Abstract
Background
Particle exposure is a risk factor for cardiovascular diseases. Mitochondrial DNA (mtDNA) is a primary target for oxidative stress generated by particle exposure. We aimed to elucidate the effects of occupational exposure to particle-containing welding fumes on different biomarkers of mtDNA function, and in turn, explore if they modify the association between particle exposure and cardiovascular response, measured as blood pressure.
Methods
We investigated 101 welders and 127 controls (all non-smoking males) from southern Sweden. Personal sampling of the welders’ exposure to respirable dust was performed during work hours (average sampling time: 6.8 h; range: 2.4-8.6 h) and blood pressure was measured once for each subject. We measured relative mtDNA copy number by quantitative PCR and methylation of the mitochondrial regulatory region D-loop and the tRNA encoding gene MT-TF by bisulfite-pyrosequencing. We calculated the relative number of unmethylated D-loop and MT-TF as markers of mtDNA function to explore the modification of mtDNA on the association between particle exposure and blood pressure. General linear models were used for statistical analyses.
Results
Welders had higher mtDNA copy number (β = 0.11, p = 0.003) and lower DNA methylation of D-loop (β = −1.4, p = 0.002) and MT-TF (β = −1.5, p = 0.004) than controls. Higher mtDNA copy number was weakly associated with higher personal respirable dust exposure among welders with exposure level above 0.7 mg/m3 (β = 0.037, p = 0.054). MtDNA function modified the effect of welding fumes on blood pressure: welders with low mtDNA function had higher blood pressure than controls, while no such difference was found in the group with high mtDNA function.
Conclusion
Increased mtDNA copy number and decreased D-loop and MT-TF methylation were associated with particle-containing welding fumes exposure, indicating exposure-related oxidative stress. The modification of mtDNA function on exposure-associated increase in blood pressure may represent a mitochondria-environment interaction.
Electronic supplementary material
The online version of this article (doi:10.1186/s12940-017-0234-4) contains supplementary material, which is available to authorized users.
Keywords: Particle, Mitochondria, Copy number, DNA methylation, Blood pressure
Background
In work environments, the welding process is an important emission source of fine and ultrafine particles with a mass median diameter of 200–300 nm [1]. The reaction between vaporized metals and air during welding produces different types of metal oxides, including iron (Fe), manganese (Mg), chromium (Cr), and nickel (Ni) that result in the complex chemical properties of welding fume [2]. Studies have found associations between exposure to welding fumes and chronic obstructive pulmonary disease [3, 4], lung cancer [5] and various cardiovascular diseases [6, 7]. In the current project, we have reported higher blood pressure in the welders than in the controls, and that years of working as a welder were associated with increased blood pressure [8]. Growing evidence suggested that oxidative stress could be an intermediate step linking welding fumes exposure and disease [9–11]. Oxidative stress induced by particles with metal components (e.g. Cr, Ni, Fe) has been consistently shown to alter the methylation level of nuclear DNA and cause DNA damage [12–15].
Mitochondria, located in all types of cells except in red blood cells, are unique organelles with the primary biological function of generating energy [16]. They carry their own extranuclear, closed circular double-strand DNA: mitochondrial DNA (mtDNA). MtDNA is more susceptible to oxidative stress than nuclear DNA due to a lack of histones for protection, less adequate DNA repair capacity, and close proximity to the electron transport chain [17]. The copy numbers of mtDNA vary in each mitochondrion, as well as in different cells, different tissues and individuals. Alteration of mtDNA copy number has been observed as a response to oxidative stress in vitro and in vivo [18–20]. Moreover, integrity of the mitochondrial genome can affect mitochondrial function [21]. MtDNA encodes 13 respiratory chain polypeptides, 22 transfer RNAs (tRNA) and 2 ribosomal RNAs (rRNA) [22], and has a noncoding control region called the displacement loop (D-loop). The presence of mtDNA methylation has been debated for decades. An in vivo study of mtDNA – protein interaction observed methylation in the mitochondrial genome [23]. Then, Shock et al. demonstrated an enrichment of 5-methylcytosine and 5-hydroxymethylcytosine together with the presence of DNA methyltransferases 1 inside mitochondria [24]. More recently, Bellizzi et al. confirmed that mtDNA is indeed methylated, particularly in the D-loop region [25]. The D-loop contains three promoters required for transcription initiation and nearly the entire mitochondrial genome transcribes from this region [26]. Another gene of interest for mtDNA epigenetics is the transfer RNA phenylalanine (MT-TF) gene that encodes a tRNA involved in intra-mitochondrial translation and essential for protein synthesis [22]. Mitochondria play a crucial role for regulation of energy generation, and redox signaling of cells in the cardiovascular system [27], and thus, indicate that mitochondrial function is important for cardiovascular health. Still, there is limited knowledge how environmental exposures modify mtDNA epigenetics. It is also reasonable to infer that the mtDNA function might modify the relationship between exposure to particles and adverse cardiovascular effects.
In the present study, we aimed to elucidate the effects of occupational exposure to welding fumes on mtDNA copy number, methylation in the D-loop region and MT-TF gene. Furthermore, we also attempted to explore if these mitochondrial markers can modify the association between welding fumes and cardiovascular response, measured as blood pressure.
Methods
Study participants
Details of study participants’ recruitment have been reported previously [8]. In short, we investigated 101 welders and 127 controls (male and currently non-smoking) from southern Sweden. All participants had been non-smoking for more than one year. The welders used the same gas metal arc welding method; therefore they were exposed to relatively homogenous compositions of welding fumes. Fifty-five % of the welders reported that they used local exhaust ventilation, 56% reported that they use only welding shields as personal protection and 44% reported that they used powered air purifying respirators. All the welders also used protective clothing. The non-exposed controls were mainly workers in storage rooms, uploading and offloading products, and thus, their physical workload was comparable to the welders in our study.
All participants went through a structured face-to-face interview regarding potential particle exposure (e.g. particle from wood burning at home, traffic intensity outside their house windows), disease history (personal and family), and daily life (e.g. smoking history, daily diet, and activity and/or training). The answers were categorized into several groups. After the interviews, the participants went through blood pressure measurements and blood sampling. The study was approved by the Regional Ethical Committee of Lund University. All study participants gave fully informed and signed consent for their participation.
Exposure assessment
We used respirable dust as the indicator of exposure to particles from welding fumes [28]. Personal sampling of respirable dust was performed in the workers’ breathing zone for 70 welders (among them, 17 welders were not participants in the medical part of the study) at the work places of 10 welding companies in the manufacturing industry by an occupational hygienist by use of pre-weighed 37 mm mixed cellulose ester filters (0.8 μm pore size) fitted in conductive cassettes attached to cyclones (BGI4L, BGI Inc., USA; 50% cutoff at an aerodynamic equivalent particle diameter of 4 μm). The air flow was set at 2.2 L/min, and was regularly checked with a primary calibrator (TSI Model 4100 Series, TSI Inc., Shoreview, MN, USA) before, during, and after the sampling. The personal sampling was also performed for 19 controls from two control companies. Real-time measurements of background particle mass concentrations were conducted in other four control companies with direct reading instrument (Sidepak Model AM510, TSI Inc., MN USA).
Most of the personal sampling was performed during full-shift work with an average 6.9 h sampling time (range 2.4-8.6 h, only 5 out of 70 welders had sampling time shorter than 4 h). The filters were analyzed gravimetrically for respirable dust therefore the concentrations were the accumulation of the full-shift work. If the welders used powered air purifying respirators, the air outside the respirators was sampled, and the measured concentrations were reduced by a correction factor of 3 to get a better estimation of the exposure inside the respirator [29]. For the 48 welders with no exposure measurements, the exposure to respirable dust was estimated from the personal exposure data of welders (n = 70) with similar tasks.
Blood pressure and blood sampling
Each participant was asked to be seated during the 15 min structured interview. Blood pressure was then measured once by the skilled occupational health nurse using a mercury sphygmomanometer, with an adjustable cuff corresponding to different arm circumference in supine position. Peripheral blood was obtained afterwards, transported to the laboratory on dry ice, and stored at −20 °C until extraction of DNA.
Analysis of relative mitochondrial DNA copy number (RmtDNAcn)
DNA was isolated from whole peripheral blood by Qiagen DNA Blood Midi kit (Qiagen, Heidelberg, Germany). An assay based on real-time quantitative polymerase chain reaction (PCR) and SYBR® Green technology was adopted to determine mtDNA copy number relative to the single copy hemoglobin beta (HBB) gene using two independent PCRs. Master mixes for mtDNA copy number and HBB were prepared with KAPA SYBR FAST qPCR Kit Master Mix (2X) ABI Prism (Kapa Biosystems, Woburn, MA, USA) and corresponding primers (0.20 μM for each primer). Primers for mtDNA were: forward 5′-CAC CCA AGA ACA GGG TTT GT-3′ and reverse 5′-TGG CCA TGG GTA TGT TGT TA-3′; and primers for the HBB gene were: forward 5′-TGT GCT GGC CCA TCA CTT TG-3′ and reverse 5′-ACC AGC CAC CAC TTT CTG ATA GG-3′, as previously described [30]. PCR was performed on a real-time PCR machine (7900HT, Applied Biosystems, Foster City, CA, USA). Each reaction (end volume 10 μL) consisted of 2.5 μL of DNA (4 ng/μL) and 7.5 μL master mix. The thermal cycle profile was 95 °C for 3 min, followed by 95 °C for 3 s and 60 °C for 20 s for 25 cycles (mtDNA) or 35 cycles (HBB). A standard curve and a blank were included in each run. For the standard curve, one reference DNA sample (a pool of 20 samples randomly picked) was diluted serially by twofold per dilution to produce 5 concentrations of 1 – 16 ng/μL. A control sample was also included in each run to monitor the variance between runs. All samples and standard curve points were run in triplicates. R2 for each standard curve was >0.99. Standard deviations of triplicates <0.1 were accepted for the Ct values. SDS 2.4.1 software (Life Technologies) calculated the relative quantity of mtDNA and HBB for each sample, based on the standard curve. The relative mtDNA copy number (RmtDNAcn) was the quotient of the relative quantity of mtDNA and HBB, thus, it is an arbitrary value. The coefficient of variation (CV) of the control sample was 14% based on 3 runs. The raw data of individual RmtDNAcn is provided as Additional file 1.
Analysis of mitochondrial DNA methylation
Bisulfite modification was performed on 500 ng of peripheral blood DNA with EZ-96 DNA Methylation-Gold kit (Catalogue number D5008; Zymoresearch, Irvine, CA) according to the manufacturer’s instructions. We used 0.6 μl bisulfite-treated DNA in a 15 μl PCR reaction using the Pyromark PCR kit (Qiagen). The customer designed PCR primers and sequencing primers for the assay of D-loop and MT-TF methylation were described by Byun et al., and reverse PCR primers were biotinylated [31]. PCR was performed using PyroMark PCR reagents (Qiagen, catalog nr 972807). Detailed PCR conditions and primer sequences for mtDNA methylation assays are listed in Additional file 2: Table S1. The PCR products of 24 randomly picked samples were tested by gel and no non-specific binding was noticed. The PCR products were purified using Streptavidin Sepharose High Performance beads (Amersham Biosciences, Uppsala, Sweden). The Sepharose beads with the immobilized PCR products were purified, washed, and denatured with 0.2 M NaOH and washed again using a vacuum prep tool (Pyrosequencing Inc., Westborough, MA, USA). We performed pyrosequencing using the PSQ HS96 Pyrosequencing System (Qiagen). Negative PCR control (without DNA template) was included in four test plates and no signal of DNA methylation was measured in any of the negative controls. The degree of DNA methylation was expressed as the percentage of methylated cytosines over the sum of methylated and unmethylated cytosines (% DNA methylation). Three CpG sites from the D-loop region and one CpG site from the MT-TF gene were measured. We took an average percentage of methylation of D-loop for the analyses since the three sites were highly correlated (r S range 0.71-0.78). The raw methylation data of four CpG sites is provided as Additional file 1. We repeated eight samples and found the coefficient of variation as 12% for D-loop and 22% for MT-TF.
Statistical analysis
Basic characteristics like age, BMI, and blood pressure, as well as mtDNA biomarkers were symmetrically distributed for welders and controls and compared by t-test, while the exposure to respirable dust was skewed between the two groups and compared by Mann–Whitney U tests. Tobacco use (proportions of previous smokers, use of the smokeless tobacco “snus”, a moist crushed tobacco placed under the upper lip), and mtDNA function group (for details of calculation see below) were compared by Fisher’s exact tests. Pearson correlation coefficients were used to evaluate the correlations between RmtDNAcn, D-loop, and MT-TF methylation.
Exposure, copy number and methylation of mtDNA
General linear models were used to analyze the effects of occupational group (welders vs. controls), or exposure level (continuous variables: personal exposure to respirable dust, and years of working as a welder) on RmtDNAcn, D-loop and MT-TF methylation. Potential confounders (age, BMI, variables regarding smoking history, family history of disease, potential particle exposure, daily diet, alcohol consumption and activity and/or training) were chosen based on published studies and general knowledge, and tested one by one in the models. Only the confounders which changed the β-estimate of exposure indexes by more than 10% remained in further analyses. Therefore, age, BMI, previous smoking years, smokeless tobacco “snus” status and current residence were included in the adjusted models.
Calculation of mtDNA function
To investigate the modification of mtDNA function, we considered markers representing both the copy number and the methylation of mtDNA. We calculated the relative number of unmethylated mtDNA in the D-loop region and MT-TF gene. This was performed by calculating unmethylated D-loop and unmethylated MT-TF separately by subtracting the percentage of methylation of each region from 1. Then, we multiplied the products of RmtDNAcn and unmethylated D-loop (relative number of unmethylated D-loop), or RmtDNAcn and unmethylated MT-TF (relative number of unmethylated MT-TF), respectively, and used the products as the markers of mtDNA function. The formulas were:
Therefore, higher mtDNA function represented more copies of active (unmethylated) mtDNA, showing as greater relative number of unmethylated D-loop or MT-TF.
Modification of mtDNA function
First, interaction terms of mtDNA function markers and occupational group/exposure level were introduced into the general linear models to see if there was a modifying effect on mtDNA function. Then, data was stratified in two subgroups by the median value of mtDNA function (low and high mtDNA function groups). Age, BMI, family history of cardiovascular disease, smoking history, and current residence were selected as adjustments based on the same criteria of inclusion.
The residuals from each linear regression model were examined and all showed symmetric distribution. All statistical analyses were completed by SPSS (Version 22.0; IBM SPSS Statistics for Windows, NY, USA).
Results
Characteristics and biomarkers of study participants
Age (p = 0.94) and BMI (p = 0.70) were similar in the welders and the controls, as were the proportions of previous smoking (p = 0.21) and the average years of previous smoking (p = 0.52). The smokeless tobacco “snus” status was not different between two occupational groups (p = 0.15, 28% in the welders and 19% in the controls). The average concentration of respirable dust was 1.1 mg/m3 in the welders, while no participant in the control group was exposed to respirable dust above 0.1 mg/m3. RmtDNAcn was higher in the welders than in the controls (p = 0.0049), and D-loop and MT-TF methylation were lower in the welders than the controls (p = 0.0012 for D-loop, p = 0.0015 for MT-TF; Table 1). When stratifying study participants into low/high mtDNA function groups based on the median value of all participants, we found more welders in the high mtDNA function group, while more controls in the low mtDNA function group (p = 0.026 for D-loop, p < 0.001 for MT-TF; Table 1).
Table 1.
Basic characteristics and mitochondrial DNA biomarkers in welders (n = 101) and controls (n = 127)
| Welder | Control | p g | |
|---|---|---|---|
| Agea | 41 (23 – 60) | 43 (23 – 56) | 0.94 |
| BMI (kg/m2)a | 28 (22 – 34) | 27 (22 – 34) | 0.70 |
| Respirable dust (mg/m3)a,b | 1.1 (0.2 – 8.4) | 0.1 (0.0 – 0.1) | <0.001h |
| Working years as welder (year)a | 15 (1 – 38) | --- | -- |
| Systolic blood pressure (mm Hg)a | 130 (115 – 155) | 125 (105 – 145) | <0.001 |
| Diastolic blood pressure (mm Hg)a | 75 (60 – 85) | 70 (60 – 85) | <0.001 |
| Previous smoking (yes/no (%)) | 43/58 (43%) | 43/83 (34%) | 0.21i |
| Previous smoking years (year)a | 10 (3 – 32) | 14 (2 – 31) | 0.52 |
| Smokeless tobacco “snus” (yes/no (%)) | 28/73 (28%) | 24/103 (19%) | 0.15i |
| Relative mtDNA copy numbera | 1.13 (0.84 – 1.52) | 1.00 (0.74 – 1.50) | 0.0049 |
| D-loop methylationa, c | 13.4 (10.5 –20.8) | 15.6 (10.9 – 21.1) | 0.0012 |
| MT-TF methylationa, c | 3.4 (0 – 9.5) | 4.5 (0 – 12.1) | 0.0015 |
| mtDNA function group (high/low (%)) (D-loop)d, f | 57/37 (60%) | 51/66 (44%) | 0.026i |
| mtDNA function group (high/low (%)) (MT-TF)e, f | 63/37 (63%) | 47/73 (39%) | <0.001i |
aPresented as median (5 – 95 percentile)
bMedian of respirable dust in welder group was based on 70 welders with measured respirable dust
cMedian of D-loop methylation was based on 211 participants (94 welders and 117 controls); median of MT-TF methylation was based on 220 participants (100 welders and 120 controls)
dDerived from unmethylated D-loop and relative mtDNA copy number: Relative number of unmethylated D-loop = (100% - % methylated D-loop) × relative mtDNA copy number
eDerived from unmethylated MT-TF and relative mtDNA copy number: Relative number of unmethylated MT-TF = (100% - % methylated MT-TF) × relative mtDNA copy number
fStratified into low and high groups based on the median. Here, we present the count (%) of participants in each function group
g p value obtained from t test unless marked otherwise
h p values obtained from Mann–Whitney U test
i p values obtained from Fisher’s exact test
D-loop methylation was positively correlated with MT-TF methylation (r = 0.31, p < 0.001; Fig. 1a), and inversely correlated with RmtDNAcn, (r = −0.25, p < 0.001; Fig. 1b). No correlation between MT-TF methylation and RmtDNAcn was found (r = −0.11, p = 0.092; Fig. 1c).
Fig. 1.
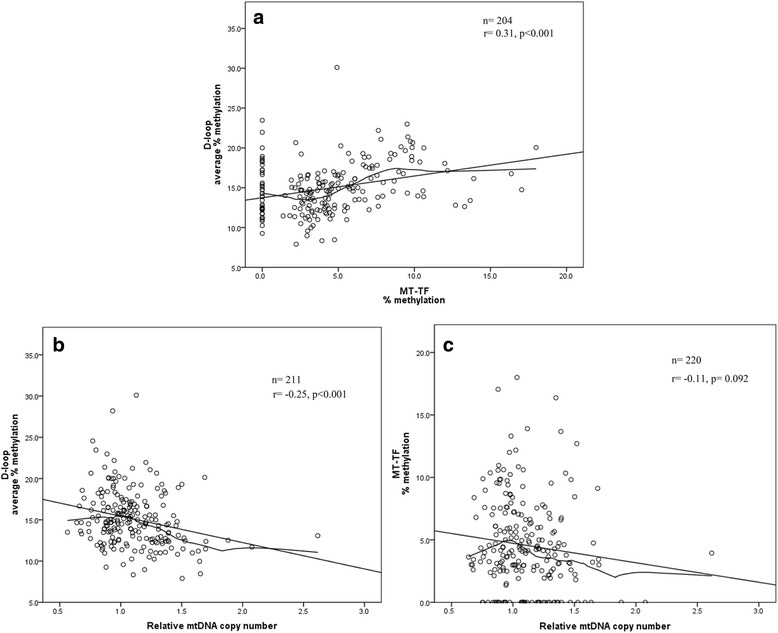
Correlations between methylation and relative mtDNA copy number. Scatterplots with linear and loess fit lines showing the correlations between mitochondrial gene methylation and relative mtDNA copy number: a D-loop methylation and MT-TF methylation; b D-loop methylation and relative mtDNA copy number; c MT-TF methylation and relative mtDNA copy number
Associations between exposure to welding fumes and mtDNA copy number and methylation
The differences of RmtDNAcn and mtDNA methylation status between welders and controls remained significant after adjustment for age, BMI, previous smoking years, smokeless tobacco “snus” status, and current residence (Table 2, unadjusted model see Additional file 3: Table S2). In order to elucidate dose-effect relationships of welding fume particles on RmtDNAcn and mtDNA methylation, analyses were performed in welders only. Figure 2 shows the associations between RmtDNAcn and personal respirable dust, indicating a slightly different pattern of dose-effect relationship among welders with different exposure levels: RmtDNAcn was weakly positively associated with respirable dust among welders with personal exposure level above 0.7 mg/m3 (β = 0.037, p = 0.054; Table 2), however, no clear association was found among welders with low exposure level (below 0.7 mg/m3). No associations were observed for personal respirable dust concentrations and D-loop methylation or MT-TF methylation (figures not shown). No association between RmtDNAcn, mtDNA methylation and years of working as a welder was found (Table 2).
Table 2.
Associations between exposure and relative mtDNA copy number, D-loop and MT-TF methylation in adjusted modelsa
| mtDNA copy number | D-loop methylation | MT-TF methylation | |||||||
|---|---|---|---|---|---|---|---|---|---|
| N | Beta (95% CI) | p | N | Beta (95% CI) | p | N | Beta (95% CI) | p | |
| Occupational group | |||||||||
| Welders vs. controlsb | 228 | 0.11 (0.037, 0.18) | 0.0032 | 211 | −1.4 (−2.3, −0.5) | 0.0021 | 220 | −1.5 (−2.5, −0.48) | 0.0038 |
| Exposure level | |||||||||
| Respirable dustc | 101 | 0.017 (−0.018, 0.052) | 0.33 | 92 | 0.012 (−0.47, 0.50) | 0.96 | 97 | 0.097 (−0.32, 0.51) | 0.64 |
| Respirable dust (<=0.7 mg/m3)c, d | 57 | −0.031 (−0.47, 0.41) | 0.89 | 54 | −0.42 (−4.76, 3.9) | 0.85 | 56 | 2.9 (−2.2, 8.1) | 0.26 |
| Respirable dust (>0.7 mg/m3)c, d | 41 | 0.037 (−0.00075, 0.075) | 0.054 | 38 | −0.043 (−0.85, 0.76) | 0.92 | 41 | 0.17 (−0.29, 0.63) | 0.46 |
| Working yearse | 100 | −0.0027 (−0.010, 0.0049) | 0.49 | 93 | 0.061 (−0.053, 0.16) | 0.29 | 99 | −0.013 (−0.10, 0.076) | 0.77 |
aThe adjusted model included age, BMI, previous smoking years, smokeless tobacco “snus” status and current residence as adjustments
bEffect estimates presented are β-values for occupation (welders compared with control) derived from general linear models
cEffect estimates presented are β-values for personal respirable dust (only welders included) derived from general linear models
dThe cut-off was based on median value of welders with measured and estimated respirable dust
eEffect estimates presented are β-values for years working as welder (only welders included) derived from general linear models
Fig. 2.
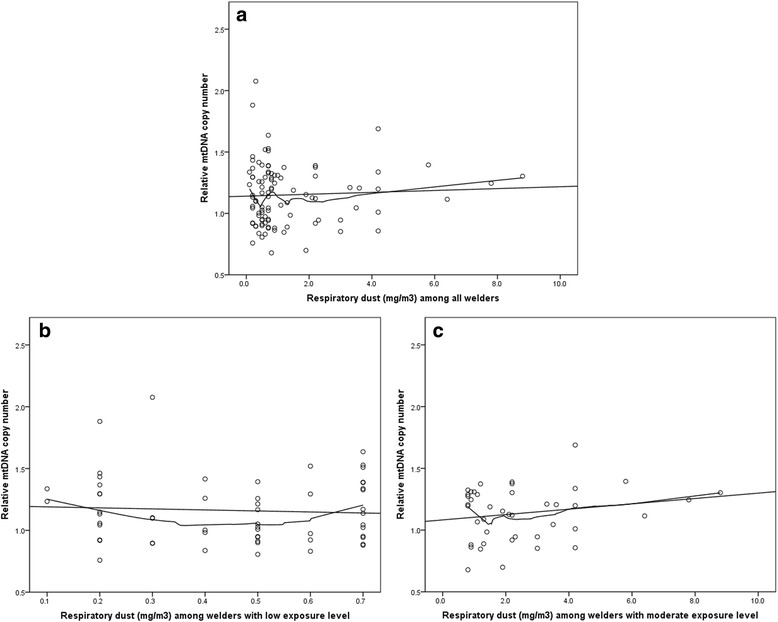
Associations between relative mtDNA copy number and personal respirable dust. Scatterplots with linear and loess fit lines showing the associations between respirable dust and relative mtDNA copy number. Associations are (a) among all welders, (b) among welders with exposure level lower than 0.7 mg/m3 (median concentration = 0.5 mg/m3) and (c) among welders with exposure level higher than 0.7 mg/m3 (median concentration = 2.0 mg/m3). The cut-off of exposure level was based on median value of respirable dust from 101 welders (including measured and estimated exposures)
Interaction of mtDNA function with exposure on blood pressure
There was some indication of interaction between the occupational groups and the mtDNA function marker (relative number of unmethylated D-loop) on blood pressure (for systolic blood pressure: interaction p = 0.15, for diastolic blood pressure, interaction p = 0.20). Similarly, interactions were suggested between the occupational groups and relative number of unmethylated MT-TF (p = 0.073 for systolic blood pressure, p = 0.25 for diastolic blood pressure). The suggested interactions may indicate a modifying effect of mtDNA function on the associations between occupational groups and blood pressure. Thus, we stratified the study participants into low and high mtDNA function groups by the median of relative number of unmethylated D-loop. The high mtDNA function group represented more copies of active (unmethylated) D-loop than the low function group. After stratifying the data, we found higher systolic blood pressure in the welders than the controls (β = 11.3, p < 0.001) in the group with low mtDNA function, but not in the high group (β = 3.3, p = 0.15, Table 3). Similarly, after stratifying the data by the relative number of unmethylated MT-TF, higher systolic blood pressure in the welders than in the controls (β = 12.3, p < 0.001, Table 3) was only found in the group with low mtDNA function. However, no modifying effects of mtDNA function on the association between blood pressure and years of working as a welder, or between blood pressure and respirable dust, were found (data not shown).
Table 3.
Associations between occupational groups and blood pressure in different mtDNA function groupsa
| mtDNA function group (D-loop)b |
mtDNA function group (MT-TF) c |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Low | High | Low | High | |||||||||
| Outcome | N | Beta (95% CI) | p | N | Beta (95% CI) | p | N | Beta (95% CI) | p | N | Beta (95% CI) | p |
| Systolic blood pressure | 100 | 11.32 (6.61, 15.91) | <0.001 | 106 | 3.34 (−1.18, 7.85) | 0.15 | 107 | 12.31 (7.93, 16.69) | <0.001 | 108 | 1.11 (−3.51, 5.73) | 0.63 |
| Diastolic blood pressure | 100 | 3.53 (0.18, 6.88) | 0.039 | 106 | 3.64 (0.28, 7.00) | 0.034 | 107 | 3.00 (−0.38, 6.38) | 0.081 | 108 | 2.39 (−0.96, 5.73) | 0.16 |
aThe model included age, BMI, family history of cardiovascular disease, smoking history, and current residence as adjustments
bDerived from unmethylated D-loop and relative mtDNA copy number. Two groups stratified by median
cDerived from unmethylated MT-TF and relative mtDNA copy number. Two groups stratified by median
Discussion
In this study, we hypothesized that occupational exposure to particles could be associated with alteration in mtDNA function, and mtDNA function may interact with exposure on adverse cardiovascular effects, measured as blood pressure. We found that the occupation as welder was associated with changes in mtDNA, both measured as copy number and the methylation status of key regions of the mtDNA, indicating an effect of welding fumes on the mitochondrial function. There was, however, no strong dose-effect relationship between personal respirable dust or working years as a welder and biomarkers of mtDNA function, and these findings need therefore to be interpreted cautiously. The changes in mtDNA may affect mitochondrial biogenesis and cellular function, and in turn contribute to higher risk of cardiovascular diseases through interaction with particle exposures.
There is a widespread interest in exploring the response of mtDNA to environmental exposures. Recent studies have shown a linkage between particle exposure and alteration in mtDNA in peripheral blood, but the results are inconsistent [30–34]. Hou et al. found particle-related increased mtDNA copy number in 63 steel workers at time-weighted PM1 concentration around 9 μg/m3 [30]. However, the same research group reported that exposure to elemental carbon at around 15 μg/m3 and ambient PM10 at around 120 μg/m3 during work hours, was related to decreased mtDNA copy number. No association was found between personal exposure to PM2.5 and mtDNA copy number [32]. One cross-sectional study involving 20 steel workers and 20 controls reported a positive association between metal-rich particle exposure and mtDNA methylation in MT-TF and MT-RNR1 region encoding 12 s rRNA but they did not find any association between particle exposure and D-loop methylation [31]. Yet, a recent paper involving 48 healthy male workers reported a negative association between PM2.5 and D-loop methylation, but not in the MT-TF and MT-RNR1 region [34].
In the present study, we observed higher RmtDNAcn together with lower methylation levels in the mtDNA D-loop region in peripheral blood in the welders than the controls, and a weak positive dose-effect relationship between personal respirable dust and RmtDNAcn among the welders with moderate exposure level. The inconclusive results between the studies might be due to the different size and compositions of particles, different exposure concentrations in each study, as well as small study size resulting in low power. The mtDNA synthesis can be stimulated by mild oxidative stress, resulting in an increase of mtDNA copy number to supply energy for cell survive, while immoderate oxidative stress may lead to decrease in mtDNA synthesis because of defect mitochondria and result in apoptosis and cell death [30, 35]. Since the median particle concentration in our study (1.1 mg/m3) is not very high (the occupational exposure limit is set as 5 mg/m3 by the Swedish Work Environment Authority [28]), we concluded that the increase in RmtDNAcn could be a feedback response to compensate defect mitochondria with an impaired respiratory chain, or mutated mtDNA caused by oxidative damage from welding fume [18, 35]. The methylation level change in the D-loop could be another molecular event related to particle induced oxidative stress, which can damage methylation of nucleotides [36]. Classic epigenetic theory suggests that hypermethylation in the promoter region are generally transcriptionally silent, while demethylated DNA is usually associated with active gene expression [37, 38]. Based on the finding that demethylated mtDNA D-loop was associated with increased ND2 expression (a subunit of NADH, a rate-limiting enzyme of oxidative phosphorylation), which can regulate the generation of ATP [39], our findings of demethylation of D-loop in the welders could be interpreted as mtDNA response to cope with the increased oxidative stress by increasing energy production related gene expression to maintain normal cellular function. Thus, both the increase in RmtDNAcn and the decrease in methylation level in welders could be the compensatory mechanisms of mtDNA responses to particle exposure. Recently, Janssen et al. studied mtDNA methylation and mtDNA content in placental tissues in the context of the early life exposure to environmental airborne particles, and they observed opposite effects of particle exposure on mtDNA: i.e. an inverse association between PM2.5 and mtDNA content, as well as a positive association with mtDNA methylation in D-loop and MT-RNR1 [40]. The contradictory finding between our study and theirs could be due to the duration of exposure that the mothers in their study were exposed to ambient PM2.5 during the entire pregnancy, while the exposure to respirable dust was only measured once in our study. Another possibility is that the different origin of DNA matters. Given the fact that the placenta is an organ that requires a constant and abundant source of energy, mtDNA in placenta may response more readily to particle exposure compared to mtDNA in blood, and therefore, produce more by-product reactive oxygen species that in turn more rapidly causes mtDNA damage.
Despite the noticeable findings of occupational group, the effect of exposure levels on mtDNA was not statistically certain. One explanation about the small effect estimates of respirable dust could be that other unmeasured exposures, like ozone production in gas phase during the welding process [2], might have been responsible for the increase in RmtDNAcn. We also failed to identify dose–response associations among welders with low exposure, which could suggest a threshold of particle exposure for mtDNA effects. However this hypothesis remains unconfirmed as we only carried out respirable dust sampling once, and it is dangerous to assume that one measurement can represent the long-term exposure dose.
The association between particle exposure and higher blood pressure has been suggested to be caused by the generation of reactive oxygen species, which interrupt the redox state, cause systemic inflammation and induce endothelial dysfunction and vascular injury in hypertension development [41, 42]. Given the fact that mitochondria are important to cellular function, mtDNA function may play a critical role in the etiology of particle-related hypertension. Our finding of possible modification of mtDNA function on exposure and blood pressure indicated that individuals carrying lower copies of active mtDNA may be a susceptible group, possibly because of insufficient energy generation and antioxidant capacity. The vicious cycle of decreased mitochondrial antioxidant capacity and continuous oxidative stress could induce endothelial dysfunction and vascular smooth muscle cell hypertrophy [43], which are the main characteristic alterations of vascular wall in hypertension [44]. We are aware that our grouping of low and high mtDNA function was rather crude; further follow-up studies are required to determine the effects of changes in mtDNA on mitochondrial function.
The primary limitation of the study was the cross-sectional study design, which precludes conclusions with regard to causality. However, the information from the structured interview allowed proper control of possible confounders in the statistical analysis to avoid biased associations. The single measurement of RmtDNAcn, mtDNA methylation did not permit us to determine intrapersonal variation over time. In the study, we only measured the blood pressure once for each subject, due to the practical circumstance that workers were busy and could not spare more time for medical examination. However, blood pressure measurement in this study served as an indicator of adverse cardiovascular effects, rather than a clinical diagnosis of hypertension. We cannot confirm the direct linkage between particle exposure, methylation and its functional significance on RNA expression since RNA was not available. This would be an interesting topic for further research. The relatively higher CV for MT-TF methylation is probably due to the low methylation of this CpG, which may be influenced by noise of the assay. Therefore, the changes in MT-TF methylation needs to be validated in further studies.
We also recognize that mtDNA methylation in this paper is likely overestimated. One recently published paper revealed that measurement of mtDNA methylation level is affected by its form: circular mtDNA appears to have higher methylation level than linear mtDNA, due to incomplete bisulfite conversion when mtDNA is circular [45]. In our study, we did not break the circular DNA into linear, and obtained an average D-loop methylation around 13 - 16%, which is similar to Liu et al. (11.65% ± 0.95%) [45] when circular mtDNA was not broken. However, we do not consider it is likely that the difference in methylation between circular and linear mtDNA would influence our conclusions. Both controls and welders were sampled in the same way and analyzed, in a randomized set-up, by the same mtDNA method; therefore the systematic overestimation apply to all study participants in the study, and should not change the relative comparison between exposed group and control group.
Conclusion
In summary, we observed a potential effect of occupational exposure to particles on mtDNA in welders, although the occupational exposure level to particles was only low to moderate. The modification of mtDNA function on the welding-associated increase in blood pressure, may represent a mitochondria-environment interaction, and may indicate that mtDNA plays a critical role in the etiology of particle-related cardiovascular disease. Studies further disentangling these associations may be an important step in understanding the mechanisms behind particle-induced cardiovascular disease.
Acknowledgements
We would like to thank Eva Assarsson for performing the recruitment of the study participants and Mary Partington for language editing. We also would like to express our gratitude to the volunteers who participated in the investigation.
Funding
This work was supported by The Swedish Council for Working Life and Social Research [2009–1888]. KB received the funding.
Availability of data and materials
The dataset of individual RmtDNAcn and mtDNA methylation is provided as Additional file 1. The other dataset analyzed during the current study are not publicly available since it contains sensitive health-related data, but it is available from the corresponding author on reasonable request.
Authors’ contributions
HL, HT and KB designed the project. MH measured and provided exposure characteristics. YX and HL measured mitochondrial DNA copy number. YX and MBH measured mitochondrial DNA methylation. YX and HL analyzed the data and interpreted the results. The manuscript was written by YX and revised critically by KB and MA. All authors read, corrected and approved the manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participants
The study was approved by the Regional Ethical Committee of Lund University. All study participants gave fully informed and signed consent for their participation.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- Cr
Chromium
- D-loop
Displacement loop
- Fe
Iron
- HBB
Hemoglobin beta
- Mg
Manganese
- mtDNA
Mitochondrial DNA
- MT-TF
Transfer RNA phenylalanine
- Ni
Nickel
- PCR
Polymerase chain reaction
- RmtDNAcn
Relative mitochondrial DNA copy number
Additional files
Raw data of mtDNA copy number and methylation in D-loop and MT-TF in Excel. (XLSX 21 kb)
Detailed PCR conditions and primer sequences for mtDNA methylation assays. (DOCX 17 kb)
The associations between occupational group/exposure level and relative mtDNA copy number, D-loop and MT-TF methylation in unadjusted models. (DOCX 18 kb)
Contributor Information
Yiyi Xu, Phone: +46-222 1637, Email: yiyi.xu@med.lu.se.
Huiqi Li, Email: huiqi.li@med.lu.se.
Maria Hedmer, Email: maria.hedmer@med.lu.se.
Mohammad Bakhtiar Hossain, Email: bakhtiar.hossain@med.lu.se.
Håkan Tinnerberg, Email: hakan.tinnerberg@skane.se.
Karin Broberg, Email: karin.broberg@ki.se, Email: karin.broberg_palmgren@med.lu.se.
Maria Albin, Email: maria.albin@med.lu.se.
References
- 1.Isaxon C, Pagels J, Gudmundsson A, Asbach C, John A, Kuhlbusch T, Karlsson J, Kammer R, Tinnerberg H, Nielsen J. Characteristics of welding fume aerosol investigated in three Swedish workshops. Inhaled Particles X. (Vol.151). IOP Publishing; 2009. doi:10.1088/1742-6596/151/1/012059.
- 2.Antonini JM. Health effects of welding. Crit Rev Toxicol. 2003;33(1):61–103. doi: 10.1080/713611032. [DOI] [PubMed] [Google Scholar]
- 3.Koh DH, Kim JI, Kim KH, Yoo SW. 0371 the relationship between welding fume exposure and chronic obstructive pulmonary disease in shipyard welders in Korea. Occup Environ Med. 2014;71(Suppl 1):A46. [Google Scholar]
- 4.Cullinan P. Occupation and chronic obstructive pulmonary disease (COPD) Br Med Bull. 2012;104:143–161. doi: 10.1093/bmb/lds028. [DOI] [PubMed] [Google Scholar]
- 5.Sorensen AR, Thulstrup AM, Hansen J, Ramlau-Hansen CH, Meersohn A, Skytthe A, Bonde JP. Risk of lung cancer according to mild steel and stainless steel welding. Scand J Work Environ Health. 2007;33(5):379–386. doi: 10.5271/sjweh.1157. [DOI] [PubMed] [Google Scholar]
- 6.Ibfelt E, Bonde JP, Hansen J. Exposure to metal welding fume particles and risk for cardiovascular disease in Denmark: a prospective cohort study. Occup Environ Med. 2010;67(11):772–777. doi: 10.1136/oem.2009.051086. [DOI] [PubMed] [Google Scholar]
- 7.Mocevic E, Kristiansen P, Bonde JP. Risk of ischemic heart disease following occupational exposure to welding fumes: a systematic review with meta-analysis. Int Arch Occup Environ Health. 2014. [DOI] [PubMed]
- 8.Li H, Hedmer M, Kåredal M, Björk J, Stockfelt L, Tinnerberg H, Albin M, Broberg K. A cross-sectional study of the cardiovascular effects of welding fumes. PLoS One. 2015;10(7) doi: 10.1371/journal.pone.0131648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li GJ, Zhang LL, Lu L, Wu P, Zheng W. Occupational exposure to welding fume among welders: alterations of manganese, iron, zinc, copper, and lead in body fluids and the oxidative stress status. J Occup Environ Med. 2004;46(3):241–248. doi: 10.1097/01.jom.0000116900.49159.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erdely A, Antonini JM, Young SH, Kashon ML, Gu JK, Hulderman T, Salmen R, Meighan T, Roberts JR, Zeidler-Erdely PC. Oxidative stress and reduced responsiveness of challenged circulating leukocytes following pulmonary instillation of metal-rich particulate matter in rats. Part Fibre Toxicol. 2014;11:34. doi: 10.1186/s12989-014-0034-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han SG, Kim Y, Kashon ML, Pack DL, Castranova V, Vallyathan V. Correlates of oxidative stress and free-radical activity in serum from asymptomatic shipyard welders. Am J Respir Crit Care Med. 2005;172(12):1541–1548. doi: 10.1164/rccm.200409-1222OC. [DOI] [PubMed] [Google Scholar]
- 12.Keyhani E, Abdi-Oskouei F, Attar F, Keyhani J. DNA strand breaks by metal-induced oxygen radicals in purified Salmonella typhimurium DNA. Ann N Y Acad Sci. 2006;1091:52–64. doi: 10.1196/annals.1378.054. [DOI] [PubMed] [Google Scholar]
- 13.Jomova K, Valko M. Advances in metal-induced oxidative stress and human disease. Toxicology. 2011;283(2–3):65–87. doi: 10.1016/j.tox.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Kile ML, Fang S, Baccarelli AA, Tarantini L, Cavallari J, Christiani DC. A panel study of occupational exposure to fine particulate matter and changes in DNA methylation over a single workday and years worked in boilermaker welders. Environ Health. 2013;12(1):47. doi: 10.1186/1476-069X-12-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Hedmer M, Wojdacz T, Hossain MB, Lindh CH, Tinnerberg H, Albin M, Broberg K. Oxidative stress, telomere shortening, and DNA methylation in relation to low-to-moderate occupational exposure to welding fumes. Environ Mol Mutagen. 2015. [DOI] [PMC free article] [PubMed]
- 16.Desagher S, Martinou J-C. Mitochondria as the central control point of apoptosis. Trends Cell Biol. 2000;10(9):369–377. doi: 10.1016/S0962-8924(00)01803-1. [DOI] [PubMed] [Google Scholar]
- 17.Lee HC, Wei YH. Mitochondrial role in life and death of the cell. J Biomed Sci. 2000;7(1):2–15. doi: 10.1007/BF02255913. [DOI] [PubMed] [Google Scholar]
- 18.Lee H, Yin P, Lu C, Chi C, Wei Y. Increase of mitochondria and mitochondrial DNA in response to oxidative stress in human cells. Biochem J. 2000;348:425–432. doi: 10.1042/bj3480425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pavanello S, Dioni L, Hoxha M, Fedeli U, Mielzynska-Svach D, Baccarelli AA. Mitochondrial DNA copy number and exposure to polycyclic aromatic hydrocarbons. Cancer Epidemiol Biomarkers Prev. 2013;22(10):1722–1729. doi: 10.1158/1055-9965.EPI-13-0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gebhard D, Mahler B, Matt K, Burger K, Bergemann J. Mitochondrial DNA copy number–but not a mitochondrial tandem CC to TT transition–is increased in sun-exposed skin. Exp Dermatol. 2014;23(3):209–211. doi: 10.1111/exd.12327. [DOI] [PubMed] [Google Scholar]
- 21.Miller FJ, Rosenfeldt FL, Zhang C, Linnane AW, Nagley P. Precise determination of mitochondrial DNA copy number in human skeletal and cardiac muscle by a PCR-based assay: lack of change of copy number with age. Nucleic Acids Res. 2003;31(11):e61. doi: 10.1093/nar/gng060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson S, Bankier AT, Barrell BG, De Bruijn M, Coulson AR, Drouin J, Eperon I, Nierlich D, Roe BA, Sanger F. Sequence and organization of the human mitochondrial genome. 1981. [DOI] [PubMed] [Google Scholar]
- 23.Rebelo AP, Williams SL, Moraes CT. In vivo methylation of mtDNA reveals the dynamics of protein–mtDNA interactions. Nucleic Acids Res. 2009;37:6701–6715. doi: 10.1093/nar/gkp727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shock LS, Thakkar PV, Peterson EJ, Moran RG, Taylor SM. DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc Natl Acad Sci. 2011;108(9):3630–3635. doi: 10.1073/pnas.1012311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bellizzi D, D’Aquila P, Scafone T, Giordano M, Riso V, Riccio A, Passarino G. The control region of mitochondrial DNA shows an unusual CpG and non-CpG methylation pattern. DNA Res. 2013;20(6):537–547. doi: 10.1093/dnares/dst029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taanman J-W. The mitochondrial genome: structure, transcription, translation and replication. BBA-Bioenergetics. 1999;1410(2):103–123. doi: 10.1016/S0005-2728(98)00161-3. [DOI] [PubMed] [Google Scholar]
- 27.Ballinger SW. Mitochondrial dysfunction in cardiovascular disease. Free Radic Biol Med. 2005;38(10):1278–1295. doi: 10.1016/j.freeradbiomed.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 28.The Swedish Work Environment Authority. Occupational exposure limit values, AFS 2011:18. Stockholm; 2011. Available at: http://www.branschutbildarna.com/wp-content/uploads/2015/09/occupational-exposure-limit-values-provisions-afs2011-18.pdf.
- 29.Hedmer M, Karlsson J-E, Andersson U, Jacobsson H, Nielsen J, Tinnerberg H. Exposure to respirable dust and manganese and prevalence of airways symptoms, among Swedish mild steel welders in the manufacturing industry. Int Arch Occup Environ Health. 2014;87(6):623–634. doi: 10.1007/s00420-013-0896-3. [DOI] [PubMed] [Google Scholar]
- 30.Hou L, Zhu ZZ, Zhang X, Nordio F, Bonzini M, Schwartz J, Hoxha M, Dioni L, Marinelli B, Pegoraro V, et al. Airborne particulate matter and mitochondrial damage: a cross-sectional study. Environ Health. 2010;9:48. doi: 10.1186/1476-069X-9-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Byun H-M, Panni T, Motta V, Hou L, Nordio F, Apostoli P, Bertazzi PA, Baccarelli AA. Effects of airborne pollutants on mitochondrial DNA methylation. Part Fibre Toxicol. 2013;10(1):18. doi: 10.1186/1743-8977-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hou L, Zhang X, Dioni L, Barretta F, Dou C, Zheng Y, Hoxha M, Bertazzi PA, Schwartz J, Wu S, et al. Inhalable particulate matter and mitochondrial DNA copy number in highly exposed individuals in Beijing, China: a repeated-measure study. Part Fibre Toxicol. 2013;10:17. doi: 10.1186/1743-8977-10-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xia Y, Chen R, Wang C, Cai J, Wang L, Zhao Z, Qian J, Kan H. Ambient air pollution, blood mitochondrial DNA copy number and telomere length in a panel of diabetes patients. Inhal Toxicol. 2015;27(10):481–487. doi: 10.3109/08958378.2015.1075090. [DOI] [PubMed] [Google Scholar]
- 34.Byun HM, Colicino E, Trevisi L, Fan T, Christiani DC, Baccarelli AA. Effects of air pollution and blood mitochondrial DNA methylation on markers of heart rate variability. J Am Heart Assoc. 2016;5(4) doi: 10.1161/JAHA.116.003218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee H-C, Wei Y-H. Mitochondrial biogenesis and mitochondrial DNA maintenance of mammalian cells under oxidative stress. Int J Biochem Cell Biol. 2005;37(4):822–834. doi: 10.1016/j.biocel.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 36.Cline SD. Mitochondrial DNA damage and its consequences for mitochondrial gene expression. BBA-Gene Regul Mech. 2012;1819(9):979–991. doi: 10.1016/j.bbagrm.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Cold Spring Harbor Monograph Archive. 1996;32:639–645. [PubMed] [Google Scholar]
- 38.Schübeler D. Function and information content of DNA methylation. Nature. 2015;517(7534):321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- 39.Feng S, Xiong L, Ji Z, Cheng W, Yang H. Correlation between increased ND2 expression and demethylated displacement loop of mtDNA in colorectal cancer. Mol Med Rep. 2012;6(1):125–130. doi: 10.3892/mmr.2012.870. [DOI] [PubMed] [Google Scholar]
- 40.Janssen BG, Byun HM, Gyselaers W, Lefebvre W, Baccarelli AA, Nawrot TS. Placental mitochondrial methylation and exposure to airborne particulate matter in the early life environment: an ENVIRONAGE birth cohort study. Epigenetics. 2015;10(6):536–544. doi: 10.1080/15592294.2015.1048412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brook RD, Rajagopalan S. Particulate matter, air pollution, and blood pressure. J Am Soc Hypertens. 2009;3(5):332–350. doi: 10.1016/j.jash.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 42.Touyz R, Schiffrin E. Reactive oxygen species in vascular biology: implications in hypertension. Histochem Cell Biol. 2004;122(4):339–352. doi: 10.1007/s00418-004-0696-7. [DOI] [PubMed] [Google Scholar]
- 43.Zalba G, San José G, Moreno MU, Fortuño MA, Fortuño A, Beaumont FJ, Díez J. Oxidative stress in arterial hypertension role of NAD (P) H oxidase. Hypertension. 2001;38(6):1395–1399. doi: 10.1161/hy1201.099611. [DOI] [PubMed] [Google Scholar]
- 44.Zalba G, Beaumont J, San José G, Fortuno A, Fortuño MA, Diez J. Vascular oxidant stress: molecular mechanisms and pathophysiological implications. J Physiol Biochem. 2000;56(1):57–64. doi: 10.1007/BF03179777. [DOI] [PubMed] [Google Scholar]
- 45.Liu B, Du Q, Chen L, Fu G, Li S, Fu L, Zhang X, Ma C, Bin C. CpG methylation patterns of human mitochondrial DNA. Sci Rep. 2016;6. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The dataset of individual RmtDNAcn and mtDNA methylation is provided as Additional file 1. The other dataset analyzed during the current study are not publicly available since it contains sensitive health-related data, but it is available from the corresponding author on reasonable request.


