PD, PRL, and VPP remain unclear. More gene clusters must be discovered to help unravel the evolutionary relationships. Moreover, some intermediates that form during nicotine metabolism have pharmacological value (Roduit et al., 1997; Wang et al., 2005; Goetz and Garg, 2013), necessitating additional genetic and metabolic resources for industrial applications. In this work, we studied a novel nicotine-degrading gene cluster, ndp, in S. melonis TY. This 31-kb ndp in strain TY exhibited a different genetic organization with the vpp cluster in strains SJY1 and S33. Genes in vpp were separated by a 20-kb interval sequence, while genes in ndp were localized together. Half of the homolog genes were in different locus in ndp and vpp. Moreover, there was a gene encoding putative transporter of nicotine or other critical metabolite in ndp, while missing in vpp. The amino acid sequence identity between two key common enzymes was low, 45 and 54% to NdpAL and NdpB, respectively. All these differences showed that ndp was special and had a different evolution process with that of vpp. Nicotine dehydrogenase ndpA, 6-hydroxynicotine oxidase ndpB, 6-hydroxypseudooxynicotine oxidase ndpC and 6-hydroxy-3-succinoyl-pyridine 3-monooxygenase ndpD were identified. In a word, ndp was found to be an integrated and compact nicotine-degrading gene cluster with a different genetic organization and coding sequence compared with vpp in strains SJY1 and S33. We analyzed the three nicotine-degrading gene clusters in the VPP and found that all of them seemed to evolve via HGT. Analysis of the origin of the three main pathways involved in nicotine degradation revealed that the VPP was more similar to PRL.
Materials and methods
Chemicals and reagents
(S)-Nicotine (>99%) was obtained from Chemsky international Co., Ltd (Shanghai, China). 6HN, 6-hydroxypseudooxynicotine (6HPON), and HSP were prepared as previously described (Ma et al., 2013). TransStart® FastPfu DNA Polymerase for fragment amplification was purchased from TransGen Biotech (Beijing, China). Restriction enzymes used for plasmid construction and premixed protein marker for protein electrophoresis were purchased from Takara Biotechnology Co., Ltd. (Dalian, China). Antibiotics, isopropyl β-D-1-thiogalactopyranoside (IPTG) and other reagents were purchased from Shanghai Sangon Biotech Co., Ltd. (Shanghai, China). The plasmid extraction, gel extraction, and DNA purification kits were obtained from Omega Bio-tek, Inc. (Norcross, GA, USA). Bacterial genomic DNA was extracted using the TIANamp Bacteria DNA Kit from Tiangen Biotech co., Ltd. (Beijing, China). All reagents and solvents were of analytical or chromatographic grade.
Bacterial strains, plasmids, and growth conditions
The bacterial strains and plasmids used in this study are listed in Table S1 and the primers are shown in Table S2. The wild-type strain S. melonis TY (deposited in China General Microbiological Culture Collection Center, collection number CGMCC1.15791) can use nicotine as a sole carbon, nitrogen, and energy source to grow (Wang et al., 2011). The wild-type strain TY and its derivatives were cultured aerobically in LB medium or inorganic salt medium (ISM) supplemented with nicotine at 30°C as described previously (Wang et al., 2011). Escherichia coli strains were grown in LB broth at 37°C. When necessary, ampicillin, kanamycin, and tetracycline were used at final concentrations of 100, 50, and 10 μg/mL, respectively, excluding Origami B(DE3) (tetracycline and kanamycin were used at final concentrations of 12.5 and 15 μg/mL, respectively). IPTG was used as an inducer at a given concentration. The 2, 6-diaminopimelic acid (2, 6-DAP) was used at a final concentration of 0.3 mM for E. coli WM3064. Competent E. coli WM3064 were prepared according to standard methods using 0.1 M CaCl2 in 20% (v/v) glycerol (Ausbel et al., 1995).
Genome sequence analysis and prediction of nicotine metabolism-related genes in strain TY
To identify putative genes involved in nicotine degradation in strain TY, we conducted a BLAST analysis against the draft genome sequence of strain S. melonis TY (the report about the draft genome sequence of strain TY is under reviewed in Frontiers in Microbiology, and the accession number of the genome is LQCK00000000) using known nicotine metabolic genes, such as ndhLSM and 6hlno in A. nicotinovorans (Grether-Beck et al., 1994) and hspA, hspB, hpo, nfo, ami, and iso in Pseudomonas putida S16 (Tang et al., 2008, 2011, 2012). The hits were designated as putative nicotine metabolism-related genes in strain TY and used for reverse transcription quantitative PCR (RT-qPCR) analysis. Subsequently, some of them were chosen for gene disruption experiments.
RT-qPCR analysis of ndpA, ndpB, ndpC, and ndpD
ISM with 1 g/L glucose and 1 g/L (NH4)2SO4 was used as the control medium for the RT-qPCR experiments. ISM supplemented with 1 g/L nicotine was established as the experimental group. Cultures in the presence and absence of nicotine were cultured in triplicate at 30°C to mid-exponential phase. Total RNA were extracted using the RNAprep Pure Bacteria Kit (Tiangen Biotech, Beijing, China) and reverse-transcribed into cDNA using the random hexamer primers and PrimeScript RT Reagent Kit with gDNA Eraser (Perfect Real Time; Takara, Dalian, China). The respective cDNA fragments were applied as templates in the PCR using the gene-specific primers shown in Table S2 designed using Beacon Designer 7 software, Premier Biosoft International (Palo Alto, CA, USA). qPCR was performed on a Rotor-Gene Q real-time PCR detection system (Qiagen, Germany) using TransStart Top Green qPCR SuperMix (TransGen Biotech, Beijing, China). The genome of strain TY was used as a positive control, and untranscribed RNA was used as a negative control. Melting curves and agarose gel analyses were used to confirm the specificity of the PCR products. The threshold cycle (CT) values for each target gene were normalized to the 16S rRNA reference gene. The 2ΔΔCT method was used to calculate the relative expression level, where ΔΔCT = (CT, target − CT, 16S)induction − (CT, target − CT, 16S)control, whereas the theoretical efficiency value 2 was replaced with the estimated PCR efficiency value (Livak and Schmittgen, 2001; Ramakers et al., 2003; Ruijter et al., 2009; Tuomi et al., 2010). In brief, the PCR data were saved in “LinReg Export Format” in the Rotor-Gene Q series software program and imported into the LinRegPCR program. Individual samples were then checked sequentially, and the mean PCR efficiency values were used for each amplicon group as a substitute for the theoretical efficiency value 2.
Gene knockout and complementation
All the vectors constructed in this work were first simulated using Vector NTI Advance 11.5.1 (Invitrogen, USA). Specific primers for fragment amplification were annotated in this software and used as output for synthesis, as were the primers used to sequence the constructed vectors. The sequencing results obtained from the biotechnology company were assembled and checked for mutations with simulative construction by Vector NTI Advance 11.5.1. In-frame disruption of ndpAL, ndpB, ndpC, and ndpD in strain S. melonis TY was performed using the suicide plasmid pEX18Tc and a two-step homologous recombination method (Chen et al., 2014). Plasmids pEX18Tc-ndpAL, pEX18Tc-ndpB, pEX18Tc-ndpC, and pEX18Tc-ndpD for gene knockout were constructed by fusing the PCR products of the kanamycin resistance gene and two upstream and downstream fragments of the target gene, amplified with the primers shown in Table S2, to Sac I and Hind III-digested pEX18Tc using the In-Fusion® HD Cloning Kit (Takara, Dalian, China). Constructed plasmids were transformed into E. coli DH5α and sequenced, and accurate constructions were preserved and used for subsequent analyses. These plasmids were then transformed into E. coli WM3064 (2, 6-DAP auxotroph; Dehio and Meyer, 1997; Saltikov and Newman, 2003) before being conjugated to strain TY as described previously (Saltikov and Newman, 2003). The TYΔndpAL, TYΔndpB, TYΔndpC, and TYΔndpD recombinants were screened on LB plates containing kanamycin and then verified using specific primers for PCR and sequencing. If double-crossover recombinants were not acquired during the first cycle of screening, the single crossover recombinants were cultured in liquid LB containing kanamycin and 10% sucrose (w/v) for several generations and then screened on LB plates supplemented with kanamycin and 10% sucrose (w/v) to obtain single colonies. Verification was performed as mentioned above until double-crossover recombinants were obtained.
Plasmids pRK415-ndpAL, pRK415-ndpB, pRK415-ndpC, and pRK415-ndpD for gene complementation were constructed by fusing the PCR products corresponding to the full-length ndpAL, ndpB, ndpC, and ndpD amplified with the primers shown in Table S2, to Hind III and EcoR I-digested pRK415. After sequencing and obtaining the desired construction, four plasmid constructs were transformed into E. coli WM3064 and then mated into the TYΔndpAL, TYΔndpB, TYΔndpC, and TYΔndpD strain by conjugation to obtain TYΔndpAL(pRK415-ndpAL), TYΔndpB(pRK415-ndpB), TYΔndpC(pRK415-ndpC), and TYΔndpD(pRK415-ndpD), respectively.
Cell growth and resting cell reactions of TY and its derivatives
The four mutants TYΔndpAL, TYΔndpB, TYΔndpC, and TYΔndpD were examined for their ability to grow in the presence of nicotine. After gene complementation, all the complementary strains were inoculated in ISM medium supplemented with nicotine to determine whether they restored nicotine-degrading ability. After preliminarily confirming the importance of these four genes in nicotine degradation of strain TY, biotransformation tests were conducted using TYΔndpAL, TYΔndpB, TYΔndpC, TYΔndpD, wild type TY, and an inactivated strain of wild type TY (heated at 80°C for 5 min to exclude nicotine absorption; Harwood et al., 1994) and a control group containing only nicotine, to determine the intermediate product of nicotine produced by these four mutant strains. The cells were harvested by centrifugation at 6,000 × g for 5 min and washed twice with 12 mM phosphate-buffered saline (PBS, pH 7.4). Subsequently, the cells pellets were resuspended in ISM (resting cells). The biotransformation test was performed in a 150-mL beaker flask containing 60 mg dry cell weight of resting cells (with an optical density at 600 nm (OD600nm) of 5.0, in which one OD600nm unit was equivalent to 0.40 g/L dry cell weight) and 0.5 mg/mL nicotine in 30 mL of sterilized ISM and incubated at 30°C and 200 rpm for 2 days. The reactions were stopped by centrifugation at 6,000 × g for 5 min, and the supernatant was subjected to spectrum scanning and liquid chromatography-mass spectrometry (LC-MS) analysis. Biotransformation tests were conducted as described for TYΔndpAL, TYΔndpB, TYΔndpC, and TYΔndpD with 6HN, 6HPON, and HSP. Additionally, growth ability was examined using these four mutants with nicotine-degrading intermediates (6HN, 6HPON, and HSP) in strain TY.
Heterologous expression of ndpA, ndpB, ndpC, and ndpD
To verify the function of ndpA, heterologous expression of NdpA was performed according to the method described for the heterologous expression of vppA (Yu et al., 2015). P. putida KT2440 and Sphingomonas aquatilis JSS7T were chosen as the expression host (details concerning the heterologous expression of ndpA, ndpB, and ndpD are provided in the supporting information).
pET-28a(+) and pET-22b(+) were used as the expression vectors for ndpB, and E. coli BL21(DE3) and Origami B(DE3) were used as the expression hosts. Origami B(DE3) was selected because it carries glutathione reductase (gor) and thioredoxin reductase (trxB) mutations to enhance the formation of disulfide bonds in the E. coli cytoplasm (Prinz et al., 1997; Aslund et al., 1999).
For heterologous expression of NdpB, pRK415-ndpB was mated into P. putida KT2440 and Sphingomonas aquatilis JSS7T through E. coli WM3064 to generate P. putida KT-ndpB and Sphingomonas-ndpB, respectively. The method applied for whole cell transformation of 6HN by NdpB was the same as that described for NdpA. For homologous expression of NdpB, His6-tagged fusion protein under the promoter of pRK415 was expressed in strain TYΔndpB and purified by Wuhan GeneCreate Biological Engineering Co., Ltd. (Wuhan, China) under native conditions. The obtained protein was evaluated by western blot analysis and detection of activity.
Heterologous expression of NdpC was conducted as described for NdpA and NdpB, with the transfer of pRK415-ndpC into P. putida KT2440 and Sphingomonas aquatilis JSS7T to obtain P. putida KT-ndpC and Sphingomonas-ndpC; the substrate was substituted with 6HPON when performing whole cell transformation to detect the enzyme activity.
C-terminal and N-terminal His6-tagged expressions of NdpD were performed by fusing the ndpD (full-length gene or without the stop codon) product to Nco I and Hind III-digested pET-28a(+) or Nde I and Xho I-digested pET-22b(+) to obtained pET28a-ndpD-C and pET22b-ndpD-N, respectively, using E. coli BL21(DE3) as the expression host. The crude enzyme activity of NdpD was detected according to Yu et al. (2014).
NdpD was also heterologously expressed in P. putida KT2440 and Sphingomonas aquatilis JSS7T with pRK415-ndpD as described above. Whole cell transformation of HSP by P. putida KT-ndpD and Sphingomonas-ndpD was also conducted as described for NdpA.
Analytical methods
Biotransformation of nicotine by resting cells was analyzed by spectrum scanning using a Lambda25 UV/VIS spectrometer (PerkinElmer, USA). LC-MS analysis was performed using liquid chromatography (Agilent 1200, USA) equipped with Sielc Obelisc_N column (5 μm, 2.1 × 150 mm) and a LCQ Deca XP Max MS instrument (Thermo Finigan) with an electrospray interface (Turbo Ion Spray). The iron spray voltage was set at 4,500 V. Nitrogen was used as the sheath gas (55 arb) and auxiliary gas (5 arb). The capillary temperature was set at 275°C, and the capillary voltage was set at 41 V. Mobile phase A was 50 mM ammonium acetate, pH 5.0, adjusted with formic acid, and mobile phase B was acetonitrile. The system was run as follows: 0–5 min, 5% A+95% B; 5–35 min, from 5% A+95% B to 70% A+30% B; 35–45 min, 70% A+30% B. The total flow rate was 0.3 mL/min, and 15 μl of the sample was injected. The column was set at 35°C, and the detection was performed at 254 nm. The obtained data were analyzed using Xcalibur software (Thermo Electron Corporation). The HPLC analysis was performed according to Ruan et al. (2005).
Genetic characteristics of genes in the VPP
The G+C content of genome of TY, SJY1, and S33 were searched at NCBI (http://www.ncbi.nlm.nih.gov/), and the G+C content of the nicotine-degrading gene cluster was calculated at DNA/RNA GC Content Calculator in EndMemo (http://www.endmemo.com/bio/gc.php). CAIcal (Puigbò et al., 2008) and condonW (http://codonw.sourceforge.net/index.html) were used to calculate the nucleotide composition and relative synonymous codon usage (RSCU) of the nicotine-degrading genes, and condonW was also used to calculated the effective number of codons (ENC) value. BLASTp was used to search for closely related taxa of the nicotine-degrading gene of strain TY, SJY1, and S33 (https://blast.ncbi.nlm.nih.gov/Blast.cgi). HGT remnants were checked by searching the annotation of contiguous genes of nicotine-degrading gene clusters.
Analysis of evolutionary relationships among nicotine degradation pathways
Hydroxylation at C6 of the pyridine ring and dehydrogenation at the C2–C3 bond of the pyrrolidine ring were common reactions in the VPP, PRL, and PD pathways. Therefore, all members of the corresponding hydroxylases and dehydrogenases identified in previous experiments were used as queries in BLAST searches against a local genomes database that included 2806 predicted prokaryotic proteomes (downloaded from the NCBI FTP server, ftp://ftp.ncbi.nih.gov/). The genomes with homologs of both NdpAL and NdpB were selected, and the corresponding protein sequences were retrieved for subsequent analyses. The NdpHFEG proteins were also determined using the BLAST method. The protein sequences of NdpAL and NdpB were aligned using ClustalW (Larkin et al., 2007), and the resulting alignments of individual proteins were used to infer the organismal phylogeny with the maximum likelihood algorithm (ML) in the MPI-parallelized version of RAxML version 7.3.0 (Stamatakis, 2006). Ambiguous alignments were removed using the Gblocks method in SEAVIEW (Gouy et al., 2010) with options for a less stringent selection. The LG model (Le and Gascuel, 2008) with a proportion of invariable sites (+I), a gamma-shaped distribution of rates across sites (+G) and observed amino acid frequencies (+F) was used for the phylogeny inference. The topologies of the phylogenetic trees were evaluated using the bootstrap resampling method of Felsenstein (Felsenstein, 1985) with 100 replicates.
Accession number of nucleotide sequence
The sequence of the ndp cluster from strain TY is available in GenBank under accession number LQCK02000019.1.
Results
A putative nicotine metabolism gene cluster is present in the genome of strain TY
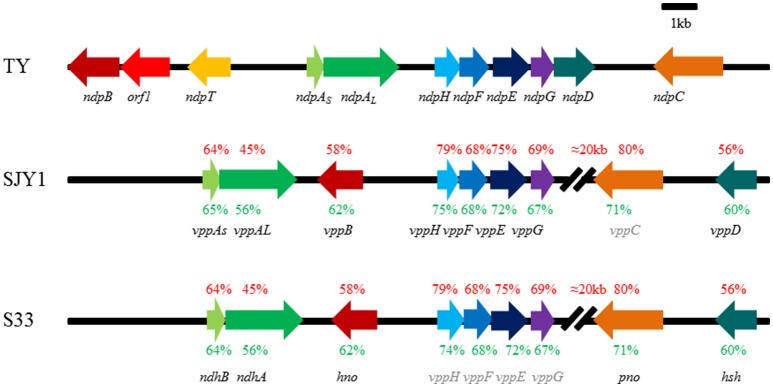
After performing a BLAST analysis against the genome of strain TY using previously known nicotine metabolism genes, we found that the genes including ndhLSM and 6hlno in A. nicotinovorans pAO1, hspB, hpo, nfo, ami, and iso in Pseudomonas putida S16 all had hits in one 31-kb scaffold of the genome of strain TY, demonstrating a compact arrangement in this scaffold (Figure 1). Among these putative genes were two genes, ndpAS (54%) and ndpAL (37%), which showed amino acid sequence similarities to ndhS and ndhL, respectively. ndpB (43%) displayed similarities to 6hlno, and ndpH (74%), ndpF (64%), ndpE (72%), ndpG (68%), and ndpD (54%) all shared considerable similarities with iso, nfo, hpo, ami, and hspB, respectively. This gene cluster was considered to be responsible for nicotine degradation and designated ndp. Until now, the underlying molecular mechanism has been the most well-studied in strain SJY1, excluding the enzyme that catalyzes the reaction from 6HPON to HSP (Yu et al., 2014, 2015); this enzyme was recently found in strain S33 (Li et al., 2016). Analysis of the whole genome sequence of strains SJY1 (accession number AZRT00000000) and S33 (CP014259.1 and CP014260.1; Yu et al., 2014; Li et al., 2016) showed that the gene organization and protein sequences of the nicotine-degrading gene clusters in these two strains were nearly identical (Figure 1). However, the sequence identity of ndp was significantly lower at the amino acid sequence (45–80%) and nucleotide sequence (56–75%) levels compared with strains SJY1 and S33. Moreover, the genetic organization of ndp was clearly different compared with the vpp gene cluster in strains SJY1 and S33. There was almost a 20-kb interval sequence between 6-hydroxypseudooxynicotine oxidase and maleamate amidase in the vpp gene cluster. In the 20-kb interval sequence, there were 6 and 11 mobile element protein genes in strains S33 and SJY1, respectively. However, the ndp gene cluster exhibited a compact arrangement without too much irrelevant sequence and no mobile element protein genes between the nicotine-degrading genes. In addition, we inferred that the undiscovered 6-hydroxypseudooxynicotine oxidase in SJY1 occurred at the same position as pno (Figure 1) due to their identical coding sequence.
Figure 1.
Genetic organization and amino acid sequence identity of nicotine degradation gene clusters in Sphingomonas melonis TY, Ochrobactrum rhizosphaerae SJY1, and Agrobacterium tumefaciens S33. The arrows indicate the size and direction of the transcription of each gene, and genes with the same fill color are isoenzymes. The gene name in gray represents speculated genes in the corresponding strain. The corresponding amino acids sequence identity between TY and S33, TY and SJY1 are shown in red text, and the nucleotide sequence identity is shown in green. ndpAs, nicotine hydroxylase, subunit S; ndpAL, nicotine hydroxylase, subunit L; ndpB, 6-hydroxy-L-nicotine oxidase; ndpC, 6-hydroxypseudooxynicotine oxidase; ndpD, HSP monooxygenase; ndpE, 2,5-DHP dioxygenase; ndpF, N-formylmaleamic acid deformylase; ndpG, maleamate amidase; ndpH, maleate isomerase; Percent amino acid sequences identity of genes compared with orthologous gene product from TY were labeled above the gene cluster in SJY1 and S33, respectively. TY, Sphingomonas melonis TY; SJY1, Ochrobactrum rhizosphaerae SJY1; and S33, Agrobacterium tumefaciens S33.
Transcription levels of putative nicotine-degrading genes in ndp are upregulated by nicotine
To elucidate the correlation between nicotine degradation and putative nicotine-degrading genes in ndp, the mRNA expression levels of nine putative target genes involved in the nicotine degradation of S. melonis TY were estimated using RT-qPCR and the 2ΔΔCT method with or without nicotine supplementation in the growth medium. The results showed that the levels of these genes were significantly upregulated in the presence of nicotine (Figure 2), suggesting that the transcription of these putative nicotine-degrading genes in ndp was induced by nicotine or other nicotine degradation intermediates. As shown in Figure 2, ndpAL, ndpB, ndpC, ndpD, and ndpH displayed more than 50-times higher than the levels of the corresponding genes in the control group, while ndpAS, ndpF, and ndpG exhibited relatively lower levels of transcription.
Figure 2.
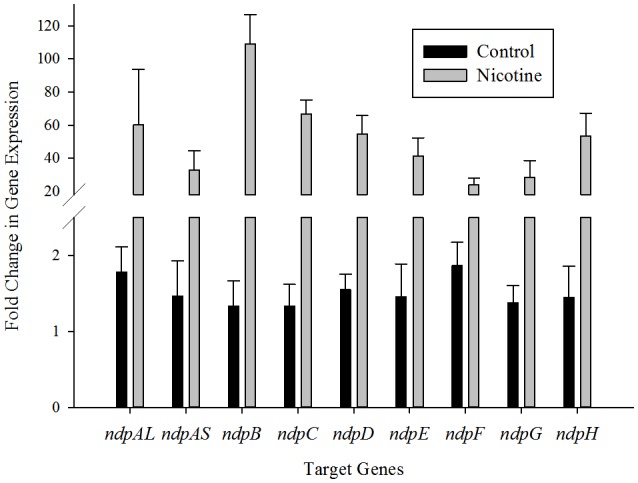
Transcriptional analysis of the ndp gene cluster. RT-qPCR analysis of target gene transcripts produced in Sphingomonas melonis TY grown with (gray bars) or without (black bars) nicotine. The expression levels of the ndp genes were normalized to the 16S rRNA and are expressed as fold changes relative to the expression level in cells. The results presented in these histograms are the means of three independent experiments, and error bars indicate the standard error.
NdpA catalyzes the reaction from nicotine to 6HN
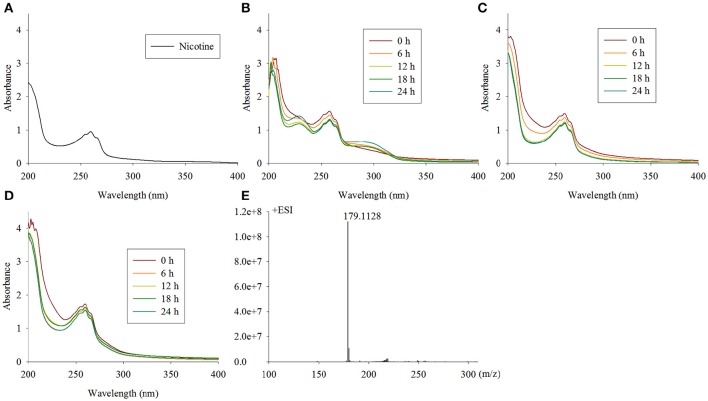
The first step in the VPP is hydroxylation of the C6 at the pyridine ring to generate 6HN. The mutant TYΔndpAL lost the ability to grow on nicotine, and after complementation of the full-length gene, it regained its ability to degrade nicotine. A biotransformation test with TYΔndpAL revealed that no intermediate was accumulated by TYΔndpAL, according to UV scanning with a maximum absorbance at 259 nm (equivalent to nicotine) and LC-MS analysis. Additionally, TYΔndpAL could transform 6HN, 6HPON and HSP but not nicotine, as expected (Table 1). The phenotypic traits of TYΔndpAL suggested that NdpA was responsible for catalyzing the reaction from nicotine to 6HN. Heterologous expression of NdpA in P. putida KT2440 provided negative results, irrespective of the presence of pRK415ndpA or pRK415ndpAplus. NdpA was successfully expressed in Sphingomonas aquatilis JSS7T, and a decrease in the nicotine substrate resulted in the gradual production of the product 6HN (Figure 3).
Table 1.
Characteristics of mutant and complementary strains.
| Biotransformation | ||||||
|---|---|---|---|---|---|---|
| Strains | Growth with nicotine | Accumulated intermediate metabolite | Nicotine | 6HN | 6HPON | HSP |
| TY | +a | NAb | + | + | + | + |
| TYΔndpAL | −c | Nicotine | − | + | + | + |
| TYΔndpB | − | 6HN | + | − | + | + |
| TYΔndpC | − | 6HPON | + | + | − | + |
| TYΔndpD | − | HSP | + | + | + | − |
| TYΔndpAL(pRK415-ndpAL) | + | NA | NA | NA | NA | NA |
| TYΔndpB(pRK415-ndpB) | + | NA | NA | NA | NA | NA |
| TYΔndpC(pRK415-ndpC) | + | NA | NA | NA | NA | NA |
| TYΔndpD(pRK415-ndpD) | + | NA | NA | NA | NA | NA |
means have corresponding ability;
means not applicable;
means have no corresponding ability.
Figure 3.
Intermediate accumulated by TYΔndpAL and biotransformation of nicotine by Sphingomonas-ndpA, KT-ndpA and KT-ndpAplus. (A) Nicotine with a maximum absorbance at 259 nm by UV scanning was accumulated by TYΔndpAL; (B–D) biotransformation of nicotine by Sphingomonas-ndpA, KT-ndpA, and KT-ndpAplus, shows that product was only formed in Sphingomonas-ndpA (product should be 6-hydroxynicotine with a maximum absorbance at 232 and 295 nm); (E) LC-MS analysis of the 6HN produced in (B).
The ndpB gene encodes 6-hydroxynicotine oxidase
The second enzymatic step in the VPP of nicotine degradation is the transformation of 6HN to 6-hydroxy-N-methylmyosmine (6HMM). After the disruption of ndpB, the mutant strain TYΔndpB lost the ability to grow on nicotine, and complementation restored nicotine-degrading ability similar to the wild-type strain. A biotransformation test illustrated that the intermediate that accumulated in TYΔndpB was 6HN, according to UV scanning (maximum absorbances at 232 and 295 nm) and LC-MS analysis. It also showed that TYΔndpB could effectively transform nicotine, 6HPON and HSP (Table 1). The phenotypic traits of TYΔndpB and the complementary experiment suggested that ndpB catalyzed the transformation from 6HN to 6HMM. Recombinant NdpB formed in inclusion bodies following the overexpression in E. coli BL21(DE3). Additionally, renaturation of NdpB failed in the absence of active enzyme.
To avoid inclusion body formation and obtain soluble protein, pET-22b(+) was selected for secretive expression of NdpB, and the expression host was changed to Origami B(DE3). Unfortunately, the NdpB still formed in inclusion bodies (data not shown).
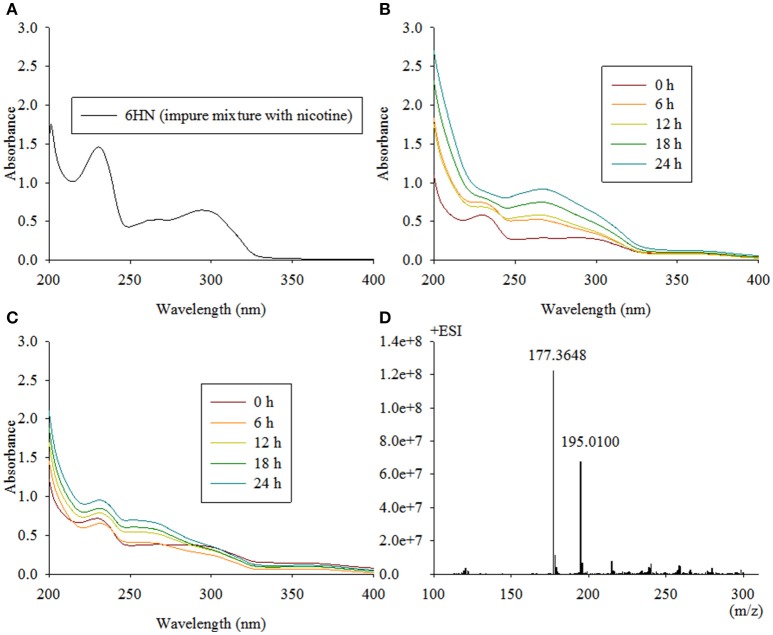
Because E. coli strains were not suitable for the expression of NdpB, we placed pRK415ndpB in Sphingomonas aquatilis JSS7T and detected the catalysis of 6HN. Interestingly, NdpB expressed in Sphingomonas aquatilis JSS7T was active and transformed 6HN to 6HPON, whereas active expression in P. putida KT2440 failed (Figure 4). We obtained 100 μg of purified NdpB from the expression of NdpB in strain TYΔndpB, and the western blot analysis is shown in Figure 5. The results suggested that the correct protein was acquired with the expected molecular weight and with high purity. However, very weak activity was detected at the primary stage when conducting the enzymatic reaction, and the product was found to be 6HMM by LC-MS (Figure 4).
Figure 4.
Intermediate accumulated by TYΔndpB and biotransformation of 6HN by Sphingomonas-ndpB and KT-ndpB. (A) 6HN with a maximum absorbance at 232 and 295 nm by UV scanning was accumulated by TYΔndpB; (B,C) biotransformation of 6HN by Sphingomonas-ndpB and KT-ndpB, respectively, showed that no product was formed in strain KT-ndpB, whereas Sphingomonas-ndpB exhibited catalytic activity based on 6-hydroxy-N-methylmyosmine formation. LC-MS analysis of the product shown in (D), and 6HMM formed by purified NdpB demonstrates the same peak as in (D).
Figure 5.

Western blot analysis of purified NdpB expressed in Sphingomonas TYΔndpB. Western blot analysis of the samples using a His tag-specific antibody. The contents of the lanes are as follows: M, marker; a1, original sample; a2, parallel sample with a2; b, 10-fold dilution of the original sample; and c, 100-fold dilution of the original sample.
NdpC is responsible for the transformation of 6HPON
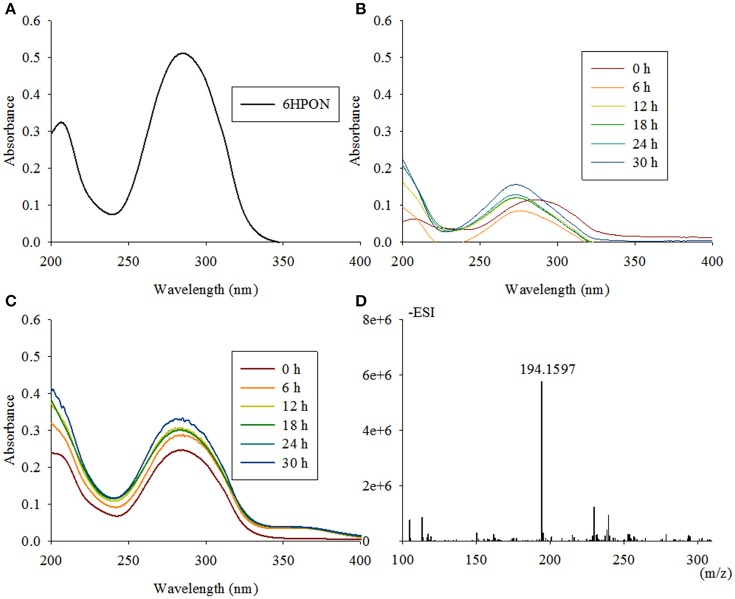
The reaction from 6HPON to HSP is one of the key steps in the VPP, causing the degradation pathway that starts at the PD to switch to the PRL. TYΔndpC, as well as TYΔndpB and TYΔndpAL, lost the ability to grow with nicotine as the sole carbon and nitrogen source. After gene complementation, the complementary strain recovered the ability to degrade nicotine. A biotransformation test showed that 6HPON was the accumulated intermediate produced by TYΔndpC, according to UV scanning (maximum absorbance at 289 nm, pH < 8) and LC-MS analysis. Additionally, TYΔndpC could convert nicotine, 6HN and HSP (Table 1). The phenotypic characteristics of TYΔndpC indicated that ndpC might be responsible for transforming 6HPON to HSP. Heterologous expression of ndpC in P. putida KT2440 provided negative results, whereas the transfer of Sphingomonas aquatilis JSS7T with ndpC provided the ability to transform 6HPON into HSP (Figure 6). However, it was considered that there was a 6-hydroxy-3-succinoylsemialdehyde-pyridine formed between 6HPON and HSP (Ma et al., 2013), but this dehydrogenation step may be performed by another non-specific semialdehyde dehydrogenase in strain.
Figure 6.
Intermediate accumulated by TYΔndpC and biotransformation of 6HPON by Sphingomonas-ndpC and KT-ndpC. (A) 6HPON with a maximum absorbance at 289 nm was accumulated by TYΔndpC based on UV scanning; (B,C) biotransformation of 6HPON by Sphingomonas-ndpC and KT-ndpC showed that no product was formed in strain KT-ndpC, whereas Sphingomonas-ndpC had catalytic activity based on the formation of 6-hydroxy-3-succinoyl-pyridine. LC-MS analysis of the product shown in (D).
NdpD is 6-hydroxy-3-succinoyl-pyridine 3-monooxygenase
The fourth enzymatic step in the VPP is the formation of 2, 5-dihydroxypyridine (2, 5-DHP), which is an intermediate that is generated during the degradation of many pyridine derivatives by aerobic microorganisms (Yao et al., 2013). The mutant strain TYΔndpD lost the ability to grow on nicotine, which was restored after gene complementation. A biotransformation test with TYΔndpD revealed an accumulation of the intermediate HSP, according to UV scanning with a maximum absorbance at 276 nm and LC-MS analysis. Additionally, it was anticipated that TYΔndpD could catalyze the transformations of nicotine, 6HN, and 6HPON (Table 1). Recombinant NdpD was primarily overexpressed in E. coli BL21(DE3) as a C-terminal His6-tagged fusion protein in pET-28a(+). A band at an apparent molecular mass of 45.1 kDa was detected by SDS-PAGE in the precipitate (data not shown), which corresponded to the molecular weight of His6-tagged NdpD. To express soluble NdpD, the N-terminal His6-tagged fusion protein in pET-28a(+) was selected for expression in E. coli BL21(DE3). Fortunately, a small amount of soluble NdpD protein in supernatant (data not shown), but no product formed during detection of the crude enzyme activity. However, heterologous expression of NdpD in P. putida KT2440 provided negative results, but the ability of NdpD to catalyze the transformation of HSP to 2, 5-DHP was successfully detected in Sphingomonas aquatilis JSS7T (Figure 7).
Figure 7.
Intermediate accumulated by TYΔndpD and biotransformation of 6-hydroxy-3-succinoyl-pyridine by Sphingomonas-ndpD and KT-ndpD. (A) 6-hydroxy-3-succinoyl-pyridine with a maximum absorbance at 276 nm was accumulated by TYΔndpD based on UV scanning; (B,C) biotransformation of 6-hydroxy-3-succinoyl-pyridine by Sphingomonas-ndpD and KT-ndpD showed that no product was formed in strain KT-ndpC, whereas Sphingomonas-ndpC displayed catalytic activity based on 2, 5-dihydroxy-pyridine formation. LC-MS analysis of the product shown in (D).
ndp has different genetic characteristics compared with vpp
The G+C content of nicotine-degrading gene clusters in TY, SJY1, and S33 compared with the G+C content of their genomes revealed no significant differences (Table 2). The RSCU analysis showed that strains SJY1 and S33 have nearly the same codon usage bias as well as %G3s+C3s for each corresponding nicotine-degrading gene, whereas strain TY has a distinct RSCU value and significantly higher %G3s+C3s (Figure 8, Table 2). A BLASTp search revealed that homologous genes of nicotine-degrading genes in strain TY are present in Sphingomonas sp. Ant20, Sphingobium xenophagum, Sphingobium chungbukense, Sphingobium sp. KK22, Sphingopyxis sp. H080, and other closely related species, all of which belong to the family Sphingomonadaceae. While homologous genes of nicotine-degrading genes in SJY1 and S33 are distributed in much more distantly related taxa, such as species of Sphingomonas and Sphingobium for strain SJY1 and species of Rhizobium, Shinella, Paramesorhizobium, Sphingopyxis, and Sphingomonas for strain S33, all the species that displayed similarity to strain SJY1 or S33 belonged to a different order or class. Several clusters of mobile element protein genes were found in nicotine-degrading genes or adjacent sequences of strain SJY1 and S33, but none were found in ndp. In addition to all the necessary nicotine metabolism genes, a transmembrane protein was encoded by ndp and was probably related to the transport of nicotine or other critical metabolite of nicotine degradation in strain TY. The mean ENC in TY was 35.67, and the mean ENC in ndp was 34.34. In general, based on the clustered genetic organization and integrity of ndp and considering the above results, ndp had different genetic characteristics compared with vpp, and vpp in strains SJY1 and S33 appeared to evolve from HGT. Nevertheless, these results didn't rule out the possibility that ndp was originated from HGT. On the contrary, from the results of RSCU analysis of 3867 genes in draft genome of strain TY (Figure S2), it seems that there was a considerable possibility that some genes in ndp were originated from HGT.
Table 2.
Comparison of the G+C content and nucleotide composition of the genome, gene cluster, and encoded genes.
| TY | %G+C | %G3s+C3s | SJY1 | %G+C | %G3s+C3s | S33 | %G+C | %G3s+C3s |
|---|---|---|---|---|---|---|---|---|
| Genome | 67.10 | / | Genome | 54.2 | Genome | 59.16 | / | |
| Gene cluster | 66 | / | Gene cluster | 55.70 | / | Gene cluster | 55 | / |
| ndpAL | 67.21 | 85.93 | vppAL | 55.46 | 60.9655 | ndhA | 55.28 | 60.68 |
| ndpAS | 64.90 | 87.5 | vppAS | 56.41 | 59.7315 | ndhB | 56.19 | 59.06 |
| ndpB | 66.39 | 88.18 | vppB | 53.34 | 56.5012 | hno | 53.42 | 56.97 |
| ndpC | 65.73 | 83.38 | vppCa | 53.22 | 50.463 | pno | 53.32 | 50.77 |
| ndpD | 65.89 | 87.63 | vppD | 53.74 | 56.3342 | hsh | 53.74 | 56.06 |
| ndpE | 65.90 | 85.49 | vppE | 58.79 | 73.27044 | S33vppE | 58.69 | 72.95 |
| ndpF | 67.51 | 83.26 | vppF | 64.62 | 71.6599 | S33vppF | 64.62 | 71.65 |
| ndpG | 66.82 | 82.5 | vppG | 55.76 | 55.3922 | S33vppG | 55.29 | 53.69 |
| ndpH | 65.95 | 84.61 | vppH | 62.73 | 77.1739 | S33vppH | 61.06 | 75.74 |
Gene name in gray text means they were putative by analysis this work.
Figure 8.
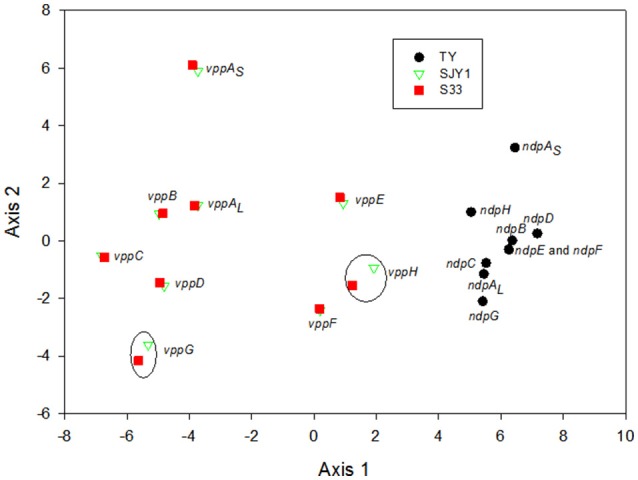
Principal component analysis of the codon usage of nicotine-catalyzing genes in strains TY, SJY1, and S33. Black dots, genes from strain TY; red squares, genes from strain S33; green triangles, genes from strain SJY1. Most of the homologous genes in SJY1 and S33 were overlapping, and the genes in black circles were homologous genes in strains SJY1 and S33. ndpE and ndpF almost overlapped with each other in strain TY.
Discussion
Microbial degradation plays an important role in the elimination of nicotine pollution in the environment. In this study, a novel nicotine degradation gene cluster, ndp, was identified in strain TY, which degraded nicotine efficiently via the VPP. The molecular mechanisms of nicotine degradation in this strain and the functions of four genes, ndpA, ndpB, ndpC, and ndpD, were studied. NdpA is responsible for the hydroxylation of nicotine to 6HN, NdpB catalyzes the conversion of 6HN to 6HMM, NdpC transforms 6HPON to HSP, and NdpD converts HSP to 2, 5-DHP. Elucidation of the molecular mechanism of nicotine degradation in strain TY provides a new resource for industrial applications and the management of nicotine-polluted environments.
Analysis based on vpp and ndp shows that the homologous genes of nicotine-degrading genes show a scattered phylogenetic distribution in strains SJY1 and S33, whereas the homologous genes of nicotine-degrading genes in TY are distributed all in closely related taxa and species. There were no distinct difference in G+C content between the nicotine-degrading gene cluster and the genome in strains S33 and SJY1; however, this may be due to genetic homogenization during long-term evolution following HGT of these genes. Moreover, there were no distinct differences in the G+C content of the genome and cluster in strain TY. The RSCU values are useful for comparing codon usage among genes or sets of genes (Andersson and Sharp, 1996). The RSCU value and percentage of G3s+C3s of nicotine-degrading genes in strains SJY1 and S33 were highly consistent, despite the very distant relationship of these strains. In contrast, the nicotine-degrading genes in strain TY had a remarkably different codon usage bias and nucleotide composition compared with strains SJY1 and S33. Additionally, the regions contiguous to the genes that were confirmed to be horizontally transferred were observed for the nicotine-degrading gene cluster in strains SJY1 and S33. However, there was no trace of mobile elements in ndp in strain TY, may be due to the deletion of intervening genes (such as mobile elements) that do not provide a selectable function in certain environments, according to the Selfish Operon Model of gene clustering in prokaryotes and eukaryotes, thus facilitating the evolution of clustered, energy efficient, dissemination (both by vertical transmission and by horizontal transfer) and functional gene clusters (Lawrence and Roth, 1996; Lawrence, 1999). Meanwhile, it may be due to the genome rearrangement of orthologous pathway (Periwal and Scaria, 2014).
Substrate transport-related transporter genes are often contiguous with catabolism genes in the bacterial genome to save energy and respond rapidly to environmental stress. For example, MhbT, a specific transporter for the uptake of 3-hydroxybenzoate in Klebsiella pneumoniae M5a1, is encoded by a cluster, mhbRTDHIM, containing enzymes that convert 3-hydroxybenzoate to pyruvate and fumarate via gentisate and a gene activator, mhbR (Xu et al., 2012). MhpT, a 3-(3-hydroxyphenyl) propionate (3HPP) transporter in Escherichia coli K-12, is encoded by cluster mhpRABCDFET, and the other genes in this cluster are responsible for regulatory functions and 3HPP catabolism (Ferrández et al., 1997; Torres et al., 2003; Xu et al., 2013). GabPCg, a γ-aminobutyric acid (GABA) transporter, is adjacent to succinic semialdehyde dehydrogenase (GabD) and GABA oxoglutarate aminotransferase (GabT) in the genome of Corynebacterium glutamicum (Zhao et al., 2012). Interestingly, we identified a transmembrane protein, NdpT, in ndp (Figure 1). Additionally, ndpT was tentatively proposed to encode a potential transporter involved in the uptake of nicotine, its metabolic intermediates, or both. Furthermore, this gene was absent in strains SJY1 and S33. Combining the scattered phylogenetic distribution of nicotine-degrading genes in strains SJY1 and S33, the concentrated structure of nicotine-degrading genes in strain TY and the above-described results, we considered that the vpp in strains SJY1 and S33 resulted directly from HGT and that ndp was the gene cluster result from genome rearrangement of orthologous pathways or from Selfish Operon Model, but with a high degree of homogenization. The specificity of the ndp gene cluster may explain why P. putida KT2440 is unable to express active nicotine-degrading genes in strain TY, in contrast to Sphingomonas aquatilis JSS7T. However, VppA can be successfully expressed in P. putida KT2440 (Yu et al., 2015), further supporting the differences between ndp and vpp clusters.
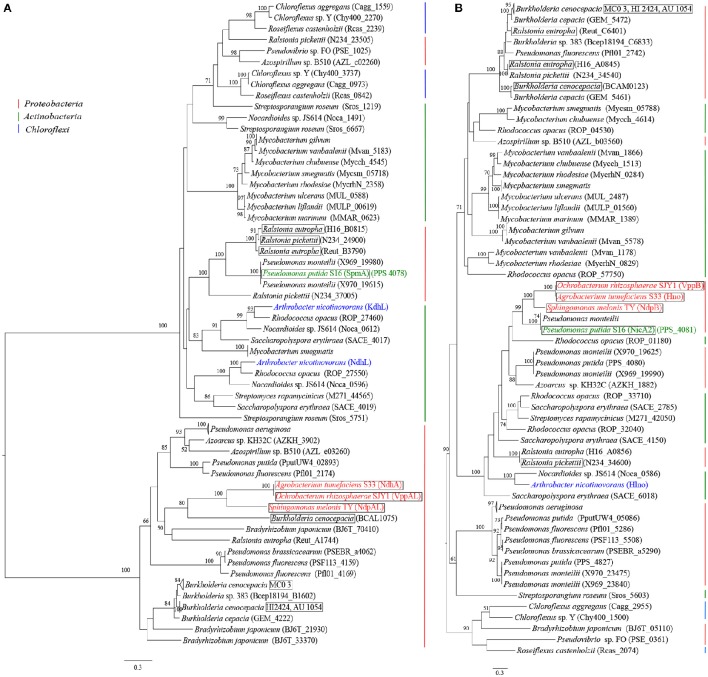
The evolutionary relationships analysis showed that the nicotine hydroxylase large subunit (NdpAL) in the VPP, 3-succinoylpyridine monooxygenase alpha subunit (SpmA) in the PRL, and the nicotine dehydrogenase (NdhL) and ketone dehydrogenase (KdhL) in the PD belonged to the same protein family (Figure 9). Evidently, the phylogenies of the NdpAL and NdpB proteins were completely different (Figure 9). This result indicated that although they were recruited in the same metabolic pathway, they had different evolutionary patterns. In particular, NdpAL resided in two clear phylogenetic clades. Interestingly, although the VPP and PRL pathways were more similar in terms of metabolic mechanisms, NdpAL and SpmA, respectively, belonged to different protein subfamilies with potentially different origins that had undergone convergent evolution. However, 6-hydroxy-L-nicotine oxidase, NdpB, and NicA2 (PRL) had relative closer evolutionary relationships. According to the patchwork hypothesis, the ancient gene could have encoded a primitive enzyme with low substrate specificity to catalyze distinct but similar reactions, and it was recruited into various biological pathways (Jensen, 1976; Lazcano and Miller, 1996). Our results based on the phylogenies of the NdpAL and NdpB proteins demonstrated that the VPP and PRL pathways recruited different genotypes. We also found that the ndpHFEG genes were linked in some strains (Figure 9). The linkages among these four genes might represent a common genetic structure in the VPP and PRL pathways.
Figure 9.
Phylogenetic trees of the homologs of the hydroxylase of the pyridine ring and dehydrogenase of the pyrrolidine ring. (A) Phylogenetic tree of the homologs of the hydroxylase of the pyridine ring; (B) Phylogenetic tree of the homologs of the dehydrogenase of the pyrrolidine ring. The trees were constructed using homologs of the corresponding enzymes from 50 bacterial genomes (the information of these homologs is listed in Table S4). The colored text denotes reported nicotine-degrading strains, and the strains marked by a square frame have linked ndpHFEG.
In the PD, we found that KdhL and NdhL had a closer evolutionary relationship and that these proteins can also be identified in other closely related species such as Rhodococcus opacus, Nocardioides sp. JS614 and Saccharopolyspora erythraea. We thought that the PD would be distributed in Gram-positive bacteria and that the VPP and PRL pathways would be distributed in Gram-negative bacteria (Table S3). To further study this hypothesis, we evaluated the gram staining of the strains with of ndpHFEG. As expected, the linked homologs of ndpHFEG identified by BLAST searches against 2806 predicted prokaryotic proteomes were in Gram-negative strains (Table S3). Moreover, one of the three degradation pathways of nicotinic acid gone through 2, 5-DHP was only found in Gram-negative strains (Table S3). Surprisingly, we found that Streptomyces rapamycinicus might have an entire nicotine-degrading pathway that is not a PD. Although S. rapamycinicus only had linked ndpHFG genes, one protein exhibited 34.02% similarity to the NdpE of S. melonis TY. However, further experimental evidence is needed to support this result. In summary, in terms of both evolution and metabolic mechanism, the VPP was more similar to the PRL. The linkage of ndpHFEG genes shared by both pathways indicated that these two pathways might have the same origin; however, variants occurred in some bacteria.
In addition to the common reactions of hydroxylation at C6 of the pyridine ring and dehydrogenation at the C2–C3 bond of the pyrrolidine ring, there are three other similar enzyme-catalyzed reactions (cleavage of the pyrrolidine residue in the pyridine ring, deamination and dehydrogenation of the pyrrolidine residue) and one step of autohydrolysis of the pyrrolidine ring that are, in fact, shared by the VPP, PRL, and PD pathways (Wang et al., 2012). Moreover, the codon usage of representative strains of the VPP, PRL, and PD pathways was distinct (Figure S1). We speculated that a series of ancient genes could have encoded primitive enzymes with low substrate specificity to catalyze distinct but similar reactions, and these genes were recruited to various biological pathways to form distinct pathways over long-term evolution, according to the patchwork hypothesis (Jensen, 1976; Lazcano and Miller, 1996). Together with the analysis of evolution and relationship of the metabolic mechanisms of the VPP and PRL pathways described above, it was reasonable to consider that the VPP, PRL, and PD pathways may have experienced independent but interrelated evolutionary events.
The hypothesis that the PD is distributed in Gram-positive bacteria and that the VPP and PRL pathway is distributed in Gram-negative bacteria was based on current research and was an innovative inference. This hypothesis can be verified by a simple experiment that is theoretically feasible. The representative intermediate product of the VPP and PRL pathways can be used as a sole carbon and/or nitrogen source to screen Gram-positive bacterial growth, and conversely, we can screen the growth of Gram-negative bacteria on the pyridine pathway-specific metabolite. After analyzing the intermediates produced in the pyridine, VPP and PRL pathways, we considered selecting 2, 5-DHP as the representative metabolic product in the VPP and PRL pathways and 2, 6-dihydroxy-pseudohydroxypyridine as the product in the PD. The 2, 5-DHP is an intermediate product that is shared by the nicotine and nicotinic acid degradation pathways (Jiménez et al., 2008), and all the strains collected using the 2, 5-DHP pathway were Gram-negative, supporting our speculation. A second strategy to support or dismiss our hypothesis is to examine whether certain homologs of ndpHFEG are active in specific Gram-positive strains. If these homologs become active, then our hypothesis is incorrect, and natural Gram-positive strain that utilize the VPP or PRL pathway will eventually be obtained. Conversely, our hypothesis is reasonable and has research value. Of course, the Gram-positive strain S. rapamycinicus, which carries the homologs of ndpAL, ndpB, ndpE, and ndpHFG, can be assessed for nicotine degradation ability and the associated pathway. Strain, Pusillimonas sp. T2, which contained nicotine metabolites from both the VPP and PD pathways (2, 6-dihydroxypyridine), may be explained by the substrate ambiguity of nicotine hydroxylase. After hydroxylation of C6 of the pyridine ring, some of the 6HPON intermediate was hydroxylated again at C2 of pyridine to generate 2, 6-dihydroxy-pseudooxynicotine, which was then hydroxylated by a non-specific C-C hydrolase to produce 2, 6-dihydroxypyridine. Otherwise, the portion of the vpp cluster in Pusillimonas sp. T2 was different with what we were discussed about. Four nicotine degradation intermediates produced by Gram-negative Achromobacter nicotinophagum have been mentioned above and are related to the PD (6HN), PRL (pseudooxynicotine, 3-succinoyl-pyridine and HSP), and VPP (6HN and HSP). We thought that this strain might contain a complete PRL and that due to the substrate ambiguity of the hydroxylase of pyridine ring, the strain might be capable of forming 6HN. On the other hand, this strain may contain both the PD and the PRL pathway.
In summary, we discovered a new nicotine degradation gene cluster, ndp, and characterized four genes that catalyze the first four enzymatic steps in the VPP in S. melonis TY. These results provide relatively comprehensive evidence for the molecular mechanism of nicotine degradation in S. melonis TY. We also formulated an inference that both the evolutionary features and metabolic mechanisms of the VPP were more similar to the PRL. These findings provide a deeper understanding of the evolution of nicotine metabolism in Sphingomonas. The hypothesis that the PD is distributed in Gram-positive bacteria while the VPP and PRL pathways are distributed in Gram-negative bacteria based on the result of comprehensive homologs searching and phylogenetic tree construction and that the evolutionary relationships among the VPP, PRL and PD pathways will provide critical information that will improve our understanding of the evolution of nicotine-degrading gene clusters.
Ethics statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Author contributions
Performed experiments: HW and XZ. Analyzed data: HW, XZ, JQ, and LS. Conceived and designed experiments, wrote the paper and approved the final manuscript: All authors.
Funding
This work was financially supported by the Natural Science Foundation for Distinguished Young Scholars of Zhejiang Province (LR14D030001), and the National Natural Science Foundation of China (No. 31170115 and 31422003).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Professor Ning-Yi Zhou from Shanghai Jiao Tong University for experimental suggestion. We also thank Professor Min Wu and Yuhua Zhao from Zhejiang University for providing several strains and plasmids used in this study.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00337/full#supplementary-material
References
- Andersson G. E., Sharp P. M. (1996). Codon usage in the Mycobacterium tuberculosis complex. Microbiology 142, 915–925. 10.1099/00221287-142-4-915 [DOI] [PubMed] [Google Scholar]
- Aslund F., Bessette P. H., Georgiou G., Beckwith J. (1999). Efficient production of disulfide bonded proteins in the cytoplasm in “oxidizing” mutants of E. coli. Innovations 10, 11–12. [Google Scholar]
- Ausbel F., Brent R., Kingston R., Moore D., Seidman J., Smith J., et al. (1995). Short Protocols in Molecular Biology. New York, NY: JohnWiley. [Google Scholar]
- Baitsch D., Sandu C., Brandsch R., Igloi G. L. (2001). Gene cluster on pAO1 of Arthrobacter nicotinovorans involved in degradation of the plant alkaloid nicotine: cloning, purification, and characterization of 2,6-dihydroxypyridine 3-hydroxylase. J. Bacteriol. 183, 5262–5267. 10.1128/JB.183.18.5262-5267.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandsch R. (2006). Microbiology and biochemistry of nicotine degradation. Appl. Microbiol. Biotechnol. 69, 493–498. 10.1007/s00253-005-0226-0 [DOI] [PubMed] [Google Scholar]
- Brühmüller M., Möhler H., Decker K. (1972). Covalently bound flavin in d-6-hydroxynicotine oxidase from Arthrobacter oxidans. Eur. J. Biochem. 29, 143–151. 10.1111/j.1432-1033.1972.tb01968.x [DOI] [PubMed] [Google Scholar]
- Chen Y. F., Chao H., Zhou N. Y. (2014). The catabolism of 2,4-xylenol and p-cresol share the enzymes for the oxidation of para-methyl group in Pseudomonas putida NCIMB 9866. Appl. Microbiol. Biotechnol. 98, 1349–1356. 10.1007/s00253-013-5001-z [DOI] [PubMed] [Google Scholar]
- Chiribau C. B., Mihasan M., Ganas P., Igloi G. L., Artenie V., Brandsch R. (2006). Final steps in the catabolism of nicotine. FEBS J. 273, 1528–1536. 10.1111/j.1742-4658.2006.05173.x [DOI] [PubMed] [Google Scholar]
- Chiribau C. B., Sandu C., Fraaije M., Schiltz E., Brandsch R. (2004). A novel gamma-N-methylaminobutyrate demethylating oxidase involved in catabolism of the tobacco alkaloid nicotine by Arthrobacter nicotinovorans pAO1. Eur. J. Biochem. 271, 4677–4684. 10.1111/j.1432-1033.2004.04432.x [DOI] [PubMed] [Google Scholar]
- Civilini M., Domenis C., Sebastianutto N., de Bertoldi M. (1997). Nicotine decontamination of tobacco agro-industrial waste and its degradation by micro-organisms. Waste Manage. Res. 15, 349–358. 10.1177/0734242X9701500403 [DOI] [Google Scholar]
- Cobzaru C., Ganas P., Mihasan M., Schleberger P., Brandsch R. (2011). Homologous gene clusters of nicotine catabolism, including a new ω-amidase for α-ketoglutaramate, in species of three genera of Gram-positive bacteria. Res. Microbiol. 162, 285–291. 10.1016/j.resmic.2011.01.001 [DOI] [PubMed] [Google Scholar]
- Cosic I., Vukovic M., Gomzi Z., Briski F. (2012). Comparison of various kinetic models for batch biodegradation of leachate from tobacco waste composting. Rev. Chim. 63, 967–971. [Google Scholar]
- Dang V. D., Decker K., Sund H. (1968). Purification and properties of l-6-hydroxynicotine oxidase. Eur. J. Biochem. 4, 95–102. 10.1111/j.1432-1033.1968.tb00177.x [DOI] [PubMed] [Google Scholar]
- Dehio C., Meyer M. (1997). Maintenance of broad-host-range incompatibility group P and group Q plasmids and transposition of Tn5 in Bartonella henselae following conjugal plasmid transfer from Escherichia coli. J. Bacteriol. 179, 538–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeTraglia M. C., Tometsko A. M. (1980). Separation of d-(+)-nicotine from a racemic mixture by stereospecific degradation of the l-(−) isomer with Pseudomonas putida. Appl. Environ. Microbiol. 39, 1067–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. 10.2307/2408678 [DOI] [PubMed] [Google Scholar]
- Ferrández A., Garciá J. L., Díaz E. (1997). Genetic characterization and expression in heterologous hosts of the 3-(3-hydroxyphenyl)propionate catabolic pathway of Escherichia coli K-12. J. Bacteriol. 179, 2573–2581. 10.1128/jb.179.8.2573-2581.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganas P., Sachelaru P., Mihasan M., Igloi G., Brandsch R. (2008). Two closely related pathways of nicotine catabolism in Arthrobacter nicotinovorans and Nocardioides sp. strain JS614. Arch. Microbiol. 189, 511–517. 10.1007/s00203-007-0340-8 [DOI] [PubMed] [Google Scholar]
- Goetz A. E., Garg N. K. (2013). Regioselective reactions of 3,4-pyridynes enabled by the aryne distortion model. Nat. Chem. 5, 54–60. 10.1038/nchem.1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M., Guindon S., Gascuel O. (2010). SeaView Version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27, 221–224. 10.1093/molbev/msp259 [DOI] [PubMed] [Google Scholar]
- Gray M. W. (1992). The endosymbiont hypothesis revisited. Int. Rev. Cytol. 141, 233–357. 10.1016/S0074-7696(08)62068-9 [DOI] [PubMed] [Google Scholar]
- Grether-Beck S., Igloi G. L., Pust S., Schilz E., Decker K., Brandsch R. (1994). Structural analysis and molybdenum-dependent expression of the pAO1-encoded nicotine dehydrogenase genes of Arthrobacter nicotinovorans. Mol. Microbiol. 13, 929–936. 10.1111/j.1365-2958.1994.tb00484.x [DOI] [PubMed] [Google Scholar]
- Gurusamy R., Natarajan S. (2013). Current status on biochemistry and molecular biology of microbial degradation of nicotine. Sci. World J. 2013:125385. 10.1155/2013/125385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood C. S., Nichols N. N., Kim M. K., Ditty J. L., Parales R. E. (1994). Identification of the pcaRKF gene cluster from Pseudomonas putida: involvement in chemotaxis, biodegradation, and transport of 4-hydroxybenzoate. J. Bacteriol. 176, 6479–6488. 10.1128/jb.176.21.6479-6488.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstein L. I., Rittenberg S. C. (1959a). The bacterial oxidation of nicotine: I. nicotine oxidation by cell-free preparations. J. Biol. Chem. 234, 151–155. [PubMed] [Google Scholar]
- Hochstein L. I., Rittenberg S. C. (1959b). The bacterial oxidation of nicotine: II. the isolation of the first oxidative product and its identification as (l)-6-hydroxynicotine. J. Biol. Chem. 234, 156–160. [PubMed] [Google Scholar]
- Hylin J. W. (1958). Microbial degradation of nicotine I. Morphology and physiology of Achromobacter nicotinophagumn sp. J. Bacteriol. 76, 36–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hylin J. W. (1959). The microbial degradation of nicotine. II. The mode of action of Achromobacter nicotinophagum. Arch. Biochem. Biophys. 83, 528–537. 10.1016/0003-9861(59)90061-X [DOI] [PubMed] [Google Scholar]
- Jensen R. A. (1976). Enzyme recruitment in evolution of new function. Annu. Rev. Microbiol. 30, 409–425. 10.1146/annurev.mi.30.100176.002205 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Tang H., Wu G., Xu P. (2015). Functional identification of a novel gene, moaE, for 3-succinoylpyridine degradation in Pseudomonas putida S16. Sci. Rep. 5:13464. 10.1038/srep13464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez J. I., Canales A., Jiménez-Barbero J., Ginalski K., Rychlewski L., García J. L., et al. (2008). Deciphering the genetic determinants for aerobic nicotinic acid degradation: the nic cluster from Pseudomonas putida KT2440. Proc. Natl. Acad. Sci. U.S.A. 105, 11329–11334. 10.1073/pnas.0802273105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S., Campbell A. M., Mrazek J. (1998). Comparative DNA analysis across diverse genomes. Annu. Rev. Genet. 32, 185–225. 10.1146/annurev.genet.32.1.185 [DOI] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Lawrence J. (1999). Selfish operons: the evolutionary impact of gene clustering in prokaryotes and eukaryotes. Curr. Opin. Genet. Dev. 9, 642–648. 10.1016/S0959-437X(99)00025-8 [DOI] [PubMed] [Google Scholar]
- Lawrence J. G., Ochman H. (1998). Molecular archaeology of the Escherichia coli genome. Proc. Natl. Acad. Sci. U.S.A. 95, 9413–9417. 10.1073/pnas.95.16.9413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence J. G., Roth J. R. (1996). Selfish operons: horizontal transfer may drive the evolution of gene clusters. Genetics 143, 1843–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazcano A., Miller S. L. (1996). The origin and early evolution of life: prebiotic chemistry, the pre-RNA world, and time. Cell 85, 793–798. 10.1016/S0092-8674(00)81263-5 [DOI] [PubMed] [Google Scholar]
- Le S. Q., Gascuel O. (2008). An improved general amino acid replacement matrix. Mol. Biol. Evol. 25, 1307–1320. 10.1093/molbev/msn067 [DOI] [PubMed] [Google Scholar]
- Lemay S., Chouinard S., Blanchet P., Masson H., Soland V., Beuter A., et al. (2004). Lack of efficacy of a nicotine transdermal treatment on motor and cognitive deficits in Parkinson's disease. Prog. Neuropsychopharmacol. Biol. Psychiatry 28, 31–39. 10.1016/S0278-5846(03)00172-6 [DOI] [PubMed] [Google Scholar]
- Li H., Xie K., Yu W., Hu L., Huang H., Xie H., et al. (2016). Nicotine dehydrogenase complexed with 6-hydroxypseudooxynicotine oxidase involved in the hybrid nicotine-degrading pathway in Agrobacterium tumefaciens S33. Appl. Environ. Microbiol. 82, 1745–1755. 10.1128/AEM.03909-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Ma Y., Wei Y., Qiu J., Wen R., Hong J., Liu W. (2013). Isolation, transposon mutagenesis, and characterization of the novel nicotine-degrading strain Shinella sp. HZN7. Appl. Microbiol. Biotechnol. 98, 2625–2636. 10.1007/s00253-013-5207-0 [DOI] [PubMed] [Google Scholar]
- Ma Y., Wen R., Qiu J., Hong J., Liu M., Zhang D. (2014). Biodegradation of nicotine by a novel strain Pusillimonas. Res. Microbiol. 166, 67–71. 10.1016/j.resmic.2014.12.009 [DOI] [PubMed] [Google Scholar]
- Meng X. J., Lu L. L., Gu G. F., Xiao M. (2010). A novel pathway for nicotine degradation by Aspergillus oryzae 112822 isolated from tobacco leaves. Res. Microbiol. 161, 626–633. 10.1016/j.resmic.2010.05.017 [DOI] [PubMed] [Google Scholar]
- Mihasan M., Chiribau C. B., Friedrich T., Artenie V., Brandsch R. (2007). An NAD(P)H-nicotine blue oxidoreductase is part of the nicotine regulon and may protect Arthrobacter nicotinovorans from oxidative stress during nicotine catabolism. Appl. Environ. Microbiol. 73, 2479–2485. 10.1128/AEM.02668-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A., Osawa S. (1987). The guanine and cytosine content of genomic DNA and bacterial evolution. Proc. Natl. Acad. Sci. U.S.A. 84, 166–169. 10.1073/pnas.84.1.166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotny T. E., Zhao F. (1999). Consumption and production waste: another externality of tobacco use. Tob. Control 8, 75–80. 10.1136/tc.8.1.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H., Lawrence J. G., Groisman E. A. (2000). Lateral gene transfer and the nature of bacterial innovation. Nature 405, 299–304. 10.1038/35012500 [DOI] [PubMed] [Google Scholar]
- Oldendorf W., Braun L., Cornford E. (1979). pH dependence of blood-brain barrier permeability to lactate and nicotine. Stroke 10, 577–581. 10.1161/01.STR.10.5.577 [DOI] [PubMed] [Google Scholar]
- Oldendorf W. H. (1974). Lipid solubility and drug penetration of the blood brain barrier. Exp. Biol. Med. 147, 813–816. 10.3181/00379727-147-38444 [DOI] [PubMed] [Google Scholar]
- O'neill M., Murray T., Lakics V., Visanji N., Duty S. (2002). The role of neuronal nicotinic acetylcholine receptors in acute and chronic neurodegeneration. Curr. Drug Targets CNS Neurol. Disord. 1, 399–411. 10.2174/1568007023339166 [DOI] [PubMed] [Google Scholar]
- Periwal V., Scaria V. (2014). Insights into structural variations and genome rearrangements in prokaryotic genomes. Bioinformatics 31, 1–9. 10.1093/bioinformatics/btu600 [DOI] [PubMed] [Google Scholar]
- Prinz W. A., Aslund F., Holmgren A., Beckwith J. (1997). The role of the thioredoxin and glutaredoxin pathways in reducing protein disulfide bonds in the Escherichia coli cytoplasm. J. Biol. Chem. 272, 15661–15667. 10.1074/jbc.272.25.15661 [DOI] [PubMed] [Google Scholar]
- Puigbò P., Bravo I. G., Garcia-Vallve S. (2008). CAIcal: a combined set of tools to assess codon usage adaptation. Biol. Direct 3:38. 10.1186/1745-6150-3-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J., Ma Y., Chen L., Wu L., Wen Y., Liu W. (2011). A sirA-like gene, sirA2, is essential for 3-succinoyl-pyridine metabolism in the newly isolated nicotine-degrading Pseudomonas sp. HZN6 strain. Appl. Microbiol. Biotechnol. 92, 1023–1032. 10.1007/s00253-011-3353-9 [DOI] [PubMed] [Google Scholar]
- Ramakers C., Ruijter J. M., Deprez R. H., Moorman A. F. (2003). Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339, 62–66. 10.1016/S0304-3940(02)01423-4 [DOI] [PubMed] [Google Scholar]
- Raman G., Mohan K., Manohar V., Sakthivel N. (2013). Biodegradation of nicotine by a novel nicotine-degrading bacterium, Pseudomonas plecoglossicida TND35 and its new biotransformation intermediates. Biodegradation 25, 95–107. 10.1007/s10532-013-9643-4 [DOI] [PubMed] [Google Scholar]
- Roduit J. P., Wellig A., Kiener A. (1997). Renewable functionalized pyridines derived from microbial metabolites of the alkaloid (S)-nicotine. Heterocycles 9, 1687–1702. [Google Scholar]
- Ruan A., Min H., Peng X., Huang Z. (2005). Isolation and characterization of Pseudomonas sp. strain HF-1, capable of degrading nicotine. Res. Microbiol. 156, 700–706. 10.1016/j.resmic.2005.02.010 [DOI] [PubMed] [Google Scholar]
- Ruijter J. M., Ramakers C., Hoogaars W. M. H., Karlen Y., Bakker O., van den Hoff M. J., et al. (2009). Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 37:e45. 10.1093/nar/gkp045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachelaru P., Schiltz E., Brandsch R. (2006). A functional mobA gene for molybdopterin cytosine dinucleotide cofactor biosynthesis is required for activity and holoenzyme assembly of the heterotrimeric nicotine dehydrogenases of Arthrobacter nicotinovorans. Appl. Environ. Microbiol. 72, 5126–5131. 10.1128/AEM.00437-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachelaru P., Schiltz E., Igloi G. L., Brandsch R. (2005). An alpha/beta-fold C–C bond hydrolase is involved in a central step of nicotine catabolism by Arthrobacter nicotinovorans. J. Bacteriol. 187, 8516–8519. 10.1128/JB.187.24.8516-8519.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltikov C. W., Newman D. K. (2003). Genetic identification of a respiratory arsenate reductase. Proc. Natl. Acad. Sci. U.S.A. 100, 10983–10988. 10.1073/pnas.1834303100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk S., Hoelz A., Krauss B., Decker K. (1998). Gene structures and properties of enzymes of the plasmid-encoded nicotine catabolism of Arthrobacter nicotinovorans. J. Mol. Biol. 284, 1323–1339. 10.1006/jmbi.1998.2227 [DOI] [PubMed] [Google Scholar]
- Sindelar R. D., Rosazza J. P., Barfknecht C. F. (1979). N-demethylation of nicotine and reduction of nicotine-1′-N-oxide by Microsporum gypseum. Appl. Environ. Microbiol. 38, 836–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Sueoka N. (1962). On the genetic basis of variation and heterogeneity of DNA base composition. Proc. Natl. Acad. Sci. U.S.A. 48:582. 10.1073/pnas.48.4.582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Wang L., Meng X., Ma L., Wang S., He X., et al. (2009). Novel nicotine oxidoreductase-encoding gene involved in nicotine degradation by Pseudomonas putida strain S16. Appl. Environ. Microbiol. 75, 772–778. 10.1128/AEM.02300-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Wang L., Wang W., Yu H., Zhang K., Yao Y., et al. (2013). Systematic unraveling of the unsolved pathway of nicotine degradation in Pseudomonas. PLoS Genet. 9:e1003923. 10.1371/journal.pgen.1003923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Wang S., Ma L., Meng X., Deng Z., Zhang D., et al. (2008). A novel gene, encoding 6-hydroxy-3-succinoylpyridine hydroxylase, involved in nicotine degradation by Pseudomonas putida strain S16. Appl. Environ. Microbiol. 74, 1567–1574. 10.1128/AEM.02529-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Yao Y., Wang L., Yu H., Ren Y., Wu G., et al. (2012). Genomic analysis of Pseudomonas putida: genes in a genome island are crucial for nicotine degradation. Sci. Rep. 2:377. 10.1038/srep00377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Yao Y., Zhang D., Meng X., Wang L., Yu H., et al. (2011). A novel NADH-dependent and FAD-containing hydroxylase is crucial for nicotine degradation by Pseudomonas putida. J. Biol. Chem. 286, 39179–39187. 10.1074/jbc.M111.283929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres B., Porras G., García J. L., Díaz E. (2003). Regulation of the mhp cluster responsible for 3-(3-hydroxyphenyl)propionic acid degradation in Escherichia coli. J. Biol. Chem. 278, 27575–27585. 10.1074/jbc.M303245200 [DOI] [PubMed] [Google Scholar]
- Tuomi J. M., Voorbraak F., Jones D. L., Ruijter J. M. (2010). Bias in the Cq value observed with hydrolysis probe based quantitative PCR can be corrected with the estimated PCR efficiency value. Methods 50, 313–322. 10.1016/j.ymeth.2010.02.003 [DOI] [PubMed] [Google Scholar]
- Uchida S., Maeda S., Kisaki T. (1983). Conversion of nicotine into nornicotine and N-methylmyosmine by fungi. Agric. Biol. Chem. 47, 1949–1953. [Google Scholar]
- Wada E., Yamasaki K. (1953). Mechanism of microbial degradation of nicotine. Science 117, 152–153. 10.1126/science.117.3033.152 [DOI] [PubMed] [Google Scholar]
- Wang M., Yang G., Wang X., Yao Y., Min H., Lu Z. (2011). Nicotine degradation by two novel bacterial isolates of Acinetobacter sp. TW and Sphingomonas sp. TY and their responses in the presence of neonicotinoid insecticides. World J. Microbiol. Biotechnol. 27, 1633–1640. 10.1007/s11274-010-0617-y [DOI] [Google Scholar]
- Wang S., Huang H., Xie K., Xu P. (2012). Identification of nicotine biotransformation intermediates by Agrobacterium tumefaciens strain S33 suggests a novel nicotine degradation pathway. Appl. Microbiol. Biotechnol. 95, 1567–1578. 10.1007/s00253-012-4007-2 [DOI] [PubMed] [Google Scholar]
- Wang S. N., Liu Z., Xu P. (2009). Biodegradation of nicotine by a newly isolated Agrobacterium sp. strain S33. J. Appl. Microbiol. 107, 838–847. 10.1111/j.1365-2672.2009.04259.x [DOI] [PubMed] [Google Scholar]
- Wang S. N., Xu P., Tang H. Z., Meng J., Liu X. L., Huang J., et al. (2004). Biodegradation and detoxification of nicotine in tobacco solid waste by a Pseudomonas sp. Biotechnol. Lett. 26, 1493–1496. 10.1023/B:BILE.0000044450.16235.65 [DOI] [PubMed] [Google Scholar]
- Wang S. N., Xu P., Tang H. Z., Meng J., Liu X. L., Ma C. Q. (2005). “Green” route to 6-hydroxy-3-succinoyl-pyridine from (S)-nicotine of tobacco waste by whole cells of a Pseudomonas sp. Environ. Sci. Technol. 39, 6877–6880. 10.1021/es0500759 [DOI] [PubMed] [Google Scholar]
- Xu Y., Chen B., Chao H., Zhou N. Y. (2013). mhpT encodes an active transporter involved in 3-(3-hydroxyphenyl) propionate catabolism by Escherichia coli K-12. Appl. Environ. Microbiol. 79, 6362–6368. 10.1128/AEM.02110-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Gao X., Wang S. H., Liu H., Williams P. A., Zhou N. Y. (2012). MhbT is a specific transporter for 3-hydroxybenzoate uptake by Gram-negative bacteria. Appl. Environ. Microbiol. 78, 6113–6120. 10.1128/AEM.01511-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., Tang H., Ren H., Yu H., Wang L., Xu P. (2012). Genome sequence of a nicotine-degrading strain of Arthrobacter. J. Bacteriol. 194, 5714–5715. 10.1128/JB.01370-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., Tang H., Ren H., Yu H., Wang L., Zhang W., et al. (2013). Iron (II)-dependent dioxygenase and N-formylamide deformylase catalyze the reactions from 5-hydroxy-2-pyridone to maleamate. Sci. Rep. 3:3235. 10.1038/srep03235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., Tang H., Su F., Xu P. (2015). Comparative genome analysis reveals the molecular basis of nicotine degradation and survival capacities of Arthrobacter. Sci. Rep. 5:8642. 10.1038/srep08642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Tang H., Li Y., Xu P. (2015). Molybdenum-containing nicotine hydroxylase genes in a nicotine degradation pathway that is a variant of the pyridine and pyrrolidine pathways. Appl. Environ. Microbiol. 81, 8330–8338. 10.1128/AEM.02253-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Tang H., Zhu X., Li Y., Xu P. (2014). Molecular mechanism of nicotine degradation by a newly isolated strain Ochrobactrum sp. SJY1. Appl. Environ. Microbiol. 81, 272–281. 10.1128/AEM.02265-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Ding J. Y., Ma W. H., Zhou N. Y., Liu S. J. (2012). Identification and characterization of γ-aminobutyric acid uptake system GabPCg (NCgl0464) in Corynebacterium glutamicum. Appl. Environ. Microbiol. 78, 2596–2601. 10.1128/AEM.07406-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.