Summary
The nuclear factor‐κB (NF‐κB) family of transcription factors play an essential role for the regulation of inflammatory responses, immune function and malignant transformation. Aberrant activity of this signalling pathway may lead to inflammation, autoimmune diseases and oncogenesis. Over the last two decades great progress has been made in the understanding of NF‐κB activation and how the response is counteracted for maintaining tissue homeostasis. Therapeutic targeting of this pathway has largely remained ineffective due to the widespread role of this vital pathway and the lack of specificity of the therapies currently available. Besides regulatory proteins and microRNAs, long non‐coding RNA (lncRNA) is emerging as another critical layer of the intricate modulatory architecture for the control of the NF‐κB signalling circuit. In this paper we focus on recent progress concerning lncRNA‐mediated modulation of the NF‐κB pathway, and evaluate the potential therapeutic uses and challenges of using lncRNAs that regulate NF‐κB activity.
Keywords: cancer, inflammation, long non‐coding RNA, nuclear factor‐κB signalling, regulation
Introduction
Nuclear factor‐κB (NF‐κB) is a transcription factor that plays its most important and evolutionarily conserved role in coordinating immune and inflammatory responses. Moreover, NF‐κB influences the expression of genes that function in cell differentiation, proliferation and survival in almost all multicellular organisms.1 As a result, both activation of the NF‐κB signalling pathway and termination of the NF‐κB response are tightly regulated, and dysregulation of the NF‐κB system is known to be associated with a wide range of disorders, ranging from inflammatory and autoimmune diseases to various types of cancer.
In mammals, six transcription factors have been indentified in the NF‐κB family: RelA (p65), RelB, c‐Rel, p50 (p105 precursor), p52 (p100 precursor) and Relish.2 These proteins carry an N‐terminal Rel homology domain, which is required for dimerization, nuclear targeting, DNA binding and interaction with the inhibitory IκB proteins.2, 3 In addition, RelA, RelB and c‐Rel contain a domain at their C‐terminal that is responsible for transcriptional activation of target genes. Among the NF‐κB family members, the heterodimer p50/p65 is the most prominent and serves as the prototype of NF‐κB. In most quiescent cells, the prototypical IκB proteins (IκBα, IκBβ and IκBε) that harbour ankyrin repeats at their C‐termini, preferentially associate with different c‐Rel and p65 dimeric complexes and inhibit NF‐κB activity.4 Similar to IκB proteins, the precursors p105 and p100 also possess C‐terminal ankyrin repeats and can also bind and act as cytoplasmic inhibitors for p65, c‐Rel and RelB dimer partners.4
Activation of dormant NF‐κB in the cytoplasm occurs by releasing NF‐κB from the NF‐κB/IκB complex or by cleaving the inhibitory ankyrin repeat domains of p100 and p105.5 In the canonical (classical) activation pathway, a variety of structurally diverse signals originating from antigen receptors, pattern‐recognition receptors and tumour necrosis factor (TNF) and interleukin‐1 (IL‐1) receptors trigger activation of a serine‐specific IκB kinase complex termed IKK.6 The IKK complex consists of two catalytic subunits, IKKα (IKK1) and IKKβ (IKK2), together with at least one non‐catalytic regulatory protein (NF‐κB Essential Modulator, NEMO or IKKγ).2, 5 Phosphorylation of IκB by activated IKK2 is a prerequisite for Lys48‐linked polyubiquitination and proteasomal degradation of IκB. In the non‐canonical (alternative) activation pathway, a subset of TNF superfamily receptors, including CD40, lymphotoxin β receptor, B‐cell activating factor receptor, receptor activator of NF‐κB, Fn14 and CD27, activate NF‐κB.7 In this pathway, NF‐κB‐inducing kinase is accumulated following receptor ligation, and then phosphorylates IKK1. Activated IKK1 induces p100 phosphorylation leading to its ubiquitination and partial degradation to p52.5 After the release of various NF‐κB dimers by either pathway, NF‐κB migrates to the nucleus and activates target gene transcription by binding to NF‐κB elements in the promoter region.
As an important regulator of immunity and inflammation, activation of NF‐κB signalling is influenced by multiple regulatory mechanisms. Numerous post‐translational modifications of p65, including ubiquitination, phosphorylation, acetylation, sumoylation and nitrosylation and, more recently, methylation,3 have been shown to have positive or negative effects on transcriptional responses of NF‐κB. In addition, NF‐κB signalling components, including NF‐κB itself, have been reported to interact with chromatin‐modifying enzymes such as histone deacetylases or acetyltransferases (p300/CBP),5 with other transcription factors,5, 8 and with phosphatases9, 10 to fine‐tune the NF‐κB response. Notably, many NF‐κB target genes encode inhibitors of the NF‐κB response. Among them are IκBα, IκBε, A20, CYLD and several microRNAs that are induced by NF‐κB‐dependent transcription.11
Growing evidence suggests that NF‐κB control is also regulated through long non‐coding RNAs (lncRNAs), which is the topic of this review. Unlike protein‐coding genes, most lncRNA do not show sequence conservation across species. This could be due to current methods for sequence comparison not being suited for finding homology between lncRNAs, a lack of functionality of large portions of a given lncRNA, or the functional dependence of a secondary structure for lncRNA action.12, 13 So far, how well lncRNA sequences, secondary structures and their functions are conserved is unknown. Nevertheless, certain human lncRNAs with poor evolutionary conservation beyond primates have been demonstrated to be functional and possess therapeutic potential.13 Although the precise mechanisms by which lncRNAs carry out their roles remain poorly understood, accumulating data suggest that through specific interactions with proteins, DNA and other types of RNA, lncRNAs may regulate their neighbouring genes in cis, or distant genes in trans.12 Moreover, a considerable number of lncRNAs, particularly those derived from enhancer regions (eRNAs), have been found to locally regulate gene expression through the act of transcription rather than lncRNAs themselves,14, 15 although it is evident that some eRNA transcripts (e.g. HOTTIP) do contribute to gene regulation.16
The lncRNAs function in multiple cellular processes, and dysregulated expression of lncRNAs has been found to be associated with a diverse set of human ailments, including cancer.17 Recently, the roles of lncRNAs in innate immunity and inflammation have attracted much attention.18, 19, 20 Below we focus on individual lncRNAs that have been shown to influence NF‐κB signalling under pathological and physiological conditions (Table 1).
Table 1.
Long non‐coding RNAs (lncRNAs) involved in the regulation of nuclear factor‐κB (NF‐κB) signalling
| lncRNA | System | Function | Mechanism | Reference |
|---|---|---|---|---|
| lncRNAs that interact with NF‐κB or its transcripts | ||||
| PACER | Primary human mammary epithelial cells; human monocyte/macrophage cell line U937; primary human osteoarthritis chondrocytes | Activates COX2 in cis | Interacts and sequesters excess p50 from COX2 promoter | 22, 23 |
| lincRNA‐Cox2 | Mouse RAW264.7 macrophages and mouse BV2 microglia | Activates the transcription of NF‐κB‐regulated late‐primary inflammatory genes stimulated by lipopolysaccharide (LPS) | Interacts with SWI/SNF and p65/p50 to facilitate NF‐κB binding to DNA by modifying epigenetic marks and chromatin accessibility | 25 |
| Lethe | Mouse embryonic fibroblast (MEF) cells; human 293T cells | Represses NF‐κB target gene expression | Binds to NF‐κB and prevents it from interacting with DNA | 26 |
| NKILA | Human breast cancer cell lines | Inhibits NF‐κB activation and breast cancer metastasis | Blocks IκB phosphorylation via interacting with NF‐κB/IκB complex | 29 |
| MALAT1 | Human monocyte/macrophage cell line THP1 | Negatively regulates LPS‐induced inflammatory response | Associates with p50/p65 and occludes NF‐κB from the promoter | 32 |
| lincRNA‐p21 | Patients with rheumatoid arthritis; human Jurkat T cell and THP1 monocyte lines | Enhances anti‐inflammatory properties of methotrexate by inhibiting NF‐κB | Interacting with p65 mRNA to inhibit its translation | 37 |
| lncRNAs that interfere with signalling components or related molecules upstream of NF‐κB | ||||
| HOTAIR | Ovarian cancer cell lines; patients with ovarian cancer | Activates NF‐κB target genes implicated in DNA damage response and chemoresistance | Decreases IκBα protein level most likely by chromatin‐mediated repression | 38 |
| HOTAIR | Mouse cardiomyocytes | Activates the expression of TNFa, an inducer of myocardial dysfunction during LPS‐induced sepsis | Promotes NF‐κB activation through increasing LPS‐induced p65 phosphorylation | 39 |
| MIR31HG | Human adipose‐derived stem cells | Suppresses osteogenic differentiation and enhances inflammation‐induced inhibition of osteogenesis | Activates NF‐κB through interacting with IκBα | 41 |
| C2dat1 | Murine model of ischaemia/reperfusion (I/R); mouse neuroblastoma (N2a) cells in response to in vitro ischaemia | Promotes neuron survival following ischaemia | Activates NF‐κB through enhancing CAMK2D expression | 42 |
| Arid2‐IR | Mouse unilateral ureteral obstructive (UUO) kidney and mouse tubular epithelial cells (mTECs) | Promotes NF‐κB‐driven renal inflammation | Promotes interleukin‐1β‐induced NF‐κB signalling through unknown mechanism | 44 |
| DLEU1 and DLEU2 | Primary human chronic lymphocytic leukaemia (CLL) cells; human cell lines including HEK293T | Represses neighbouring genes in cis; play a role in tumorigenesis | Transcriptional down‐regulation of its neighbouring candidate tumour suppressor genes that are regulators of NF‐κB | 45 |
| lncRNAs that are induced by NF‐κB signalling and regulate NF‐κB target gene expression | ||||
| IL1β‐eRNA, IL1β‐RBT46 | Primary human monocytes | Enhance LPS‐induced transcription of IL1β in cis and other inflammatory genes in trans | Unknown | 46 |
| AS‐IL1α | Mouse primary bone marrow‐derived macrophages (BMDMs) | Enhances LPS‐inducible transcription of IL‐1α in cis | Recruits RNA polymerase II to the promoter | 48 |
| ANRIL | Human umbilical vein endothelial cells | Regulates the inflammatory response related to coronary artery disease | Interacts with and facilitates binding of the transcription factor YY1 to promoters | 50 |
| THRIL | Human THP1 monocyte/macrophage cell line | Required for the expression of many immune‐response genes including TNFα | Forms a complex with hnRNPL that binds to TNFa promoter | 52 |
| lincRNA‐Cox2 | Mouse bone marrow‐derived macrophages | Represses inflammatory genes in response to TLR signalling | Forms a complex with hnRNP‐A/B and hnRNP‐A2/B1 to repress the transcription of immune genes | 24 |
| lincRNA‐Cox2 | Murine intestinal epithelial cell line | Represses TNF‐α‐induced Il12b transcription | Recruits Mi‐2/NuRD repressor complex to the Il12b promoter | 53 |
| lnc‐IL7R | Human monocyte/macrophage cell line THP1; human PBMCs; human umbilical vein endothelial cells | Diminishes LPS‐induced expression of inflammatory mediators including E‐selectin, VCAM‐1 and IL‐8 | Increases trimethylation of histone H3 at lysine 27 (H3K27me3) at target promoters | 54 |
lncRNAs interact with NF‐κB or its transcripts
PACER
Cyclo‐oxygenase 2 (COX2) participates in the biosynthesis of prostaglandin that plays a pivotal role in inflammatory processes and has also been demonstrated to have a role in tumour development.21 Using human primary mammary epithelial cells and a PMA‐driven human monocyte–macrophage differentiation system, Krawczyk and Emerson detected the transcript of a nuclear lncRNA, in the upstream region of COX2, named PACER (P50‐Associated COX‐2 Extragenic RNA).22 PMA induction strongly induced both COX2 and PACER expression in monocytes, and similarly, treatment of macrophages with lipopolysaccharide (LPS) markedly up‐regulated PACER and COX2 expression; however, in PACER knockdown cells, PMA‐induced or LPS‐stimulated COX2 expression was severely attenuated.22 Hence, PACER appears to mediate COX2 transcription.
The underlying mechanism involves direct association of PACER with p50, the inhibitory NF‐κB subunit.22 In the absence of stimuli such as LPS, p50/p50 homodimers associate with the COX2 promoter to repress its transcription. However, upon LPS stimulation, LPS‐induced PACER may restrict excess p50 from binding COX2 promoter, so promoting the formation of active p65/p50 heterodimers rather than repressive p50/p50 homodimers at the COX2 promoter. This facilitates recruitment of the histone acetyltransferase p300 to the promoter and consequently enables the assembly of transcription initiation complexes that are capable of gene activation.22 Therefore, PACER, an lncRNA transcript produced from the promoter region of COX2, functions in cis to enhance NF‐κB‐dependent COX2 transcription. In support of this notion, Pearson et al. found that following IL‐1β stimulation PACER was rapidly induced in primary human osteoarthritis chondrocytes and there was a strong positive correlation between the expression of PACER and COX2 (PTGS2).23 Unfortunately, they did not test the physical association between PACER and p50 in these cells. Whether PACER acts in trans to regulate the expression of other NF‐κB target genes is still unknown.
lincRNA‐Cox2
Besides PACER, another lncRNA, named lincRNA‐Cox2, is also located proximal to but downstream of the Cox2 gene. It is one of the most highly induced lncRNAs during the innate immune response.24 Although lincRNA‐Cox2 and its adjacent Cox2 gene displayed similar expression patterns in mouse bone‐marrow‐derived dendritic cells and macrophages following Toll‐like receptor (TLR) ligand stimulation, silencing of lincRNA‐Cox2 did not alter Cox2 expression.24 This suggests that lincRNA‐Cox2 itself may not regulate Cox2 in cis. However, it is still possible that lincRNA‐Cox2 activates Cox2 by acting through lncRNA transcription.
Work by Hu et al. indicated that increased lincRNA‐Cox2 expression was needed for the transcription of NF‐κB‐regulated late inflammatory genes in LPS‐treated murine macrophages.25 RNA immunoprecipitation and RNA pull‐down analyses revealed a direct physical association between lincRNA‐Cox2 and the switch/sucrose nonfermentable (SWI/SNF) chromatin remodelling complex.25 Furthermore, lincRNA‐Cox2 was demonstrated to participate in the assembly of NF‐κB subunits p65 and p50 into the SWI/SNF complex.25 A series of elegant experiments demonstrated the recruitment of both lincRNA‐Cox2 and the SWI/SNF complex to the promoters of late inflammatory genes such as Saa3 and Ccl5, leading to increased histone H3 methylations (H3K4Me3 and H3K36Me3) at target promoters following LPS stimulation.25 Taken together, the mechanism of action for lincRNA‐Cox2 in activated macrophages involves the formation of a lincRNA‐Cox2‐containing SWI/SNF complex. The resulting complex, which is recruited to the promoter/enhancer region of late inflammatory genes, modulates the recruitment of NF‐κB to the complex, triggers chromatin remodelling, and ultimately, results in access to sites for NF‐κB binding and gene transcription.
Lethe
Lethe, a pseudogene induced by pro‐inflammatory cytokines through NF‐κB, was one of the first lncRNAs demonstrated to be involved in modulating NF‐κB signalling.26 Lethe is expressed in mouse embryonic fibroblasts upon exposure to TNF‐α, IL‐1β and the anti‐inflammatory agent dexamethasone, but it is not responsive to TLR agonists, indicating that Lethe may have a function in inflammation, but not in native immunity.26 Expression analysis of Lethe in p65−/− cells and p65 chromatin immunoprecipitation revealed that Lethe is a direct transcriptional target of p65.26 Lethe is largely located in the nucleus and is preferentially associated with chromatin, suggesting that it is directly involved in gene regulation by interacting with the chromatin.
Further characterization using human 293T cells indicated that Lethe physically associates with p65 to block the DNA binding activity of NF‐κB.26 Therefore, Lethe, which is induced in a p65‐dependent fashion, appears to act as a negative feedback regulator of NF‐κB to help fine tune inflammatory responses.26 Although the best example of a pseudogene functioning as an lncRNA may come from X‐inactive specific transcript, which is essential for dosage compensation and X chromosome inactivation in female mammals,26, 27 Lethe provides the first evidence that a pseudogene is capable of functioning as an lncRNA to regulate inflammatory signalling. Interestingly, Lethe can also be induced by signal transducer and activator of transcription 3 (STAT3), and elevated expression of Lethe promotes hepatitis C virus replication by down‐regulating type I interferon response.28 This seems logical considering the fact that the p65 subunit of NF‐κB could physically associate with STAT3, facilitating NF‐κB recruitment to STAT3 promoters and vice versa.5
NKILA
Abnormal NF‐κB activation has been implicated in many cancers. To probe the role of lncRNA in cancer development, Liu et al. used microarray to comprehensively analyse lncRNA expression level in human breast cancer cells and found that an lncRNA, which they named NKILA (NF‐κB interacting lncRNA), was up‐regulated by inflammatory cytokines via NF‐κB signalling.29 NKILA, located primarily in the cytoplasm, inhibits both basal and cytokine‐stimulated NF‐κB activation in breast cancer cells.29 This inhibition is mediated through an interaction between p65 and NKILA, leading to the formation of a stable NKILA/NF‐κB/IκBα complex and subsequent veiling of phosphorylation sites of IκB, thereby suppressing IKK‐triggered IκB phosphorylation, NF‐κB release, and nuclear localization.29 Based on detailed analysis of the cell lines and primary cells of breast cancer with different metastatic potentials, the study confirmed that reduced NKILA expression in human breast cancer significantly correlates with metastatic dissemination and poor prognosis.29 Although NKILA runs antisense to a cancer‐associated gene PMEPA1 and might indirectly exert its effect on NF‐κB function through interference with PMEPA1 expression,30 the elaborate work by Liu et al. convincingly indicates that NF‐κB‐induced NKILA participates in the negative feedback regulation of NF‐κB, and may serve as a tumour suppressor‐like lncRNA.
MALAT1
Metastasis‐associated lung adenocarcinoma transcript 1 (MALAT1) is a highly conserved lncRNA whose abnormal expression is considered to correlate with the development, progression and metastasis of multiple cancer types.31 Recently we reported the role of MALAT1 in regulating the production of cytokines in macrophages.32 Using PMA‐differentiated macrophages derived from the human THP1 monocyte cell line, we showed that following stimulation with LPS, a ligand for the innate pattern recognition receptor TLR4, MALAT1 expression is increased in an NF‐κB‐dependent manner.32 In the nucleus, MALAT1 interacts with both p65 and p50 to suppress their DNA binding activity and consequently attenuates the expression of two NF‐κB‐responsive genes, TNF‐a and IL‐6.32 This finding is in agreement with a report based on in silico analysis predicting that MALAT1 could influence NF‐κB/RelA activity in the context of epithelial–mesenchymal transition.32, 33 Therefore, in LPS‐activated macrophages MALAT1 is engaged in the tight control of the inflammatory response through interacting with NF‐κB, demonstrating for the first time its role in regulating innate immunity‐mediated inflammation. As MALAT1 is capable of binding hundreds of active chromatin sites throughout the human genome,34 the function and mechanism of action so far uncovered for this evolutionarily conserved lncRNA may be just the tip of an iceberg.
lincRNA‐p21
Initially reported by Huarte et al., lincRNA‐p21 resides 15 kb upstream of the p21 (Cdkn1a) gene.35 This lncRNA has been demonstrated to participate in a variety of biological processes through multiple mechanisms that range from influencing transcription of p53‐regulated genes to modulating mRNA translation and protein stability.35, 36 The anti‐inflammatory properties of p53 triggered Spurlock et al. to investigate the relationship between p53, lincRNA‐p21 and NF‐κB in the context of rheumatoid arthritis and methotrexate therapy.37 They observed that lincRNA‐p21 levels were decreased whereas NF‐κB activity was increased in rheumatoid arthritis, and moreover, methotrexate treatment could correct both defects in vivo.37 The existence of complementary regions between lincRNA‐p21 and p65 mRNA and the results of RNA pull‐down suggest that lincRNA‐p21 associates with p65 mRNA.37 Notably, reducing the level of lincRNA‐p21 did not alter the level of p65 transcript, indicating a role for lincRNA‐p21 by inhibiting NF‐κB activity through inhibition of p65 mRNA translation. Although the possibility of additional regulatory mechanisms cannot be ruled out,37 collectively, their results suggest that lincRNA‐p21 suppresses NF‐κB activity and lincRNA‐p21 induction may contribute to the anti‐inflammatory effects of methotrexate.
lncRNAs interfere with signalling components or related molecules upstream of NF‐κB
HOTAIR
In addition to inhibiting IκB phosphorylation, lncRNA can also regulate NF‐κB activity by down‐regulating IκBα expression. Hox antisense intergenic RNA (HOTAIR) functions in chromatin remodelling and transcription, and is correlated with metastasis and poor prognosis in a range of cancers. Studies in human ovarian cancer cell lines indicated that up‐regulation of HOTAIR induces platinum resistance and leads to persistent activation of the DNA damage response following treatment with platinum.38 The correlation between HOTAIR expression and chemoresistance was further confirmed in patients with ovarian cancer with distinct platinum sensitivity. Mechanistically, HOTAIR decreases IκBα protein levels, most likely by recruiting the polycomb repressive complex 2 (PRC2) to the IκBα gene. This results in prolonged activation of NF‐κB and the expression of its target genes that promote cellular senescence and resistance to DNA‐damaging agents.38 Based on these data the authors propose that the NF‐κB–HOTAIR pathway modulates the DNA damage response and contributes to chemoresistance and cellular senescence in ovarian and other types of cancer.38
Besides its inhibitory effect on the expression of NF‐κB inhibitor IκBα, HOTAIR can regulate NF‐κB activity by post‐translational modification of its subunit p65. Wu et al. showed that HOTAIR expression was significantly up‐regulated in cardiomyocytes from mice with LPS‐induced sepsis as well as in LPS‐treated cardiomycytes from normal mice.39 Up‐regulation of HOTAIR in these cells was found to be associated with elevated levels of TNF‐α and p65 phosphorylation. The association between HOTAIR expression, TNF‐α production and p65 phosphorylation was confirmed by HOTAIR knockdown or overexpression studies using murine HL‐1 cardiomyocytes in vitro and LPS‐induced septic mice in vivo. Hence, in cardiomyocytes, HOTAIR can promote NF‐κB activation through increasing LPS‐induced p65 phosphorylation. HOTAIR has both nuclear and cytoplasmic distribution, and p65 phosphorylation is catalysed either by cAMP‐dependent protein kinase, which is constitutively associated with cytosolic p65‐containing complexes, or by nuclear kinases such as MSK1 and MSK2.1 Whether HOTAIR interacts with these kinases and the location of the interaction, if it occurs, awaits further investigation.
MIR31HG
MIR31HG is an lncRNA previously demonstrated to be associated with tumour development. As the MIR31HG promoter contains three potential NF‐κB binding sites,40 Jin et al. examined whether MIR31HG plays a role in inflammation or bone formation by interacting with the NF‐κB pathway.41 Their results showed that MIR31HG suppressed osteoblast differentiation of human adipose‐derived stem cells, and that knockdown of MIR31HG inhibited NF‐κB signalling, reversed pro‐inflammatory cytokine‐induced inhibition of osteogenesis in human adipose‐derived stem cells, and enhanced heterotopic bone formation in vivo.41 Mechanistically, MIR31HG interacts with the cytoplasmic NF‐κB:IκB complex through IκBα, participates in IκBα phosphorylation, and hence activates NF‐κB signalling.41 Because binding of p65 to MIR31HG promoter was experimentally demonstrated, MIR31HG‐ NF‐κB is believed to constitute a positive feedback loop that inhibits osteoblast differentiation and bone formation under inflammatory conditions.
C2dat1
CaMKII, including CaMKIIα, β, γ and δ, is the most abundant protein kinase in the brain. The lncRNA C2dat1 overlaps the CaMKIIδ coding gene, implying potential regulation of C2dat1 on CAMK2D expression.42 Indeed, Xu et al. found that C2dat1 was induced both in an in vivo murine model of ischaemia/reperfusion and in an in vitro ischaemic model in which neurons were subjected to ischaemia‐like injury.42 They demonstrated that in mouse neuronal cells C2dat1 up‐regulated CAMK2D expression and enhanced neuron death under ischaemic conditions. By examination of several signalling pathways potentially implicated in ischaemia/reperfusion‐induced neuronal cell death, the level of phosphorylated IKKα/β of the NF‐κB pathway was shown to be consistently altered upon C2dat1 knockdown.42 The mechanism of action of this lncRNA is currently unknown. However, as knockdown of C2dat1 down‐regulated CaMKIIδ expression, suppressed IKK expression and phosphorylation, and eventually inhibited NF‐κB activity, C2dat1 appears to promote neuron survival by activating NF‐κB signalling through enhancing CAMK2D expression in cis.
Arid2‐IR
Arid2‐IR is located in the intron region of the mouse Arid2 gene (hence the name of this lncRNA). It was first identified as the most highly expressed lncRNA (np_28496) in the mouse unilateral ureteral obstructive model of chronic kidney disease.43 Further characterization of Arid2‐IR indicated that it was markedly up‐regulated in the wild‐type but down‐regulated in Smad3 knockout unilateral ureteral obstructive kidney, implying functional association of Arid2‐IR with Smad3.44 Indeed, the Arid2‐IR promoter region contained a Smad3 binding site that was experimentally verified and knockout of Smad3 abolished increased expression of Arid2‐IR in the diseased kidney.44 Knockdown and overexpression studies of Arid2‐IR in mouse tubular epithelial cells revealed that Arid2‐IR enhanced IL‐1β‐induced NF‐κB activity and the expression of pro‐inflammatory cytokines, which was further validated in vivo using a mouse unilateral ureteral obstructive kidney model.44 Overall, work by Zhou et al. is the first to have shown that a Smad3‐induced lncRNA functions to drive NF‐κB‐mediated renal inflammation.44
DLEU1 and DLEU2
DLEU1 and DLEU2 are two lncRNA genes at 13q14.3, a chromosomal region that is frequently deleted in haematopoietic and solid tumours. Characterization of the epigenetic makeup of 13q14.3 in primary human chronic lymphocytic leukaemia cells revealed histone modifications associated with active chromatin and DNA hypomethylation at the transcription start sites of DLEU1 and DLEU2.45 The 13q14.3 locus contains candidate tumour suppressor genes encoding KPNA3, RFP2, C13ORF1 and miR‐15a/16‐1, all of which, either proteins or microRNAs, are regulators of NF‐κB activity.45 In chronic lymphocytic leukaemia cells these candidate tumour suppressor genes are down‐regulated whereas DLEU1 and DLEU2 are significantly up‐regulated, suggesting co‐regulated expression between the two lncRNAs and the tumour suppressor genes within this region.45
DLEU1 and DLEU2 do not exert their function through recruiting repressors, as neither of them binds to chromatin. Rather, they probably regulate their neighbouring genes through, for instance, competition for common transcription factors.45 How the products of the candidate tumour suppressor genes activate NF‐κB activity is largely unknown. However, as down‐regulation of p65 and p50 reduced the activation of NF‐κB activity by RFP2 and loss of RFP2 ubiquitin‐ligase activity diminished RFP2‐mediated NF‐κB activation in human HEK293T cells, RFP2 has been speculated to activate canonical NF‐κB subunits p65 and p105 through its ubiquitin ligase activity.45 In essence, at the 13q14.3 locus, the two lncRNAs expressed from this region act in cis to regulate the expression of a cluster of tumour suppressor genes whose products modulate NF‐κB signalling activity.
lncRNAs that are induced by NF‐κB signalling and regulate NF‐κB target gene expression
The lncRNAs described above regulate NF‐κB activity by interacting with NF‐κB or interfering with its upstream components or related signalling molecules. However, some NF‐κB‐induced lncRNAs, presented below, regulate NF‐κB target gene transcription without interfering with NF‐κB or its upstream signal relay components.
IL1β‐eRNA and IL1β‐RBT46
IL1β‐eRNA and IL1β‐RBT46 were two lncRNAs induced in primary human monocytes upon LPS treatment.46 L1β‐eRNA is an enhancer RNA whereas IL1β‐RBT46 is within a region of bidirectional transcription surrounding the IL1β locus.46 The two nucleus‐located, NF‐κB‐regulated lncRNAs enhance the transcription and release of several pro‐inflammatory cytokines such as IL‐1β, IL‐6 and CXCL8.46 Because both of these lncRNAs are located close to the IL1β gene, they may regulate the transcription of IL1β in cis and other inflammatory genes in trans,46 although the precise mechanism is currently unknown. It is not unlikely that the process of IL1β‐eRNA and IL1β‐RBT46 transcription contributes to regulatory effects on IL1β.
AS‐IL1α
As with IL‐1β, IL‐1α is a master cytokine capable of amplifying local and systemic inflammation.47 AS‐IL1α, a natural antisense lncRNA partially complementary to IL‐1α coding gene, is up‐regulated through NF‐κB in primary mouse bone marrow‐derived macrophages after treatment with TLR ligands including LPS (TLR4), Pam3CSK4 (TLR1/2), and Poly(I:C) (TLR3).48 Functionally, AS‐IL1α is required for the recruitment of RNA polymerase II to IL‐1α promoter and acts in cis to enhance LPS‐induced transcription of IL‐1α. Therefore, suppression of IL‐1α activity by specific blockage of AS‐IL1α transcript would be beneficial to a wide spectrum of inflammatory diseases such as rheumatoid arthritis.
ANRIL
ANRIL is an lncRNA transcribed from chromosome 9 on p21.3, a locus associated with a risk of coronary artery disease.49 In human vascular endothelial cells, this lncRNA was remarkably induced in response to pro‐inflammatory factors in an NF‐κB‐dependent manner, and elevated ANRIL affected the expression of a large portion of inflammatory genes downstream of NF‐κB, such as IL6 and IL8.50 Mechanistic studies indicated that ANRIL interacts with YY1, and facilitates the binding of this transcription factor to IL‐6 and IL‐8 promoters due to direct association between ANRIL and the promoters.50 Hence, besides its role in the pathogenesis of a number of cancers, ANRIL also regulates the inflammatory response in the pathological process of coronary artery disease.
THRIL
It is well documented that activation of TLR2 signalling in response to microbial infections induces the production of various pro‐inflammatory cytokines and may cause acute and chronic inflammation.51 The TNF‐α and hnRNPL‐related immunoregulatory lncRNA (THRIL), is required for both basal and TLR2 ligand, Pam3CSK4, stimulated expression of many immune‐response genes including TNF‐α, in human THP1 macrophages.52 THRIL binds specifically to hnRNPL, forming a functional complex capable of binding to the promoter of TNF‐α and regulating its transcription.52
lincRNA‐Cox2
As mentioned above, lincRNA‐Cox2 interacts with NF‐κB and SWI/SNF to transcriptionally activate late inflammatory genes in LPS‐treated murine macrophages. Besides its positive role on the expression of certain genes, this lncRNA is capable of mediating transcriptional repression of inflammatory genes in response to TLR signalling in mouse bone‐marrow‐derived macrophages and intestinal epithelial cells.24, 53 The repressive action is dependent on interactions of lincRNA‐Cox2 with hnRNP‐A2/B1, hnRNP‐A/B or the Mi‐2/NuRD complex that is involved in histone modifications.24, 53 Therefore, rather than regulating its neighbouring gene Cox2, lincRNA‐Cox2 acts in trans and serves as repressors and activators of distinct groups of inflammatory genes by interacting with various functional complexes.
lnc‐IL7R
The lnc‐IL7R, which overlaps with the 3′ untranslated region of the human IL‐7 receptor α‐subunit gene, is another example of lncRNAs that regulate NF‐κB target gene expression in trans.54 lnc‐IL7R is induced by TLR2 or TLR4 in THP1 cells and human peripheral blood mononuclear cells. Functional characterization of this lncRNA in human umbilical vein endothelial cells indicates that lnc‐IL7R does not regulate the expression of its overlapping gene IL7R in cis. Rather, it acts in trans to inhibit LPS‐induced inflammatory responses by increasing H3K27me3 levels at the promoters of NF‐κB‐regulated inflammatory mediators.54
Therapeutic potential and concluding remarks
Dynamic changes in lncRNA expression have been demonstrated upon activation of NF‐κB signalling. These changes not only regulate NF‐κB activity through direct interaction between lncRNA and NF‐κB or its transcripts, but also regulate NF‐κB signalling activity indirectly, through upstream components. Some NF‐κB‐induced lncRNAs can even directly regulate NF‐κB target gene transcription without interfering with NF‐κB or its upstream signal relay components (Fig. 1). Of note, by interacting with different partners in respective cells, a given lncRNA may exert its regulatory impact on NF‐κB in different ways, and consequently fulfil its diverse biological functions. A well‐characterized example is lincRNA‐Cox2, which, depending on its interaction partners, is able to mediate the activation or repression of different groups of inflammatory genes.24, 25, 53 Therefore, in addition to the complex regulatory layer composed of proteins and microRNAs,55 lncRNAs represent another critical layer of the intricate modulatory architecture, for maintaining suitable levels of NF‐κB signalling activity.
Figure 1.
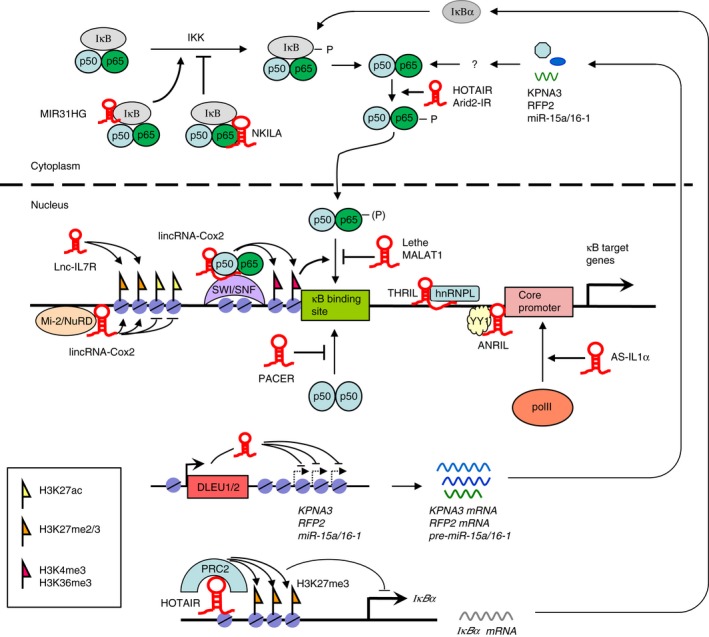
Diagram illustrating long non‐coding RNAs (lncRNAs) involved in regulating nuclear factor‐κB (NF‐κB) signalling activity. NKILA is capable of interfering with IκB phosphorylation by interacting with the NF‐κB/IκB complex. Lethe and MALAT1 inhibit while PACER facilitates the binding of p65/p50 heterodimer to target promoters. AS‐IL1α recruits RNA polymerase II to interleukin‐1α (IL‐1α) promoter whereas DLEU1 and DLEU2 suppress the expression of KPNA3, RFP2 and miR‐15a/16‐1, all of which are regulators of NF‐κB activity. lincRNA‐Cox2 recruits NF‐κB subunits p65 and p50 into the SWI/SNF complex, facilitating binding of the p65/p50 heterodimer to the κB site due to chromatin remodelling and histone modifications (H3K4me3 and H3K36me3). lincRNA‐Cox2 also represses gene transcription by interacting with the Mi‐2/NuRD complex that functions in histone modifications. lnc‐IL7R regulates NF‐κB target gene expression by up‐regulating H3K27me3 levels at the promoter region. THRIL recruits protein complexes involved in transcriptional regulation via RNA‐binding proteins (e.g. hnRNPL). ANRIL interacts and facilitates binding of transcription factor YY1 to target promoters. HOTAIR acts in trans on IκBα gene to repress its expression, at least in part, through recruitment of the chromatin modifying enzyme complex PRC2. For simplicity, several lncRNAs mentioned in the text (lincRNA‐p21, Arid2‐IR, IL1β‐eRNA and IL1β‐RBT46) are not shown in the figure because their mechanisms of action are not clearly defined.
It is well known that aberrant NF‐κB signalling is contributed to the pathogenesis of a wide range of autoimmune and inflammatory diseases wherein pro‐inflammatory cytokines stimulate NF‐κB activity, which in turn promotes pro‐inflammatory cytokine production. In addition, constitutive activation of NF‐κB in malignancies may up‐regulate the expression of genes functioning in cell proliferation and anti‐apoptosis, leading to persistent tumour survival.1 Therefore, inhibiting overactive and/or prolonged activation of the NF‐κB pathway may offer new therapeutic options for the treatment of inflammation and cancer. Unfortunately, although successfully demonstrated in some studies using animal models, so far no pharmaceutical agents designed to target the components of the NF‐κB pathway have efficacy in human diseases. This can be at least in part ascribed to adverse off‐target effects, as exemplified by small molecules targeting protein kinases of the NF‐κB pathway with multiple closely related family members.
Given the involvement of lncRNAs in NF‐κB regulation, in particular the evidence of association between lncRNA (e.g. THRIL, NKILA and lincRNA‐p21) and human diseases relevant to NF‐κB dysfunction,29, 37, 52 lncRNA‐targeted nucleic acid‐based therapies, owing to their ability to distinguish a single nucleotide mismatch and hence pronounced target specificity, may be a better alternative to current NF‐κB‐targeting small molecules, which have poor specificity and cause many unfavourable off‐target effects.56, 57 One strategy involves using antisense technology or small interfering RNA/short hairpin RNA‐mediated RNA interference to silence those lncRNAs that promote the NF‐κB‐dependent inflammatory response or cell survival. Alternatively, masking the binding sites for intermolecular interactions can functionally block lncRNAs. Conversely, introducing synthetic lncRNA mimics that structurally resemble the functional motifs of those lncRNAs that repress NF‐κB signalling could be a viable anti‐inflammatory and anticancer therapeutic option. Finally, because of its ease and efficiency, CRISPR/Cas9 can be used to introduce desired alterations in lncRNA sequences. Moreover, this technology can be used to alter lncRNA abundance via for example targeted acetylation at genomic loci containing lncRNA promoters or enhancers.58
On the other hand, as NF‐κB has broad effects on a number of physiological processes, complete, non‐selective, inhibition of NF‐κB signalling may be detrimental. Fortunately, lncRNAs are often expressed on a restricted subpopulation of cells.59 This unique specificity is another advantage of further exploring lncRNAs as therapeutic targets of the NF‐κB pathway. Notably, despite its high abundance and strong conservation, the loss of MALAT1 has no effect on mouse development and life.31 This finding suggests that in contrast to certain NF‐κB pathway components (e.g. NEMO, IKK, p65 or IκBα) whose deficiency results in lethality during embryonic development or shortly after birth, MALAT1 is dispensable outside the context of diseased cells/tissues and targeting MALAT1 may offer minimal side effects.31, 60
However, although lncRNAs might be targeted for correcting aberrant NF‐κB activity, this field is still at its early stage and faces many challenges, including poor lncRNA sequence conservation across species, poor knowledge of structural and functional elements of lncRNA, diverse roles of a given lncRNA, and different lncRNA isoforms that may or may not have common functions.
Overall, increasing evidence suggests that lncRNAs are capable of influencing NF‐κB signalling whose dysregulation has been associated with inflammatory and autoimmune diseases as well as cancer. A hallmark of these lncRNAs is that, similar to protein and miRNA regulators of NF‐κB, they are often induced by TLR/NF‐κB signalling, and so are involved in the maintenance of immune homeostasis through feedback regulation. Although lncRNAs have been shown as promising targets for NF‐κB‐mediated inflammation and cancer because of their high specificity, which could potentially result in fewer off‐target and adverse effects than current NF‐κB targeted therapies, improved knowledge is needed before lncRNA‐based therapeutics move from bench to the bedside, including the mechanisms and pathways that lncRNAs are involved in the regulatory network of NF‐κB, the isoform composition and tissue specificity of these lncRNAs, as well as more extensive pharmacological evidence of therapeutic targeting of lncRNAs for NF‐κB blockade in animal models, particularly in more clinically relevant large animal models.
Disclosures
The authors have no competing interests to declare.
Acknowledgement
This work was supported by Natural Science Foundation of China (81071445, 31670798).
References
- 1. Hayden MS, Ghosh S. NF‐κB, the first quarter‐century: remarkable progress and outstanding questions. Genes Dev 2012; 26:203–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Napetschnig J, Wu H. Molecular basis of NF‐κB signaling. Ann Rev Biophy 2013; 42:443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lu T, Stark GR. NF‐κB: regulation by methylation. Cancer Res 2015; 75:3692–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gerondakis S, Fulford TS, Messina NL, Grumont RJ. NF‐κB control of T cell development. Nat Immunol 2014; 15:15–25. [DOI] [PubMed] [Google Scholar]
- 5. Hoesel B, Schmid JA. The complexity of NF‐κB signaling in inflammation and cancer. Mol Cancer 2013; 12:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vallabhapurapu S, Karin M. Regulation and function of NF‐κB transcription factors in the immune system. Annu Rev Immunol 2009; 27:693–733. [DOI] [PubMed] [Google Scholar]
- 7. Razani B, Reichardt AD, Cheng G. Non‐canonical NF‐κB signaling activation and regulation: principles and perspectives. Immunol Rev 2011; 244:44–54. [DOI] [PubMed] [Google Scholar]
- 8. Liu Q, Chen Y, Auger‐Messier M, Molkentin JD. Interaction between NFκB and NFAT coordinates cardiac hypertrophy and pathological remodeling. Circ Res 2012; 110:1077–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shinzawa M, Konno H, Qin J, Akiyama N, Miyauchi M, Ohashi H et al Catalytic subunits of the phosphatase calcineurin interact with NF‐κB‐inducing kinase (NIK) and attenuate NIK‐dependent gene expression. Sci Rep 2015; 5:10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hadweh P, Habelhah H, Kieff E, Mosialos G, Hatzivassiliou E. The PP4R1 subunit of protein phosphatase PP4 targets TRAF2 and TRAF6 to mediate inhibition of NF‐κB activation. Cell Signal 2014; 26:2730–7. [DOI] [PubMed] [Google Scholar]
- 11. Ruland J. Return to homeostasis: downregulation of NF‐κB responses. Nat Immunol 2011; 12:709–14. [DOI] [PubMed] [Google Scholar]
- 12. Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell 2013; 154:26–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Johnsson P, Lipovich L, Grandér D, Morris KV. Evolutionary conservation of long non‐coding RNAs; sequence, structure, function. Biochim Biophys Acta 2014; 1840:1063–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Paralkar VR, Taborda CC, Huang P, Yao Y, Kossenkov AV, Prasad R et al Unlinking an lncRNA from its associated cis element. Mol Cell 2016; 62:104–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, et alLocal regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 2016; 539:452–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang KC, Yang YW, Liu B, Sanyal A, Corces‐Zimmerman R, Chen Y et al A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011; 472:120–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huarte M. The emerging role of lncRNAs in cancer. Nat Med 2015; 21:1253–61. [DOI] [PubMed] [Google Scholar]
- 18. Cheng HS, Njock M‐S, Khyzha N, Dang LT, Fish JE. Noncoding RNAs regulate NF‐κB signaling to modulate blood vessel inflammation. Front Genet 2014; 5:422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang Y, Cao X. Long noncoding RNAs in innate immunity. Cell Mol Immunol 2016; 13:138–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Heward JA, Lindsay MA. Long non‐coding RNAs in the regulation of the immune response. Trends Immunol 2014; 35:408–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hoellen F, Kelling K, Dittmer C, Diedrich K, Friedrich M, Thill M. Impact of cyclooxygenase‐2 in breast cancer. Anticancer Res 2011; 31:4359–67. [PubMed] [Google Scholar]
- 22. Krawczyk M, Emerson BM. p50‐associated COX‐2 extragenic RNA (PACER) activates COX‐2 gene expression by occluding repressive NF‐κB complexes. Elife 2014; 3:e01776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pearson MJ, Philp AM, Heward JA, Roux BT, Walsh DA, Davis ET et al Long intergenic noncoding RNAs mediate the human chondrocyte inflammatory response and are differentially expressed in osteoarthritis cartilage. Arthritis Rheumatol 2016; 68:845–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL et al A long noncoding RNA mediates both activation and repression of immune response genes. Science 2013; 341:789–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hu G, Gong A‐Y, Wang Y, Ma S, Chen X, Chen J et al LincRNA‐Cox2 promotes late inflammatory gene transcription in macrophages through modulating SWI/SNF‐mediated chromatin remodeling. J Immunol 2016; 196:2799–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rapicavoli NA, Qu K, Zhang J, Mikhail M, Laberge R‐M, Chang HY. A mammalian pseudogene lncRNA at the interface of inflammation and anti‐inflammatory therapeutics. Elife 2013; 2:e00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Duret L, Chureau C, Samain S, Weissenbach J, Avner P. The Xist RNA gene evolved in eutherians by pseudogenization of a protein‐coding gene. Science 2006; 312:1653–5. [DOI] [PubMed] [Google Scholar]
- 28. Xiong Y, Yuan J, Zhang C, Zhu Y, Kuang X, Lan L et al The STAT3‐regulated long non‐coding RNA Lethe promote the HCV replication. Biomed Pharmacother 2015; 72:165–71. [DOI] [PubMed] [Google Scholar]
- 29. Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X et al A cytoplasmic NF‐κB interacting long noncoding RNA blocks IκB phosphorylation and suppresses breast cancer metastasis. Cancer Cell 2015; 27:370–81. [DOI] [PubMed] [Google Scholar]
- 30. Dijkstra JM, Alexander DB. The “NF‐κB interacting long noncoding RNA”(NKILA) transcript is antisense to cancer‐associated gene PMEPA1. F1000Res 2015; 4:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gutschner T, Hämmerle M, Diederichs S. MALAT1—a paradigm for long noncoding RNA function in cancer. J Mol Med 2013; 91:791–801. [DOI] [PubMed] [Google Scholar]
- 32. Zhao G, Su Z, Song D, Mao Y, Mao X. The long noncoding RNA MALAT1 regulates the lipopolysaccharide‐induced inflammatory response through its interaction with NF‐κB. FEBS Lett 2016; 590:2884–95. [DOI] [PubMed] [Google Scholar]
- 33. Li X, Zhu M, Brasier AR, Kudlicki AS. Inferring genome‐wide functional modulatory network: a case study on NF‐κB/RelA transcription factor. J Comput Biol 2015; 22:300–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. West JA, Davis CP, Sunwoo H, Simon MD, Sadreyev RI, Wang PI et al The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol Cell 2014; 55:791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann‐Broz D et al A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010; 142:409–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dimitrova N, Zamudio JR, Jong RM, Soukup D, Resnick R, Sarma K et al LincRNA‐p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol Cell 2014; 54:777–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Spurlock CF, Tossberg JT, Matlock BK, Olsen NJ, Aune TM. Methotrexate inhibits NF‐κB activity via long intergenic (noncoding) RNA–p21 induction. Arthritis Rheumatol 2014; 66:2947–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Özeş A, Miller D, Özeş O, Fang F, Liu Y, Matei D et al NF‐κB‐HOTAIR axis links DNA damage response, chemoresistance and cellular senescence in ovarian cancer. Oncogene 2016; 35:5350–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu H, Liu J, Li W, Liu G, Li Z. LncRNA‐HOTAIR promotes TNF‐α production in cardiomyocytes of LPS‐induced sepsis mice by activating NF‐κB pathway. Biochem Biophys Res Commun 2016; 471:240–6. [DOI] [PubMed] [Google Scholar]
- 40. Rajbhandari R, McFarland BC, Patel A, Gerigk M, Gray GK, Fehling SC et al Loss of tumor suppressive microRNA‐31 enhances TRADD/NF‐κB signaling in glioblastoma. Oncotarget 2015; 6:17805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jin C, Jia L, Huang Y, Zheng Y, Du N, Liu Y et al Inhibition of lncRNA MIR31HG promotes osteogenic differentiation of human adipose‐derived stem cells. Stem Cells 2016; 34:2707–20. [DOI] [PubMed] [Google Scholar]
- 42. Xu Q, Deng F, Xing Z, Wu Z, Cen B, Xu S et al Long non‐coding RNA C2dat1 regulates CaMKIIδ expression to promote neuronal survival through the NF‐κB signaling pathway following cerebral ischemia. Cell Death Dis 2016; 7:e2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhou Q, Chung AC, Huang XR, Dong Y, Yu X, Lan HY. Identification of novel long noncoding RNAs associated with TGF‐β/Smad3‐mediated renal inflammation and fibrosis by RNA sequencing. Am J Pathol 2014; 184:409–17. [DOI] [PubMed] [Google Scholar]
- 44. Zhou Q, Huang XR, Yu J, Yu X, Lan HY. Long noncoding RNA Arid2‐IR is a novel therapeutic target for renal inflammation. Mol Ther 2015; 23:1034–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Garding A, Bhattacharya N, Claus R, Ruppel M, Tschuch C, Filarsky K et al Epigenetic upregulation of lncRNAs at 13q14. 3 in leukemia is linked to the in cis downregulation of a gene cluster that targets NF‐κB. PLoS Genet 2013; 9:e1003373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ilott NE, Heward JA, Roux B, Tsitsiou E, Fenwick PS, Lenzi L et al Long non‐coding RNAs and enhancer RNAs regulate the lipopolysaccharide‐induced inflammatory response in human monocytes. Nat Commun 2014; 5:3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dinarello CA, van der Meer JW. Treating inflammation by blocking interleukin‐1 in humans Semin Immunol 2013; 25:469–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chan J, Atianand M, Jiang Z, Carpenter S, Aiello D, Elling R et al Cutting edge: a natural antisense transcript, AS‐IL1α, controls inducible transcription of the proinflammatory cytokine IL‐1α. J Immunol 2015; 195:1359–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Broadbent HM, Peden JF, Lorkowski S, Goel A, Ongen H, Green F et al Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum Mol Genet 2008; 17:806–14. [DOI] [PubMed] [Google Scholar]
- 50. Zhou X, Han X, Wittfeldt A, Sun J, Liu C, Wang X et al Long non‐coding RNA ANRIL regulates inflammatory responses as a novel component of NF‐κB pathway. RNA Biol 2016; 13:98–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell 2010; 140:805–20. [DOI] [PubMed] [Google Scholar]
- 52. Li Z, Chao T‐C, Chang K‐Y, Lin N, Patil VS, Shimizu C et al The long noncoding RNA THRIL regulates TNFα expression through its interaction with hnRNPL. Proc Natl Acad Sci U S A 2014; 111:1002–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tong Q, Gong A‐Y, Zhang X‐T, Lin C, Ma S, Chen J et al LincRNA‐Cox2 modulates TNF‐α‐induced transcription of Il12b gene in intestinal epithelial cells through regulation of Mi‐2/NuRD‐mediated epigenetic histone modifications. FASEB J 2016; 30:1187–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cui H, Xie N, Tan Z, Banerjee S, Thannickal VJ, Abraham E et al The human long noncoding RNA lnc‐IL7R regulates the inflammatory response. Eur J Immunol 2014; 44:2085–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sun S‐C, Chang J‐H, Jin J. Regulation of nuclear factor‐κB in autoimmunity. Trends Immunol 2013; 34:282–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chery J, Näär A. RNA therapeutics: RNAi and antisense mechanisms and clinical applications. Postdoc J 2016; 4:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhou Y, Zhang C, Liang W. Development of RNAi technology for targeted therapy—a track of siRNA based agents to RNAi therapeutics. J Controlled Release 2014; 193:270–81. [DOI] [PubMed] [Google Scholar]
- 58. Hilton IB, D'Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE et al Epigenome editing by a CRISPR‐Cas9‐based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol 2015; 33:510–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A et al Landscape of transcription in human cells. Nature 2012; 489:101–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mendell JT. Targeting a long noncoding RNA in breast cancer. N Engl J Med 2016; 374:2287–9. [DOI] [PubMed] [Google Scholar]


