Abstract
Introduction:
Streptococcus suis serotype 2 is an important swine pathogen and emerging zoonotic agent causing meningitis and septicemia/septic shock. Strains are usually virulent (Eurasia) or of intermediate/low virulence (North America). Very few data regarding human and swine isolates from South America are available.
Case presentation:
Seventeen new human S. suis cases in Argentina (16 serotype 2 strains and a serotype 5 strain) are reported. Alongside, 14 isolates from pigs are analyzed: 12 from systemic disease, one from lungs and one from tonsils of a healthy animal. All human serotype 2 strains and most swine isolates are sequence type (ST) 1, as determined by multilocus sequence typing and present a mrp+/epf+/sly+ genotype typical of virulent Eurasian ST1 strains. The remaining two strains (recovered from swine lungs and tonsils) are ST28 and possess a mrp+/epf−/sly− genotype typical of low virulence North American strains. Representative human ST1 strains as well as one swine ST28 strain were analyzed by whole-genome sequencing and compared with genomes from GenBank. ST1 strains clustered together with three strains from Vietnam and this cluster is close to another one composed of 11 strains from the United Kingdom.
Conclusion:
Close contact with pigs/pork products, a good surveillance system, and the presence of potentially virulent Eurasian-like serotype 2 strains in Argentina may be an important factor contributing to the higher number of human cases observed. In fact, Argentina is now fifth among Western countries regarding the number of reported human cases after the Netherlands, France, the UK and Poland.
Keywords: Streptococcus suis serotype 2, swine, zoonosis, virulence factors, multilocus sequence typing, sequencing
Introduction
Streptococcus suis is an important swine pathogen mainly causing septicemia, meningitis and arthritis (Gottschalk, 2012). In addition, it is an emerging zoonotic agent responsible for septicemia with or without septic shock and meningitis (Gottschalk, 2012). During the last decade, the number of human cases due to S. suis has increased, and while most sporadic human cases of infection occur following close occupational contact with pigs/pork products, particularly in Western countries, important outbreaks have been recorded in Asia. In the latter, the general population is at risk due to consumption of raw pork products as part of traditional dishes. Indeed, S. suis infections are considered to be among the most frequent causes of adult meningitis in Asia (Wertheim et al., 2009).
Although different serotypes have been described, serotype 2 is the most commonly isolated from diseased pigs; it also represents more than 95 % of human cases worldwide (Goyette-Desjardins et al., 2014). However, important differences in virulence of isolates within this serotype have been described (Auger et al., 2016). It has been shown that Eurasian strains generally present a higher virulence potential than those from Canada and the USA, two countries that together are the second most important swine producers worldwide after China (Gottschalk, 2012).
Multilocus sequence typing (MLST) has been used worldwide to determine the sequence types (STs) of S. suis strains, thus allowing the gathering of further information regarding their genetic diversity and evolution (Goyette-Desjardins et al., 2014). More recently, studies have begun combining data obtained from MLST with the presence or absence of different S. suis serotype 2 virulence-associated markers such as muramidase released protein (mrp), extracellular protein factor (epf), and suilysin (sly), to compare ST data with pathotypes. ST1 strains, which are usually more virulent and possess a genotypic mrp+/epf+/sly+pathotype profile, are mostly present in Eurasia, whereas ST25 (intermediate virulence) and ST28 (low virulence) strains with a mrp−/epf+/−/sly− profile predominate in North America (Goyette-Desjardins et al., 2014; Lopreto et al., 2005). Interestingly, human cases reported in North America represent less than 0.5 % of described cases worldwide, compared with more than 8 % in European countries, confirming differences in virulence of S. suis serotype 2 isolates (Goyette-Desjardins et al., 2014; Auger et al., 2016). Very few data are available concerning serotype 2 strains recovered from diseased pigs in South America (Doto et al., 2016; Martinez et al., 2003), where the swine industry is either well-developed (such as in Brazil) or in clear expansion (such as in Argentina).
Herein, we describe new human cases of S. suis recovered from diseased patients in Argentina. We have also studied strains isolated from diseased pigs in this country. MLST and pathotype characteristics of strains were determined and representative strains recovered from humans were further analyzed by whole-genome sequencing (WGS).
Case Report
We present the characteristics of 32 S. suis strains isolated in Argentina (Table 1). We report and analyze 17 human cases of S. suis in Argentina isolated between 1995 and 2016. An additional strain from a previously reported human patient (Lopreto et al., 2005) was also included. All but one of the human strains were isolated from cases of meningitis; the remaining strain was recovered from a patient presenting septic arthritis. Of the 17 cases of meningitis, 7 strains were isolated exclusively from the cerebrospinal fluid (CSF), while the remaining strains were isolated from both CSF and blood. In the latter cases, only one isolate per patient was further characterized. Three isolates were recovered from female patients and 13 patients, from rural areas, recalled having had contact with swine, pork or pork-derived products. Four patients did not recall this information, whereas one patient assured us of having had no such contact.
Table 1. S. suis strains isolated from humans or pigs in Argentina included in this study.
| Strain | Source* | Clinical manifestation |
Serotype | Host | Date | Gender | Swine/pork contact |
Pathotype | ST |
|---|---|---|---|---|---|---|---|---|---|
| 285† | CSF, blood | Meningitis | 2 | H | 1995 | M | Yes | mrp+/epf+/sly+ | 1 |
| 284 | CSF, blood | Meningitis | 2 | H | 1995 | M | Yes | mrp+/epf+/sly+ | 1 |
| 263† | CSF, blood | Meningitis | 2 | H | 2003 | M | Yes | mrp+/epf+/sly+ | 1 |
| 178† | CSF, blood | Meningitis | 2 | H | 2003 | M | Yes | mrp+/epf+/sly+ | 1 |
| 247† | CSF, blood | Meningitis | 2 | H | 2003 | M | Yes | mrp+/epf+/sly+ | 1 |
| 2376‡ | CSF | Meningitis | 2 | H | 2004 | F | Yes | mrp+/epf+/sly+ | 1 |
| 12† | CSF, blood | Meningitis | 2 | H | 2009 | M | Yes | mrp+/epf+/sly+ | 1 |
| 83 | CSF, blood | Meningitis | 2 | H | 2009 | M | Unknown | mrp+/epf+/sly+ | 1 |
| 88 | CSF | Meningitis | 2 | H | 2009 | M | Yes | mrp+/epf+/sly+ | 1 |
| 245† | CSF | Meningitis | 2 | H | 2012 | M | Yes | mrp+/epf+/sly+ | 1 |
| 486† | CSF, blood | Meningitis | 2 | H | 2012 | M | Unknown | mrp+/epf+/sly+ | 1 |
| 371† | CSF, blood | Meningitis | 2 | H | 2013 | M | Yes | mrp+/epf+/sly+ | 1 |
| 473† | CSF | Meningitis | 2 | H | 2013 | F | Unknown | mrp+/epf+/sly+ | 1 |
| 42 | CSF, blood | Meningitis | 2 | H | 2014 | F | Unknown | mrp+/epf+/sly+ | 1 |
| 130/15 | CSF | Meningitis | 2 | H | 2015 | M | Yes | mrp+/epf+/sly+ | 1 |
| 695-15 | CSF | Meningitis | 2 | H | 2015 | M | Yes | mrp+/epf+/sly+ | 1 |
| 136-16 | Joint | Arthritis | 2 | H | 2016 | M | No | mrp+/epf+/sly+ | 1 |
| 15§ | CSF | Meningitis | 5 | H | 2014 | M | Yes | nd | nd |
| P156 | Spleen | Septicemia | 2 | S | 2000 | na | na | mrp+/epf+/sly+ | 1 |
| P232 | Joint | Arthritis | 2 | S | 2001 | na | na | mrp+/epf+/sly+ | 1 |
| 050798 | Brain | Meningitis | 2 | S | 2001 | na | na | mrp+/epf+/sly+ | 1 |
| P421 | Joint | Arthritis | 2 | S | 2002 | na | na | mrp+/epf+/sly+ | 1 |
| 130387 | Brain | Meningitis | 2 | S | 2002 | na | na | mrp+/epf+/sly+ | 1 |
| P517 | Brain | Meningitis | 2 | S | 2003 | na | na | mrp+/epf+/sly+ | 1 |
| 477 | Heart | Endocarditis | 2 | S | 2003 | na | na | mrp+/epf+/sly+ | 1 |
| P613 | Brain | Meningitis | 2 | S | 2005 | na | na | mrp+/epf+/sly+ | 1 |
| P655 | Heart | Endocarditis | 2 | S | 2005 | na | na | mrp+/epf+/sly+ | 1 |
| P574 | Joint | Arthritis | 2 | S | 2005 | na | na | mrp+/epf+/sly+ | 1 |
| P611 | Brain | Meningitis | 2 | S | 2005 | na | na | mrp+/epf+/sly+ | 1 |
| P706† | Lungs | Pneumonia | 2 | S | 2006 | na | na | mrp+/epf−/sly− | 28 |
| 50703-8 | Brain | Meningitis | 2 | S | 2010 | na | na | mrp+/epf−/sly− | 1 |
| BE 21¶ | Tonsils | None | 2 | S | 2014 | na | na | mrp+/epf−/sly− | 28 |
*When two sources of isolates are mentioned for the same patient, only one strain was characterized; †Strains studied by whole-genome sequencing (see Table 2); ‡(Lopreto et al., 2005); §Fatal case; ¶Clinically healthy pig. ST, sequence type (as evaluated by multilocus sequence typing); CSF, cerebrospinal fluid; nd, not determined; na, not applicable.
We also studied 14 S. suis serotype 2 strains recovered from pigs, as pure cultures, from non-related farms: 12 were from systemic disease (meningitis, arthritis or endocarditis). In addition, a strain isolated from the lungs of an animal with pneumonia and another strain isolated from the tonsils of a clinically healthy animal were included.
Investigations
Identification of S. suis by PCR and multiplex PCR was performed as previously described (Okura et al., 2014). Strains positive for serotype 2 were further differentiated from those positive for serotype 1/2 by coagglutination test (Gottschalk et al., 1993). All but one strain from human cases of infection were serotype 2; the remaining strain (the only fatal case) was characterized as serotype 5. No further characterization of this strain was completed.
To evaluate pathotype based on the presence or absence of the traditional virulence factor genes mrp, epf and sly, specific PCRs were performed on serotype 2 strains as previously described (Silva et al., 2006). Interestingly, all serotype 2 strains isolated from humans present the mrp+/epf+/sly+ profile usually associated with virulent Eurasian serotype 2 strains (Goyette-Desjardins et al., 2014). The epf+ genotype obtained refers to the variant encoding the 110 kDa protein (Smith et al., 1993). All but two strains recovered from pigs also presented this pathotype. The two exceptions were the strain isolated from lungs and that recovered from tonsils of a clinically healthy animal, which both presented the mrp+/epf−/sly−profile usually found in North America.
Results from MLST studies, performed as previously described (King et al., 2002), confirm such results. All strains presenting the mrp+/epf+/sly+ pathotype were ST1, similar to most virulent Eurasian strains (Goyette-Desjardins et al., 2014). The two swine strains presenting the mrp+/epf−/sly− profile (one recovered from lungs and the other from tonsils of a clinically healthy animal) were ST28, which is usually associated with low virulence strains in Canada and the USA (Goyette-Desjardins et al., 2014).
Since almost all Argentinean serotype 2 strains from humans and diseased pigs included in this study were ST1, we further compared nine available human ST1 strains and one porcine ST28 strain by WGS. Phylogenetic analyses were performed and compared with the available assembled GenBank genome sequence read data of 26 ST1 strains from the United Kingdom (13 strains), the Netherlands (one strain, reference strain), PR China (three strains), and Vietnam (nine strains) (Table 2). In addition, a Chinese serotype 8 strain was also included as an outsider control. The sequences of strain P1/7 (UK), GZ1 (PR China), BM407 (Vietnam) and R735 (the Netherlands) are completely finished. The methodology used was that previously described (Chen et al., 2013). For each strain, a 500 bp library was constructed and then sequenced using the Hiseq 4000 system (Illumina) to produce 150 bp paired-end reads. The high-throughput read data were mapped to the reference genome of S. suis ST1 strain P1/7 (Accession number: NC_012925) using SOAP2 and SNPs detected using SOAPsnp v1.03 (Li et al., 2008). For the assembled genome sequences, SNPs were called using MUMmer v3.23 (Delcher et al., 2003). These SNP patterns were distilled and concatenated using an automatic pipeline as previously described (Chen et al., 2013). Based on this, a phylogenetic tree was reconstructed using the maximum-likelihood method and GTRGAMMA substitution model via raxml software (version 7.2.8) with 1000 bootstrap replications (Stamatakis, 2006). (Fig. 1). The sequencing data were deposited in the GenBank database (Accession number SRP079937).
Table 2. List of strains used for comparison of the whole-genome sequencing of S. suis from Argentina (see Table 1).
| Strain number | Country | ST | Year | Host | Clinical signs |
|---|---|---|---|---|---|
| S90C | UK | 1 | 2010 | Pig | Diseased |
| S11N | UK | 1 | 2010 | Pig | Diseased |
| S98Y | UK | 1 | 2010 | Pig | Diseased |
| S13B | UK | 1 | 2010 | Pig | Diseased |
| S16E | UK | 1 | 2010 | Pig | Diseased |
| S15U | UK | 1 | 2010 | Pig | Diseased |
| S13F | UK | 1 | 2010 | Pig | Diseased |
| P1/7 | UK | 1 | 1980 | Pig | Diseased |
| S92D | UK | 1 | 2010 | Pig | Diseased |
| S17G | UK | 1 | 2010 | Pig | Diseased |
| S16D | UK | 1 | 2010 | Pig | Diseased |
| S16B | UK | 1 | 2010 | Pig | Diseased |
| S12U | UK | 1 | 2010 | Pig | Diseased |
| R735 | Netherlands | 1 | 1963 | Pig | Meningitis |
| YS1 | China | 1 | 2011 | Pig | Healthy |
| GZ1 | China | 1 | 2005 | Human | Meningitis |
| ZGST1 | China | 1 | 2015 | Human | Meningitis |
| RC1* | China | 308 | 2005 | Pig | Healthy |
| BM237a | Vietnam | 1 | 2001 | Human | Meningitis |
| BM407 | Vietnam | 1 | 2004 | Human | Meningitis |
| BM190a | Vietnam | 1 | 2000 | Human | Meningitis |
| BM264a | Vietnam | 1 | 2002 | Human | Meningitis |
| BM346B | Vietnam | 1 | 2003 | Human | Meningitis |
| BM478 | Vietnam | 1 | 2014 | Human | Meningitis |
| BM224C | Vietnam | 1 | 2001 | Human | Meningitis |
| BM461 | Vietnam | 1 | 2014 | Human | Meningitis |
| BM424a | Vietnam | 1 | 2004 | Human | Meningitis |
*Serotype 8 (outgroup)
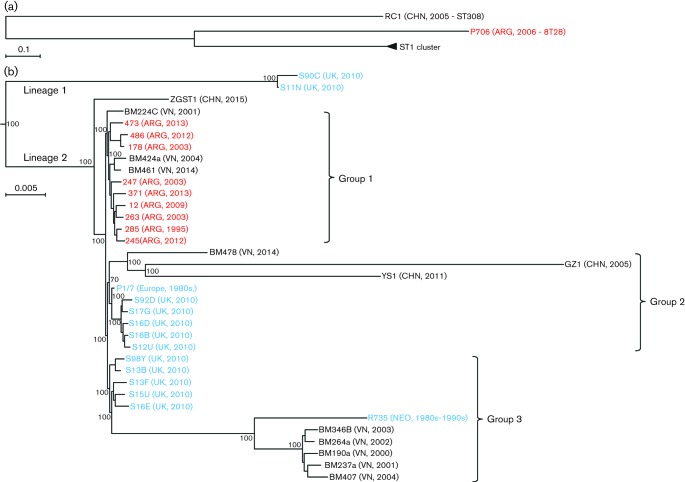
Fig. 1.
Phylogenetic relationship of ST1 strains. (a) Phylogenetic tree of all strains (including sequence type) used in this study. The ST1 cluster was compressed and is indicated by a triangle. (b) Phylogenetic tree of ST1 strains included in the present study compared with available data from GenBank. The numbers on the branches correspond to the bootstrap values. Strains from different areas are represented by different colors: Asia (black), Europe (blue) and Argentina (red). Geographical origins and year of isolation are included, in parentheses, after the strain name.
Approximately 1.36–1.63 Gb high-quality read data was obtained for each strain and covered 680–858-fold (769±45) of the complete genome of P1/7. A phylogenetic study showed that all of the ST1 strains clustered tightly together when compared with the ST28 strain P706 and ST308 (RC1, PR China) strain (Fig. 1a). Within the ST1 cluster, the nine Argentinean ST1 strains tested clustered together with three other strains from Vietnam, with this cluster being close to another one composed of 11 strains from the UK (Fig. 1b). These two clusters are relatively far away from other European (UK and the Netherlands) and Asian (China and Vietnam) strains.
Discussion
It has been clearly shown that serotype 2 strains isolated in North America (Canada and the USA) are phenotypically and genotypically highly different from those recovered in Europe and Asia (Fittipaldi et al., 2011). Preliminary results with archetypal strains (Fittipaldi et al., 2011), as well as more recent results with field strains (Auger et al., 2016; Athey et al., 2015), showed that North American S. suis serotype 2 strains are, in general, of low virulence. It has been strongly suggested that the relatively low number of human cases in both Canada and the USA (which together represent the second largest swine producing region worldwide after China) are probably due to the lower virulence properties of S. suis ST25 and ST28 strains that predominate in these countries.
Very few data are available regarding strains isolated in South America. A few reports on serotype 2 strains recovered from diseased pigs in Brazil have been published (Costa et al., 2005; Doto et al., 2016; Rocha et al., 2012), though no human case has yet been reported. Human cases have been reported in Argentina (Lopreto et al., 2005; Callejo et al., 2014; Nagel et al., 2008) and, only once, in Chile (Alarcon et al., 2013), but strains were not further characterized. Although not a traditional pork-producing country, Argentina has significantly increased its swine production in the last few years, having presently more than 4 000 000 pigs (http://www.porcinos.org.ar/). In this study, we analyzed 18 S. suis strains isolated from humans, 17 of which were serotype 2. With the addition of another case for which the strain was not available (Nagel et al., 2008) and a serotype 21 strain previously described by us (Callejo et al., 2014), this number is considerably higher than in the important swine producing countries of Canada and the USA. Three different factors may explain these differences: (a) the presence of backyard types of swine production; (b) a good surveillance system in rural hospitals and (c) the presence of potentially virulent strains, when compared with those found in North America.
Relatively close contact with pigs/pork products and animals slaughtered at home cannot be ruled out as a possible factor contributing to this atypically high prevalence. More than 75 % of farms in Argentina have fewer than 10 sows (http://www.porcinos.org.ar/), indicating a relatively high number of backyard family types of production with which human S. suis infections have been traditionally associated (Huong et al., 2014). Most patients described in this study had contact with swine/pork products and/or live in rural areas. However, it is important to note that typical dishes using raw blood or meat (such as are commonly consumed in some Asian countries) do not exist in the Argentinean culture (Gottschalk et al., 2010). In addition, and since the first human case of S. suis has been described more than 10 years ago (Lopreto et al., 2005), a good surveillance system for the characterization of alpha-hemolytic bacteria recovered from cases of meningitis in rural hospitals has been established. Each suspicious isolate is immediately sent to the National Institute of Microbiology for further identification. Consequently, close contact with pigs/pork and a good surveillance system may have contributed to the proper identification of these cases.
However, in the present study, we also demonstrated that serotype 2 strains recovered from either ill patients or systemically diseased pigs in Argentina are mostly ST1 strains with a typical mrp+/epf+/sly+genotype that characterizes virulent Eurasian strains. This markedly differs from lower virulence ST25/ST28 North American strains, which have a mrp+/−/epf−/sly− genotype (Fittipaldi et al., 2011). WGS results indicated that two strains from the UK (S90C and S11N) are clustered together in lineage 1 and significantly diverged from other strains of lineage 2 (Fig. 1b). These two lineages may have evolved from the ancestor of ST1 before separating, with lineage 2 having become dominant. Within this lineage, most strains could be clustered into three main groups with the exception of the Chinese strain ZGST1. Argentinean strains and three strains from Vietnam were clustered into group 1. Strains from the UK, two Chinese strains, other Vietnamese strains, and the old reference strain from the Netherlands were clustered into groups 2 and 3. A reasonable hypothesis is that the ST1 originated in Europe (UK) where a dominant lineage evolved before spreading to other countries maybe via the introduction of animals from genetic companies. In many countries, including China, Vietnam and Argentina, the advanced breeds of pigs have been introduced from European countries like the UK and Denmark. Interestingly, the strains in these three groups of lineage 2 were all isolated in more than one country from different continents. It would appear that these groups diverged before the spreading and were transmitted, in parallel, to different countries.
Interestingly, all strains from diseased pigs in this study were also ST1. Since some genetic breeders from North America have also been incorporated in Argentina, it is possible that North-American-like serotype 2 strains are also present. In fact, one strain isolated from a diseased pig (pneumonia, lungs) and another recovered from tonsils of a clinically healthy pig were typical ST28 strains with a mrp+/epf−/sly− genotype profile identical to that found in Canada and the USA (Fittipaldi et al., 2011). In this study, the strain isolated from lungs grouped far from all ST1 strains (Fig. 1a). These strains are most probably of low virulence; it has been previously reported that S. suis is not a primary cause of pneumonia and isolates recovered from lungs are often low-virulence (Gottschalk, 2012). It is important to note that these strains are also able to induce serious disease in Canada and the USA. The most important difference between these countries and Argentina, from the disease status point of view, is the absence of the most important swine virus from this South American country: the porcine reproductive and respiratory syndrome virus, which is considered one of the most important predisposing factors for S. suis infection (Gottschalk, 2012). In the absence of this virus, only virulent S. suis strains are usually able to cause important disease in swine. Finally, transmission of ST1 strains from South America to North America is probably low since pig flow from genetic companies and breeders is usually from Europe and North America to South America.
Finally, we report the first human case of serotype 5 in Argentina (the only fatal case in the present study). Although this is the first report of this serotype in South America, one case of septic arthritis and one case of peritonitis have been previously described in Sweden and Thailand, respectively (Kerdsin et al., 2011; Gustavsson et al., 2014). In addition, one human case of arthroplasty infection with streptococcal toxic shock-like syndrome, was caused by a non-encapsulated strain belonging to this serotype in the USA (Gomez et al., 2014).
In conclusion, and in addition to the probably close contact with pigs/pork products and a good surveillance system, the presence of potentially virulent Eurasian-like serotype 2 strains in Argentina may be an important risk factor contributing to the higher number of human cases observed. In fact, and with this report, Argentina is now among the Western countries with the highest number of reported human cases after the Netherlands, France and the UK, with a similar number of cases to Poland (Bojarska et al., 2016). However, the three former European countries began the identification of S. suis in humans more than 15 years before Argentina, so the total number of human cases in this South American country may have been underestimated. Further studies in other South American countries, such as Brazil, where swine production is very important, should be performed.
Acknowledgements
We thank all physicians from the different hospitals in Argentina who were involved in the diagnosis of human cases. We also thank Sonia Lacouture for technical assistance. This study was funded by Natural Science and Engineering Research Council of Canada (NSERC) 04435 and by the Canadian Institutes of Health Research (CIHR) 125684 to M. G. The authors declare no conflicts of interest.
Abbreviations:
- CSF
cerebrospinal fluid
- MLST
multilocus sequence typing
- ST
sequence type
- WGS
whole-genome sequencing
References
- Alarcón L P., Araya R P., Aguayo C., Fernández J., Illesca V., Zaror A., Vaquero A.(2013). [Laboratory confirmation of Streptococcus suis in Chile]. Rev Chilena Infectol 30539–540. 10.4067/S0716-10182013000500011 [DOI] [PubMed] [Google Scholar]
- Athey T. B., Auger J. P., Teatero S., Dumesnil A., Takamatsu D., Wasserscheid J., Dewar K., Gottschalk M., Fittipaldi N.(2015). Complex population structure and virulence differences among serotype 2 Streptococcus suis strains belonging to sequence type 28. PLoS One 10e0137760. 10.1371/journal.pone.0137760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auger J.-P., Fittipaldi N., Benoit-Biancamano M.-O., Segura M., Gottschalk M.(2016). Virulence studies of different sequence types and geographical origins of Streptococcus suis serotype 2 in a mouse model of infection. Pathogens 548. 10.3390/pathogens5030048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojarska A., Molska E., Janas K., Skoczyńska A., Stefaniuk E., Hryniewicz W., Sadowy E.(2016). Streptococcus suis in invasive human infections in Poland: clonality and determinants of virulence and antimicrobial resistance. Eur J Clin Microbiol Infect Dis 35917–925. 10.1007/s10096-016-2616-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callejo R., Prieto M., Salamone F., Auger J. P., Goyette-Desjardins G., Gottschalk M.(2014). Atypical Streptococcus suis in man, Argentina, 2013. Emerg Infect Dis 20500–502. 10.3201/eid2003.131148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Zhang W., Zheng H., Lan R., Wang H., Du P., Bai X., Ji S., Meng Q., et al. (2013). Minimum core genome sequence typing of bacterial pathogens: a unified approach for clinical and public health microbiology. J Clin Microbiol 512582–2591. 10.1128/JCM.00535-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A. T., Lobato F. C., Abreu V. L., Assis R. A., Reis R., Uzal F. A.(2005). Serotyping and evaluation of the virulence in mice of Streptococcus suis strains isolated from diseased pigs. Rev Inst Med Trop Sao Paulo 47113–115. 10.1590/S0036-46652005000200012 [DOI] [PubMed] [Google Scholar]
- Delcher A. L., Salzberg S. L., Phillippy A. M.(2003). Using MUMmer to identify similar regions in large sequence sets. Curr Protoc Bioinformatics 103. [DOI] [PubMed] [Google Scholar]
- Doto D. S., Moreno L. Z., Calderaro F. F., Matajira C. E., de Moura Gomes V. T., Ferreira T. S., Mesquita R. E., Timenetsky J., Gottschalk M., et al. (2016). Genetic diversity of Streptococcus suis serotype 2 isolated from pigs in Brazil. Can J Vet Res 80106–111. [PMC free article] [PubMed] [Google Scholar]
- Fittipaldi N., Xu J., Lacouture S., Tharavichitkul P., Osaki M., Sekizaki T., Takamatsu D., Gottschalk M.(2011). Lineage and virulence of Streptococcus suis serotype 2 isolates from North America. EmergInfect Dis 172239–2244. 10.3201/eid1712.110609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez E., Kennedy C. C., Gottschalk M., Cunningham S. A., Patel R., Virk A.(2014). Streptococcus suis-related prosthetic joint infection and streptococcal toxic shock-like syndrome in a pig farmer in the United States. J Clin Microbiol 522254–2258. 10.1128/JCM.02934-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk M.(2012). Streptococcocis. Diseases of Swine. Edited by Straw B. E., Zimmerman J. J., D’Allaire S., Taylor D. J.Ames, Iowa, USA: Blackwell Publishing. [Google Scholar]
- Gottschalk M., Higgins R., Boudreau M.(1993). Use of polyvalent coagglutination reagents for serotyping of Streptococcus suis. J Clin Microbiol 312192–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk M., Xu J., Lecours M.-P., Grenier D., Fittipaldi N., Segura M.(2010). Streptococcus suis infections in humans: what is the prognosis for Western countries? (Part II). Clin Microbiol Newsl 3297–102. 10.1016/j.clinmicnews.2010.06.001 [DOI] [Google Scholar]
- Goyette-Desjardins G., Auger J.-P., Xu J., Segura M., Gottschalk M.(2014). Streptococcus suis, an important pig pathogen and emerging zoonotic agent—an update on the worldwide distribution based on serotyping and sequence typing. Emerg Microb Infect 3e45 10.1038/emi.2014.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustavsson C., Rasmussen C.(2014). Septic arthritis caused by serotype 5 in pig farmer. Emerg Infect Dis 20489–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huong V. T., Hoa N. T., Horby P., Bryant J. E., Van Kinh N., Toan T. K., Wertheim H. F., Horby V. T., Bryant V. T., Wertheim V. T.(2014). Raw pig blood consumption and potential risk for Streptococcus suis infection, Vietnam. Emerg Infect Dis 201895–1898. 10.3201/eid2011.140915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerdsin A., Dejsirilert S., Sawanpanyalert P., Boonnark A., Noithachang W., Sriyakum D., Simkum S., Chokngam S., Gottschalk M., et al. (2011). Sepsis and spontaneous bacterial peritonitis in Thailand. Lancet 378960. 10.1016/S0140-6736(11)60923-9 [DOI] [PubMed] [Google Scholar]
- King S. J., Leigh J. A., Heath P. J., Luque I., Tarradas C., Dowson C. G., Whatmore A. M.(2002). Development of a multilocus sequence typing scheme for the pig pathogen Streptococcus suis: identification of virulent clones and potential capsular serotype exchange. J Clin Microbiol 403671–3680. 10.1128/JCM.40.10.3671-3680.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Li Y., Kristiansen K., Wang J.(2008). SOAP: short oligonucleotide alignment program. Bioinformatics 24713–714. 10.1093/bioinformatics/btn025 [DOI] [PubMed] [Google Scholar]
- Lopreto C., Lopardo H. A., Bardi M. C., Gottschalk M.(2005). [Primary Streptococcus suis meningitis: first case in humans described in Latin America]. Enferm Infecc Microbiol Clin 23110. [DOI] [PubMed] [Google Scholar]
- Martinez G., Pestana de Castro A. F., Ribeiro Pagnani K. J., Nakazato G., Dias da Silveira W., Gottschalk M.(2003). Clonal distribution of an atypical MRP+, EF*, and suilysin+ phenotype of virulent Streptococcus suis serotype 2 strains in Brazil. Can J Vet Res 6752–55. [PMC free article] [PubMed] [Google Scholar]
- Nagel A., Manias V., Busquets N., Sniadowsky S., Anzardi J., Méndez E. L.(2008). [Streptococcus suis meningitis in an immunocompetent patient]. Rev Argent Microbiol 40158–160. [PubMed] [Google Scholar]
- Okura M., Lachance C., Osaki M., Sekizaki T., Maruyama F., Nozawa T., Nakagawa I., Hamada S., Rossignol C., et al. (2014). Development of a two-step multiplex PCR assay for typing of capsular polysaccharide synthesis gene clusters of Streptococcus suis. J Clin Microbiol 521714–1719. 10.1128/JCM.03411-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha D. L., Santos L. F., Santos D. L., Costa W. M. T., Santos J. L.(2012). Sorotipos de Streptococcus suis identificados em suínos com meningite no estado do Paraná. Arq Bras Med Vet Zootec 64488–490. 10.1590/S0102-09352012000200032 [DOI] [Google Scholar]
- Silva L. M., Baums C. G., Rehm T., Wisselink H. J., Goethe R., Valentin-Weigand P.(2006). Virulence-associated gene profiling of Streptococcus suis isolates by PCR. Vet Microbiol 115117–127. 10.1016/j.vetmic.2005.12.013 [DOI] [PubMed] [Google Scholar]
- Smith H. E., Reek F. H., Vecht U., Gielkens A. L., Smits M. A.(1993). Repeats in an extracellular protein of weakly pathogenic strains of Streptococcus suis type 2 are absent in pathogenic strains. Infect Immun 613318–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A.(2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 222688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Wertheim H. F., Nguyen H. N., Taylor W., Lien T. T., Ngo H. T., Nguyen T. Q., Nguyen B. N., Nguyen H. H., Nguyen H. M., et al. (2009). Streptococcus suis, an important cause of adult bacterial meningitis in northern Vietnam. PLoS One 4e5973. 10.1371/journal.pone.0005973 [DOI] [PMC free article] [PubMed] [Google Scholar]