Abstract
Barth syndrome (BTHS) is a rare X-linked disease characterized by dilated cardiomyopathy, proximal skeletal myopathy and cyclic neutropenia. It is caused by various mutations in the tafazzin (TAZ) gene located on Xq28 that results in remodeling of cardiolipin and abnormalities in mitochondria stability and energy production. Here we report on a novel c.285-1G>C splice site mutation in intron 3 of the TAZ gene that was detected prenatally.
Keywords: Barth Syndrome (BTHS), cardiomyopathy, Neutropenia, 3-Methylglutaconin aciduria, Tafazzin (TAZ) gene
Introduction
Barth syndrome (BTHS, OMIM 302060) is a rare X-linked disease characterized by dilated cardiomyopathy, proximal skeletal myopathy and cyclic neutropenia and was first described in 1983 by Barth et al. [1]. The incidence of BTHS is about 1 in 300,000-400,000 births [2]. It is also presented with organic aciduria, particularly excess of 3-methylglutaconic acid [1,3]. The excretion of 3-methylglutaconic acid in urine can be highly variable and is often intermittent. At the moment, 3-methylglutaconic acid is a biochemical marker for mitochondrial dysfunction of still unknown origin [4]. Less common features of the disease include hypertrophic cardiomyopathy, isolated left ventricular noncompaction, ventricular arrhythmia, motor delay, poor appetite, fatigue and exercise intolerance, hypoglycemia, lactic acidosis, hyperammonemia and growth delay [5,6]. The main cause of death in infants with BTHS is either heart failure or sepsis due to neutropenia [1,2,5].
Barth syndrome is caused by various mutations in the tafazzin ( TAZ, previously termed G4.5) gene [7], comprising 11 exons and located on Xq28 [8,9]. This gene encodes a protein tafazzin that plays an important role in remodelling of cardiolipin and phosphatidylglycerol structure [1]. Cardiolipin is a component of the inner mitochondrial membrane. It stabilizes highly ordered respiratory chain supercomplexes and optimises energy production in mitochondria [10]. Studies carried out on a BTHS zebrafish model suggest that the expression of tafazzins is both tissue-specific and age-dependent, and plays an essential role in cardiac development and function [11,12]. We report a novel splice site mutation in intron 3 of the TAZ gene in this study.
Case Report
The proband, a 22-year-old primigravida, was referred to the Center for Medical Genetics, Vilnius University Hospital Santariškių Klinikos, Vilnius, Lithuania, at 13 weeks of gestation for genetic counseling because of a familial history of cardiomyopathy. She had three biological brothers. The first brother was healthy. The second brother was born after an uncomplicated pregnancy; on the 9th day after birth, the boy became febrile and was referred to the intensive care unit because of impaired cardiac function, where he was diagnosed with endocardial fibroelas-tosis. Later, the diagnosis was changed to myocarditis. At 4 months of age the boy repeatedly showed symptoms of fever and worsened symptoms of myocarditis. He subsequently died due to cardiac failure at 6 months of age.
The third brother of the proband was also born after a normal pregnancy and was diagnosed with cardiac insufficiency due to endocardial fibroelastosis on the 3rd week of life after the episode of fever and cyanosis. Since the heart function was improving significantly, the diagnosis was changed to myocarditis. He then developed dilated cardio-myopathy at 5 months of age. Subsequently, at 1 year and 4 months of age the boy presented with an episode of impaired consciousness and convulsion. At that time the boy was diagnosed with hypoglycemia, cardiac insufficiency, repeated episodes of neutropenia, growth retardation and hypotonia. Congenital metabolic disorder was suspected. Biochemical findings included excess amounts of 3-meth-ylglutaric and 3-methylglutaconic acids in urine and low free carnitine in blood. As a result, based on clinical, biochemical findings and family history, BTHS was diagnosed. Genetic testing was not performed at that time. The boy died at 11 years of age of cardiopulmonary insufficiency due to severe pulmonary infection.
The first trimester ultrasound performed on our proband demonstrated the male sex of the fetus and revealed no markers of chromosomal abnormalities and no fetal pathology. Chorionic villus sampling was performed and molecular tests were carried out. There were no aneuploidies detected and sex chromosomes were XY.
As the clinical diagnosis of BTHS was defined for our proband’s third brother and the family history highly suggested an X-linked disorder, genetic analysis was performed on the proband’s fetus. Genetic tests were initiated in this order because the purpose of genetic counseling was to investigate if the proband’s fetus had BTHS. Furthermore, as mentioned previously, the third brother of our proband died 10 years ago and it was not possible to start genetic testing on the deceased patient. Informed consent was obtained from all family members who participated in the molecular analyses study.
Materials and Methods
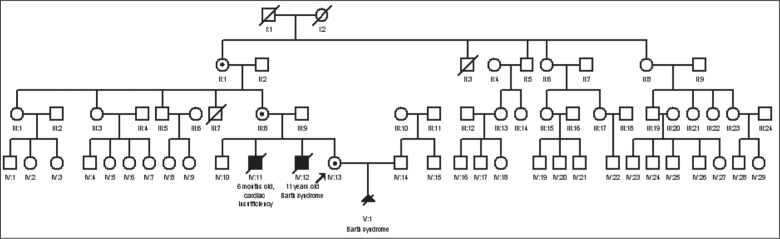
A detailed genealogy of the family was constructed. Inheritance in the presented pedigree was consistent with an X-linked recessive pattern (Figure 1). The DNA of the proband’s fetus (V:1) was extracted from chorionic villi using the InstaGene™ Matrix (Bio-Rad Laboratories, Hercules, CA, USA). The DNA of the proband’s sibling (IV: 12) was extracted from a dried blood spot sample (Guthrie card; Newborn Bloodspot Screening, Wales, Cardiff, UK; http://www.newbornbloodspotscreening.wales.nhs.uk/) using the InstaGene™ Matrix (Bio-Rad Laboratories). The DNA of other family members (IV:13, III:8, II:1) was isolated from the venous peripheral blood samples using the phenol-chloroform extraction method. Amplification using specific primers for X-linked TAZ gene (NM_ 000116) was performed for the proband’s fetus (V:1). Polymerase chain reaction (PCR) primers for exons 1-11 exons and adjacent intronic regions of the TAZ gene were designed using the Primer3 (http://bioinfo.ut.ee/primer3-0.4.0/primer3/) program [13,14]. Primer sequences are available from the authors upon request. Family members were tested only for the mutation that was observed in the fetus (i.e., c.285-1G>C). Amplification products were electrophoresed with TBE 2.0% agarose gel and sequenced using a BigDye Terminator version 3.1 cycle sequence kit (Applied Biosystems, Waltham, MA, USA) on an ABI PRISM™ 3130xl sequencer.
Figure 1.
Pedigree of the studied family
Results of Mutational Analyses
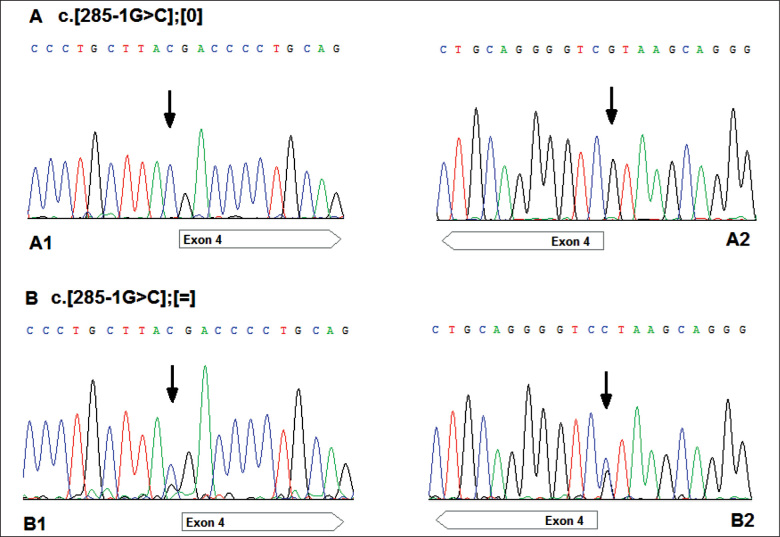
Sequence analysis of the TAZ gene identified a hemizygous c.285-1G>C substitution (c.[285-1G>C];[0]) in intron 3 in chorionic villi DNA of the proband’s fetus (V:1). The same hemizygous c.285-1G>C mutation was subsequently identified in the deceased sibling of the proband (IV:12). The mutation was detected in proband’s DNA (IV:13) in the heterozygous form (c.[285-1G>C];[=]) and the carrier status was confirmed. The mother (III:8) and maternal grandmother (II:1) of the proband also carries the mutation (heterozygous genotype for this mutation) [Figure 2].
Figure 2.
The TAZ sequencing electrophoregrams showing position c.285-1 of the TAZ sequence (NM_000116) (indicated by an arrow).
(A) The results in the fetus and proband’s sibling: hemizygous mutation (c.[285-1G>C];[0]); A1: forward strand; A2: reverse strand.
(B) The fragments of the TAZ gene sequences of the proband, mother and maternal grandmother: heterozygous form (c.[285-1G>C];[=]); B1: forward strand; B2: reverse strand.
Discussion
To date, more than 220 different TAZ gene mutations in all exons have been identified [Human Tafazzin (TAZ ) Gene Mutation and Variation Database (last updated March 28, 2015; http://www.barthsyndrome.org/home)], 94 of which were found in patients with diagnosed BTHS (Human Gene Mutation Database Professional 2015.4; http://www.hgmd.cf.ac.uk/ac/index.php). Only 13.0% of boys with BTHS carry de novo mutations [15]. Phenotype-genotype correlations have not yet been identified [16,17]. It has been observed that mutations in the TAZ gene also result in non syndromic left ventricular non compaction, endocardial fibroelastosis, X-linked infantile cardiomyopathy and dilated cardiomyopathy. Thus, mutations in the TAZ gene can result in a broad spectrum of clinical phenotypes including, but not limited to classical BTHS.
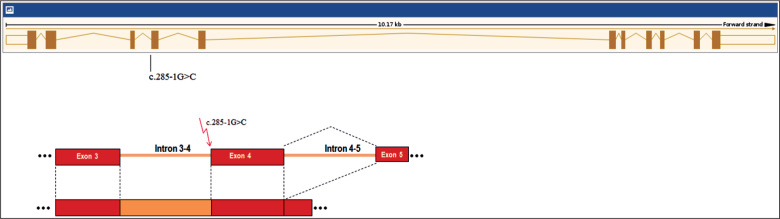
The c.285-1G>C mutation has not been previously reported in the Human Tafazzin (TAZ) Gene Mutation and Variation Database (last updated March 28, 2015; http://www.barth syndrome.org/home). The mutation is predicted to alter the wild type constitutive acceptor splice site. Most probably the presence of mutation affects splicing in tafazzin [18]. Family studies showed that the proband, her mother and maternal grandmother carried the same mutation. Our proband’s fetus as well as her brother was affected with BTHS, suggesting that this newly discovered splice site mutation is an anomaly strongly affecting the normal function of the tafazzin protein, and thus, is likely to be pathogenic (Figure 3). An intronic TAZ gene mutation (aberrant splicing and elongation of exon 3 because of the insertion of 106 bases between exons 3 and 4) has been reported in a Japanese patient with BTHS, who had very similar symptoms and course of the disease as the third brother of our patient [19]. Our proband decided to terminate the pregnancy. Fetal autopsy was inconclusive.
Figure 3.
The location of the c.285-1G>C mutation of the TAZ gene and its effect on splicing.
Evidence is accumulating that the disorder is substantially underdiagnosed. Historically regarded as a cardiac disease, BTHS is now considered a multi-system disorder that may be first seen by many different specialists. Phenotypic variability raises a major challenge, as some children with BTHS have never showed neutropenia, others lack increased 3-methylglutaconic acid and a minority has occult or absent cardiomyopathy [20]. Furthermore, BTHS was described in 2010 as an unrecognized cause of fetal death. It is recommended that investigation for BTHS should now be seriously considered in male neonates, babies and young boys presenting with idiopathic dilated cardiomyopathy or left ventricular non compaction, and in males with unexplained ventricular arrhythmia or sudden death [20].
Female carriers are usually healthy and have no cardiovascular pathology. It is however theoretically possible for a female to develop symptoms of the disease because of impaired X chromosome inactivation. The only female ever described with the disease had abnormalities of both X chromosomes [21]. In this case, the proband, her mother and maternal grandmother had normal electrocardiograms and no history of cardiac disease.
Conclusions
A novel mutation in the TAZ gene was identified prenatally in a family with a clinical diagnosis of BTHS. The diagnosis for the proband’s third brother was confirmed by molecular genetic testing and the second brother’s underlying cause of early death was revealed. Mutational analysis offers the possibility of prenatal genetic counseling and preimplantation genetic diagnosis for the family.
Declaration of Interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
References
- 1.Barth PG, Scholte HR, Berden JM, van der Klei-Van Moorsel JM, Luyt Houwen IE, van’t Veer Korthof ET. et al. An X-linked mitochondrial disease affecting cardiac muscle, skeletal muscle and neutrophil leukocytes. J Neurol Sci 1983. 62(1-3):327–355. doi: 10.1016/0022-510x(83)90209-5. [DOI] [PubMed] [Google Scholar]
- 2.Barth Syndrome Foundation Website: Frequently Asked Questions. 2006 http://www.barthsyndrome.org [Google Scholar]
- 3.Kelley RI, Cheatham JP, Clark BJ, Nigro MA, Powell BR, Sherwood GW. et al. X-linked dilated cardiomyopathy with neutropenia, growth retardation, and 3-methylglutaconic aciduria. J Pediatr. 1991;119(5):738–747. doi: 10.1016/s0022-3476(05)80289-6. [DOI] [PubMed] [Google Scholar]
- 4.Wortmann SB, Kluijtmans LA, Engelke UFH, Wevers RA, Morava E. The 3-methylglutaconic acidurias: What’s new? J Inherit Metab Dis. 2012;35(1):13–22. doi: 10.1007/s10545-010-9210-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steward CG, Newbury-Ecob RA, Hastings R, Smithson SF, Tsai-Goodman B, Quarrell OW. et al. Barth syndrome: An X-linked cause of fetal cardiomyopathy and stillbirth. Prenatal Diag. 2010;30(10):970–976. doi: 10.1002/pd.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spencer CT, Bryant RM, Day J, Gonzalez IL, Colan SD, Thompson WR. et al. Cardiac and clinical phenotype in Barth syndrome. Pediatrics. 2006;118(2):e337–e346. doi: 10.1542/peds.2005-2667. [DOI] [PubMed] [Google Scholar]
- 7.Bione S, Dadamo P, Maestrini E, Gedeon AK, Bolhuis PA, Toniolo D. A novel X-linked gene, G4.5. is responsible for Barth syndrome. Nat Genet. 1996;12(4):385–389. doi: 10.1038/ng0496-385. [DOI] [PubMed] [Google Scholar]
- 8.Ades LC, Gedeon AK, Wilson MJ, Latham M, Partington MW, Mulley JC. et al. Barth syndrome – Clinical features and confrmation of gene localization to distal Xq28. Am J Med Genet. 1993;45(3):327–334. doi: 10.1002/ajmg.1320450309. [DOI] [PubMed] [Google Scholar]
- 9.Bolhuis PA, Hensels GW, Hulsebos TJM, Baas F, Barth PG. Mapping of the locus for X-linked cardioskeletal myopathy with neutropenia and abnormal mitochondria (Barth Syndrome) to Xq28. Am J Hum Genet. 1991;48(3):481–485. [PMC free article] [PubMed] [Google Scholar]
- 10.Klingenberg M. Cardiolipin and mitochondrial carriers. Biochim Biophys Acta. 2009;1788(10):2048–2058. doi: 10.1016/j.bbamem.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 11.Ferri L, Donati MA, Funghini S, Malvagia S, Catarzi S, Lugli L. et al. New clinical and molecular insights on Barth syndrome. Orphanet J Rare Dis. 2013;8(1):27. doi: 10.118/1750-1172-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khuchua Z, Yue Z, Batts L, Strauss AW. A zebrafish model of human Barth syndrome reveals the essential role of tafazzin in cardiac development and function. Circ Res. 2006;99(2):201–208. doi: 10.1161/01.RES.0000233378.95325.ce. [DOI] [PubMed] [Google Scholar]
- 13.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M. et al. Primer3 – new capabilities and interfaces. Nucleic Acids Res. 2012;40(15):e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koressaar T, Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23(10):1289–1291. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez IL. Human tafazzin (TAZ) gene mutation and variation database. 2012 http://www.barthsyndrome.org Science and research section of. [Google Scholar]
- 16.Chen R, Tsuji T, Ichida F, Boules KR, Yu X, Watanabe S. et al. Mutation analysis of the G4.5 gene in patients with isolated left ventricular noncompaction. Mol Genet Metab. 2002;77(4):319–325. doi: 10.1016/s1096-7192(02)00195-6. [DOI] [PubMed] [Google Scholar]
- 17.Roberts AE, Nixon C, Steward CG, Gauvreau K, Maisenbacher M, Fletcher M. et al. The Barth Syndrome Registry: Distinguishing disease characteristics and growth data from a longitudinal study. Am J Med Genet Part A. 2012;158A(11):2726–2732. doi: 10.1002/ajmg.a.35609. [DOI] [PubMed] [Google Scholar]
- 18.Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C. Human Splicing Finder: An online bioinformatics tool to predict splicing signals. Nucleic Acid Res. 2009;37(9):e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakamoto O, Ohura T, Katsushima Y, Fujiwara I, Ogawa E, Miyabayashi S. et al. A novel intronic mutation of the TAZ (G4.5) gene in a patient with Barth syndrome: Creation of a 5-prime splice donor site with variant GC consensus and elongation of the upstream exon. Hum Genet. 2001;109(5):559–563. doi: 10.1007/s00439-001-0612-3. [DOI] [PubMed] [Google Scholar]
- 20.Clarke SL, Bowron A, Gonzalez IL, Groves SJ, Newbury-Ecob R, Clayton N. et al. Barth syndrome. Orphanet J Rare Dis. 2013;8:23. doi: 10.118/61760-1172-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cosson L, Toutain A, Simard G, Kulik W, Matyas G, Guichet A. et al. Barth syndrome in a female patient. Mol Genet Metab. 2012;106(1):115–120. doi: 10.1016/j.ymgme.2012.01.015. [DOI] [PubMed] [Google Scholar]