Abstract
Calcium signaling controls many key processes in neurons, including gene expression, axon guidance, and synaptic plasticity. In contrast to calcium influx through voltage- or neurotransmitter-gated channels, regulatory pathways that control store-operated calcium entry (SOCE) in neurons are poorly understood. Here, we report a transcriptional control of Stim1 (stromal interaction molecule 1) gene, which is a major sensor of endoplasmic reticulum (ER) calcium levels and a regulator of SOCE. By using a genome-wide chromatin immunoprecipitation and sequencing approach in mice, we find that NEUROD2, a neurogenic transcription factor, binds to an intronic element within the Stim1 gene. We show that NEUROD2 limits Stim1 expression in cortical neurons and consequently fine-tunes the SOCE response upon depletion of ER calcium. Our findings reveal a novel mechanism that regulates neuronal calcium homeostasis during cortical development.
Keywords: calcium, genomics, Neurod2, store-operated calcium entry, transcription factor
Significance Statement
Store-operated calcium entry (SOCE) is a major source of neuronal calcium influx. Although SOCE controls key neurodevelopmental processes, the gene expression programs that regulate this mode of calcium entry in neurons remain poorly understood. In this study, we conducted an in vivo, genome-wide target gene analysis of the neurogenic transcription factor NEUROD2. We find that NEUROD2 controls the Stim1 gene, which encodes a major ER calcium sensor and an essential component of SOCE. Importantly, we demonstrate that NEUROD2 is a critical regulator of neuronal SOCE levels. Our findings present important implications for understanding transcriptional programs that control neuronal calcium homeostasis, as well as for disease mechanisms in which deranged SOCE is observed, such as epilepsy and Alzheimer’s disease.
Introduction
By a remarkable series of regulated gene expression programs, neural progenitor cells, and eventually neurons, steadily transition from one cellular state to the next in terms of their proliferative capacities, migratory behavior, axonal growth, and dendritogenic and synaptogenic capabilities (Kohwi and Doe, 2013; Pataskar et al., 2016; Telley et al., 2016). This series of regulated transitions depends on the correct spatiotemporal expression of critical transcription factors (TFs) that allow the generation of different classes of mature neurons at the correct time and place (Leone et al., 2008; Kwan et al., 2012; Greig et al., 2013). While the phenotypes emerging from knockout mouse models of these TFs have been extensively analyzed, their genome-wide binding sites and the biological implications of such binding events are still largely unknown.
Proneural and neurogenic basic helix-loop-helix (bHLH) TFs are key players for controlling the emergence of a wide range of neuronal subtypes each with unique connectivities, and physiologic and morphologic properties (Mattar et al., 2008; Wilkinson et al., 2013; Imayoshi and Kageyama, 2014). In the developing mammalian neocortex, these TFs include proneural Neurogenins (Neurog1/2) and neurogenic NeuroDs (Neurod1/2/4/6), for which ectopic gain of function is sufficient to induce neurogenesis (Farah et al., 2000; Noda et al., 2006) and for which loss-of-function mutations result in a wide-range of neurodevelopmental abnormalities (Wilkinson et al., 2013; Yuan and Hassan, 2014). At the onset of mouse cortical neurogenesis, a transient burst of expression of Neurogenins specifies neural progenitors into cortical excitatory neurons (Fode et al., 2000; Mattar et al., 2008; Kim et al., 2011). Specifically, the Neurogenins trigger a cascade of downstream TF expression, including the NeuroD family, which then simultaneously activate genetic programs to drive excitatory neurogenesis and suppress alternative cell fates (Jo et al., 2007; Roybon et al., 2010; Pataskar et al., 2016). The importance of the Neurogenin–NeuroD gene regulatory network for human neocortical development is evidenced by a recent study demonstrating that, during the evolution of the human fetal neocortex, neural progenitor populations expressing Neurog2 had specifically expanded relative to other progenitor classes (Johnson et al., 2015). In fact, the Neurog1/2-driven network is being exploited for the in vitro production of cortical excitatory neurons from human induced pluripotent stem cells, aided by remarkable developments in cellular reprogramming and high-throughput gene expression technologies (Busskamp et al., 2014).
NEUROD2 is one of the key members of the Neurogenin–NeuroD gene network. Within the neocortex, Neurod2 expression is triggered as progenitors exit the cell cycle and is sustained throughout the lifetime of cortical excitatory neurons (McCormick et al., 1996; Olson et al., 2001). Neurod2 regulates several essential features of brain development, as mice lacking Neurod2 exhibit morphologic and physiologic defects in thalamocortical connections, hippocampal synaptogenesis, axonal guidance of callosal axons, and development of amygdalar nuclei (Olson et al., 2001; Lin et al., 2005; Ince-Dunn et al., 2006; Wilke et al., 2012; Bormuth et al., 2013; Chen et al., 2016). In gain-of-function experiments, the overexpression of Neurod2 in cortical neural progenitors induces premature exit from the cell cycle and differentiation (Telley et al., 2016). These studies clearly reveal that NEUROD2 controls a wide-range of neurodevelopmental and physiologic processes in different developmental stages and brain regions. In fact, recent target gene analyses and gene expression studies have suggested that NEUROD2 regulates components of radial migration and neuritogenesis during embryonic development (Bayam et al., 2015; Telley et al., 2016). However, questions remain regarding the genome-wide binding sites of NEUROD2 at various spatiotemporal settings and the biologically relevant effects of such binding events.
In this study, we performed a chromatin immunoprecipitation and sequencing (ChIP-Seq) analysis of NEUROD2 from postnatal cerebral cortical tissue, with the goal of identifying target genes and pathways regulating processes important for postnatal cortical development. Our in vivo analysis identified Stim1 (stromal interaction molecule 1) as a primary target of NEUROD2. Stim1 encodes a major sensor of endoplasmic reticulum (ER) calcium levels and is an important regulator of store-operated calcium entry (SOCE; Kraft, 2015; Moccia et al., 2015). Contrary to previous research describing NEUROD2 as a transcriptional activator, our data suggest that NEUROD2 restrains Stim1 expression via binding to an intronic element within intron 2 of Stim1. The NEUROD2 binding site is phylogenetically conserved and harbors clustered consensus E-box elements. Knockdown of Neurod2 expression in cultured cortical neurons increased STIM1 protein expression and consequently caused an upregulation in SOCE. Conversely, Neurod2 overexpression resulted in depression of SOCE response. Collectively, our data point to a NEUROD2-dependent gene regulatory mechanism that controls neuronal SOCE via fine-tuning STIM1 abundance.
Materials and Methods
Chromatin immunoprecipitation and sequencing
Cortices were recovered from five littermate BALB/c postnatal day 0 (P0) mice of either sex. Cortical tissue was dissected, pooled, and cross-linked for 10 min in 1% formaldehyde. Cross-linked tissue was lysed in RIPA buffer (0.05 m Tris-HCl, pH 7.5, 0.15 m NaCl, 1% Triton X-100, 1% Na-DOC, 0.1% SDS) and sonicated to achieve 200–250 bp fragments. Ten percent of the input was used to isolate input chromatin, and the remainder was used for ChIP. NEUROD2–chromatin complexes were immunoprecipitated using three separate antibodies (ab168932, ab104430, and ab109406, Abcam). Chromatin immunoprecipitated with an unrelated GFP antibody was used as a negative control (sc-8334, Santa Cruz Biotechnology). Beads used for immunoprecipitation were carried through a series of stringent wash steps (Buffer 1: 1× PBS, 0.1% SDS, 0.5% Na-DOC, 0.5% NP-40; Buffer 2: 5× PBS, 0.1% SDS, 0.5% Na-DOC, 0.5% NP-40; Buffer 3: 15 mm Tris-HCl, pH 7.5, 5 mm EDTA, 2.5 mm EGTA, 1% Triton X-100, 1% Na-DOC, 0.1% SDS, 120 mm NaCl, 25 mm KCl; Buffer 4: 15 mm Tris-HCl, pH 7.5, 5 mm EDTA, 2.5 mm EGTA, 1% Triton X-100, 1% Na-DOC, 0.1% SDS, 1M NaCl; Buffer 5: 15 mm Tris-HCl, pH 7.5, 5 mm EDTA; Buffer 6: 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm MgCl2, 0.05% NP-40; Buffer 7: 50 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 0.5% NP-40). After the washes, protein–DNA crosslinks were reversed at 65°C, RNase A and proteinase K treatments were conducted, and ChIP DNA was isolated by standard phenol-chloroform extraction and ethanol precipitation. Library preparation and 50 bp single end sequencing (HiSeq 2500 platform, Illumina) were performed at Genewiz (South Plainfield, NJ).
Bioinformatics analysis
Raw sequences from three independent NEUROD2 ChIP-Seq experiments were mapped onto the mouse genome build mm10 using Bowtie for Illumina (version 1.1.2; Langmead et al., 2009; Langmead and Salzberg, 2012). Peak locations were determined using MACS (Model-based analysis of ChIP-Seq, version 1.0.1; Zhang et al., 2008). Sequence reads from GFP ChIP-Seq were used as control files for MACS analysis. A p-value of 1 × 10−5 was used as cutoff value for peak calling. Analyses with Bowtie and MACS tools were conducted through the Galaxy interface (usegalaxy.org; Giardine et al., 2005). Overlapping peaks identified in all three NEUROD2 ChIP-Seq datasets were selected as high confidence binding sites, and all subsequent analyses were conducted using these binding sites. Individual p-values and false discovery rates (FDRs) were calculated for each of the overlapping peak regions. Processed files are available at Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE84895. Midpoints of NEUROD2 binding sites were overlapped with Ensembl annotations and labeled as intergenic, exonic, intronic, or promoter [±1000 bp of a transcription start site (TSS)]. ChIP-Seq data for various histone codes were acquired from The ENCODE Project (encodeproject.org; ENCODE Project Consortium, 2012). All histone data were generated by the Bing Ren Laboratory at the University of California, San Diego (San Diego, CA), and accession numbers are as follows: H3K9me3 (ENCFF676DBG); H3K27me3 (ENCFF102IIL); H3K27ac (ENCFF145FVU); H3K36me3 (ENCFF091JOV); H3K4me3 (ENCFF875CQU); and H3K4me1 (ENCFF152TUF). CCCTC-binding factor (CTCF) ChIP-Seq data were generated by the Richard Myers Laboratory at HudsonAlpha Institute of Biotechnology (Huntsville, AL), and the accession number is: CTCF (ENCSR677HXC). Histone-binding locations and scores obtained from the ENCODE database were overlapped with NEUROD2 binding regions, yielding six histone binding scores (one for each histone type) for each NEUROD2 binding region. Mean histone-binding scores were calculated separately for NEUROD2 binding regions located in promoters, introns, exons, and intergenic regions. The genome-wide average of the histone scores was plotted as a baseline control. Gene ontology (GO) analysis was conducted as described previously (Ashburner et al., 2000). All ChIP-Seq data were visualized either in Trackster (Goecks et al., 2012) embedded in The Galaxy Project (Giardine et al., 2005) or The UCSC Genome Browser (Kent et al., 2002).
Data access
Raw ChIP-Seq data and processed files are available at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE84895.
ChIP followed by quantitative PCR (ChIP-qPCR)
Ten percent of the cross-linked lysate was used to prepare input DNA by standard phenol-chloroform extraction followed by ethanol precipitation. Input and ChIP DNA were dissolved in 300 μl of sterile water, and 1 μl from each was used as template for quantitative PCR (qPCR). The adjusted input Ct value (representing 10% of input chromatin) was calculated by subtracting 3.32 (log210) from the input Ct value. The following formula was then used to calculate the input normalized ChIP DNA amount: 100 × 2(Adjusted input Ct − ChIP Ct). The normalized NEUROD2 ChIP DNA amounts were divided by normalized GFP ChIP DNA. The primer sequences used in ChIP-qPCR experiments were as follows: Stim1-int2-ChIP-F (gtcctgctgctgactatgtg); Stim1-int2-ChIP-R (ctaaccctttgcccctaacc); Stim1-int1-ChIP-F (gaagttctttcgtgtagtagtcatgc); Stim1-int1-ChIP-R (cagaaaggcacacctgaacaccaag); Stim1-int3-ChIP-F (aggaagggaacctcttagacaactcag); Stim1-int3-ChIP-R (ggcagtagagatggttcagtggttaag); Dlx2-ChIP-F (gacggttgcctcctttcttg); Dlx2-ChIP-R (gtcgagtgcatatcagccac); Gsx2-ChIP-F (caaaagccagttctctcccg); Gsx2-ChIP-R (ggctggtgatggtgatgatg); Gad1-ChIP-F (ccagggatcgtgcaagcaa); Gad1-ChIP-R (gtggtcttggggtctctacg); Calb2-ChIP-F (atgcgggtaggtatgcttcg); Calb2-ChIP-R (cagggcgttagcttgaagga); Npy-ChIP-F (tcacttgctggactcaggttc); Npy-ChIP-R (atgcaatctgggttcctggt); Neurod6-ChIP-F (aacagttgcaccattggcag); Neurod6-ChIP-R (gcactgatcatctggcatcc); Bhlhe22-ChIP-F (gccacacatgtcaagctaaag); Bhlhe22-ChIP-R (gccgcgagtctgaatagtttc); Nrcam-ChIP-F (aagcttcggaaacacgcac); Nrcam-ChIP-R (ggctccttgttctgctccag); and Cux1-ChIP-F (ggtgaccgatagcttgcatc); Cux1-ChIP-R (agtctccttacagtccagcg).
Cloning
shND2-1, shND2-2, and nonsilencing (NS) short hairpin RNAs (shRNA) were cloned into pSUPER-neo-EGFP (www.oligoengine.com) as described in the manual. For calcium imaging and immunofluorescent staining experiments, the EGFP cassette was removed, and an mCherry-expressing cassette was subcloned into the AgeI and NotI sites of pSUPER-neo. shND2-1-resistant cDNA (resND2) was created by site-directed mutagenesis in the pcDNA4 backbone vector by introducing three silent mutations (C1077A, G1080A, T1083A) within the shND2-1 target sequence (aagacaagagattctcgga). The primer sequences were as follows: shND2-1_F (gatccccaagacaagagattctcggattcaagagatccgagaatctcttgtcttttttta); shND2-1_R (agcttaaaaaaagacaagagattctcggatctcttgaatccgagaatctcttgtcttggg); shND2-2_F (gatcccctgccgttgagacagagcggttcaagagaccgctctgtctcaacggcattttta); shND2-2_R (agcttaaaaatgccgttgagacagagcggtctcttgaaccgctctgtctcaacggcaggg); NS_F (gatccccgcgcgatagcgctaataatttttcaagagaaaattattagcgctatcgcgcttttta); NS_R (agtctaaaaagcgcgatagcgctaataattttctcttgaaaaattattagcgctatcgcgcggg); resND2_F (ttcaccacgatcggggccccatgtac); and resND2_R (ggtgcatatcgtatgataatagattctcgga). For luciferase assays, a 570 bp fragment encompassing the NEUROD2 binding site in Stim1 intron 2 was amplified by PCR, subcloned into the KpnI and HindIII sites of pXPG backbone vector (Bert et al., 2000). All four E-boxes were destroyed by site-directed mutagenesis using the following primers: Stim1MutE1_F (cactcaagcaagggtccca); Stim1MutE1_R (ggtgaacagaatgtatcttccc); Stim1MutE2_F (cagacacatggagctacac); Stim1MutE2_R (ggtacacggacccttgcttg); Stim1MutE3_F (agctacacatttcagagagtaggc); Stim1MutE3_R (gattgactctgggtacacggac); Stim1MutE4_F (cctccgtcatgcttccagga); and Stim1MutE4_R (gattcacggggagctgctgcctc). All constructs created were confirmed by Sanger sequencing.
Reverse Transcription and quantitative PCR (RT-qPCR)
Total RNA from primary cortical cultures was prepared using the Absolutely RNA Microprep Kit (Agilent Technologies) and reverse-transcribed using Transcriptor High Fidelity cDNA Synthesis Kit (Roche). The transcripts were quantified by qPCR with Luminaris HiGreen qPCR Master Mix (Thermo Scientific) using a CFX Connect Real-Time PCR Detection System (Bio-Rad). Gapdh RNA was used for normalization. The primer sequences were as follows: Stim1-qPCR-F (cctctcttgactcggcataatc); Stim1-qPCR-R (gaccttctctacttccacagttc); Gapdh-qPCR-F (cgacttcaacagcaactcccactcttcc); Gapdh-qPCR-R (tgggtggtccagggtttcttactcctt); Stim2-qPCR-F (ctactgtgctttcttcgccc); and Stim2-qPCR-R (aactccataccgcattgctg)
Luciferase assays
Transfection of HEK293T cells for luciferase assay was performed in 24-well plates using Lipofectamine 2000 (Invitrogen). Each well was cotransfected with 200 ng of pXPG luciferase reporter construct (Bert et al., 2000), 200 ng of empty or NEUROD2-expressing pcDNA4 plasmid and 100 ng of Renilla pRL-null plasmid (Promega). Twenty-four hours after transfection, the cells were lysed in PLB buffer (Promega), and the activity of Firefly and Renilla luciferases were assessed using Dual-Luciferase Reporter Assay System (Promega).
Primary cortical cultures, immunoblotting, and immunofluorescence staining
Primary cortical cultures were prepared from embryos of either sex derived from pregnant BALB/c mice on embryonic day 14.5 (E14.5). Briefly, cortices were dissected in ice-cold 1× HBSS and digested in 20 U/ml papain enzyme for 10 min at 37ºC. Digestion was terminated by treatment with 10 mg/ml trypsin inhibitor for 1 min. Tissue was triturated two to four times and plated onto plates (pre-coated with laminin and poly-d-lysine) in Basal Media Eagle supplemented with 5% FBS, 1 mm l-glutamine, penicillin/streptomycin, 1× N-2, and 1× B27 supplements (Invitrogen). For immunoblotting and RT-qPCR experiments, shRNAs were transfected by nucleofection immediately before plating (P3 primary cell 4-D nucleofector C kit, program #CU-133, Lonza). Protein lysates were collected at 5 days in vitro (DIV) in RIPA buffer supplemented with protease inhibitors. For immunofluorescence staining, neurons were transfected with Lipofectamine 2000 transfection reagent (Invitrogen) at 2 DIV and fixed with 4% paraformaldehyde at 5 and 8 DIV. All images were collected with Nikon 90i Eclipse confocal microscope, and analysis was performed with ImageJ software. Antibodies used were as follows: myc (sc-40, Santa Cruz Biotechnology), STIM1 (5668, Cell Signaling Technlogy), GFP (H0612, Santa Cruz Biotechnology), BETA-ACTIN (PA5-16914, ThermoFisher Scientific), NEUROD2 (ab168932, ab104430, and ab109406, Abcam), and Histone H3 (9715S, Cell Signaling Technology).
Calcium imaging
Primary neuronal cultures were prepared from mouse E14.5 embryos as described above. At 2 DIV, pSUPER-neo-mCherry expressing either Neurod2 shRNA or nonsilencing shRNA was transfected. For rescue experiments, resND2 in pcDNA4 backbone vector was cotransfected with Lipofectamine 2000. Calcium-imaging protocol was conducted at ∼7–9 DIV. Briefly, calcium-sensitive dye Fluo-3 dissolved in Ringer’s solution (155 mm NaCl, 10 mm d-glucose, 5 mm HEPES, 4.5 mm KCl, 2 mm CaCl2, and 1 mm MgCl2, pH 7.4) was loaded onto cells at a final concentration of 4 µm. Before imaging, cultures were treated with 1 µm TTX to prevent spontaneous activity. All live imaging experiments were performed using an XcellencePro inverted microscope (Olympus), where the imaging chamber was kept at 37°C and 5% CO2. ER Ca2+ was depleted by treatment with 5 µm thapsigargin in Ca2+-free Ringer’s solution. Images were acquired every 6 s for a total duration of 10 min using 40× magnification. The images were quantified with ImageJ software, based on mean signal intensity measured on neuronal somas. The quantified images were plotted as ΔF/Fo after background subtraction. Fo represented the minimum signal obtained in the images after background subtraction was performed. This minimum signal was set as a value of 1 and all other measurements (ΔF) were plotted as a ratio of this minimum signal.
Use of animals
All animal experiments were done in accordance with guidelines provided by the Koç University, Ministries of Food, Agriculture and Live Stock, Forestry and Water Management, Turkey and the European Union. Ethics approval was obtained from the Institutional Animal Care and Use Committee of Koç University (no. 2013-1).
Results
ChIP-Seq reveals NEUROD2 targets in postnatal cerebral cortical tissue
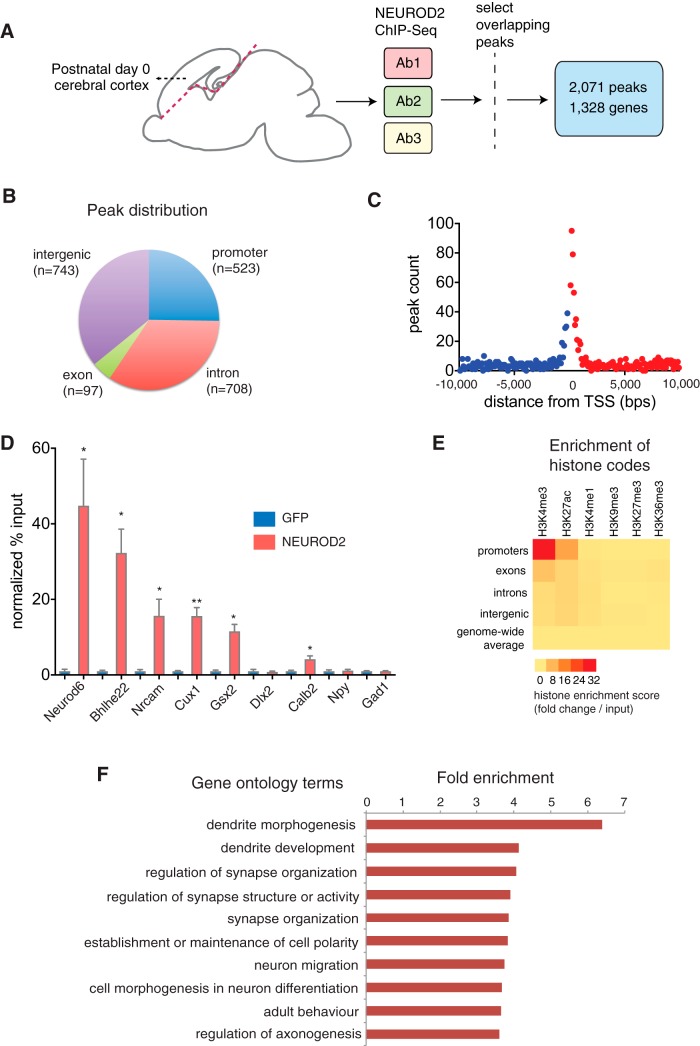
In the mouse, cortical neurogenesis and the process of neuronal migration largely come to an end by birth, and early postnatal days coincide with the initiation of a period of intense dendritic growth, synaptogenesis, and axonal myelination (Semple et al., 2013). Since Neurod2 is highly expressed in the cerebral cortex during embryonic and postnatal development (Lin et al., 2004; Ince-Dunn et al., 2006) and Neurod2 knockout mice display defects in neocortical and hippocampal dendritogenesis and synaptogenesis (Ince-Dunn et al., 2006; Wilke et al., 2012; Chen et al., 2016), we decided to identify its genome-wide targets within the cortex during this developmental time period. We dissected cerebral cortex tissue from P0 wild-type mice and conducted ChIP-Seq analysis using three separate NEUROD2 antibodies (Fig. 1A). We normalized our ChIP-Seq signals to a dataset acquired from a ChIP-Seq experiment conducted with an unrelated GFP antibody using the same tissue. Our computational analyses from three separate NEUROD2 ChIP-Seq experiments collectively yielded a total of 19,562 unique binding sites (p < 1 × 10−5). To minimize false-positive hits, we filtered for the 2,071 peaks that overlapped in all three experiments and represented highly significant binding regions (p < 1 × 10−164; FDR <0.004; Tables 1, Fig. 1A). All subsequent analyses were conducted using these 2,071 overlapping binding sites.
Figure 1.
Identification of genome-wide NEUROD2 binding sites at postnatal day 0 cerebral cortex. A, NEUROD2 ChIP-Seq was performed on cerebral cortex tissue using three separate antibodies. Selecting for overlapping peaks in all three datasets revealed 2,071 high confidence binding sites mapping to 1,328 annotated genes. B, Distribution of midpoints of NEUROD2 binding sites based on mouse Ensembl transcripts. Midpoints mapping within ± 1000 bp of TSSs are accepted as promoter binding. C, The number of NEUROD2 binding regions is plotted as a function of the distance of their midpoints to the closest TSS. A clear binding preference for NEUROD2 within ± 1000 bp is observed. D, Quantification of NEUROD2 binding to target and nontarget regions by ChIP-qPCR. Template DNA is immunoprecipitated with NEUROD2 antibody or an unrelated GFP antibody as a negative control. Amount of DNA immunoprecipitated is expressed as percentage of input DNA (% input). NEUROD2 % input values are normalized to GFP % input values as described in Materials and Methods. Enrichment of NEUROD2 is detected at target regions located on Neurod6, Bhlhe22, Nrcam, and Cux1 genes, but not at nontarget regions on Dlx2, Npy, and Gad1 genes. Slight enrichment is also observed in nontarget genes Gsx2 and Calb2. Bars represent SEM. p < 0.0001 determined by one-way ANOVA followed by unpaired t test, *p < 0.05, **p < 1 × 10−4 (Table 2). E, Enrichment of histone marks within NEUROD2 peaks located within different genomic regions is represented as a heat map. Genome-wide enrichment of histone marks, including NEUROD2 target and nontarget sequences, are plotted as baseline controls. F, Gene ontology analysis of all 1,328 genes identifies dendrite morphogenesis and synaptic organization as the two main NEUROD2-regulated biological processes. Significantly enriched GO categories (p < 0.01) are ranked based on their fold enrichment.
Table 1:
Top 10 NEUROD2 target-binding regions
| Gene symbol | Gene name | Ensembl ID | Genomic position of binding | Genomic region | Score | FDR (%) |
|---|---|---|---|---|---|---|
| Krtap16-1 | Keratin-associated protein 16-1 | ENSMUST00000105050 | chr11: 99985038- 99986631 | Promoter | 8439.73 | 0 |
| n/a | n/a | n/a | chr10: 84357827- 84358757 | Intergenic | 6366.16 | 0 |
| Prss36 | Polyserase-2 precursor | ENSMUST00000094026 | chr7: 127935318- 127936794 | Promoter | 6362.44 | 0 |
| Man1c1 | Mannosidase α class 1C member 1 | ENSMUST00000038628 | chr4: 134579940-134581329 | Exon | 5669.73 | 0 |
| n/a | n/a | n/a | chr12: 6638634- 6639508 | Intergenic | 5018.18 | 0 |
| Tecr | Synaptic glycoprotein SC2 (very-long-chain enoyl-CoA reductase) | ENSMUST00000165740 | chr8: 83584694-83585269 | Intron | 5011.04 | 0 |
| Stim1 | Stromal interaction molecule 1 | ENSMUST00000033289 | chr7: 102369602-102370206 | Intron | 4889.58 | 0 |
| Hlcs | Holocarboxylase synthetase | ENSMUST00000163193 | chr16: 94313192-94313996 | Promoter | 4595.15 | 0 |
| Btbd17 | BTB/POZ domain-containing protein 17 | ENSMUST00000156192 | chr11: 114795002- 114795886 | Promoter | 4541.48 | 0 |
| Erg | Erythroblast transformation-specific transcription factor | ENSMUST00000077773 | chr16: 9539059-95391295 | Intron | 4390.44 | 0 |
Score = −10log10(p value). n/a, Not applicable.
Table 2:
Statistical table
| Figure | Data structure | Type of test | Power |
|---|---|---|---|
| 1D | Quantification of DNA by qPCR (n = 4–6, 2 biological replicates with each having 2–3 technical replicates) | One-way ANOVA followed by Student’s t test (two-tailed, type 2) | *p < 0.05, **p < 1 × 10−4 |
| 3A | Quantification of DNA by qPCR (n = 18, 6 biological replicates with each having 3 technical replicates) | One-way ANOVA followed by Student’s t test (two-tailed, type 2) | *p < 0.05, **p < 1 × 10−4, ***p < 1 × 10−5 |
| 3B | Measurement of luciferase reporter activity (n = 9, 3 biological replicates with each having 3 technical replicates) | Gaussian distribution was confirmed by D’Agostino–Pearson normality test (α = 0.05); one-way ANOVA followed by post hoc Tukey’s test | ****p < 0.0001 |
| 4B | Quantification of mRNA by RT-qPCR (n = 9, 3 biological replicates, with each having 3 technical replicates) | Gaussian distribution was confirmed by D’Agostino–Pearson normality test (α = 0.05); one-way ANOVA followed by post hoc Tukey’s test | *p = 0.023 |
| 4D | Quantification of protein levels by immunoblotting (n = 3 biological replicates) | Unpaired t test (two-tailed, type 2) | p = 0.057 |
| 4G,H | Quantification of protein levels by immunofluorescence (n = 30, from 2 biological replicates) | Nonparametric Kruskal–Wallis test followed by Dunn’s multiple-correction analysis | *p < 0.02,**p = 0.0012, ****p < 0.0001 |
| 6B–F | Quantification of calcium levels by measuring Fluo-3 signal (n = 20–50 per condition) | Nonparametric Kruskal–Wallis test followed by Dunn’s multiple-correction analysis | **p = 0.0073, ***p = 0.0008 |
| 7B–F | Quantification of calcium levels by measuring Fluo-3 signal (n = 20–40 per condition) | Unpaired t test (two-tailed, type 2) | *p < 0.05 |
Initially, we mapped our list of genome-wide NEUROD2 binding sites onto annotated mouse transcripts from the Ensembl database (Hubbard et al., 2002). We discovered that ∼25% (n = 516) of all binding sites mapped onto promoter sequences, as defined by a window flanking both sides of TSSs by 1000 bp, 39% (n = 805) mapped onto intronic or exonic regions outside of the 2000 bp window, and another 36% (n = 743) mapped onto intergenic regions (Fig. 1B). While a majority of NEUROD2 binding sites did not map onto promoter sequences, when they were plotted relative to all TSSs in the mouse genome, a distinct binding preference was observed proximal to TSSs (Fig. 1C).
Next, we validated a number of selected binding sites on four different genes from our dataset, which were associated with a range of statistical confidence intervals [Neurod6 (785.52), Bhlhe2 (531.7), Nrcam (1045.99), Cux1 (706.74); score = −10*log10(p-value)]. For quantifying NEUROD2 binding, we conducted ChIP-qPCR. As a negative control, we used template DNA, which had been chromatin-immunoprecipitated with an unrelated GFP antibody. In additional negative controls, we also performed ChIP-qPCR using primers specific to sequences on other non-target genes (Gad1, Dlx2, Npy, Gsx2, and Calb2). As a result, we observed a significant enrichment of NEUROD2 at all target genes tested, but not at non-target genes Gad1, Dlx2, and Npy (Fig. 1D). Unexpectedly, we also observed a low but significant enrichment at promoters of Gsx2 and Calb2; whether these are biologically relevant or nonfunctional binding events remains to be elucidated.
To gain insight into the biological relevance of all 1,328 genes, which harbored NEUROD2 binding sites within their promoters or gene bodies, we conducted a GO analysis. In agreement with previous reports focused on Neurod2 mutant phenotypes (Ince-Dunn et al., 2006; Wilke et al., 2012; Chen et al., 2016), our analysis revealed dendrite development and synapse organization as the most enriched functional categories (Fig. 1F). Importantly, many of the previously confirmed NEUROD2 targets, such as Dlg4/Psd95 (Wilke et al., 2012) and Cntn2 (Bormuth et al., 2013), as well as previously unknown targets that are key regulators of dendritogenesis and synaptogenesis, such as Grip1, Nrxn3, Nrxn1, and Camk2a, were among those genes enriched in identified GO categories.
Next, we asked whether NEUROD2 binding sites within different locations in the genome corresponded to either transcriptionally active or repressed chromatin. We acquired ChIP-Seq data collected from P0 mouse forebrain using antibodies against specific histone modifications associated with active transcription or silenced chromatin (ENCODE Project Consortium, 2012; www.encodeproject.org). Specifically, we used H3K4me3 as a marker for promoters; H3K4me1, H3K27ac and H3K36me3 as markers of active enhancers; and H3K9me3 and H3K27me3 as markers of repressed chromatin (Zhang et al., 2015). We calculated peak intensities corresponding to different histone modifications within NEUROD2 binding sequences. As a baseline value, we calculated the genome-wide average of peaks corresponding to different histone modifications (Fig. 1E). Our results demonstrated that NEUROD2 binding sequences mapping onto promoter regions were enriched with markers of actively transcribed promoters (H3K4me3) and enhancers (H3K4me1 and H3K27ac), (Fig. 1E). However, binding sites that mapped onto non-promoter gene bodies or onto intergenic regions exhibited only a low level of association with markers of active transcription or enhancers, and many NEUROD2 binding sites did not overlap with markers of either active or repressed transcription (Fig. 1E). Since many of the peaks mapping onto these non-promoter binding sites were among the highest ranking of all NEUROD2 peaks, we argued that at least a subset likely represented functionally relevant binding events as opposed to nonfunctional interactions (Fig. 1B).
NEUROD2 binds to a conserved intronic element within the Stim1 gene
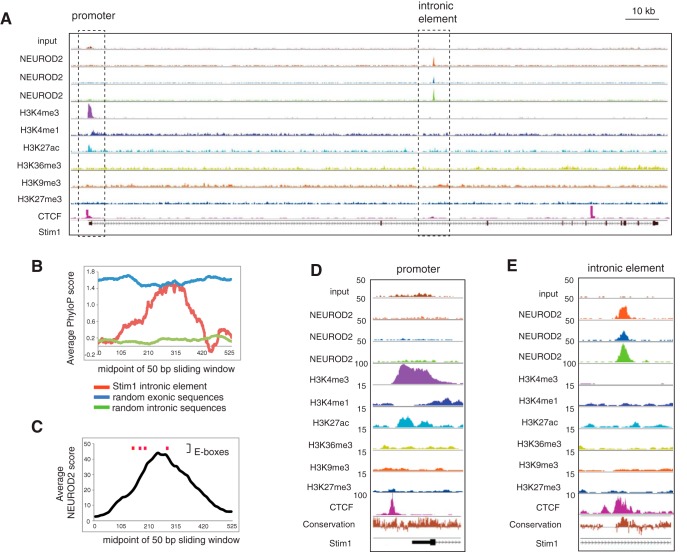
Genome-wide studies of regulatory sequences have demonstrated that development and physiology of the cerebral cortex rely on the regulation of enhancer activity (Malik et al., 2014; Nord et al., 2015) and that neocortical expansion in mammals is substantially driven by changes in cis-regulatory elements (Reilly et al., 2015; Emera et al., 2016; Silver, 2016). However, many of the trans-acting factors that control the activity of these cis-elements and the functional outcomes of such interactions at the cellular and organismal level are largely unknown. Our observation that the majority of NEUROD2 binding sites were located outside of promoter regions suggested that at least a subset of these sites may represent important regulatory sequences that can potentially play critical roles in developmental gene expression regulation in the cortex. Therefore, next we decided to focus on how one of these potential regulatory elements was impacted by NEUROD2 binding. A binding site mapping to the second intron of the Stim1 gene immediately attracted our attention as one of the highest scoring NEUROD2 binding sites (Table 1; Fig. 2A). Stim1 gene encodes for a protein that regulates store-operated calcium entry (Liou et al., 2005). Since in the past it had been suggested that NEUROD2 is a calcium influx activated TF (Ince-Dunn et al., 2006), we decided to investigate the potential regulation of calcium influx by NEUROD2 and therefore selected Stim1 gene for further investigation.
Figure 2.
NEUROD2 binds to a conserved intronic element within the Stim1 gene. A, Input DNA or ChIP-Seq tracks acquired from three separate NEUROD2 antibodies or various histone modifications along the Stim1 gene are plotted. B, The midpoint of a 50 bp sliding window across a 550 bp stretch is plotted as a function of its average evolutionary conservation score (PhyloP score). Blue and green traces represent the average of 20 randomly selected exonic or intronic sequences within the Stim1 gene, respectively. The red trace represents the NEUROD2 binding sequence within intron 2. C, The midpoint of a 50 bp sliding window encompassing the NEUROD2 binding sequence within intron 2 is plotted as a function of NEUROD2 ChIP-Seq score (MACS score from NEUROD2 ChIP-Seq with antibody 2). Red lines denote the locations of E-boxes. D, E, A closer view of Stim1 promoter and intronic element are presented. Enrichment of promoter-associated histone modifications H3K4me3 and H3K27ac are observed proximal to Stim1 TSS. While no NEUROD2 binding is observed at the Stim1 promoter, all three antibodies reveal strong enrichment at a specific sequence within intron 2.
An examination of the Stim1 intronic element revealed, within a 130 bp stretch, a cluster of four E-box elements (CANNTG), which is the consensus sequence for bHLH TFs like NEUROD2. We calculated the frequency of E-box distributions in all intronic sequences found in the mouse genome and found that on average one E-box is observed approximately every ∼180 bp. Therefore, four E-boxes within ∼130 bp represented approximately fivefold enrichment above random chance. Next, we examined evolutionary conservation across the Stim1 intronic element using a tool (PhyloP) that provides a phylogenetic conservation score for each base within a target region, by comparing sequences of placental species (Cooper et al., 2005). We calculated average conservation scores within a 50 bp sliding window across a 550 bp stretch that either spanned the NEUROD2 binding site or randomly selected exonic or intronic sequences. As expected, we observed a much higher level of sequence conservation of randomly selected exons as opposed to introns (Fig. 2B). Surprisingly, Stim1 intronic element also exhibited a high degree of conservation, which was comparable to random exonic sequences. Finally, we also calculated average NEUROD2 ChIP-Seq scores (MACS score) in a similar manner within a 50 bp sliding window across the same 550 bp stretch. As a result we observed a clear correlation between levels of NEUROD2 binding and evolutionary conservation (Fig. 2B,C).
To better understand the potential functional implications of NEUROD2 binding to Stim1 intronic element, we asked whether this location was enriched in histone modifications that are associated with either transcriptional activation or repression. In contrast with our previous global analysis of histone modifications associated with all NEUROD2 binding sites, here we focused only on the Stim1 gene. An analysis of peaks for different histone modifications uncovered a prominent peak for H3K4me3 (promoter marker) and less prominent but still significant peaks for H3K4me1 (enhancer marker) and H3K27ac (marker of transcriptional activity), all of which overlapped with the TSS of Stim1 (Fig. 2A,D). Interestingly, however, the Stim1 intronic element was not associated with histone modifications linked to promoters, enhancers, or actively transcribed or repressed chromatin (Fig. 2A,E). Moreover, NEUROD2 binding was not detected at the Stim1 promoter, as defined by the promoter-specific histone modification H3K4me3 (Fig. 2A,D). Since this prominent NEUROD2 binding site was not associated with markers of either promoters or enhancers, next we decided to investigate the binding profile of a marker for another cis-regulatory element. Toward this aim, we acquired ChIP-Seq data representing binding sites of insulator protein CTCF (CCCTC-binding factor) in P0 mouse forebrain tissue (ENCODE Project Consortium, 2012; encodeproject.org). CTCF is a transcription factor commonly associated with chromatin loop formation (Hnisz et al., 2016). Evidence from recent studies have suggested that chromatin loops represent topologically associated domains, in which promoter activities are regulated by enhancers located within a loop and insulated from outside enhancers (Hnisz et al., 2016; Long et al., 2016). We observed a CTCF binding site that overlapped with NEUROD2 binding site within Stim1 intron 2 (Fig. 2A,E). Our observation raised the possibility that NEUROD2 may function by binding to an insulator element and consequently shielding the Stim1 promoter from the influences of inappropriate enhancer activity.
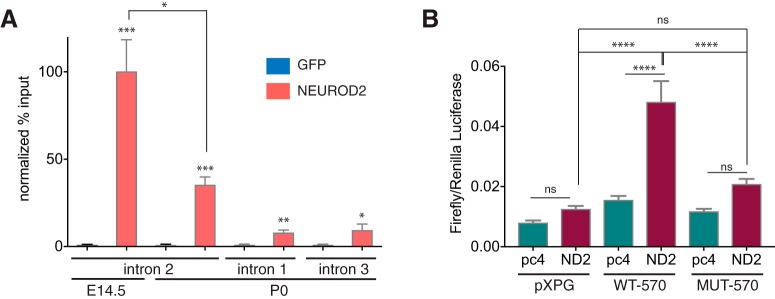
Next, we confirmed NEUROD2 binding to the Stim1 intronic element both in embryonic (E14.5) and postnatal (P0) developmental stages by ChIP-qPCR. As in our previous ChIP-qPCR analyses, as a negative control we used template chromatin DNA immunoprecipitated with a GFP antibody. In additional negative controls, we also performed ChIP-qPCR using primers specific to nontarget Stim1 intronic sequences (introns 1 and 3). As a result, we observed a robust and highly significant enrichment of NEUROD2 at the target Stim1 intronic element within intron 2 (100-fold for E14.5 samples and 35-fold for P0 samples; p < 1 × 10−5; Fig. 3A, Table 2). Notably, reduced levels of enrichment and statistical confidence were detected for sequences located on Stim1 introns 1 and 3, most likely due to the proximity of these sequences to the strong NEUROD2 peak located on intron 2. In summary, our data support a specific binding of NEUROD2 to a conserved Stim1 intronic element that is highly enriched in E-box elements.
Figure 3.
Verification of NEUROD2 binding to the conserved element within Stim1 intron 2. A, NEUROD2 binding to Stim1 intronic element is confirmed in E14.5 and P0 cortices by ChIP-qPCR. ChIP DNA acquired with an unrelated GFP antibody is used as a negative control. Amount of DNA immunoprecipitated with either a NEUROD2 antibody (NEUROD2 ChIP DNA) or GFP antibody (GFP ChIP DNA) is expressed as percentage of input DNA (% input). NEUROD2 % input values are then normalized to GFP % input values. Strong enrichment of NEUROD2 is detected at the NEUROD2 binding element located in Stim1 intron 2 both in E14.5 and P0 cortices. Slight enrichments are observed for Stim1 introns 1 and 3. Data are representative of six biological replicates each composed of three technical replicates. Bars represent SEM. p < 0.0001 determined by one-way ANOVA followed by unpaired t test, *p < 0.05, **p < 1 × 10−4, ***p < 1 × 10−5 (Table 2). B, Luciferase activity is measured from HEK 293T cell lysates that are transfected either with an empty luciferase reporter plasmid (pXPG) or with a luciferase reporter downstream of a wild-type (WT-570) or mutated 570 bp fragment (MUT-570) Stim1 intronic element. In addition, cells are also cotransfected with either an empty (pc4) or NEUROD2 expressing (ND2) pcDNA4 vector. Firefly luciferase activity is normalized to Renilla luciferase signal. Data represent three independent experiments with each sample measured in triplicates. Bars represent SEM. D’Agostino–Pearson test showed normal distribution of the data (α = 0.05). One-way ANOVA and post hoc Tukey’s multiple-comparison analysis was performed, ****p < 0.0001 (Table 2).
Next, we tested whether the four clustered E-boxes localized to the Stim1 intronic element were required for NEUROD2 recruitment. Toward this aim, we cloned a 570 bp fragment composed of the NEUROD2 binding site and encompassing all four E-boxes, upstream of the TSS of the luciferase gene in a reporter construct (WT-570; Bert et al., 2000). We cotransfected this reporter construct along with an empty or a NEUROD2-expressing plasmid into HEK293T cells and measured luciferase activity 24 h later. We observed that when recruited to a site immediately upstream of the TSS of a reporter gene, NEUROD2 significantly activated gene expression (Fig. 3B). This result was not unexpected since numerous previous studies had reported NEUROD2 acting as a transcriptional activator when bound to either endogenous promoter sequences or promoters cloned immediately upstream of reporter genes (McCormick et al., 1996; Ince-Dunn et al., 2006; Fong et al., 2012; Bayam et al., 2015). When we used a mutant reporter construct in which all four E-box elements within the 570 bp fragment were destroyed by site-directed mutagenesis (MUT-570), the capacity of NEUROD2 to influence luciferase gene expression was significantly reduced (Fig. 3B). These results suggested that the E-boxes are required for NEUROD2 recruitment to this particular sequence. However, given that the NEUROD2 binding site on the Stim1 gene was located in an intron, which was far removed from the TSS and was also bound by the chromatin insulator element-associated protein CTCF, we argued that this reporter construct most likely did not faithfully recapitulate the effect of NEUROD2 on endogenous Stim1 expression. Therefore, next we decided to investigate the effect of NEUROD2 on Stim1 expression from its genomic locus.
NEUROD2 limits Stim1 gene expression in cortical neurons
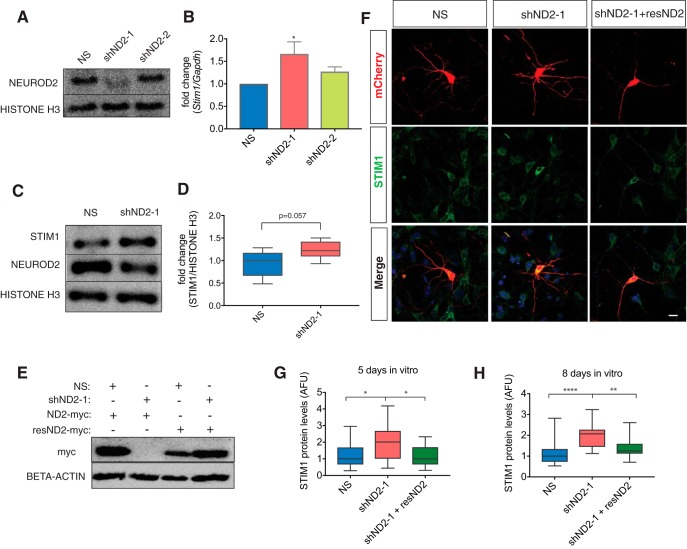
To determine the effect of NEUROD2 on Stim1 expression, we knocked down Neurod2 in mouse primary cortical neurons via transfection of shRNA and quantified Stim1 expression. To achieve a high efficiency of transfection with primary neurons, we used an electroporation-based method (nucleofection) and achieved ∼80–90% transfection efficiency. Initially, we tested two separate shRNAs and found that shND2-1 knocked down Neurod2 expression much more efficiently compared with shND2-2 (Fig. 4A). Interestingly, compared with a NS shRNA, Stim1 mRNA, and protein levels increased ∼1.3- to 1.5-fold on knockdown of Neurod2 with the more effective shND2-1 and changed minimally with the less effective shND2-2 (Fig. 4B–D). Therefore, for the remaining experiments we decided to use shND2-1, which knocked down NEUROD2 levels by ∼65% of the control and subsequently resulted in aberrant Stim1 upregulation (Fig. 4A–D).
Figure 4.
NEUROD2 suppresses Stim1 expression. A, Immunoblotting analysis reveals that two separate shRNAs (shND2-1 and shND-2) can suppress Neurod2 expression compared with a nonsilencing shRNA (NS) in primary cortical cultures with different efficiencies. B, Neurod2 mRNA levels normalized to Gapdh mRNA is measured by RT-qPCR in cortical cultures transfected with either shND2-1 or shND2-2. While both shRNAs induce an upregulation of Stim1 mRNA, the effect of the more potent shND2-1 is greater. Data represent three biological replicates, each with three technical replicates. One-way ANOVA followed by post hoc Tukey’s test, *p = 0.023. C, D, Primary cortical cultures were transfected with NS shRNA and shND2-1. STIM1 protein levels were quantified by immunoblotting and normalized to histone H3 loading control. Data are presented as bar graphs; the line marks the median; the box represents the 25th and 75th percentiles; top and bottom whiskers mark minima and maxima, respectively. Unpaired t test, p = 0.057. E, resND2-myc, a cDNA resistant to shND2-1, was generated. HEK293T cells were transfected with NS shRNA or shND2-1, along with either ND2-myc or resND2-myc cDNAs. Immunoblotting analysis against the myc epitope revealed that while shND2-1 completely knocked down the expression of ND2-myc, resND2-myc expression was not affected. F, Primary cortical neurons were transfected at low efficiency with shND2-1 either alone or together with resND2-myc and immunofluorescently stained against STIM1 protein. Transfected cells were identified based on their coexpression of mCherry from the shRNA-expressing plasmid. G, H, Quantification of STIM1 immunofluorescence signals from experiments presented in F. Experimenter was blinded to all sample identity during staining and quantification. n = 30 for each condition from two independent experiments. Scale bar, 20 µm. Data are presented as bar graphs; the line marks the median; the box represents the 25th and 75th percentiles; top and bottom whiskers mark minima and maxima, respectively. Nonparametric Kruskal–Wallis test was followed by Dunn’s multiple-comparison analysis, *p < 0.02, **p = 0.0012, ****p < 0.0001 (Table 2).
Next, we questioned whether knocking down NEUROD2 expression in the majority of neurons in culture could potentially cause an unusual culture environment and result in secondary, cell-nonautonomous effects on Stim1 expression. To eliminate this possibility, we transfected shND2-1 at a low efficiency into neurons and quantified STIM1 protein levels by immunofluorescent staining with the experimentalist blinded to sample identity (Fig. 4F). Consistent with our RT-qPCR and immunoblotting results, we observed a significant increase in STIM1 expression after Neurod2 knock down (Fig. 4G,H). Importantly, we were able to rescue STIM1 levels by cotransfection of an shRNA-resistant mouse Neurod2 cDNA, which was identical in its amino acid sequence to wild-type NEUROD2 (resND2; Fig. 4E–H). Finally, we also determined whether or not the mRNA level of Stim2, another sensor of ER Ca2+ and regulator of SOCE, was also affected after Neurod2 suppression, and we observed a slight enrichment that was not statistically significant (1.5-fold enrichment; p > 0.05). Together, we concluded that NEUROD2 binding to the Stim1 intronic element is correlated with a NEUROD2-dependent decrease in Stim1 expression.
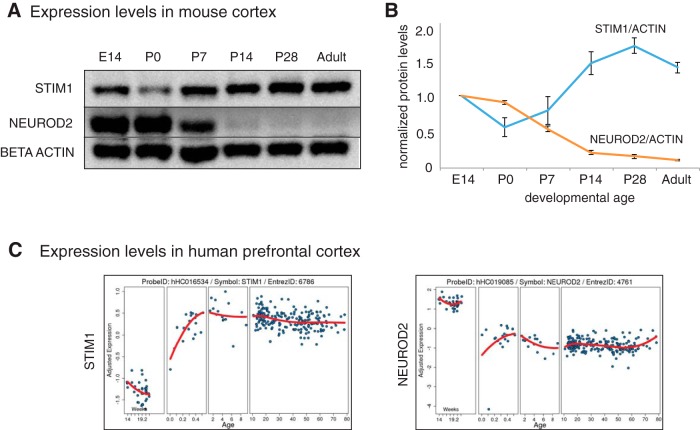
Based on our results, we argued that a model in which NEUROD2 limited Stim1 expression would be consistent with an inverse correlation in the levels of these two proteins across cortical development. Therefore, we determined NEUROD2 and STIM1 protein levels in various developmental ages in cortical tissue in mice (Fig. 5A). Our results demonstrated that, throughout development, a downregulation of NEUROD2 was correlated with an upregulation of STIM1 levels (Fig. 5A,B). Interestingly, a similar inverse correlation between Neurod2 and Stim1 mRNA levels was observed in prefrontal cortical tissue obtained across the human lifespan (Fig. 5C; braincloud.jhmi.edu/; Colantuoni et al., 2011). Together, our results are consistent with NEUROD2 acting as a brake on STIM1 expression in cortical neurons during development. However, since Neurod2 is expressed only in excitatory neurons and Stim1 is expressed in progenitors, neurons, as well as astrocytes (Kraft, 2015), the protein levels quantified by bulk analysis of cortical tissue reflected cumulative STIM1 levels from all these cell types. Future experimentation will help to clarify cell type-specific regulation of Stim1 expression by NEUROD2 across cortical development.
Figure 5.
NEUROD2 and STIM1 expression are inversely correlated across cortical development. A, B, Immunoblotting analysis and quantification of protein levels across development in cerebral cortical tissue revealed an inverse correlation between NEUROD2 and STIM1 protein expression. STIM1 protein levels are normalized to the amount of β-actin. Data represents two biological replicates, each quantified as duplicates. Bars represent the SEM. C, Stim1 and Neurod2 mRNA levels were plotted as a function of age in human prefrontal cortex using postmortem tissue. Plots were acquired from braincloud.jhmi.edu (Colantuoni et al., 2011). Similar to mouse data, an inverse correlation in Neurod2 and Stim1 expression was observed in humans as well.
NEUROD2 is required for fine-tuning of SOCE in cortical neurons
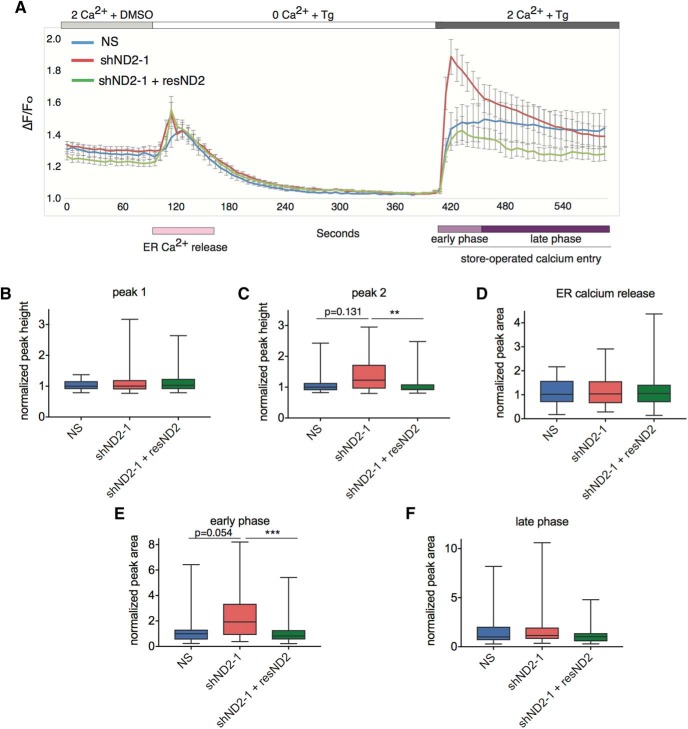
SOCE constitutes an important source of calcium entry and signaling in neurons. Briefly, depletion of ER Ca2+ stores causes the ER Ca2+ sensor STIM proteins (STIM 1 and STIM2) to interact with and activate cell surface Ca2+ release-activated Ca2+ (CRAC) channels, thereby resulting in a second wave of cytoplasmic Ca2+ rise (Moccia et al., 2015). Genetic suppression of Stim1 or CRAC channels, or pharmacological blockade of CRAC channels, results in complete abrogation of this second wave of calcium rise that constitutes SOCE (Somasundaram et al., 2014). Since our data suggested that knockdown of Neurod2 caused an upregulation in STIM1 levels, we next asked whether this effect also translated into deranged neuronal SOCE. We transfected primary cortical neurons with either NS shRNA or shND2-1 and measured cytoplasmic Ca2+ levels with the Ca2+-sensitive dye Fluo-3. We imaged transfected neurons selected by the coexpression of the marker gene mCherry. ER Ca2+ stores were released by treatment with 5 µm thapsigargin, a blocker of SERCA (sarcoplasmic/endoplasmic reticulum Ca2+ ATPase) pump. As expected, on thapsigargin treatment of neurons bathed in Ca2+-free buffer, we observed an initial wave of cytoplasmic Ca2+ increase both in control and NEUROD2-depleted neurons at comparable levels (Fig. 6A,B,D). As long as neurons were kept in Ca2+-free buffer, the ER stores remained empty, a situation that was presumably sensed by the Ca2+ sensor STIM1. On switching to a buffer containing 2 mM Ca2+, an immediate SOCE response was observed as a second wave of cytoplasmic Ca2+ rise. Consistent with a role for NEUROD2 in Stim1 regulation, we observed an upregulation of SOCE in neurons depleted of NEUROD2, a response that was rescued by coexpression of shRNA-resistant resND2 (Fig. 6A,C,E). Specifically, on induction of SOCE, a significant upregulation of Ca2+ influx above control levels was detected during the early time periods (∼50 s) following NEUROD2 depletion, although this difference was attenuated during the later phase of SOCE (Fig. 6E,F).
Figure 6.
Suppression of Neurod2 expression results in increased SOCE response. A, Primary cortical cultures were transfected with NS shRNA or shND2-1 together with either an empty or shRNA-resistant Neurod2 (resND2) expressing pcDNA4 vector. On the day of imaging, cultures were loaded with calcium-sensitive dye Fluo-3 and imaged by live imaging. Baseline signal was acquired by bathing the cells in Ringer’s buffer containing 2 µm Ca2+. Upon treatment with thapsigargin (Tg) and withdrawal of extracellular Ca2+, a first wave of rise in signal was observed that corresponded to emptying of ER Ca2+ stores (at ∼100 s). A second wave of signal was observed on providing Ca2+ containing Ringer’s buffer that corresponded to store-operated calcium entry (at ∼400 s). B, C, Quantification of initial peak heights for first wave (ER Ca2+ release) and second wave (SOCE) of Ca2+ signals unveiled an increase in SOCE on Neurod2 knockdown that was rescued by coexpression of resND2. D–F, Measurement of the total area under the peaks revealed that ER Ca2+ release was not affected; however, the early phase (∼50 s) but not the late phase of SOCE was significantly upregulated upon Neurod2 suppression. Traces are color coded as follows: NS shRNA (blue); shND2-1 (red); and shND2-1 + resND2 (green). Data are presented as bar graphs; the line marks the median; the box represents the 25th and 75th percentiles; top and bottom whiskers mark minima and maxima, respectively. Neurod2 is abbreviated as ND2. Nonparametric Kruskal–Wallis test was followed by Dunn’s multiple-comparison analysis, **p = 0.0073, ***p = 0.0008 (Table 2).
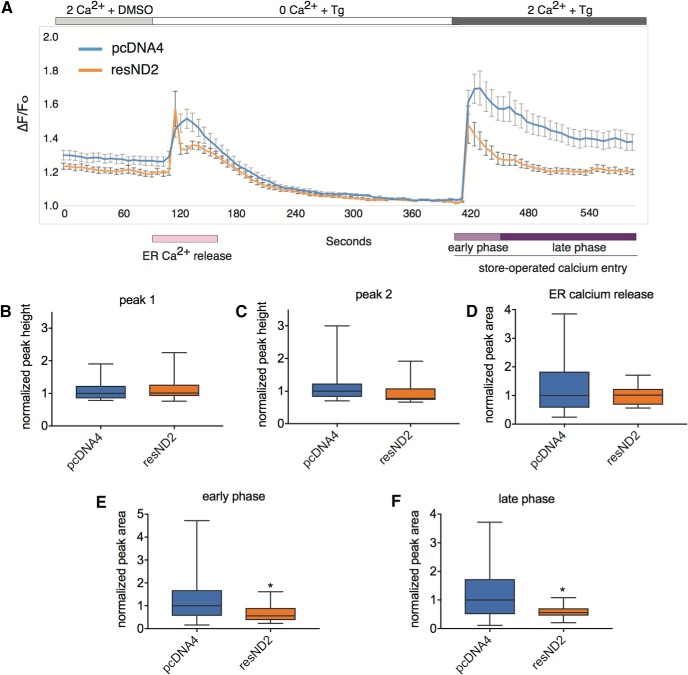
Next we tested the effects of Neurod2 overexpression on SOCE response. We transfected primary cortical neurons with an empty vector or a vector overexpressing resND2 in otherwise wild-type neurons. In agreement with our prediction, we observed a significant suppression of SOCE (Fig. 7A,E,F) but not of steady-state ER Ca2+ levels (Fig. 7A,B,D) in neurons overexpressing NEUROD2 protein. Together, our results suggest that NEUROD2 fine-tunes neuronal SOCE by inhibiting STIM1 expression and that the level of SOCE response is regulated by the amount of NEUROD2 protein available in the cell.
Figure 7.
Overexpression of Neurod2 reduces the SOCE response. A, Primary cortical neurons were transfected with either empty or resND2 expressing pcDNA4 vector, and calcium imaging was performed as described in Figure 6A. B, C, Measurement of peak heights of first- and second-wave of Ca2+ signals revealed that overexpression of Neurod2 in otherwise wild-type neurons causes a suppression of SOCE but does not affect steady-state levels of ER Ca2+. D–F, Calculation of total area under peaks demonstrated that both late and early phases of SOCE are downregulated upon Neurod2 overexpression. Traces are color coded as follows: pcDNA4 (blue) and resND2 (orange). Bars represent the SEM. Unpaired t test determined the p value: *p < 0.05. Data are presented as bar graphs; the line marks the median; the box represents the 25th and 75th percentiles; top and bottom whiskers mark minima and maxima, respectively (Table 2). ND2, Neurod2.
Discussion
Implications of NEUROD2-regulated store-operated calcium entry
Almost all phases of cortical development from neural progenitor proliferation to axon guidance and synaptic plasticity are regulated by a range of calcium signaling pathways activated by different sources of calcium influx, including intracellular stores and SOCE (Uhlén et al., 2015). In contrast to Ca2+ influx through voltage- and neurotransmitter-gated calcium channels, little is known about how SOCE influences neuronal differentiation during cortical development. Accumulating evidence indicates that SOCE exists in neurons, where it is linked to neuronal growth cone turning, axon guidance, synaptic plasticity, and regulation of neuronal excitability (Park et al., 2010; Kraft, 2015; Majewski and Kuznicki, 2015; Moccia et al., 2015). However, how SOCE itself is regulated in neurons, which cellular components are involved, how they interact with different signaling pathways, and the nature of neuronal gene expression programs that establish and control SOCE are poorly understood. Our genome-wide target gene analysis has identified an unanticipated role for NEUROD2 during early postnatal cortical development as a suppressor of SOCE. Our results reveal that NEUROD2 binds to the second intron of the Stim1 gene, and this association is linked to a decrease in Stim1 expression. We have demonstrated that while Neurod2 knockdown causes an increased SOCE response, its overexpression results in repressed SOCE. However, whether Stim1 is the sole NEUROD2 target or whether additional target genes that control SOCE exist remains to be determined.
STIM proteins (STIM1 and STIM2) are ER Ca2+ store sensors that interact with plasma membrane-localized CRAC channels on ER Ca2+ depletion, inducing their opening and providing a subsequent wave of Ca2+ influx to refill the ER (Liou et al., 2005; Roos et al., 2005; Zhang et al., 2005). SOCE and specifically, the roles of Stim genes (Stim1 and Stim2) are increasingly gaining attention as critical regulators of neuronal Ca2+ signaling and homeostasis. Recently, it was demonstrated that Stim1 regulates axonal branching, growth cone guidance, cell migration, and neural progenitor cell proliferation (Mitchell et al., 2012; Shim et al., 2013; Somasundaram et al., 2014; Tsai et al., 2014). In particular, it appears that SOCE is tightly regulated within spatiotemporally confined subcellular domains, at least in part by the localization of STIM1 to compartments, such as the growth cones of pathfinding axons. It is noteworthy to mention that a downregulation of SOCE and upregulation of voltage-gated Ca2+ influx is observed as neural progenitors mature toward a neuronal fate within the developing cortex (Maric et al., 2000; D'Ascenzo et al., 2006; Somasundaram et al., 2014). The functional implications for this switch in the mode of calcium influx are currently unknown. The acquisition of an excitable membrane might allow neurons to activate additional signaling pathways, such as those activated by voltage-gated Ca2+ channels (VGCCs), that would be inaccessible in other cell types. Since the initiation of Neurod2 expression also overlaps with neural progenitor cell cycle exit and differentiation to post-mitotic neurons, it will be interesting to explore whether and how NEUROD2 controls the transition from SOCE-mediated to voltage-mediated Ca2+ influx. Based on our results, we can sufficiently conclude that NEUROD2 is required for limiting Stim1 expression in postmitotic neurons; however, future experiments will be required to understand the exact stage of neuronal differentiation in which this NEUROD2-dependent effect is initially observed.
Several studies have demonstrated STIM1-dependent reciprocal regulation between SOCE- and depolarization-induced Ca2+ influx. While the inhibition of Stim1 caused a decrease in SOCE response, as expected, it also induced augmentation of Ca2+ influx through L-type VGCCs (Park et al., 2010; Wang et al., 2010). Interestingly, NEUROD2 transcriptional activity is induced on Ca2+ influx through VGCCs (Aizawa et al., 2004; Ince-Dunn et al., 2006). Together, one can imagine a scenario in which depolarization-induced calcium influx activates NEUROD2, which in turn calibrates levels of STIM1 and SOCE and, consequently, STIM1-dependent inhibition of L-type VGCCs.
To date, the role of STIM1 in radial migration of neurons in the developing cortex has not been investigated. However, a recent study investigating sheet migration of endothelial cells (human umbilical vein endothelial cells) demonstrate that STIM1 localizes to the leading edges of migrating cells (Tsai et al., 2014). Further gain-of-function and loss-of-function experiments in this study support a model where STIM1 is a negative regulator of migration speed. Interestingly, NEUROD2 is highly expressed within the cortex during the peak of radial migration (Telley et al., 2016). Although, endothelial sheet migration and neuronal radial migration most likely differ substantially from each other, we speculate that NEUROD2 might promote migration by suppressing a repressor of migration. On the other hand, additional studies investigating STIM1 impact on cell migration speed using various cancer cell lines have reported opposite results, suggesting that STIM1 is an accelerator of migration speed (Yang et al., 2009; Chen et al., 2011). While the involvement of STIM1 in the migration process is evident, its cell type-specific effects necessitate further investigations in relevant tissue and developmental stages.
NEUROD2 as a transcriptional regulator
Our data demonstrate that one of the most prominent NEUROD2 binding sites in the mouse genome maps to a conserved intronic element enriched in E-boxes within the Stim1 gene. Further, we demonstrate that shRNA-mediated suppression of Neurod2 expression is correlated with an upregulation of Stim1 mRNA and protein abundance. Our results uncover a novel mode of gene expression regulation by NEUROD2. To date, NEUROD2 has been presumed to act only as a transcriptional activator. Interestingly, in equivalent tissue and developmental stages, this element is also bound by CTCF, a transcription factor associated with chromatin insulators. Collectively, our results are consistent with a model in which NEUROD2 can fine-tune gene expression by functioning as part of an insulator complex and preventing the promoter from spuriously associating with enhancers located outside of the insulated neighborhood. Consequently, a downregulation of NEUROD2 protein levels may render the Stim1 promoter vulnerable to the influence of an enhancer that can now ectopically promote transcription. It is generally acknowledged that, while not all CTCF peaks function as insulators, a large majority of insulators are bound by CTCF (Hnisz et al., 2016). Therefore, whether this Stim1 intronic element is functioning as an insulator is currently unknown. In future experiments, we are very interested in further pursuing this possibility and testing whether this intronic element is required for limiting Stim1 gene expression in a NEUROD2-dependent manner in neurons.
Our findings add to a series of recent discoveries of several neuronally expressed TFs limiting gene expression by binding to intronic elements (Wiegreffe et al., 2015; Rannals et al., 2016; Soldner et al., 2016). Most relevant to our findings is a study exploring TCF4, a ubiquitously expressed bHLH factor known to heterodimerize with tissue-specific bHLH TFs, including NEUROD2 (Ravanpay and Olson, 2008). The authors demonstrate that TCF4 binding to intronic elements within the Scn10a and Kcnq1 genes, both encoding ion channels, induces a suppression of gene expression, which is associated with a deregulation of intrinsic excitability of cortical neurons (Rannals et al., 2016). In light of the finding that TCF4 biochemically interacts with NEUROD2 (Ravanpay and Olson, 2008), it is of future interest to test whether NEUROD2 and TCF4 functionally interact on specific sites to regulate gene expression.
In summary, our genome-wide target gene analysis of the neurogenic TF NEUROD2 has uncovered a novel gene expression mechanism by which differentiating cortical excitatory neurons fine-tune the extent of SOCE. We believe that future analyses of our results in the context of other high-throughput datasets representing TF binding, gene expression, chromatin accessibility, and conformation will provide valuable insights into how gene regulatory circuits function in guiding the differentiation of cortical excitatory neurons.
Acknowledgments
Acknowledgments: We thank Cory D. Dunn for feedback on this manuscript. We also thank Ali C. Taskin, Ahmet Kocabay, and Mehmet Yucel for their help in providing timed pregnant mice. In addition, we thank Tamer Onder and Gulben Gurhan for assistance in usage of the nucleofector.
Synthesis
Reviewing Editor: Orly Reiner, Weizmann Institute of Science
Decisions are customarily a result of the Reviewing Editor and the peer reviewers coming together and discussing their recommendations until a consensus is reached. When revisions are invited, a fact-based synthesis statement explaining their decision and outlining what is needed to prepare a revision will be listed below. The following reviewer(s) agreed to reveal their identity: Jean-Bernard Manent, Takeshi Kawauchi.
Overall the reviewers have found that your study is interesting and worthy. They note that this is an interesting study suggesting that NeuroD2 may contribute to regulating calcium signaling and homeostasis in maturing neurons through controlling expression of the Stim1 gene, which encodes an ER calcium sensor.
However, they do find that several issues need to be addressed prior to publication, they believe that their comments can be resolved within two months of work. Their specific comments are summarized below:
Reviewer 1
In this study, the authors suggest that NeuroD2 may regulate expression of the Stim1 gene, which encodes an ER calcium sensor essential for controlling store-operated calcium entry. These are interesting novel findings, although several comments have to be addressed and additional evidences provided, especially for addressing comments #2 and #3.
1) From the provided material and text (‘we selected one of the highest scoring NEUROD2 binding sites’ pg15 ln7), it is unclear why the authors decided to focus their study on the second intron of the Stim1 gene among many other potential binding sites. What were the criteria and scoring method? How was the scoring cutoff defined? Etc. Clarification would help the reader a lot, and may be accompanied with a table showing other genes/binding sites.
2) Because they claim that NEUROD2 acts as a transcriptional repressor on Stim1 expression through binding E-box elements on intronic sequences of the Stim1 gene, the authors may wish to provide evidences that altering the sequence of these E-boxes (or removing them) results in increased Stim1 expression. If feasible, this would provide with more direct evidences than those of the NeuroD2 knockdown experiments.
3) Conclusions of the NeuroD2 experiments are somewhat overstated (‘NEUROD2 binding to Stim1 intronic element is associated with suppression of gene expression’ pg17, ln15). From the conducted experiments, the authors can reasonably conclude than knocking down NeuroD2 results in increased Stim1 mRNA and protein levels, but could hardly claim that this effect is a direct consequence of a reduced binding to the Stim1 gene intronic element (‘we concluded that NEUROD2 binding to the Stim1 intronic element is associated with a NEUROD2-dependent suppression of Stim1 expression’, pg 18 ln20-21). Direct evidences such as those suggested in the above comment are missing.
4) There is a general trend toward improving data representation in the scientific literature and Neurosciences so that distributional information regarding individual data points is available for the reader, especially for small samples. Distributional differences between conditions would be much more convincing if boxplots or scatterplots are utilized in the figures, especially for the Ca imaging experiments, as suggested in (Allen et al., Neuron 2012; Weissgerber et al., Plos Biol 2015).
5) The authors may wish to cite and discuss the results of a recent study utilizing a similar ChIP-seq analysis of NeuroD2 targets from murine embryonic neocortex (Bayam et al BMC Genomics 2015). Their results are complementary to those of the Bayam et al study.
Reviewer 2
The manuscript describes an upstream regulator for store-operated calcium entry (SOCE). The authors show that NeuroD2, a bHLH transcription factor, negatively regulates the expression of Stim1, a major ER calcium sensor, via the direct binding to the intron 2 of a Stim1 gene. They also show that knockdown or overexpression of NeuroD2 increases or decreases SOCE response, respectively. This study also provides evidence that NeuroD2 functions as a transcriptional repressor (in addition to an activator as previously described). The findings described in this manuscript are informative to many developmental neuroscientists, but there are several issues that should be improved.
[Major points]
1) Neuronal differentiation occurs in a stepwise fashion and a developing brain (e.g. E14.5 cerebral cortex) contains cells with several differentiation stages, including radial glial progenitors (Pax6+), intermediate progenitors (Tbr1+) and newly generated neurons (Tuj1+). As the authors mentioned, SOCE is downregulated in a subset of neuron-committed progenitors, but it is unclear whether these progenitors are Neurogenin-positive or NeuroD-positive or Tbr2-positive. To clarify it, immunohistochemistry experiments for NeuroD2 and Stim1 (and markers for neural progenitors or neurons) in the developing cerebral cortex are required.
2) It is strange that the culture periods are different in Fig. 4A-C (4 div), Fig. 4D-F (5 div) and Figs. 6 and 7 (7-9 div). The authors should consolidate the same culture period in all experiments. If there is a special reason, they should explain it and provide data of the expression levels of NeuroD2 and Stim1 in each time point. Particularly, the experiments in Figs. 6 and 7 should use the same stage of primary neurons.
3) In Fig. 4A-C, shRNAs were transfected immediately prior to plating, which may lead to defects in neuronal differentiation. Do these cells exhibit normal morphologies? The representative images are required. If neurite length would be short in the knockdown cells, actin (or tubulin) would not be appropriate for the loading control.
4) Neurons in the lower panels of Fig. 4E do not seem to be healthy. They do not extend their neurites.
[Minor points]
5) Student's t-test is not appropriate for statistical analyses of rescue experiments in Figs. 4F and 6B-F. Multiple comparison analyses should be used.
6) No error bars in Fig. 5B.
7) In Fig. 7, wild type-NeuroD2, but not rNeuroD2, should be used.
References
- Aizawa H, Hu SC, Bobb K, Balakrishnan K, Ince G, Gurevich I, Cowan M, Ghosh A (2004) Dendrite development regulated by CREST, a calcium-regulated transcriptional activator. Science 303:197–202. 10.1126/science.1089845 [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25:25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayam E, Sahin GS, Guzelsoy G, Guner G, Kabakcioglu A, Ince-Dunn G (2015) Genome-wide target analysis of NEUROD2 provides new insights into regulation of cortical projection neuron migration and differentiation. BMC Genomics 16:681 10.1186/s12864-015-1882-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bert AG, Burrows J, Osborne CS, Cockerill PN (2000) Generation of an improved luciferase reporter gene plasmid that employs a novel mechanism for high-copy replication. Plasmid 44:173–182. 10.1006/plas.2000.1474 [DOI] [PubMed] [Google Scholar]
- Bormuth I, Yan K, Yonemasu T, Gummert M, Zhang M, Wichert S, Grishina O, Pieper A, Zhang W, Goebbels S, Tarabykin V, Nave KA, Schwab MH (2013) Neuronal basic helix-loop-helix proteins Neurod2/6 regulate cortical commissure formation before midline interactions. J Neurosci 33:641–651. 10.1523/JNEUROSCI.0899-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busskamp V, Lewis NE, Guye P, Ng AH, Shipman SL, Byrne SM, Sanjana NE, Murn J, Li Y, Li S, Stadler M, Weiss R, Church GM (2014) Rapid neurogenesis through transcriptional activation in human stem cells. Mol Syst Biol 10:760 10.15252/msb.20145508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Moran JT, Zhang Y, Ates K, Yu D, Schrader LA, Das PM, Jones FE, Hall BJ (2016) The transcription factor NeuroD2 coordinates synaptic innervation and cell intrinsic properties to control excitability of cortical pyramidal neurons. J Physiol 594:3729–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YF, Chiu WT, Chen YT, Lin PY, Huang HJ, Chou CY, Chang HC, Tang MJ, Shen MR (2011) Calcium store sensor stromal-interaction molecule 1-dependent signaling plays an important role in cervical cancer growth, migration, and angiogenesis. Proc Natl Acad Sci U S A 108:15225–15230. 10.1073/pnas.1103315108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, Colantuoni EA, Elkahloun AG, Herman MM, Weinberger DR, Kleinman JE (2011) Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature 478:519–523. 10.1038/nature10524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, Stone EA, Asimenos G, Green ED, Batzoglou S, Sidow A (2005) Distribution and intensity of constraint in mammalian genomic sequence. Genome Res 15:901–913. 10.1101/gr.3577405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Ascenzo M, Piacentini R, Casalbore P, Budoni M, Pallini R, Azzena GB, Grassi C (2006) Role of L-type Ca2+ channels in neural stem/progenitor cell differentiation. Eur J Neurosci 23:935–944. [DOI] [PubMed] [Google Scholar]
- Emera D, Yin J, Reilly SK, Gockley J, Noonan JP (2016) Origin and evolution of developmental enhancers in the mammalian neocortex. Proc Natl Acad Scie U S A 113:E2617–E2626. 10.1073/pnas.1603718113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489:57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farah MH, Olson JM, Sucic HB, Hume RI, Tapscott SJ, Turner DL (2000) Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development 127:693–702. [DOI] [PubMed] [Google Scholar]
- Fode C, Ma Q, Casarosa S, Ang SL, Anderson DJ, Guillemot F (2000) A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev 14:67–80. [PMC free article] [PubMed] [Google Scholar]
- Fong AP, Yao Z, Zhong JW, Cao Y, Ruzzo WL, Gentleman RC, Tapscott SJ (2012) Genetic and epigenetic determinants of neurogenesis and myogenesis. Dev Cell 22:721–735. 10.1016/j.devcel.2012.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, Miller W, Kent WJ, Nekrutenko A (2005) Galaxy: a platform for interactive large-scale genome analysis. Genome Res 15:1451–1455. 10.1101/gr.4086505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, Coraor N, Nekrutenko A, Taylor J (2012) NGS analyses by visualization with Trackster. Nat Biotechnol 30:1036–1039. 10.1038/nbt.2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig LC, Woodworth MB, Galazo MJ, Padmanabhan H, Macklis JD (2013) Molecular logic of neocortical projection neuron specification, development and diversity. Nat Rev Neurosci 14:755–769. 10.1038/nrn3586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Day DS, Young RA (2016) Insulated neighborhoods: structural and functional units of mammalian gene control. Cell 167:1188–1200. 10.1016/j.cell.2016.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, Cox T, Cuff J, Curwen V, Down T, Durbin R, Eyras E, Gilbert J, Hammond M, Huminiecki L, Kasprzyk A, Lehvaslaiho H, Lijnzaad P, Melsopp C, Mongin E, et al. (2002) The Ensembl genome database project. Nucleic Acids Res 30:38–41. 10.1093/nar/30.1.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imayoshi I, Kageyama R (2014) bHLH factors in self-renewal, multipotency, and fate choice of neural progenitor cells. Neuron 82:9–23. 10.1016/j.neuron.2014.03.018 [DOI] [PubMed] [Google Scholar]
- Ince-Dunn G, Hall BJ, Hu SC, Ripley B, Huganir RL, Olson JM, Tapscott SJ, Ghosh A (2006) Regulation of thalamocortical patterning and synaptic maturation by NeuroD2. Neuron 49:683–695. 10.1016/j.neuron.2006.01.031 [DOI] [PubMed] [Google Scholar]
- Jo AY, Park CH, Aizawa S, Lee SH (2007) Contrasting and brain region-specific roles of neurogenin2 and mash1 in GABAergic neuron differentiation in vitro. Exp Cell Res 313:4066–4081. 10.1016/j.yexcr.2007.08.026 [DOI] [PubMed] [Google Scholar]
- Johnson MB, Wang PP, Atabay KD, Murphy EA, Doan RN, Hecht JL, Walsh CA (2015) Single-cell analysis reveals transcriptional heterogeneity of neural progenitors in human cortex. Nat Neurosci 18:637–646. 10.1038/nn.3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D (2002) The human genome browser at UCSC. Genome Res 12:996–1006. 10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EJ, Hori K, Wyckoff A, Dickel LK, Koundakjian EJ, Goodrich LV, Johnson JE (2011) Spatiotemporal fate map of neurogenin1 (Neurog1) lineages in the mouse central nervous system. J Comp Neurol 519:1355–1370. 10.1002/cne.22574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi M, Doe CQ (2013) Temporal fate specification and neural progenitor competence during development. Nat Rev Neurosci 14:823–838. 10.1038/nrn3618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft R (2015) STIM and ORAI proteins in the nervous system. Channels 9:245–252. 10.1080/19336950.2015.1071747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KY, Sestan N, Anton ES (2012) Transcriptional co-regulation of neuronal migration and laminar identity in the neocortex. Development 139:1535–1546. 10.1242/dev.069963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone DP, Srinivasan K, Chen B, Alcamo E, McConnell SK (2008) The determination of projection neuron identity in the developing cerebral cortex. Curr Opin Neurobiol 18:28–35. 10.1016/j.conb.2008.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, Hansen S, Wang Z, Storm DR, Tapscott SJ, Olson JM (2005) The dosage of the neuroD2 transcription factor regulates amygdala development and emotional learning. Proc Natl Acad Sci U S A 102:14877–14882. 10.1073/pnas.0506785102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, Stoeck J, Ravanpay AC, Guillemot F, Tapscott SJ, Olson JM (2004) Regulation of neuroD2 expression in mouse brain. Devel Biol 265:234–245. [DOI] [PubMed] [Google Scholar]
- Liou J, Kim ML, Heo WD, Jones JT, Myers JW, Ferrell JE Jr, Meyer T (2005) STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr Biol 15:1235–1241. 10.1016/j.cub.2005.05.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long HK, Prescott SL, Wysocka J (2016) Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell 167:1170–1187. 10.1016/j.cell.2016.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski L, Kuznicki J (2015) SOCE in neurons: signaling or just refilling? Biochim Biophys Acta 1853:1940–1952. 10.1016/j.bbamcr.2015.01.019 [DOI] [PubMed] [Google Scholar]
- Malik AN, Vierbuchen T, Hemberg M, Rubin AA, Ling E, Couch CH, Stroud H, Spiegel I, Farh KK, Harmin DA, Greenberg ME (2014) Genome-wide identification and characterization of functional neuronal activity-dependent enhancers. Nat Neurosci 17:1330–1339. 10.1038/nn.3808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maric D, Maric I, Barker JL (2000) Developmental changes in cell calcium homeostasis during neurogenesis of the embryonic rat cerebral cortex. Cereb Cortex 10:561–573. 10.1093/cercor/10.6.561 [DOI] [PubMed] [Google Scholar]
- Mattar P, Langevin LM, Markham K, Klenin N, Shivji S, Zinyk D, Schuurmans C (2008) Basic helix-loop-helix transcription factors cooperate to specify a cortical projection neuron identity. Mol Cell Biol 28:1456–1469. 10.1128/MCB.01510-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick MB, Tamimi RM, Snider L, Asakura A, Bergstrom D, Tapscott SJ (1996) NeuroD2 and neuroD3: distinct expression patterns and transcriptional activation potentials within the neuroD gene family. Mol Cell Biol 16:5792–5800. 10.1128/MCB.16.10.5792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell CB, Gasperini RJ, Small DH, Foa L (2012) STIM1 is necessary for store-operated calcium entry in turning growth cones. J Neurochem 122:1155–1166. 10.1111/j.1471-4159.2012.07840.x [DOI] [PubMed] [Google Scholar]
- Moccia F, Zuccolo E, Soda T, Tanzi F, Guerra G, Mapelli L, Lodola F, D'Angelo E (2015) Stim and Orai proteins in neuronal Ca(2+) signaling and excitability. Front Cell Neurosci 9:153. 10.3389/fncel.2015.00153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Kawamura R, Funabashi H, Mie M, Kobatake E (2006) Transduction of NeuroD2 protein induced neural cell differentiation. J Biotechnol 126:230–236. 10.1016/j.jbiotec.2006.04.021 [DOI] [PubMed] [Google Scholar]
- Nord AS, Pattabiraman K, Visel A, Rubenstein JL (2015) Genomic perspectives of transcriptional regulation in forebrain development. Neuron 85:27–47. 10.1016/j.neuron.2014.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson JM, Asakura A, Snider L, Hawkes R, Strand A, Stoeck J, Hallahan A, Pritchard J, Tapscott SJ (2001) NeuroD2 is necessary for development and survival of central nervous system neurons. Developmental Biology 234:174–187. 10.1006/dbio.2001.0245 [DOI] [PubMed] [Google Scholar]
- Park CY, Shcheglovitov A, Dolmetsch R (2010) The CRAC channel activator STIM1 binds and inhibits L-type voltage-gated calcium channels. Science 330:101–105. 10.1126/science.1191027 [DOI] [PubMed] [Google Scholar]
- Pataskar A, Jung J, Smialowski P, Noack F, Calegari F, Straub T, Tiwari VK (2016) NeuroD1 reprograms chromatin and transcription factor landscapes to induce the neuronal program. EMBO J 35:24–45. 10.15252/embj.201591206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rannals MD, Hamersky GR, Page SC, Campbell MN, Briley A, Gallo RA, Phan BN, Hyde TM, Kleinman JE, Shin JH, Jaffe AE, Weinberger DR, Maher BJ (2016) Psychiatric risk gene transcription factor 4 regulates intrinsic excitability of prefrontal neurons via repression of SCN10a and KCNQ1. Neuron 90:43–55. 10.1016/j.neuron.2016.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravanpay AC, Olson JM (2008) E protein dosage influences brain development more than family member identity. J Neurosci Res 86:1472–1481. 10.1002/jnr.21615 [DOI] [PubMed] [Google Scholar]
- Reilly SK, Yin J, Ayoub AE, Emera D, Leng J, Cotney J, Sarro R, Rakic P, Noonan JP (2015) Evolutionary genomics. Evolutionary changes in promoter and enhancer activity during human corticogenesis. Science 347:1155–1159. 10.1126/science.1260943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos J, DiGregorio PJ, Yeromin AV, Ohlsen K, Lioudyno M, Zhang S, Safrina O, Kozak JA, Wagner SL, Cahalan MD, Veliçelebi G, Stauderman KA (2005) STIM1, an essential and conserved component of store-operated Ca2+ channel function. J Cell Biol 169:435–445. 10.1083/jcb.200502019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roybon L, Mastracci TL, Ribeiro D, Sussel L, Brundin P, Li JY (2010) GABAergic differentiation induced by Mash1 is compromised by the bHLH proteins Neurogenin2, NeuroD1, and NeuroD2. Cereb Cortex 20:1234–1244. 10.1093/cercor/bhp187 [DOI] [PubMed] [Google Scholar]
- Semple BD, Blomgren K, Gimlin K, Ferriero DM, Noble-Haeusslein LJ (2013) Brain development in rodents and humans: identifying benchmarks of maturation and vulnerability to injury across species. Prog Neurobiol 106-107:1–16. 10.1016/j.pneurobio.2013.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim S, Zheng JQ, Ming GL (2013) A critical role for STIM1 in filopodial calcium entry and axon guidance. Mol Brain 6:51. 10.1186/1756-6606-6-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver DL (2016) Genomic divergence and brain evolution: how regulatory DNA influences development of the cerebral cortex. Bioessays 38:162–171. 10.1002/bies.201500108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F, Stelzer Y, Shivalila CS, Abraham BJ, Latourelle JC, Barrasa MI, Goldmann J, Myers RH, Young RA, Jaenisch R (2016) Parkinson-associated risk variant in distal enhancer of α-synuclein modulates target gene expression. Nature 533:95–99. 10.1038/nature17939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somasundaram A, Shum AK, McBride HJ, Kessler JA, Feske S, Miller RJ, Prakriya M (2014) Store-operated CRAC channels regulate gene expression and proliferation in neural progenitor cells. J Neurosci 34:9107–9123. 10.1523/JNEUROSCI.0263-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telley L, Govindan S, Prados J, Stevant I, Nef S, Dermitzakis E, Dayer A, Jabaudon D (2016) Sequential transcriptional waves direct the differentiation of newborn neurons in the mouse neocortex. Science 351:1443–1446. 10.1126/science.aad8361 [DOI] [PubMed] [Google Scholar]
- Tsai FC, Seki A, Yang HW, Hayer A, Carrasco S, Malmersjö S, Meyer T (2014) A polarized Ca2+, diacylglycerol and STIM1 signalling system regulates directed cell migration. Nat Cell Biol 16:133–144. 10.1038/ncb2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén P, Fritz N, Smedler E, Malmersjö S, Kanatani S (2015) Calcium signaling in neocortical development. Dev Neurobiol 75:360–368. 10.1002/dneu.22273 [DOI] [PubMed] [Google Scholar]
- Wang Y, Deng X, Mancarella S, Hendron E, Eguchi S, Soboloff J, Tang XD, Gill DL (2010) The calcium store sensor, STIM1, reciprocally controls Orai and CaV1.2 channels. Science 330:105–109. 10.1126/science.1191086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegreffe C, Simon R, Peschkes K, Kling C, Strehle M, Cheng J, Srivatsa S, Liu P, Jenkins NA, Copeland NG, Tarabykin V, Britsch S (2015) Bcl11a (Ctip1) controls migration of cortical projection neurons through regulation of Sema3c. Neuron 87:311–325. 10.1016/j.neuron.2015.06.023 [DOI] [PubMed] [Google Scholar]
- Wilke SA, Hall BJ, Antonios JK, Denardo LA, Otto S, Yuan B, Chen F, Robbins EM, Tiglio K, Williams ME, Qiu Z, Biederer T, Ghosh A (2012) NeuroD2 regulates the development of hippocampal mossy fiber synapses. Neural Dev 7:9. 10.1186/1749-8104-7-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson G, Dennis D, Schuurmans C (2013) Proneural genes in neocortical development. Neuroscience 253:256–273. 10.1016/j.neuroscience.2013.08.029 [DOI] [PubMed] [Google Scholar]
- Yang S, Zhang JJ, Huang XY (2009) Orai1 and STIM1 are critical for breast tumor cell migration and metastasis. Cancer Cell 15:124–134. 10.1016/j.ccr.2008.12.019 [DOI] [PubMed] [Google Scholar]
- Yuan L, Hassan BA (2014) Neurogenins in brain development and disease: an overview. Arch Biochem Biophys 558:10–13. 10.1016/j.abb.2014.05.028 [DOI] [PubMed] [Google Scholar]
- Zhang SL, Yu Y, Roos J, Kozak JA, Deerinck TJ, Ellisman MH, Stauderman KA, Cahalan MD (2005) STIM1 is a Ca2+ sensor that activates CRAC channels and migrates from the Ca2+ store to the plasma membrane. Nature 437:902–905. 10.1038/nature04147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Cooper S, Brockdorff N (2015) The interplay of histone modifications—writers that read. EMBO Rep 16:1467–1481. 10.15252/embr.201540945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9:R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]