Abstract
Hepatitis C virus (HCV) infection represents a serious health problem. The –174 G/C mutation in the pro inflammatory cytokine interleukin-6 (IL-6) is associated with developing liver diseases. Likewise, the S and Z mutations in the serine protease inhibitor α1-antitrypsin (A1AT) are associated with pulmonary emphysema and/or liver cirrhosis. We explored the distribution of the single nucleotide polymorphisms (SNPs) of IL-6 and A1AT genes in chronic HCV-infected patients and evaluated their impact on the progression of liver cirrhosis.
One hundred and fifty Egyptian HCV-infected patients together with 100 healthy controls were enrolled in this study. The patient groups were subdivided into chronic hepatitis patients (n = 85) and cirrhotic patients (n = 65). The SNP of IL-6 (–174 G/C, rs1800795), A1AT Z mutation (342 Glu/Lys, rs28929474) and A1AT S mutation (264 Glu/Val, rs17580) were determined using a polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method.
Cirrhotic patients exhibited significantly increased frequency of the A1AT S allele compared with the controls (34.6 vs. 5.0%), while the chronic hepatitis patients showed a higher frequency of the A1AT Z allele compared with the controls (14.7 vs. 2.5%). Remarkably, IL-6 (CC genotype) was detected only in the chronic hepatitis patients. Multivariate regression analysis showed that aspartate transaminase (AST) and the S alleles of A1AT, represented as SS+MS genotypes, were significantly independent predictors for development of liver cirrhosis. We concluded that inheritance of deficient S and Z alleles of the A1AT gene but not IL-6 (–174 G/C), were associated with progressive liver diseases.
Keywords: α1 Antitrypsin (A1AT), Cirrhosis, Inter-leukin 6 (IL-6), Hepatitis C virus (HCV) infection, Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP), Polymorphism
Introduction
Hepatitis C virus (HCV) infection represents a global health problem, especially in Egypt that showed the highest HCV epidemics worldwide. The national prevalence of HCV infection was recently estimated to be 14.7% [1]. The patients infected with HCV are always at risk of developing progressive liver diseases such as liver cirrhosis and hepatocellular carcinoma [2]. However, it is recognized that some HCV-infected patients do not develop liver cirrhosis during the course of the disease. Such prognostic variation in response to HCV infection suggests that the host genetic factors are involved in the outcome of the disease [3]. Several genetic association studies have implicated different genes such as cytokines to influence the susceptibility or the course of HCV infection [4].
Interleukin-6 (IL-6) is a multifactorial cytokine with a central role in the host immune defense. Recently, over 150 single nucleotide polymorphisms (SNPs) have been discovered at both 3’ and 5’ flanking regions of the IL-6 gene. These polymorphisms are associated with susceptibility to many different diseases such as idiopathic arthritis and some types of cancer [5]. Specifically, the SNP within the IL-6 promoter region at position –174 (IL-6, rs1800795 is associated with liver cirrhosis and hepatocellular carcinoma [6]. This SNP gives rise to two alleles, G and C, which produce two distinct phenotypes as follows: IL-6 (–174 G/G) and IL-6 (–174 G/C) genotypes that are called the high producer phenotypes because they are associated with producing higher levels of IL-6 in the blood. The IL-6 (–174 C/C) genotype, which is called the low producer phenotype, is associated with producing lower levels of IL-6 [7,8].
α1-Antitrypsin (A1AT) is an acute phase protein produced primarily in the hepatocytes. The blood concentration of A1AT increases three to four times in response to inflammation or tissue injury [9]. Interestingly, A1AT deficiency is associated with chronic liver disease and pulmonary emphysema [10]. So far, over 100 variants have been discovered in the A1AT gene [11]. The wild type M allele is associated with normal concentrations of A1AT in the serum, while the S allele (Glu264Val, rs17580) and Z allele (Glu342Lys, rs28929474) are associated with low concentrations of A1AT and therefore they are known as deficiency variants [12]. Remarkably, the homozygous ZZ genotype is associated with development of liver diseases however, only a few patients with the ZZ genotype eventually develop liver cirrhosis [13]. Importantly, it is widely accepted that IL-6 regulates the expression of acute phase proteins such as A1AT in both hepatic and extra hepatic tissues, where the plasma level of A1AT increases three- to four-fold in response to IL-6 expression [14].
In this study, we aimed to identify the distribution of the SNPs IL-6 –174 G/C, rs1800795, S (Glu264Val, rs 17580) and Z (Glu342Lys, rs28929474) mutations at the A1AT gene in Egyptian patients with chronic HCV infection. In addition, we examined the effect of these polymorphisms on the progression of liver cirrhosis.
Materials and Methods
Patients
In this study, 150 patients were recruited from a large group of 250 Egyptian HCV carriers together with 100 sex- and age-matched controls who attended the outpatient clinic at the Kasr El-Aini Hospital, Cairo University, Cairo, Egypt. The patients were divided into two subgroups: group 1 comprised 85 patients with chronic HCV infection and group 2 comprised 65 patients with liver cirrhosis.
Inclusion Criteria
All patients underwent clinical examination and routine liver function tests such as serum alanine transaminase (ALT), aspartate transaminase (AST), bilirubin, albumin, prothrombin time and viral hepatitis markers (anti-HCV antibodies, HBsAg and HBeAg). Patients were diagnosed with liver cirrhosis based on imaging studies in the form of abdominal ultrasound with Doppler as well as upper endoscopy for functional evaluation of decompensated cases. Staging of fibrosis was assessed using the Child Pugh assessment fibrosis score [15,16]. All patients provided written informed consent before participating in the study. The study was approved by the Ethics Committee of Kasr El-Aini, Faculty of Medicine, Cairo University, Cairo, Egypt and the study was carried out in accordance with the Code of Ethics of the World Medical Association (Declaration of Helsinki) for experiments in humans.
Exclusion Criteria
Patients with inflammatory diseases, cardiovascular diseases, thyroid dysfunction, diabetes mellitus (DM), alcohol intake, active schistosomiasis, co-infection with hepatitis B virus (HBV) and previously treated with interferon therapy, were excluded from the study.
Specimen Collection
A blood sample of 10 mL was drawn from each participant after overnight fasting. The blood samples were divided into two aliquots. The first aliquot was centrifuged for separation of serum to determine all routine and serological tests. The second aliquot was collected in a vacutainer containing EDTA as anticoagulant and stored at –80 °C for polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) analyses.
Routine Biochemical Tests
Serum ALT and serum AST were determined based on the method of Henry et al. [17] and Amador et al. [18] respectively using kits supplied by Roche Diagnostics GmbH (Penzberg, Germany). Serum albumin was determined using kits also supplied by Roche Diagnostics GmbH. Serum alkaline phosphatase (ALP) was determined based on the method of Bretaudiere et al. [19] using kits supplied by Bio Diagnostic (Giza, Egypt). Serum bilirubin was detected based on the method of Landis et al. [20] using kits supplied by Roche Diagnostics GmbH. Serum α fetoprotein (AFP) was detected according to the enzyme-linked-immunosorbent serologic assay (ELISA) method of Hirai [21] using kits supplied by Affymetrix (Santa Clara, CA, USA).
DNA Purification
DNA was purified using a generation capture column kit (Gentra Systems, Minneapolis, MN, USA) according to the manufacturer’s instructions. DNA was released by DNA elution buffer and heat without precipitation according to method of Glasel [22].
Detection of the Single Nucleotide Polymorphisms of α1-Antitrypsin and Interleukin 6 Genes by Polymerase Chain Reaction-Restriction Fragment Length Polymorphism
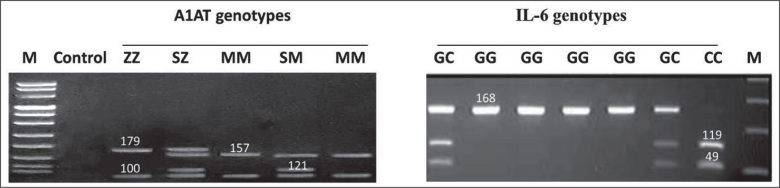
The Z mutation (342 Glu/Lys, rs28929474) and S mutation (264 Glu/Val, rs17580) on the A1AT gene in addition to the IL-6 (–174 G/C, rs1800795) mutation were identified by multiplex PCR using the thermal cycler (Techne Genius, Cambridge, Cambridgeshire, UK). Twenty pmol of each primer (described below) and 0.5 U of Taq DNA polymerase (Qiagen Ltd., Crawley, West Sussex, UK) were added to 100 ng of DNA in 30 µL (final volume) of a solution containing 20 mM Tris-HCL (pH 8.4), 50 mM KCL, 1.5 mM MgCl2, and 200 mM of each dNTP. Temperature cycling conditions were adjusted as follows for all primers used: initial denaturation for 5 min. at 94 °C; 35 cycles of 1 min. at 94 °C, 1 min. at 55 °C, and 2 min. at 72 °C; final extension for 10 min. at 72 °C. The PCR products of IL-6 were digested by 5 U of NIaIII at 37 °C for 24 hours. The presence of the –174 G/C mutation destroys a NIaIII restriction site in the respective PCR products. The generated fragments were 119 + 49 bp for the C allele and a single fragment of 168 bp for the G allele. The PCR products of the A1AT gene were digested by Taq1, where the presence of either S or Z mutation destroys a Taq1 restriction site in the respective PCR products. Fragments of (157 + 22 bp) and (100 + 21bp) were detected for the wild type M allele, a 179 bp fragment for the Z allele and a 121 bp fragment for the S allele. The analysis was carried out using a 1% agarose gel stained with ethidium bromide (Figure 1).
Figure 1.
The A1AT and IL-6 genotypes by PCR-RFLP. The left panel shows the M, S and Z alleles of the A1AT genotypes. M: DNA marker; ZZ genotype (179 and 100 bp band); SZ genotype (179, 157, 121 and 100 bp bands); MM genotype (157 and 100 bp bands); SM genotype (157, 121 and 100 bp bands). The right panel shows the G and C alleles of the IL-6 genotypes. GC genotype (168, 119 and 49 bp); GG genotype (168 bp band); CC genotype (119 and 49 bp bands); M: DNA marker.
Primer sequences of the A1AT gene: S (forward) 5’-TGA GGG GAA ACT ACA GCA CCT CG-3’; S (reverse) 5’-AGG TGT GGG CAG CTT CTT GGT CA-3’; Z (forward) 5’-ATA AGG CTG TGC TGA CCA TCG TC-3’; Z (reverse) 5’-TTG GGT GGG ATT CAC CAC TTT TC-3’. Primer sequences of the IL-6 gene: IL-6 (forward) 5’-TGA CTT CAG CTT TAC TCT TTG-3’; IL-6 (reverse) 5’-CTG ATT GGA AAC CTT ATT AAG-3’.
Statistical Analyses
The obtained data were analyzed using the Statistical Package for the Social Sciences version 12 software (SPSS Inc., Chicago, IL, USA). Mean ± standard error (SE) was used to describe continuous variables, while percentages and frequencies were used to describe categorical variables. One way repeated measure analysis of variance (ANOVA) followed by post Hoc test Scheffe’s method were used to compare the continuous variables of the groups. The differences in the frequency of A1AT and IL-6 genotypes were analyzed by the c2 test. The unpaired t-test was used to compare the continuous variables between different A1AT and IL6 genotypes.
Multiple logistic regression analysis was performed to evaluate the independent associations between liver cirrhosis and the polymorphic variants of the studied SNPs at both A1AT and IL-6 genes in addition to the other variables that may affect liver cirrhosis such as serum level of ALT, AST, ALP, total and direct bilirubin, albumin, prothrombin time and AFP. For this analysis, patients were divided as cases with absence of cirrhosis vs. cases with presence of cirrhosis (fibrosis score F0-F3 vs. F4 respectively). Odds ratio (OR), 95% confidence interval (95% CI) and p values were calculated with the SPSS Inc. software. A p value of <0.05 was considered statistically significant.
Results
The Demographic and Biochemical Parameters of the Studied Groups
In this study, we recruited two groups of chronic HCV-infected patients; the chronic hepatic patients and the cirrhotic patients in addition to the control group. We found that the chronic hepatic patients exhibited significantly higher values of ALT, AST and ALP compared with the control group (p <0.05). In the same way, the cirrhotic patient group exhibited significantly higher activities of ALT, AST and ALP in addition to higher values of total bilirubin, prothrombin time and AFP and lower albumin compared to the control group (p <0.05). Interestingly, the cirrhotic patient group exhibited significantly higher values of AST, albumin, prothrombin time, AFP and fibrosis score compared to the chronic hepatic patient group (p <0.05). These results confirm the progression of the end stage complications associated with HCV infection in the cirrhotic patients. The demographic and biochemical parameters of all studied groups are summarized in Table 1.
Table 1.
The clinical characteristics and biochemical parameters of the control group, chronic hepatitis patients and cirrhotic patients.
| Parameters | Control (n = 100) | Chronic Hepatitis Patients (n= 85) | Cirrhotic Patients (n = 65) |
|---|---|---|---|
| Sexes | M: 45; F: 55 | M: 55; F: 30 | M: 35; F: 30 |
| Age (years ± SE) | 33.10 ± 1.83 | 38.88 ± 1.92 | 38.92 ± 2.36 |
| Serum ALT (IU/mL) | 30.00 ± 1.34 | 83.53 ± 16.03a | 110.00 ± 14.63a |
| Serum AST (IU/mL) | 30.20 ± 1.36 | 76.76 ± 11.07a | 172.92 ± 27.76a,b |
| Serum ALP (IU/mL) | 43.65 ± 1.69 | 113.71 ± 6.51a | 119.85 ± 9.49a |
| Total bilirubin (mg/dL) | 0.75 ± 0.045 | 0.92 ± 0.098 | 1.09 ± 0.14a |
| Direct bilirubin (mg/dL) | 0.15 ± 0.01 | 0.25 ± 0.06 | 0.28 ± 0.05 |
| Albumin (g/dL) | 3.85 ± 0.05 | 3.69 ± 0.05 | 3.21 ± 0.11a,b |
| Prothrombin time (seconds) | 11.19 ± 0.14 | 11.53 ± 0.19 | 13.46 ± 0.51a,b |
| AFP (ng/mL) | 5.86 ± 0.45 | 8.76 ± 0.55 | 27.38 ± 6.91a,b |
| Fibrosis score (1-6) | — | 1.94 ± 0.18 | 4.46 ± 0.14a |
M: males; F: females; SE: standard error; ALT: alanine transaminase; AST: aspartate transaminase; ALP: alkaline phosphate; AFP: α fetopreotein.
significant values (p <0.05) when compared to the control group.
significant values (p <0.05) between chronic hepatitis and cirrhotic patients.
Frequency of Interleukin 6 Genotypes and Alleles of the Studied Groups
The SNP of IL-6 (–174 G/C, rs 1800795) was detected in the control, chronic hepatic and cirrhotic patients by the PCR-RFLP method. We found that the G allele was distributed at 100.0% in the control group, while the C allele was found only in the chronic hepatitis and cirrhotic patients at frequencies of 17.6 and 19.2%, respectively (Table 2). In more detail, the IL-6 (GG genotype) showed significantly lower frequency in the cirrhotic patients (61.5%) when compared to the control group (100.0%) at p <0.05. On the other hand, the IL-6 (CC genotype) was only found in the chronic hepatic patients (5.9% frequency), while the heterozygous IL-6 (GC genotype) was detected in both chronic hepatic and cirrhotic patients at 23.5 and 38.5% frequencies, respectively (Table 2).
Table 2.
The single nucleotide polymorphism pattern of the IL-6 gene in the control group, chronic hepatitis patients and cirrhotic patients.
| Parameters | Control Group n (%) | Chronic Hepatitis Patients n (%) | Cirrhotic Patients n (%) |
|---|---|---|---|
| Genotypes: | |||
| GG | 100 (100.0) | 60 (70.6) | 40 (61.5)a |
| GC | 0 (0.0) | 20 (23.5) | 25 (38.5) |
| CC | 0 (0.0) | 5 (5.9) | 0 (0.0) |
| Alleles: | |||
| G | 200 (100.0) | 140 (82.4) | 105 (80.8) |
| C | 0 (0.0) | 30 (17.6) | 25 (19.2)a |
significant values (p <0.05) when compared to the control group.
Frequency of α1-Antitrypsin Genotypes and Alleles of the Studied Groups
The S (264 Glu/Val, rs17580) and Z (342 Glu/Lys, rs28929474) mutations of the A1AT gene were detected in the control, chronic hepatic and cirrhotic patients by the PCR-RFLP method. We found that the wild type M allele was the most common allele in all groups. However, the frequency of the M allele decreased significantly in cirrhotic patients (65.4%) when compared to the control group (92.5%). Interestingly, the cirrhotic patients also showed a significantly increased frequency of the S allele (34.6%) compared to the control group (5.0%), while the Z allele was not detected in the cirrhotic patients (Table 3). The distribution of the A1AT genotypes revealed that the A1AT (SS genotype) was only detected in the cirrhotic patients (23.1% frequency) compared to control and chronic hepatic patients (both at 0.0% frequencies). In the same way, the A1AT (ZZ genotype) was only detected in the chronic hepatic patients (11.8% frequency) when compared to control and cirrhotic patients. The heterozygous MS and MZ genotypes were detected mostly in the chronic HCV patients (Table 3).
Table 3.
The single nucleotide polymorphism pattern of the A1TAT gene in the control group, chronic hepatitis patients and cirrhotic patients.
| Parameters | Control Group n (%) | Chronic Hepatitis Patients n (%) | Cirrhotic Patients n (%) |
|---|---|---|---|
| Genotypes: | |||
| MM | 85 (85.0) | 40 (47.0) | 35 (53.8)a |
| MS | 10 (10.0) | 30 (35.3) | 15 (23.1) |
| MZ | 5 (5.0) | 5 (5.9) | 0 (0.0) |
| ZZ | 0 (0.0) | 10 (11.8) | 0 (0.0) |
| SS | 0 (0.0 | 0 (0.0) | 15 (23.1) |
| Alleles: | |||
| M | 185 (92.5) | 115 (67.6) | 85 (56.4)a |
| S | 10 (5.0) | 30 (17.6) | 45 (34.6)a |
| Z | 5 (2.5) | 25 (14.7) | 0 (0.0) |
significant values (p <0.05) when compared to the control group.
The Biochemical Parameters Associated with Different Interleukin 6 Genotypes
As the low producer, IL-6 (CC genotype), was detected only in the chronic hepatitis patients, comparing the CC genotype with the high producer IL-6 (GG+GC) genotypes in the same group did not show significant increased values of ALT, AST, ALP, total and direct bilirubin or AFP (Table 4). These findings indicate that inheritance of the IL-6 (CC genotype) is not associated with increased risk of developing progressive liver diseases.
Table 4.
The biochemical parameters within different IL-6 genotypes in the control group, chronic hepatitis patients and cirrhotic patients.
| Parameters | Genotypes | ||||
|---|---|---|---|---|---|
| Control Group | Chronic Hepatitis Patients | Cirrhotic Patients | |||
| GG | GG+GC | CC | GG | GC | |
| Serum ALT (IU/mL) | 30.00±1.34 | 80.75±7.09 | 128.10±52.91 | 102.63±17.87 | 121.80±26.81 |
| Serum AST (IU/mL) | 30.20±1.36 | 73.33±5.61 | 137.45±35.74 | 150.50±28.65 | 208.80±54.69 |
| Serum ALP (IU/mL) | 43.65±1.69 | 115.93±5.45 | 78.34±19.49 | 125.25±14.21 | 111.20±10.31 |
| Total bilirubin (mg/dL) | 0.75±0.045 | 0.92±0.06 | 0.90±0.32 | 1.09±0.17 | 1.00±0.28 |
| Direct bilirubin (mg/dL) | 0.15±0.01 | 0.26±0.02 | 0.20±0.19 | 0.29±0.07 | 0.26±0.09 |
| Albumin (g/dL) | 3.85±0.05 | 3.69±0.07 | 3.60±0.06 | 3.26±0.13 | 3.12±0.23 |
| Prothrombin time (seconds) | 11.19±0.14 | 11.50±0.15 | 12.10±0.58 | 14.00±0.63 | 12.60±0.81 |
| AFP (ng/mL) | 5.86±0.45 | 8.87±0.65 | 7.00±1.05 | 25.63±8.94 | 30.20±12.04 |
| Fibrosis score (1-6) | — | 1.93±0.23 | 2.00±0.32 | 4.50±0.19 | 4.40±0.24 |
SE: standard error; ALT: alanine transaminase; AST: aspartate transaminase; ALP: alkaline phosphate; AFP: α fetopreotein.
The Biochemical Parameters Associated with Different α1-Antitrypsin Genotypes
The chronic hepatitis patients with the Z allele (MZ+ZZ genotypes) showed a significantly increased AST activity when compared to the chronic hepatitis patients with either the MM or MS genotypes (p <0.05). Interestingly, the cirrhotic patients with the S allele (MS+SS genotypes) showed significantly increased levels of AST, ALT, total bilirubin, direct bilirubin and lower albumin levels when compared with the cirrhotic liver patients with the A1AT (MM genotype) at p <0.05 (Table 5). These results indicate that inheritance of A1AT (S or Z allele) increases the severity of liver disease in either cirrhotic or chronic hepatitis patients, respectively.
Table 5.
The biochemical parameters within different A1AT genotypes in the control group, chronic hepatitis patients and cirrhotic patients.
| Parameters | Control Group | Chronic Hepatitis Patients | Cirrhotic Patients | ||||
|---|---|---|---|---|---|---|---|
| MM | MS+MZ | MM | MS | MZ+ZZ | MM | MS+SS | |
| Serum ALT (IU/mL) | 30.2±1.5 | 28.7±2.6 | 65.4±6.9 | 67.7±13.20 | 163.7±80.2 | 87.0±80.2 | 176.3±43.4a |
| Serum AST (IU/mL) | 30.1±1.6 | 31.0±2.5 | 68.0±5.6 | 57.3±10.50 | 139.0±47.9b,c | 127.7±20.1 | 300.7±65.9a |
| Serum ALP (IU/mL) | 44.5±1.9 | 38.7±0.9 | 109.3±7.4 | 115.5±5.70 | 122.0±34.5 | 114.6±10.8 | 130.7±34.8 |
| Total bilirubin (mg/dL) | 0.8±0.1 | 0.7±0.2 | 0.8±0.07 | 0.9±0.10 | 1.3±0.5 | 0.9±0.1 | 1.8±0.4a |
| Direct bilirubin (mg/dL) | 0.2±0.02 | 0.1±0.03 | 0.2±0.02 | 0.2±0.04 | 0.5±0.3 | 0.2±0.01 | 0.5±0.2a |
| Albumin (g/dL) | 3.8±0.05 | 3.9±0.2 | 3.7±0.1 | 3.7±0.10 | 3.6±0.03 | 3.3±0.1 | 2.7±0.1a |
| Prothrombin time (seconds) | 11.2±0.2 | 11.3±0.3 | 11.5±0.2 | 11.2±0.20 | 12.3±0.9 | 13.7±0.6 | 15.0±0.6 |
| AFP (ng/mL) | 6.0±0.5 | 5.0±1.0 | 8.6±0.9 | 8.7±1.00 | 9.3±1.0 | 25.6±10.3 | 48.3±11.2 |
| Fibrosis score (1-6) | — | — | 2.1±0.3 | 1.5±0.20 | 2.3±0.3 | 4.6±0.2 | 4.3±0.3 |
SE: standard error; ALT: alanine transaminase; AST: aspartate transaminase; ALP: alkaline phosphate; AFP: α fetopreotein.
significant values (p <0.05) when the MS+SS genotypes of cirrhotic patients were compared to the MM genotypes of cirrhotic patients.
significant values (p <0.05) when the MZ+ZZ genotypes of chronic hepatitis patients were compared to the MM genotypes of chronic hepatitis patients.
significant values (p <0.05) when the MZ+ZZ genotypes of chronic hepatitis patients were compared to the MS genotypes of chronic hepatitis patients.
Multivariate regression analyses showed that AST (p = 0.003, OR = 1.6, 95% CI = 1.3-2.0) and the S allele of A1AT represented as SS+MS genotypes (p = 0.001, OR = 7.7, 95% CI = 3.4-17.4) to be significantly independent predictors for development of liver cirrhosis. Although the IL-6 gene is known to regulate the expression of acute phase reactants such as A1AT, we did not find a statistically significant correlation between any of the genotypes of IL-6 (GG, GC, CC) and any of the A1AT (MM, MS, MZ, SS, ZZ) genotypes.
Discussion
In the present study, we investigated the association between IL-6 and A1AT SNPs and the progression of liver cirrhosis in Egyptian HCV-infected patients. In Egypt, numerous risk factors such as age, poverty, blood transfusion and intravenous anti-schistosomal treatment are associated with increased risk of being infected with HCV. Altogether, they render the numbers of HCV-infected patients to reach epidemic proportions [23,24].
Our analysis of the SNP IL-6 (–174 G/C) revealed that the G allele represents about 87.7% frequency in all subjects (controls and patients), while the C allele represents only a 12.3% frequency. These results are compatible with the findings that the G allele is highly distributed in non Caucasian populations compared with Caucasian population [25]. Interestingly, the CC genotype is only found in the chronic hepatitis patients but not in the control group or the cirrhotic patients. However, this CC genotype was not associated with significantly increased liver parameters in the chronic hepatitis patient group. The absence of the C allele from the control group may be attributed to the small size of our analyzed sample. However, our findings are still compatible with the few studies that examined the correlation between IL-6 polymorphisms and the progression of liver fibrosis in HCV-infected patients. For instance, Barrett et al. [26] found a positive correlation between IL-6 (–174 C/C) genotype and HCV clearance in an Irish population sample. On the other hand, Pereira et al. [27] found no difference in the frequency of IL-6 alleles at the –174 locus between controls and HCV-infected patients in the South American population. This can be explained because chronic hepatitis patients with low IL-6 producer genotypes (CC) are less susceptible to development of progressive liver diseases after HCV infection. Moreover, the chronic exposure of hepatocytes to IL-6, probably produced by the high producer genotypes (GG and GC), results in increased induction of the proapoptotic proteins: Bax, caspase 9 and cytochrome C that eventually lead to liver cell death [28]. To date, our study is the first one to describe the distribution of the IL-6 (-174 G/C) genotypes in Egyptian chronic hepatitis patients. For example, the polymorphisms of IL-4, IL-10 and IL-28B cytokines but not IL-6 were previously studied in the Egyptian HCV-infected patients to determine their impact on the response to antiviral treatments [29,30]. However, the frequency of IL-6 (–174 CC) genotype was found to be higher in Egyptian patients with other diseases such as intravascular coagulopathy in neonatal sepsis [31] and type 2 DM (T2DM) [32]. It is worth mentioning that Zekri et al. [33] measured the level of IL-6 in the sera of Egyptian patients with hepatocellular carcinoma by the ELISA method and found that they had a lower IL-6 level in comparison to asymptomatic HCV carriers.
Regarding A1AT mutations, our results showed that the highest frequency of the S allele (MS+SS genotypes) was found in the cirrhotic patients and it was associated with increased activity of liver enzymes and decreased serum albumin. Interestingly, the highest frequency of Z allele (MZ+ZZ genotypes) was found in the chronic hepatitis patients and was significantly associated with high levels of AST. We cannot exclude the existence of any of the deficient alleles in both groups due to the small number of patients in each group. Nevertheless, our results are compatible with Settin et al. [34] who found that the S allele and both MS and SS genotypes of A1AT were significantly higher in the Egyptian patients with HCV liver cirrhosis than in controls. On the other hand, there was no significant difference between the alleles or genotypes of S or Z mutations of A1AT in the Egyptian patients with hepatocellular carcinoma compared to controls [35].
It is widely accepted that the epidemiology of the A1AT gene deficiency is distributed worldwide and not restricted to Northern Europe as thought before. For example, in a study comprising around 97 countries from different geographic regions worldwide, the prevalence of the S allele in the Egyptian population was estimated at 57.1 cases/1000 population, while the Z allele was estimated at 28.6 cases/1000 population [36]. Our findings indicate that the presence of the Z allele in the chronic hepatitis patients and the S allele in the liver cirrhotic patients are associated with significantly aggressive liver disease.
Conclusions
We concluded that the inheritance of the deficient S and Z variants of the A1AT gene but not IL-6 (–174 G/C) mutation were associated with progressive liver diseases and liver cirrhosis. However, the small sample size of the current study did not allow us to extrapolate these results in the Egyptian HCV-infected patients. Even though we recommend screening the HCV-infected patients for the existence of the A1AT and IL-6 gene mutations in order to follow their future progression to liver cirrhosis.
Declaration of Interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
References
- 1.Breban R, Doss W, Esmat G, Elsayed M, Hellard M, Ayscue P. et al. Towards realistic estimates of HCV incidence in Egypt. J Viral Hepat. 2013;20(4):294–296. doi: 10.1111/j.1365-2893.2012.01650.x. [DOI] [PubMed] [Google Scholar]
- 2.Zucker S. Whomsoever ignores the natural history of the hepatitis C virus is doomed to treat it. Gastroenterology. 2002;122(2):578–579. doi: 10.1016/s0016-5085(02)80326-2. [DOI] [PubMed] [Google Scholar]
- 3.Missiha SB, Ostrowski M, Heathcote EJ. Disease progression in chronic hepatitis C: Modifiable and non-modifable factors. Gastroenterology. 2008;134(6):1699–1714. doi: 10.1053/j.gastro.2008.02.069. [DOI] [PubMed] [Google Scholar]
- 4.Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ. et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461(7262):399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 5.Cussigh A, Falleti E, Fabris C, Bitetto D, Cmet S, Fontanini E. et al. Interleukin 6 promoter polymorphisms influence the outcome of chronic hepatitis C. Immunogenetics. 2011;63(1):33–41. doi: 10.1007/s00251-010-0491-7. [DOI] [PubMed] [Google Scholar]
- 6.Giannitrapani L, Soresi M, Giacalone A, Campagna ME, Marasà M, Cervello M. et al. IL-6-174G/C polymorphism and IL-6 serum levels in patients with liver cirrhosis and hepatocellular carcinoma. OMICS. 2011;15(3):183–186. doi: 10.1089/omi.2010.0093. [DOI] [PubMed] [Google Scholar]
- 7.Fishman D, Faulds G, Jeffery R, Mohamed-Ali V, Yudkin JS, Humphries S. et al. The effect of novel poly-morphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. J Clin Invest. 1998;102(7):1369–1376. doi: 10.1172/JCI2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falleti E, Fabris C, Vandelli C, Colletta C, Cussigh A, Smirne C. et al. Genetic polymorphisms of interleu-kin-6 modulate fibrosis progression in mild chronic hepatitis C. Hum Immunol. 2010;71(10):999–1004. doi: 10.1016/j.humimm.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 9.Crystal RG, Brantly ML, Hubbard RC, Curiel DT, Holmes MD. The α1-antitrypsin gene and its mutations. Clinical consequences and strategies for therapy. Chest. 1989;95(1):196–208. doi: 10.1378/chest.95.1.196. [DOI] [PubMed] [Google Scholar]
- 10.Norman MR, Mowat AP, Hutchison D. Molecular basis, clinical consequences and diagnosis of α-1 anti-trypsin deficiency. Ann Clin Biochem. 1997;34(3):230–246. doi: 10.1177/000456329703400303. [DOI] [PubMed] [Google Scholar]
- 11.Canva V, Piotte S, Aubert J-P, Porchet N, Lecomte-Houcke M, Huet G. et al. Heterozygous M3Mmalton α1-antitrypsin deficiency associated with end-stage liver disease: Case report and review. Clin Chem. 2001;47(8):1490–1496. [PubMed] [Google Scholar]
- 12.Luisetti M, Seersholm N. α1-Antitrypsin deficiency 1: Epidemiology of α1-antitrypsin deficiency. Thorax. 2004;59(2):164–169. doi: 10.1136/thorax.2003.006494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fregonese L, Stolk J. Hereditary α1-antitrypsin deficiency and its clinical consequences. Orphanet J Rare Dis. 2008;3:16. doi: 10.1186/1750-1172-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perlmutter DH, May LT, Sehgal PB. Interferon beta 2/interleukin 6 modulates synthesis of α1-antitrypsin in human mononuclear phagocytes and in human hepatoma cells. J Clin Invest. 1989;84(1):138–144. doi: 10.1172/JCI114133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Child CG, Turcotte J. Surgery and portal hypertension. Major Probl Clin Surg. 1964;1:1–85. [PubMed] [Google Scholar]
- 16.Pugh R, Murray Lyon I, Dawson J, Pietroni M, Williams R. Transection of the oesophagus for bleeding oesophageal varices. Br J Surg. 1973;60(8):646–649. doi: 10.1002/bjs.1800600817. [DOI] [PubMed] [Google Scholar]
- 17.Henry R, Chiamori M, Golub O, Berkman S. Revised spectrophotometric methods for the determination of glutamate oxaloacetic transaminase, glutamic pyruvate transaminase and lactic acid dehydrogenase. Am J Clin Pathol. 1960;34:381–387. doi: 10.1093/ajcp/34.4_ts.381. [DOI] [PubMed] [Google Scholar]
- 18.Amador E, Wacker WE. Serum glutamic-oxaloacetic transaminase activity. A new modification and an analytical assessment of current assay technics. Clin Chem. 1962;8(4):343–350. [PubMed] [Google Scholar]
- 19.Bretaudiere J, Vassault A, Amsellem L, Pourci M, Thieu-Phung H, Bailly M. Criteria for establishing a standardized method for determining alkaline phosphatase activity in human serum. Clin Chem. 1977;23(12):2263–2274. [PubMed] [Google Scholar]
- 20.Landis JB, Pardue HL. Kinetics of the reactions of unconjugated and conjugated bilirubins with p-diazobenzenesulfonic acid. Clin Chem. 1978;24(10):1690–1699. [PubMed] [Google Scholar]
- 21.Hirai H. In: Biochemical Markers for Cancer. Chu TM, editor. New York, NY, USA: Marcel Dekker Inc; 1982. Alpha fetoprotein; pp. 23–59. [Google Scholar]
- 22.Glasel J. Validity of nucleic acid purities monitored by 260nm/280nm absorbance ratios. Biotechniques. 1995;18(1):62–63. [PubMed] [Google Scholar]
- 23.Mohamoud YA, Mumtaz GR, Riome S, Miller D, Abu-Raddad LJ. The epidemiology of hepatitis C virus in Egypt: A systematic review and data synthesis. BMC Infect Dis. 2013;13(1):288. doi: 10.1186/1471-2334-13-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guerra J, Garenne M, Mohamed M, Fontanet A. HCV burden of infection in Egypt: Results from a nationwide survey. J Viral Hepat. 2012;19(8):560–567. doi: 10.1111/j.1365-2893.2011.01576.x. [DOI] [PubMed] [Google Scholar]
- 25.Meenagh A, Williams F, Ross OA, Patterson C, Gorodezky C, Hammond M. et al. Frequency of cytokine polymorphisms in populations from western Europe, Africa, Asia, the Middle East and South America. Hum Immunol. 2002;63(11):1055–1061. doi: 10.1016/s0198-8859(02)00440-8. [DOI] [PubMed] [Google Scholar]
- 26.Barrett S, Collins M, Kenny C, Ryan E, Keane C, Crowe J. Polymorphisms in tumour necrosis factor α, transforming growth factor β, interleukin 10, interleukin 6, interferon γ, and outcome of hepatitis C virus infection. J Med Virol. 2003;71(2):212–218. doi: 10.1002/jmv.10472. [DOI] [PubMed] [Google Scholar]
- 27.Pereira FA, Silva NNPd, Rodart IF, Carmo TMAd, Lemaire DC, Reis MGd. Association of TGF-beta1 codon 25 (G915C) polymorphism with hepatitis C virus infection. J Med Virol. 2008;80(1):58–64. doi: 10.1002/jmv.21011. [DOI] [PubMed] [Google Scholar]
- 28.Jin X, Zimmers TA, Perez EA, Pierce RH, Zhang Z, Koniaris LG. Paradoxical effects of short and long term interleukin 6 exposure on liver injury and repair. Hepatology. 2006;43(3):474–484. doi: 10.1002/hep.21087. [DOI] [PubMed] [Google Scholar]
- 29.Shaker OG, Sadik NA. Polymorphisms in interleukin 10 and interleukin 28B genes in Egyptian patients with chronic hepatitis C virus genotype 4 and their effect on the response to pegylated interferon/ribavirin therapy. J Gastroenterol Hepatol. 2012;27(12):1842–1849. doi: 10.1111/j.1440-1746.2012.07273.x. [DOI] [PubMed] [Google Scholar]
- 30.Shalaby SM, Radwan MI, Abdelazim S, Nafee AM. Interleukin-4 polymorphisms and response to combination therapy in Egyptian chronic hepatitis C patients. Cell Immunol. 2012;276(1):110–113. doi: 10.1016/j.cellimm.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 31.Abdel-Hady H, El-Naggar M, El-Nady G, Badr R, El-Daker M. Genetic polymorphisms of IL-6-174 and IL-10-1082 in full term neonates with late onset blood stream infections. J Pediatr Infect Dis. 2009;4(4):357–365. [Google Scholar]
- 32.Helaly MA-H, Hatata E-SZ, Abu-Elmagd M, Ibrahem EF, Alsaid A, El-Aal IAA. et al. Association of IL-10 and IL-6 gene polymorphisms with type 2 diabetes mellitus among Egyptian patients. Eur J Gen Med. 2013;10(3):158–162. [Google Scholar]
- 33.Zekri A, Ashour MS, Hassan A, AE-D HM, El-Shehaby A, Abu-Shady MA. Cytokine profile in Egyptian hepatitis C virus genotype-4 in relation to liver disease progression. World J Gastroenterol. 2005;11(42):6624–6630. doi: 10.3748/wjg.v11.i42.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Settin A, El-Bendary M, Abo-Al-Kassem R, El Baz R. Molecular analysis of A1AT (S and Z) and HFE (C282Y and H63D) gene mutations in Egyptian cases with HCV liver cirrhosis. J Gastrointestin Liver Dis. 2006;15(2):131–135. [PubMed] [Google Scholar]
- 35.Gharib AF, Karam RA, Pasha HF, Radwan MI, Elsawy WH. Polymorphisms of hemochromatosis, and α-1 antitrypsin genes in Egyptian HCV patients with and without hepatocellular carcinoma. Gene. 2011;489(2):98–102. doi: 10.1016/j.gene.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 36.de Serres FJ, Blanco I. Prevalence of α1-antitrypsin deficiency alleles PI* S and PI* Z worldwide and effective screening for each of the five phenotypic classes PI* MS, PI* MZ, PI* SS, PI* SZ, and PI* ZZ: a comprehensive review. Ther Adv Respir Dis. 2012;6(5):277–295. doi: 10.1177/1753465812457113. [DOI] [PubMed] [Google Scholar]