Abstract
Background
Avian coccidiosis is an intracellular intestinal parasitic disease, caused by intracellular intestinal parasites from the genus Eimeria, among which Eimeria tenella is one of the most pathogenic species and causes great economic losses. Frequent applications of anticoccidial drugs have resulted in the development of drug-resistance in E. tenella. In the present study, we sought to determine the genetic diversity of E. tenella isolates prevalent in chicken farms in Hubei Province of China and examine their sensitivity to three anticoccidial drugs. The results provide useful information for the prevention and control of coccidiosis in this region.
Methods
Eimeria tenella oocysts were isolated from faecal samples collected from different commercial broiler production farms in Hubei Province, China. After oocyst sporulation and animal inoculation for expansion of the field isolates, DNA and RNA were extracted from excysted sporozoites for molecular characterization. Species identification of field isolates were performed by polymerase chain reaction (PCR) amplification of the internal transcribed spacer 1 (ITS1) region of ribosomal DNA. Random amplified polymorphic DNA (RAPD) was used for population genetic analysis. Subsequently, sequences of the major sporozoite surface antigen (SAG), micronemal protein 2 (MIC-2) and cytochrome b (cytb) genes from genomic DNA, and the Eimeria tenella cation-transport ATPase (EtCat ATPase) gene from cDNA were obtained for genotyping using multi-sequence alignments. Finally, sensitivity of the field isolates to three commonly used anticoccidial drugs (diclazuril, decoquinate and maduramycin) were tested to assess the prevalence of drug resistance in E. tenella in Hubei Province of China.
Results
Analysis of the ITS1 sequences indicated that all the isolates were E. tenella. RAPD analysis and multi-sequence alignments of the SAG, MIC-2, EtCat ATPase and cytb showed genetic diversity among these isolates. Finally, drug sensitivity tests demonstrated that all field isolates were sensitive to diclazuril but resistant to decoquinate (except for the isolates from eastern Hubei) and maduramicin.
Conclusions
Population genetic analysis indicated that genetic polymorphisms among field isolates were closely related with their regional distributions. Drug sensitivity testing demonstrated that E. tenella isolates in Hubei Province were sensitive to diclazuril, but resistant to maduramycin and decoquinate. The results presented here provide important information for the control and preventions of coccidiosis in the Hubei Province of China.
Electronic supplementary material
The online version of this article (doi:10.1186/s13071-017-2067-y) contains supplementary material, which is available to authorized users.
Keywords: Eimeria tenella, Genetic diversity, Maduramycin, Decoquinate, Diclazuril
Background
Avian coccidiosis, an intestinal parasitic disease caused by Eimeria spp., has caused great economic losses to the poultry industry worldwide [1, 2]. When chickens are infected with Eimeria tenella, clinical signs include lethargy, feather dishevelment and bloody feces. The main pathological changes include thickening of the intestinal wall and petechial hemorrhages.
Eimeria tenella is one of the most pathogenic species of Eimeria. Accurate identification is essential for the prevention and control of E. tenella. Many studies have focused on genetic diversity of E. tenella [3, 4]. As a useful molecular marker, the ITS1 fragment has been widely used for species identification of Eimeria [5]. Clark et al. used phylogenetic analysis of ITS sequence data to define species diversity between and within populations for all seven Eimeria species of chickens [6]. Schwarz et al. examined the genetic diversity of Eimeria species in Arkansas (AR) and North Carolina (NC) by analyzing ITS [7]. Williams et al. developed a RAPD technique based on the amplification of undefined targets by arbitrary primers to detect genetic polymorphisms [8], the technique has been widely used for the analysis of Eimeria genetic diversity [9, 10]. Such analysis can help us to estimate the phylogenetic relationship among different Eimeria isolates [11].
Currently, the main method to control Eimeria infection is anticoccidial drugs [12, 13]. Maduramycin, decoquinate and diclazuril are three chemotherapeutic agents. Maduramycin is thought to kill coccidium by interrupting their normal intracellular ion balance and influence of Na+-K+-ATPase activity [14]. Decoquinate interferes with the electron transport in the mitochondrial cytochrome system and CytB is an important part of the system [15]. The working mechanism of diclazuril is still unknown. Because of the prolonged use of anticoccidial drugs, resistance to such drugs has been frequently reported [16]. In addition, MIC2 and SAG genes are thought to be involved in host cell adhesion and invasion [17, 18]. Therefore, we aimed to compare the genetic diversity among field isolates using the four selected genes: EtCat ATPase and cytb genes which are drug targets; MIC2 and SAG genes which are important for interactions between parasites and host.
In the present study, the genetic diversity and drug sensitivity of E. tenella field isolates from Hubei Province of China were analyzed. The results should provide useful information for prevention and control of coccidiosis in this region.
Methods
Animals
Coccidia-free, 0-day-old chickens were purchased from Charoen Pokphand Group (Wuhan, China). Chickens were housed in a clean, coccidia-free environment in an isolated brooder room, and fed with commercial broiler feed and water [19].
Parasite material
Faecal samples were collected from eight different local commercial broiler production farms in Hubei Province between January 2012 and November 2013 (Additional file 1: Table S1). Eimeria oocysts obtained from the faecal samples were purified by saturated sodium nitrate flotation and sporulated using standard procedures [20].
Specific pathogens free (SPF) chickens (14-day-old) obtained from Charoen Pokphand Group were orally inoculated with 5 × 104 sporulated oocysts. Seven days post-inoculation, the chickens were sacrificed and necropsies were conducted. The caecal contents were collected for the isolation of Eimeria oocysts using the protease digestion method. The protease digestion and sporulation method were as follow: (i) the caeca from Eimeria oocysts infected groups were homogenized using a tissue grinder and 2 mg/ml of trypsin was added; (ii) this suspension was incubated in a water bath kettle for 1 h at 39 °C, sieved (180 diameter mesh) and transferred into a 500 ml centrifuge tube; (iii) the suspension was centrifuged at 1000× g for 5 min; (iv) the sediment was suspended in 2% w/v aqueous potassium dichromate and transferred to a 50 ml conical tube for oocysts sporulation in an aerobic incubator for 4 days at 28.6 °C. Sporulated oocysts were stored in 2.5% potassium dichromate at 4 °C [21] and were enumerated using the McMaster’s method under the microscope [22].
DNA and RNA extraction
For DNA extraction, 104 sporulated oocysts from each sample were washed with 1 mM sodium hypochlorite solution for 10 min at 4 °C, then washed three times with deionized water. The oocyst walls were ruptured using a grinding tissue homogenizer for released sporozoites (the volume 200 μl and homogenized), then 100 μl (1 mg/ml) of chicken bile, 100 μl of buffer suspension solution GA, and 20 μl (20 mg/ml) of proteinase K (Genomic DNA Kit, TIANGEN, Beijing, China) were added and the mixture was incubated at 56 °C for 2.5 h. DNA was extracted using a Genomic DNA Kit (TIANGEN), according to the manufacturer’s instructions. DNA concentration was measured using a NanoDrop 2000 nanodrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). In parallel, total RNA from sporozoites (5 × 104) was extracted using TRIzol reagent, according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA). First strand cDNA was synthesized by a reverse transcription (RT) reaction using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, USA).
ITS1 amplification and RAPD analysis
Extracted DNA was used as a template to amplify the ITS1 region of Eimeria spp., as described by Schnitzler et al. [23, 24]. RAPD random primers were synthesized by Sangon Biotech (Shanghai, China), and the conditions used for RAPD typing were the same as described by Fernandez et al. [10]. The sequences of the random primers used for RAPD amplification are listed in Additional file 1: Table S2. To examine the amplification results, all amplicons were assessed by electrophoresis through 1% agarose gels and then visualized by ethidium bromide staining.
Polymorphism analysis
Polymorphisms in cytb, MIC-2, SAG and EtCat ATPase genes were identified by sequencing the corresponding genes. The cytb and SAG genes were PCR amplified from the genomic DNA of isolated isolates using PrimeSTAR® Max DNA Polymerase (TaKaRa, Tokyo, Japan). The primers used are listed in Additional file 1: Table S3 and the following PCR program was used: initial denaturation at 94 °C for 5 min, followed by 35 cycles of 30 s denaturation at 94 °C, 30 s annealing at primer-dependent temperatures, and 60 s extension at 72 °C, followed by a final 7 min extension at 72 °C. The annealing temperatures were 60 °C for the cytb gene, 58 °C for MIC-2 and 55 °C for SAG. EtCat ATPase gene sequence is unavailable, the cation-transporting ATPase gene is highly conserved across Eimeria species, therefore, the primers used for amplifying E. tenella cation-transporting ATPase gene were designed based on the E. acervulina clone Dui-10 cation-transporting ATPase gene complete coding region (EU590120.1). The EtCat ATPase gene were amplified from the cDNA of field isolates for sequence alignments using similar PCR reaction conditions. All PCR products were subject to agarose gel electrophoresis and visualized by ethidium bromide staining. PCR products for ITS-1 amplification and polymorphism analysis were send to Sangon Biotech (Shanghai, China) for sequencing using primers in both directions.
Drug sensitivity tests
Selected field isolates were tested for their sensitivity to three coccidiostat drugs maduramycin, decoquinate and diclazuril. The design of drug-sensitivity tests in Additional file 1: Table S4. In a coccidia-free environment, 170 14-day-old SPF chickens were weighed and randomly divided into 17 groups, each containing ten chickens. All 17 groups were inoculated with E. tenella except one, which was not infected and used as a control. Infected chickens were given 5 mg/kg maduramycin, or 1 mg/kg diclazuril, or 30 mg/kg decoquinate, or no treatment as control, in fodder from the time of infection. Chicken faeces were collected from each post-infection groups between 5–7 days single droppings per day to evaluate the relative number of oocysts per gram of feces (OPG). At 21-day of age (7 days post-infection), all the chickens were individually weighed, sacrificed, and necropsied. The weight gain of chickens was recorded. The coccidial lesions present in the chickens were scored as category 0–4 following the methods described by Johnson & Reid [25]. Subsequently the anti-coccidial index (ACI) was calculated to assess drug effectiveness.
Sequence analysis
Alignment of the ITS1 sequences from Eimeria isolates was performed using MAFFT version 7 (http://mafft.cbrc.jp/alignment/server/index.html). Pairwise percentage identity was determined using BioEdit v.7.0 (http://www.mbio.ncsu.edu/bioedit/bioedit.html). RAPD results are analyzed by using SAHN program of the NTSYS-pc software (version 2.02 K,
Applied Biostatistics, Inc, NY, USA). The amino acid sequences of the EtCat ATPase gene from selected parasites were compared by the DNAMAN software (http://www.lynnon.com/dnaman.html). Phylogenetic analyses were conducted using MEGA, version 5.0.
Results
Isolation and species identification of Eimeria tenella field isolates from local farms in China
Twenty-one Eimeria field samples were isolated from different local commercial broiler production farms in Hubei Province (Fig. 1). To confirm the species identity of these Eimeria samples, seven pairs of species-specific primers (for E. acervulina, E. brunetti, E. mitis, E. necatrix, E. maxima, E. praecox and E. tenella, respectively) amplifying ITS1 region were used. The identification results of Eimeria species using species specific primers amplifying the internal transcribed spacer 1 (ITS1) region in Additional file 1: Table S5. The results indicate that all the field isolates were E. tenella since they produced a specific 278 bp band using the E. tenella-specific primer (one example is given in Fig. 2). This fragment obtained from each strain was subject to DNA sequencing and the results showed that they were 97–100% identical to the ITS1 sequence of E. tenella (GenBank No. AF026388), further confirming that these isolates are E. tenella (GenBank No. KY117132–KY117152). Pairwise comparison of ITS1 sequences from these isolates revealed high sequence identities ranging from 92.5–100% (Table 1), suggesting homology among these isolates. Phylogenetic relationship based on the ITS1 of E. tenella north (Suizhou isolate), E. tenella east (Huanggang isolate), E. tenella south (Jingzhou isolate), E. tenella middle (Tianmen isolate), E. tenella UK (GenBank: LN609779), E. tenella US (GenBank: LN609784), E. maxima (GenBank: AF065095), Neospora caninum (GenBank: AF029702) and Toxoplasma gondii (GenBank: L49390) was accomplished by MEGA v6.0 using neighbor-joining method with default setting [26], and nodal support values were indicated (%). The phylogenetic tree demonstrated the four E. tenella field isolates had close relationship with E. tenella UK (Fig. 3).
Fig. 1.
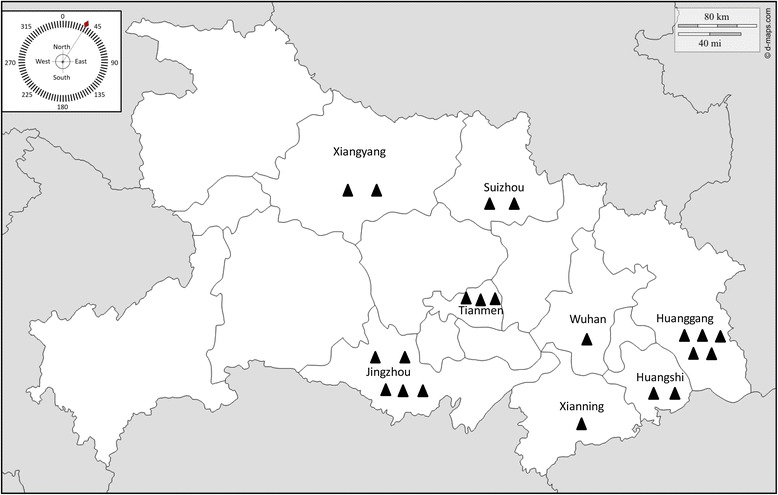
Local commercial broiler farms in Hubei Province from which Eimeria isolates were isolated. Faecal samples were collected from different local and different commercial broiler production farms (▲)
Fig. 2.
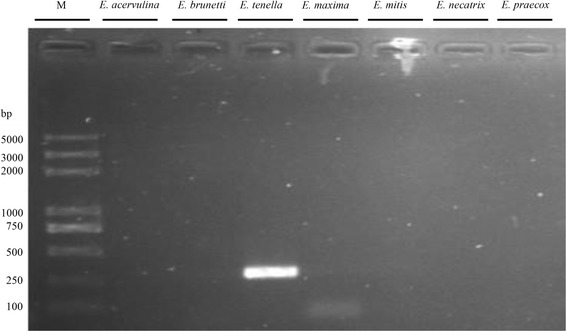
Identification of Eimeria species using species-specific primers amplifying the internal transcribed spacer 1 (ITS1) region. Seven pairs of primers specific for E. acervulina, E. brunetti, E. mitis, E. necatrix, E.maxima, E.praecox, and E.tenella were used to amplify the genomic DNA of field isolates, and the products were analyzed by agrose gel electrophoresis. One strain is shown as an example. Lane M: Trans2K Plus DNA Marker
Table 1.
Pairwise comparison between the ITS-1 sequences of different Eimeria tenella isolates
| Seq-> | TM1 | TM2 | TM3 | SZ1 | SZ2 | HS | JZ1 | JZ2 | JX | XN | XS1 | XS2 | XS3 | XS4 | XS5 | SS1 | SS2 | SS3 | XY1 | XY2 | DY |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TM1 | |||||||||||||||||||||
| TM2 | 94.8 | ||||||||||||||||||||
| TM3 | 95.6 | 94.8 | |||||||||||||||||||
| SZ1 | 96.8 | 95.6 | 97.6 | ||||||||||||||||||
| SZ2 | 96.0 | 95.6 | 97.2 | 96.8 | |||||||||||||||||
| HS | 94.8 | 96.4 | 94.8 | 95.6 | 95.2 | ||||||||||||||||
| JZ1 | 97.6 | 95.2 | 96.8 | 98.0 | 97.2 | 96.4 | |||||||||||||||
| JZ2 | 94.9 | 93.7 | 94.5 | 94.9 | 94.9 | 94.9 | 95.7 | ||||||||||||||
| JX | 96.0 | 94.4 | 95.2 | 96.0 | 96.8 | 95.6 | 97.6 | 95.3 | |||||||||||||
| XN | 95.6 | 95.2 | 96.0 | 96.0 | 96.0 | 94.5 | 96.8 | 94.9 | 95.7 | ||||||||||||
| XS1 | 94.4 | 96.4 | 96.0 | 96.4 | 95.6 | 96.0 | 95.6 | 94.5 | 94.4 | 94.9 | |||||||||||
| XS2 | 97.2 | 95.6 | 97.2 | 98.4 | 97.6 | 96.4 | 99.6 | 95.7 | 97.6 | 97.2 | 96.0 | ||||||||||
| XS3 | 97.2 | 95.2 | 97.2 | 97.2 | 97.2 | 95.6 | 98.4 | 95.7 | 96.4 | 96.8 | 95.6 | 98.8 | |||||||||
| XS4 | 96.8 | 96.0 | 96.8 | 97.2 | 98.4 | 96.0 | 98.4 | 96.4 | 97.2 | 97.2 | 96.4 | 98.8 | 98.0 | ||||||||
| XS5 | 96.8 | 94.4 | 96.0 | 96.8 | 96.4 | 95.2 | 98.8 | 95.7 | 96.4 | 96.0 | 96.0 | 98.4 | 97.6 | 97.6 | |||||||
| SS1 | 92.5 | 94.0 | 94.8 | 93.6 | 93.6 | 94.8 | 94 | 92.6 | 93.7 | 93.7 | 96.0 | 94.4 | 94.0 | 94.0 | 94.1 | ||||||
| SS2 | 94.1 | 95.2 | 95.6 | 95.2 | 95.2 | 95.6 | 95.2 | 94.9 | 94.8 | 94.9 | 96.8 | 95.6 | 95.2 | 96.0 | 94.5 | 97.2 | |||||
| SS3 | 96.8 | 95.6 | 98.3 | 97.6 | 97.2 | 94.8 | 97.2 | 94.5 | 96.4 | 96.0 | 96.0 | 97.6 | 96.8 | 96.8 | 96.0 | 94.4 | 95.6 | ||||
| XY1 | 96.8 | 95.6 | 97.2 | 98.0 | 96.4 | 95.6 | 98 | 94.5 | 96.0 | 96.0 | 95.6 | 98.4 | 97.2 | 97.2 | 96.8 | 94.0 | 94.8 | 98.0 | |||
| XY2 | 95.2 | 96.4 | 96.4 | 96.0 | 98.4 | 96.0 | 96.4 | 94.1 | 96.0 | 95.2 | 96.4 | 96.8 | 96.0 | 97.2 | 96.0 | 95.2 | 96.4 | 96.8 | 95.6 | ||
| DY | 93.3 | 94.0 | 95.2 | 94.0 | 94.4 | 95.2 | 94.4 | 94.1 | 94.0 | 93.7 | 96.0 | 94.8 | 94.4 | 95.2 | 94.5 | 97.6 | 98.0 | 94.4 | 93.6 | 96.0 |
Fig. 3.
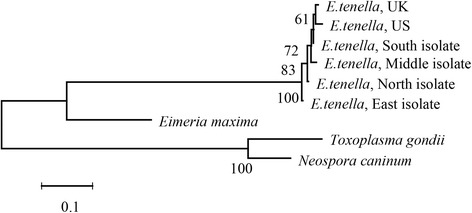
Phylogenetic relationships of E. tenella north (Suizhou isolate), E. tenella east (Huanggang isolate), E. tenella south (Jingzhou isolate) and E. tenella middle (Tianmen isolate) with E. tenella UK (GenBank: LN609779), E. tenella US (GenBank: LN609784), E. maxima (GenBank: AF065095), Neospora caninum (GenBank: AF029702) and Toxoplasma gondii (GenBank: L49390); based on ITS1 sequences by neighbor-joining analysis. Nodal support values are indicated (%). The scale-bar indicates sequence substitution per site
RAPD analysis
To optimize the primers for RAPD analysis, 70 decamer were screened using E. tenella DNA samples. After the initial screening, seven primers were found to discriminate the E. tenella species. They resulted in two to eight amplicons ranging from 250–1,500 bp using the field isolates as templates. The amplification patterns of the field isolates generated by the S2133 primer are shown in Fig. 4. NTSYS-pc software was used to convert the amplification results into data to assess the phylogenetic relationships among the E. tenella field isolates (Fig. 5). Briefly, the unweighted pair-group method with arithmetic averages (UPGMA) dendrogram of the RAPD data was calculated and analysised using SAHN program of the NTSYS-pc software. The RAPD data were generated from the DNA fingerprints of the eight E. tenella isolates using seven random primers, and the results indicated eight E. tenella isolates were separated into two branch clusters mainly based on geographical distribution, the first cluster included SZ, XY, TM and WH, and the second cluster included JZ, SS, XS and HG. The phylogenetic branch lengths ranged from 0.49 to 0.75, and the clusters may be caused by cross-regional transportation of animals.
Fig. 4.
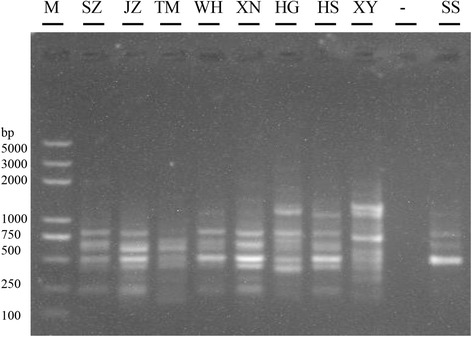
Random amplified polymorphic fragments obtained with decamer primers S2133 using DNA samples of different field isolates. Abbreviations: SZ, Suizhou; JZ, Jingzhou; TM, Tianmen; WH, Wuhan; XN, Xianning; XS, Xishui; HS, Huangshi; XY, Xiangyang; SS, Shashi; “-”, Negative control; M: Trans2K Plus DNA Marker
Fig. 5.
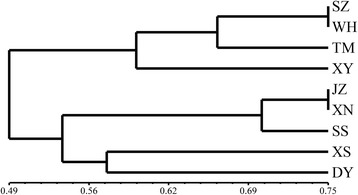
Clustering of E. tenella field isolates following RAPD analysis. Abbreviations: SZ, Suizhou; JZ, Jingzhou; TM, Tianmen; WH, Wuhan; XN, Xianning; XS, Xishui; HS, Huangshi; XY, Xiangyang; SS, Shashi
Sequence diversity of the cytochrome B gene
Amplicons of the E. tenella cytb gene (1,268 bp) derived from the field isolates were sequenced and aligned with that of the E. tenella Houghton strain (GenBank: HQ173891.1). The result revealed that the cytb gene sequences from our field isolates (GenBank: KY117213–KY117232) have 99.9% similarity with the cytb gene from the E. tenella Houghton strain. Polymorphisms were observed at three positions (position 22, C-A; position 49, A-G; position 838, T-C); these are listed in Table 2. These nucleotide changes do result in amino acid alterations in the CytB protein.
Table 2.
Mutations in the CytB gene of Eimeria tenella field isolates
| Strains | Mutations |
|---|---|
| DY | C-A(22th) A-G(49th) T-C(838th) |
| XS1 | C-A (22th) |
| XS2 | C-A (22th) |
| XS3 | 0 |
| XS4 | 0 |
| HS | 0 |
| JZ1 | C-A/C(22th) A-G/A(49th) T-C(838th) |
| JZ2 | C-A/C(22th) A-G/A(49th) T-C(838th) |
| SS1 | C-A/C(22th) A-G/A(49th) T-C(838th) |
| SS2 | C-A/C(22th) A-G/A(49th) T-C(838th) |
| SS3 | C-A(22th) A-G(49th) T-C(838th) |
| SZ1 | C-A/C(22th) A-G/A(49th) T-C(838th) |
| SZ2 | C-A/C(22th) A-G/A(49th) T-C(838th) |
| TM1 | C-A(22th) A-G(49th) T-C(838th) |
| TM2 | C-A(22th) A-G(49th) T-C(838th) |
| TM3 | C-A(22th) A-G(49th) T-C(838th)` |
| WH | C-A(22th) A-G(49th) T-C/T(838th) |
| XN | C-A(22th) A-G(49th) T-C(838th) |
| XY1 | C-A(22th) A-G(49th) T-C(838th) |
| XY2 | C-A(22th) A-G(49th) T-C(838th) |
Sequence diversity of the MIC-2 and SAG genes
Amplification of the MIC-2 gene from the E. tenella field isolates produced a specific 1,647-bp band. This fragment from each isolate was sequenced and aligned with the E. tenella MIC-2 gene (GenBank: AF111702.1). The results indicated that there is no polymorphism between field isolates (GenBank: KY117173 to KY117192) and the reference strain. Amplification of SAG gene yielded a single 1,101-bp band in all field isolates. Sequence alignment with the E. tenella surface antigen gene (GenBank: M21088.1) indicated that all of the field isolates (GenBank: KY117193–KY117212) contained two nucleotide substitutions at positions 209 (A-G) and 901 (C-G), but these nucleotide substitutions did not cause amino acid changes.
EtCat ATPase gene
PCR amplification of the EtCat ATPase gene from the field isolates produced one single band of 494 bp on agarose gels. Sequencing analysis indicate that the sequences of Et.Cat ATPase gene from all field isolates (GenBank: KY126385 to KY126404) are also identical to the E. tenella mitochondrial hypothetical protein (GenBank: KF670727.1). The amino acid sequence of Et.Cat ATPase shared 87% similarity with that of the E. acervulina cation transporter ATPase gene (GenBank: EU590120.1), 80% similarity with E. maxima Cation-transporting ATPase (GenBank: XP_013337038.1), and 34% similarity with the cation-transporting ATPase gene (GenBank: EPR61390.1) of T. gondii (Fig. 6).
Fig. 6.
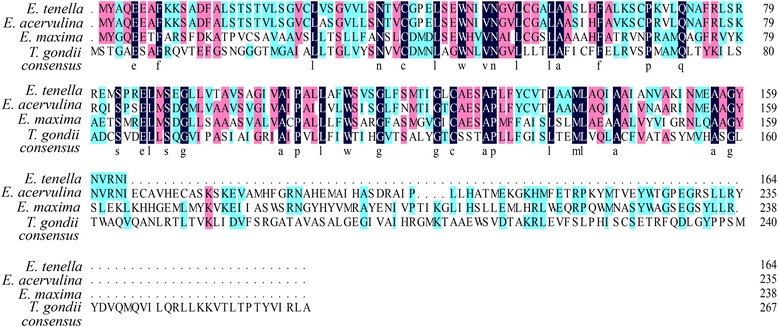
Sequence alignment of E. tenella Cation-transport ATPase with its homologs from selected parasites. Black: 100% amino acid identity; red: 75% amino acid identity; blue: 50% amino acid identity
Drug susceptibility of E. tenella isolates from Hubei Province
To test the status of drug resistance in the field, four isolates representing different geographical origins were selected and their sensitivity to commercially available drugs were tested. Diclazuril had an ACI higher than 180 in the field isolates from northern, eastern and middle regions of Hubei Province. The ACI of Diclazuril on the strain from southern part of Hubei is 168.8. These results suggest that all field isolates from these regions are sensitive to Diclazuril. The data for decoquinate drug tests showed that E. tenella field isolates from northern, southern and middle Hubei had ACI below 160, suggesting decoquinate tolerance. While for the strain from eastern Hubei, decoquinate had an ACI value of 175.8, indicating that this isolates is still sensitive to decoquinate. For maduramicin, and all tested isolates had ACI values below 160, which indicate that they were resistant to this drug (Table 3).
Table 3.
The drug susceptibility of E. tenella field isolates
| Groups | Field isolates | Weight gain (g) | Weight gain of NNCg (%) | Survival rate (%) | Lesion score | OPG (105) | Anticoccidial index |
|---|---|---|---|---|---|---|---|
| 1a | North isolated | 76 | 100 | 100 | 1.2 | 0.813 | 183 |
| 2a | East isolatee | 77 | 101.3 | 100 | 1.0 | 0.658 | 186.3 |
| 3a | South isolatef | 69 | 90.8 | 90 | 1.2 | 1.44 | 168.8 |
| 4a | Middle isolateg | 77 | 101.3 | 100 | 1.1 | 0.792 | 185.3 |
| 5b | North isolated | 54 | 71.1 | 100 | 2.6 | 1.06 | 140.1 |
| 6b | East isolatee | 66 | 86.8 | 100 | 0.6 | 0.598 | 175.8 |
| 7b | South isolatef | 47 | 61.8 | 100 | 1.6 | 1.27 | 140.8 |
| 8b | Middle isolateg | 62 | 81.6 | 70 | 3.2 | 1.3 | 129.6 |
| 9c | North isolated | 53 | 69.7 | 100 | 2.8 | 0.938 | 136.7 |
| 10c | East isolatee | 55 | 73.3 | 100 | 2.1 | 0.957 | 147.3 |
| 11c | South isolatef | 51 | 67.1 | 90 | 2.4 | 1.16 | 128.1 |
| 12c | Middle isolateg | 50 | 65.8 | 90 | 3.1 | 1.25 | 119.8 |
| 13 | North control | 39 | 51.3 | 90 | 3.5 | 6.25 | 108.8 |
| 14 | East control | 63 | 82.9 | 80 | 3.4 | 5.98 | 123.5 |
| 15 | South control | 46 | 60.5 | 90 | 3.4 | 5.52 | 114.1 |
| 16 | Middle control | 44 | 57.9 | 80 | 3.3 | 5.66 | 98.6 |
| 17h | NNC control | 76 | 100 | 100 | 0 | 0 | 200 |
aDiclazuril treatment groups
bDecoquinate treatment groups
cMaduramycin treatment groups
dSuizhou isolate
eHuanggang isolate
fJingzhou isolate
gTianmen isolate
d-g isolates control: infected dose per chicken for 5 × 104 sporulated oocysts, non-treatment control (groups, 13–16)
hNNC control: non-infected, non-treatment control (group,17)
Discussion
Chicken coccidiosis caused by Eimeria species leads to severe economic losses to the poultry industry worldwide [27, 28]. It is listed as one of the top five most devastating diseases in poultry. The morbidity of coccidiosis is estimated to be 50–70% and the disease is a major threat to 15–50 day-old chickens [29–31].
In this study, we purified Eimeria oocysts from chicken faecal samples collected from eight regions (Suizhou, Xiangyang, Huanggang, Huangshi, Xianning, Wuhan, Jingzhou, Tianmen) in Hubei Province of China. Subsequently all the isolates were identified to be E. tenella using species-specific PCR amplification of the ITS1 rDNA region.
The PCR-based RAPD technique, which was originally developed in the 1990s, can be used to genotype organisms and identify unknown sequence polymorphisms among genetically diverse isolates [32]. Here, we used random primers designed by Sangon Biotech for RAPD analysis of the E. tenella field isolates collected from different regions of Hubei, China. The result indicated that there are genetic differences among the E. tenella field isolates in this region and the differences may correlate with geographical origin of the isolates.
Many studies have investigated the development of resistance mechanisms against anti-coccidian drugs in Eimeria parasites and proposed various hypotheses [33]. However, the most popular opinion for the development of drug resistance is the endogenous mutations occurring in coccidian parasite [34]. Because of continuous and prolonged use of anticoccidial drugs, mutations have been observed among coccidian population during this continuous selection process. These mutations maybe involved in the development of drug resistance [35].
In other apicomplexan species, genetic mutations are known to be associated with drug resistance. The correlation between the acquisition of mutations and drug resistance has been verified, for example, a point mutation at position 268 of the cytb gene was reported to induce atovaquone resistance in Plasmodium falciparum [36]. In the present study, genetic mutations also existed in resistant field isolates, whether the mutations are related with drug resistance of field isolates or not need further verification.
In addition to RAPD, we also examined the genetic diversity of field isolates by checking the sequence polymorphisms in MIC-2, SAG, EtCat ATPase and cytb genes. Sequence analysis indicated that the MIC-2, SAG, EtCat ATPase genes are highly conserved among E. tenella isolates. For the cytb gene, the alignments showed that some isolates contain polymorphisms leading to missense mutations. These changes occurred at positions eight, 17 and 280 of the CytB protein.
Conclusions
In this study, we isolated and analyzed E. tenella field isolates from eight regions of Hubei Province, P. R. China. RAPD analysis and multiple sequence alignments of SAG, MIC-2, cytb and EtCat ATPase revealed genetic diversity among field isolates. The drug sensitivity tests indicated that maduramycin and decoquinate resistance are widely present in Hubei, but diclazuril is still effective towards E. tenella isolates in this region. These results are important for the selection of strategies to control chicken coccidiosis in this region.
Acknowledgements
We would like to thank professor Shijun Li for providing space for animal experiments.
Funding
This study was supported by the national key research and development program (2016YFD0501303) and the Fundamental Research Funds for the Central Universities (Grant No. 2662015PY048).
Availability of data and materials
All data generated or analyzed during this study are included in the article. The sequences are submitted to the GenBank database under accession numbers KY117132–KY117152, KY117173–KY117232 and KY126385–KY126404.
Authors’ contributions
JLZ and RF conceived and designed the study. LT and YLL wrote the manuscript with input from other coauthors. LT, XY, WQL, QYK, and MNM performed the experiments and analyzed the data. YQZ and BS assisted in study design and manuscript editing. All authors read and approved the final manuscript.
Competing interests
Not applicable.
Consent for publication
Not applicable.
Ethics approval
All animal experiments were performed under the instructions of Laboratory Animals Centre of Hubei province in P. R. China and approved by the ethical committee of Huazhong Agricultural University, according to the Regulations of the Care and Use of Laboratory Animals in China.
Abbreviations
- RAPD
Random amplified polymorphic DNA
- ITS
Internal transcribed spacer 1
- SAG
Surface antigen
- MIC-2
Micronemal protein 2
- cytb
Cytochrome b
- EtCat ATPase
Eimeria tenella cation-transport ATPase
- RT
Reverse transcription
- OPG
Oocysts per gram of faeces
- SPF
Specific pathogens free
- ACI
Anti-coccidial index
Additional file
Local commercial broiler farms in Hubei province from which Eimeria isolates were isolated. Table S2. Primers used in RAPD analysis by Sangon Biotech, Shanghai. Table S3. Primers used for PCR amplification of indicated genes. Table S4. The design of drug-sensitivity tests. Table S5. Identification results of Eimeria species using species specific primers amplifying the internal transcribed spacer 1 (ITS1) region. (DOC 126 kb)
Contributor Information
Li Tan, Email: 396322874@qq.com.
Yalin Li, Email: 275694748@qq.com.
Xin Yang, Email: 695237569@qq.com.
Qiyun Ke, Email: 1121648285@qq.com.
Weiqiang Lei, Email: 1017475188@qq.com.
Mudassar Niaz Mughal, Email: 2691629093@qq.com.
Rui Fang, Email: fangrui19810705@163.com.
Yanqin Zhou, Email: yanqinzhou@mail.hzau.edu.cn.
Bang Shen, Email: shenbang@mail.hzau.edu.cn.
Junlong Zhao, Email: zhaojunlong@mail.hzau.edu.cn.
References
- 1.McDonald V, Shirley MW. Past and future: vaccination against Eimeria. Parasitology. 2009;136(12):1477–1489. doi: 10.1017/S0031182009006349. [DOI] [PubMed] [Google Scholar]
- 2.Blake DP, Tomley FM. Securing poultry production from the ever-present Eimeria challenge. Trends Parasitol. 2014;30(1):12–19. doi: 10.1016/j.pt.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Blake DP, Clark EL, Macdonald SE, Thenmozhi V, Kundu K, Garg R, et al. Population, genetic, and antigenic diversity of the apicomplexan Eimeria tenella and their relevance to vaccine development. Proc Natl Acad Sci USA. 2015;112(38):E5343–5350. [DOI] [PMC free article] [PubMed]
- 4.Beck HP, Blake D, Darde ML, Felger I, Pedraza-Diaz S, Regidor-Cerrillo J, et al. Molecular approaches to diversity of populations of apicomplexan parasites. Int J Parasitol. 2009;39(2):175–89. [DOI] [PubMed]
- 5.Lew AE, Anderson GR, Minchin CM, Jeston PJ, Jorgensen WK. Inter- and intra-strain variation and PCR detection of the internal transcribed spacer 1 (ITS-1) sequences of Australian isolates of Eimeria species from chickens. Vet Parasitol. 2003;112(1–2):33–50. doi: 10.1016/S0304-4017(02)00393-X. [DOI] [PubMed] [Google Scholar]
- 6.Clark EL, Macdonald SE, Thenmozhi V, Kundu K, Garg R, Kumar S, et al. Cryptic Eimeria genotypes are common across the southern but not northern hemisphere. Int J Parasitol. 2016;46(9):537–544. doi: 10.1016/j.ijpara.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwarz RS, Jenkins MC, Klopp S, Miska KB. Genomic analysis of Eimeria spp. populations in relation to performance levels of broiler chicken farms in Arkansas and North Carolina. J Parasitol. 2009;95(4):871–880. doi: 10.1645/GE-1898.1. [DOI] [PubMed] [Google Scholar]
- 8.Williams JG, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18(22):6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnston DA, Fernando MA. Eimeria spp. of the domestic fowl: analysis of genetic variability between species and strains using DNA polymorphisms amplified by arbitrary primers and denaturing gradient-gel electrophoresis. Parasitol Res. 1995;81(2):91–97. doi: 10.1007/BF00931611. [DOI] [PubMed] [Google Scholar]
- 10.Fernandez S, Costa AC, Katsuyama AM, Madeira AM, Gruber A. A survey of the inter- and intraspecific RAPD markers of Eimeria spp. of the domestic fowl and the development of reliable diagnostic tools. Parasitol Res. 2003;89(6):437–445. doi: 10.1007/s00436-002-0785-2. [DOI] [PubMed] [Google Scholar]
- 11.Reid AJ, Blake DP, Ansari HR, Billington K, Browne HP, Bryant J, et al. Genomic analysis of the causative agents of coccidiosis in domestic chickens. Genome Res. 2014;24(10):1676–1685. doi: 10.1101/gr.168955.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shumard RF, Callender ME. Anticoccidial drugs: screening methods. Exp Parasitol. 1970;28(1):13–24. doi: 10.1016/0014-4894(70)90061-5. [DOI] [PubMed] [Google Scholar]
- 13.Peek HW, Landman WJ. Coccidiosis in poultry: anticoccidial products, vaccines and other prevention strategies. Vet Q. 2011;31(3):143–161. doi: 10.1080/01652176.2011.605247. [DOI] [PubMed] [Google Scholar]
- 14.Chapman HD, Jeffers TK, Williams RB. Forty years of monensin for the control of coccidiosis in poultry. Poult Sci. 2010;89(9):1788–1801. doi: 10.3382/ps.2010-00931. [DOI] [PubMed] [Google Scholar]
- 15.Guo FC, Suo X, Zhang GZ, Shen JZ. Efficacy of decoquinate against drug sensitive laboratory strains of Eimeria tenella and field isolates of Eimeria spp. in broiler chickens in China. Vet Parasitol. 2007;147(3–4):239–245. doi: 10.1016/j.vetpar.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 16.Peek HW, Landman WJ. Resistance to anticoccidial drugs of Dutch avian Eimeria spp. field isolates originating from 1996, 1999 and 2001. Avian Pathol. 2003;32(4):391–401. doi: 10.1080/0307945031000121149. [DOI] [PubMed] [Google Scholar]
- 17.Tomley FM, Bumstead JM, Billington KJ, Dunn PP. Molecular cloning and characterization of a novel acidic microneme protein (Etmic-2) from the apicomplexan protozoan parasite, Eimeria tenella. Mol Biochem Parasitol. 1996;79(2):195–206. doi: 10.1016/0166-6851(96)02662-X. [DOI] [PubMed] [Google Scholar]
- 18.Tabares E, Ferguson D, Clark J, Soon PE, Wan KL, Tomley F. Eimeria tenella sporozoites and merozoites differentially express glycosylphosphatidylinositol-anchored variant surface proteins. Mol Biochem Parasitol. 2004;135(1):123–132. doi: 10.1016/j.molbiopara.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 19.Upton SJ, McAllister CT, Brillhart DB, Duszynski DW, Wash CD. Cross-transmission studies with Eimeria arizonensis-like oocysts (Apicomplexa) in New World rodents of the genera Baiomys, Neotoma, Onychomys, Peromyscus, and Reithrodontomys (Muridae). J Parasitol. 1992;78(3):406–13. [PubMed]
- 20.Shirley MW. Eimeria spp. from the chicken: occurrence, identification and genetics. Acta Vet Hung.1997;45(3):331–47. [PubMed]
- 21.Duszynski DW, Wilber PG. A guideline for the preparation of species descriptions in the Eimeriidae. J Parasitol. 1997;83(2):333–336. doi: 10.2307/3284470. [DOI] [PubMed] [Google Scholar]
- 22.Jiang L, Zhao Q, Zhu S, Han H, Dong H, Huang B. Establishment of Eimera tenella (local isolate) in chicken embryos. Parasite. 2012;19:285–289. doi: 10.1051/parasite/2012193285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schnitzler BE, Thebo PL, Tomley FM, Uggla A, Shirley MW. PCR identification of chicken Eimeria: a simplified read-out. Avian Pathol. 1999;28(1):89–93. doi: 10.1080/03079459995091. [DOI] [PubMed] [Google Scholar]
- 24.Schnitzler BE, Thebo PL, Mattsson JG, Tomley FM, Shirley MW. Development of a diagnostic PCR assay for the detection and discrimination of four pathogenic Eimeria species of the chicken. Avian Pathol. 1998;27(5):490–497. doi: 10.1080/03079459808419373. [DOI] [PubMed] [Google Scholar]
- 25.Johnson J, Reid WM. Anticoccidial drugs: lesion scoring techniques in battery and floor-pen experiments with chickens. Exp Parasitol. 1970;28(1):30–36. doi: 10.1016/0014-4894(70)90063-9. [DOI] [PubMed] [Google Scholar]
- 26.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clark EL, Tomley FM, Blake DP: Are Eimeria genetically diverse, and does it matter?. Trends in Parasitol. 2016;33(3):231-41. [DOI] [PubMed]
- 28.Fornace KM, Clark EL, Macdonald SE, Namangala B, Karimuribo E, Awuni JA, et al. Occurrence of Eimeria species parasites on small-scale commercial chicken farms in Africa and indication of economic profitability. PLoS One. 2013;8(12):e84254. doi: 10.1371/journal.pone.0084254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang M. Veterinary parasitology. Beijing: China Agricultural Press; 2003. [Google Scholar]
- 30.Jiang JS. Aminal protozoology. Beijing: China Agricultural University Press; 2000. [Google Scholar]
- 31.Suo X, Cai J. Poultry coccidiosis. Beijing: China Agricultural Press; 2004. [Google Scholar]
- 32.Procunier JD, Fernando MA, Barta JR. Species and strain differentiation of Eimeria spp. of the domestic fowl using DNA polymorphisms amplified by arbitrary primers. Parasitol Res. 1993;79(2):98–102. doi: 10.1007/BF00932253. [DOI] [PubMed] [Google Scholar]
- 33.Jeffers TK. Avian coccidiosis. British Poultry Science. 1978;50-125.
- 34.Jeffers TK. Genetic transfer of anticoccidial drug resistance in Eimeria tenella. J Parasitol. 1974;60(6):900–904. doi: 10.2307/3278505. [DOI] [PubMed] [Google Scholar]
- 35.Weppelman RM, Battaglia JA, Wang CC. Eimeria tenella: the selection and frequency of drug-resistant mutants. Exp Parasitol. 1977;42(1):56–66. doi: 10.1016/0014-4894(77)90061-3. [DOI] [PubMed] [Google Scholar]
- 36.Teo BH, Lansdell P, Smith V, Blaze M, Nolder D, Beshir KB, et al. Delayed onset of symptoms and atovaquone-proguanil chemoprophylaxis breakthrough by Plasmodium malariae in the absence of mutation at codon 268 of pmcytb. PLoS Negl Trop Dis. 2015;9(10):e0004068. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in the article. The sequences are submitted to the GenBank database under accession numbers KY117132–KY117152, KY117173–KY117232 and KY126385–KY126404.


