Abstract
Circulating endothelial colony forming cells (ECFCs) contribute to vascular repair where they are a target for therapy. Since ECFC proliferative potential is increased in cord versus peripheral blood and to define regulatory factors controlling this proliferation, we compared the miRNA profiles of cord blood and peripheral blood ECFC-derived cells. Of the top 25 differentially regulated miRNAs selected, 22 were more highly expressed in peripheral blood ECFC-derived cells. After validating candidate miRNAs by q-RT-PCR, we selected miR-193a-3p for further investigation. The miR-193a-3p mimic reduced cord blood ECFC-derived cell proliferation, migration and vascular tubule formation, while the miR-193a-3p inhibitor significantly enhanced these parameters in peripheral blood ECFC-derived cells. Using in silico miRNA target database analyses combined with proteome arrays and luciferase reporter assays of miR-193a-3p mimic treated cord blood ECFC-derived cells, we identified 2 novel miR-193a-3p targets, the high mobility group box-1 (HMGB1) and the hypoxia upregulated-1 (HYOU1) gene products. HMGB1 silencing in cord blood ECFC-derived cells confirmed its role in regulating vascular function. Thus, we show, for the first time, that miR-193a-3p negatively regulates human ECFC vasculo/angiogenesis and propose that antagonising miR-193a-3p in less proliferative and less angiogenic ECFC-derived cells will enhance their vasculo/angiogenic function.
Endothelial colony forming cells (ECFCs) or late-outgrowth endothelial cells (OECs) are a subpopulation of endothelial precursors found circulating in human cord blood (CB) and adult peripheral blood (PB), although they are also resident in tissues including the bone marrow, umbilical cord and the placental vasculature1,2,3,4. These ECFCs generate de novo vessels in vitro and in immunodeficient murine models in vivo, and their functional potency correlates with their proliferative potential1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19. In pre-clinical models, ECFCs are reported to enhance the patency of implanted vascular grafts and to improve outcomes in ischemic diseases, such as stroke, myocardial infarction and limb ischemia20,21,22,23,24,25,26,27,28,29,30,31. Given their ability to proliferate, migrate, incorporate into vascular networks, release proangiogenic factors and promote vascular repair in preclinical models, ECFCs have attracted substantial interest as a therapeutic target for treating vascular injury.
Studies in non-human primates are reported to recapitulate those in the human, with ECFC frequency and ability to form inosculating vessels decreasing with age32. In the human, ECFCs from healthy perinatal CB are significantly more frequent than those found circulating in healthy adult PB and have a higher proliferative capacity3,5,6,11,15,16,18,19. Thus, defining strategies to enhance the content of these cells in the adult, while retaining their vasculogenic/angiogenic functions would be beneficial. In this respect, there is increasing evidence that small, non-coding microRNAs (miRNAs) regulate endothelial cell generation and functions33,34,35.
miRNAs are short non-coding RNA molecules, 21–25 nucleotides long, that function to downregulate the expression of multiple genes by such processes as mRNA cleavage or repression of translation and acceleration of target mRNA deadenylation. Interestingly, miRNA mediated knockdown impairs vasculogenesis and angiogenesis in vitro and in in vivo models36,37,38, while miRNA profiling and functional studies have identified both pro-and anti-angiogenic miRNAs33,34,37. Examples of those reported to be negative regulators of angiogenesis include the miR-17, miR-92a, miR-200, miR-221/222, and the 14q32 miR cluster (miR-329, -487b, -494, -495)34,36,38,39. In contrast, pro-angiogenic miRNAs include mir-126, let-7f, miR-93, miR-210, miR-130a and miR-1334,38.
In order to identify miRNAs, which regulate the proliferation of ECFC-derived cells, whilst also enhancing their angiogenic or vasculogenic potential, we compared the microRNA profiles of the more proliferative CB with the less proliferative PB ECFC-derived cells. In this paper, we identify miR193a-3p as an anti-proliferative and anti-angiogenic miRNA, which is preferentially expressed in human adult PB ECFC-derived cells. Subsequently, using miR-193a-3p inhibitors, we demonstrate that these PB cells exhibit improved proliferative and vasculo-/angio-genic abilities compared to control transfected cells. Based on reporter assays, we report that miR-193a-3p inhibits ECFC-derived cell proliferation and subsequently angiogenic functions via a novel target, HMGB1.
Materials and Methods
Human endothelial cell culture
Human CB units were sourced from the John Radcliffe Hospital in Oxford with ethical approval from the Oxfordshire Research Ethics Committee C and Berkshire Research Ethics Committee and with informed written consent6,11. The research was carried out with institutional R&D committee approval, and the University of Oxford and NHSBT Oxford performed all methods in accordance with the relevant guidelines and regulations set. Adult PB cells were obtained from anonymised consented donors and supplied by NHS Blood and Transplant from human leukoreduction filters collected in transparent cones within 4 hr of donation. Nine CB and 9 PB donations were collected and used for the experimental studies. Human ECFC-derived cells were next generated by culturing the CB and PB mononuclear cells (2 × 107 cells in 6 well collagen coated plates) in 4 mls complete EGM-2 media (Lonza Biologics, Cambridge, England) supplemented with 10% (v/v) Hyclone heat inactivated fetal bovine serum (FBS; ThermoScientific, Waltham, MA, USA)5,10. Endothelial colonies, which formed by 21 days of culture, were selected using cloning rings and those from individual CB or PB units were pooled and then passaged in the above medium5,10. Passage 0 was the time of the appearance of the first ECFC-derived colonies. The cells possessed a typical endothelial morphology and were confirmed by flow cytometric phenotyping as endothelial lineage cells (Supplemental Methods and Supplementary Fig. S1).
miRNA microarray analysis
Total and miRNA enriched RNA was extracted using Qiagen RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. RNA quality and abundance were determined after extraction using a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, NC, USA). miRNA profiling was done by Exiqon A/S, Vedbaek Denmark using 3 individual donations for both CB and PB ECFC-derived cells at passage 3. The samples were labeled using the miRCURY Hy3/Hy5 Power labeling kit and hybridized on the miRCURY LNA Array (version 5th Generation arrays, hsa, mmu and rno)(Exiqon A/S), which consisted of control probes, mismatch probes and 1900 capture probes that perfectly matched all of the human miRNAs registered and annotated in miRBase v15.0. The quantified signals were normalized using the global Lowess (LOcally WEighted Scatterplot Smoothing) regression algorithm, which minimizes the intensity-dependent differences between the dyes. The expression matrix contained normalized Hy3/Hy5 ratios (log2 transformed) from all hybridizations. The subset of differentially expressed miRNAs was used to construct an unsupervised hierarchical clustering of the different samples (Exiqon A/S).
Reverse transcription PCR and real time-quantitative-PCR (miRNA)
Detailed protocols are provided in the Supplemental information.
Reverse transcription PCR and real time-quantitative-PCR (mRNA)
Detailed protocols are provided in the Supplemental information.
miRNA Target Prediction
Bioinformatics prediction of target genes and miRNA binding sites was performed using a combination of TargetScan version 5.1 (http://www.targetscan.com), miRanda August 2010 release (http://www.microrna.org/), mirDB April 2009 (www.mirdb.org), RNA hybrid Version 2.1 (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/) and miRWalk 2.0 (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/).
miRNA mimic and inhibitor transfections
CB and PB ECFC-derived cells (1 × 105 cells) were plated onto 6 well plates in complete EGM-2 media (Lonza Biologics) overnight. The following day, media were removed and cells transfected with 10 nM mimic non-targeting control, and the relevant miRNA mimic or 10 and 50 nM inhibitor non-targeting control and the relevant miRNA inhibitor (Dharmacon, Lafayette, CO, USA) using Oligofectamine transfection reagent according the manufacturer’s protocol (Life Technologies). After 5 hr transfection, the transfection reagent was replaced with complete EGM-2 media (Lonza Biologics). Cells were harvested for other experiments at 48 hr after transfection.
siRNA transfection
CB ECFC-derived cells were cultured either at 500 cells/well in 96 well plates, or 0.5 × 105 cells in 12 well plates, and transfected with siRNAs at final concentrations of 5 nM using 0.2% Lipofectamine 2000 (Life Technologies). Two Silencer Select siRNAs (Life Technologies) for HMGB1 (s20254 and s20255) and HYOU1 (s20632 and s20634) were used, together with the siRNA negative control (Silencer Select Negative Control No. 2 siRNA).
Cell cycle analysis
After 48 hr transfection with miR-193a-3p, miR-34a or control mimics, CB ECFC-derived cells were stained with propidium iodide. The cells were fixed in ice-cold 70% ethanol for at least 16 hr at 4 °C. After fixation, the cells were incubated with 2 mg/ml RNase A (Sigma-Aldrich Ltd. Gillingham, Dorset, England) at 37 °C for 40 minutes. Then, 50 μg/ml propidium iodide (Sigma-Aldrich Ltd.) was added, and the cells were analyzed by flow cytometry. Single cells were selected using pulse area versus pulse height and obvious debris excluded by forward and side scatter gating of single cells, prior to propidium iodide based cell cycle analyses. The data are presented as the mean ± SEM of at least three independent experiments.
CyQUANT NF proliferation assay
To assess the proliferation of CB and PB ECFC-derived cells, the CyQUANT NF assay (Invitrogen Ltd., Paisley, Scotland) was used. Three to five different CB ECFC-derived cell batches were plated at 5 × 102 cells per well in EGM-2 media in 96 well black walled clear bottomed plates (Corning Life Sciences, Amsterdam, The Netherlands) and incubated overnight at 37 °C in 5% CO2. The following day, cells were transfected with miRNA mimics or miRNA inhibitors in parallel with respective mimic non-targeting/negative control or inhibitor non-targeting/negative control miRNAs (n = 5). Subsequently, the CyQUANT NF assay was performed on the same cells on a different plate to obtain fluorescence intensity values for Day 0 (on day of transfection). Cells were incubated with x1 CyQUANT NF dye binding solution for 60 mins at 37 °C and the fluorescence intensity was measured at excitation wavelength of 485 nm and emission detection at 530 nm using VICTOR fluorescence microplate reader (Perkin Elmer, Vienna, Austria). Supernatants from transfected cells were removed after 24 hr and replaced with fresh 10% FBS containing EGM-2 media. At Day 3, the CyQUANT NF assay was performed to obtain fluorescence intensity for Day 3. The proliferation index was calculated using the following formula: (Absorbance value Day 3/Absorbance value Day 0).
Matrigel and 3D co-culture vascular tubule assays
We used both the matrigel and co-culture assays to assess the effects of miRs on in vitro tubule formation by ECFC-derived cells. For the matrigel assay, CB and PB ECFC-derived cells were trypsinized 48 hr after transfection and 1.5 × 104 cells resuspended in complete EGM-2 media in a 96 well plate coated with 50 μl of Growth Factor Reduced Matrigel (GFR matrigel; BD Biosciences, Sunnyvale, CA, USA)40. Plates were incubated for 20 hr at 37 °C before photomicroscopy. Each image from each well was taken at x4 magnification using a Nikon Eclipse TE2000-U microscope (Nikon Ltd., London, England). Total tubule length was quantified using Wimasis Image Analysis online platform (www.wimasis.com)40.
For the tubule assay, unlabeled human bone marrow mesenchymal stromal cells (hBM MSCs) were co-cultured with transfected CB ECFC-derived cells (transduced with eGFP) in complete EGM-2 media containing VEGF and FGF-2 for up to 14 days as described previously11,18. For a standard assay, 1.6 × 103 CB ECFC-derived cells or HUVECs were co-cultured with 6.4 × 103 hBM MSCs on a 48-well plate in triplicate wells in complete EGM-2 media (Lonza Biologics). Media were changed every 3–4 days for the duration of the assay. Quantification of the resulting tubules was performed by dividing each well into quarters and photographing eGFP-expressing tubules in each quarter at x4 magnification on a Nikon Inverted TE300 microscope (Nikon UK Ltd.) fitted with a cooled CCD camera and IPLab v3.61 imaging software (Scanalytics; BD Biosciences). Images were processed in Adobe Photoshop 7 and tubule numbers, lengths, and number of junctions were quantified using Angiosys software (TCS Cellworks, Abingdon, England).
Transwell migration assay
CB and PB ECFC-derived cell migration was evaluated using a modified Boyden chamber assay15. Briefly, CB and PB ECFC-derived cells were detached using trypsin/EDTA, harvested by centrifugation, resuspended in EBM-2 media containing 0.5% FBS and counted. Then, 1 × 104 ECFC-derived cells in EBM-2 media containing 0.5% FBS were placed in the upper chamber of a modified Boyden chamber (Corning Life Sciences, Amsterdam, The Netherlands). Complete EGM-2 media were placed in the lower compartment of the chamber. After 4 hrs incubation at 37 °C, the lower side of the filter was washed with PBS and fixed with 4% paraformaldehyde. For visualisation under the fluorescence microscope, cells were stained with DAPI in mounting media (Life Technologies). Cells that migrated into the lower chamber were counted manually in four random microscopic fields (×4).
Luciferase assay
The pMirTarget-HMGB1-3′-UTR and pMirTarget-HYOU1-3′-UTR were obtained from Origene Technologies Inc. (Rockville, MD, USA). Transient transfections were performed where ~2 × 104 cells were plated in 96-well plates in alpha MEM containing 10% FBS with 1% penicillin-streptomycin 24 hr before transfection. Each vector, along with 10 nM of miR-193a-3p mimic or mimic non-targeting control (Dharmacon), was transfected into HEK293 cells with Lipofectamine 2000 (Invitrogen Ltd.) in Opti-mem media in a 96 well plates, according to the manufacturer’s instructions. Approximately 4–5 hr after transfection, alpha MEM containing 10% FBS with 1% penicillin streptomycin was added. Complete media were changed after 15 hr and incubated for an additional 24 hr, after which luciferase activity was assayed with the Dual-Glo Luciferase Reporter Assay System (Promega, Madison, WI, USA). The pMiRTarget luciferase reporter vector also encodes red fluorescent protein, which was used as a reporter for transfection monitoring and normalization. Relative luciferase activity (firefly luciferase/red fluorescent protein) from the cells transfected with miRNA mimic control was set to 1.0 as a reference. Each experiment was performed at least three times and in triplicate.
Western blot
Detailed protocols are provided in the Supplementary information.
Proteome array
Detailed protocols are provided in the Supplementary information.
Statistics
Experiments were repeated on at least three independent occasions or with at least three biological replicates. Data are presented as the mean ± S.E.M and analysed using unpaired two tailed Student’s t-test, or ANOVA with Dunnett’s post-hoc test or Sidak’s adjustment for multiple comparisons where appropriate, using Prism software version 7.0 (GraphPad software). Differences were considered significant where p ≤ 0.05 represented as *p ≤ 0.01 represented as ** and p ≤ 0.001 represented as ***.
Results
Differential miRNA expression between CB and PB ECFC-derived cells
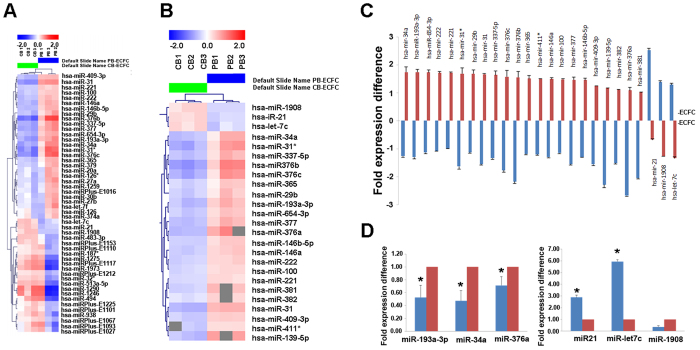
The miRNA profiling of CB and PB ECFC-derived cells identified 50 of 1273 miRNAs that passed the filtering criteria on variation across samples; standard deviation top 50 (Fig. 1A). Further statistical analysis identified twenty five miRNAs that were differentially expressed between CB and PB ECFC-derived cells and are represented in the heat map plot and graph (p < 0.01; Student’s two-tailed t-test; Fig. 1B and C). Three donors per ECFC source were used for miRNA profiling, however we further validated the expression of differentially regulated miRNAs identified in the miRCURY™ LNA Array using an additional 6 different donors of CB and 6 of PB ECFC-derived cells by real-time q-RT-PCR analysis. Three miRNAs (hsa-miR-193a-3p, hsa-miR-34a, hsa-miR-376a) upregulated in PB ECFC-derived cells from all donors tested were chosen for validation by real time q-RT-PCR based on preliminary screen of anti-proliferative activity using mimics. Three additional miRNAs (hsa-miR-21, hsa-let-7c and hsa-miR-1908) chosen from the original miRNA screen as upregulated miRNAs in CB ECFC-derived cells were also used for q-RT-PCR validation. Our results confirmed statistically significant differences between the CB and PB cells for all except one of these miRNAs, hsa-miR-1908. As illustrated in Fig. 1D, for hsa-miR-193a-3p, hsa-miR-34a, hsa-miR-376a, CB ECFC-derived cells showed reduced expression of these miRNAs (0.50 ± 0.03; 0.51 ± 0.02 and 0.39 ± 0.004 fold respectively; p < 0.01, one-way ANOVA) compared to PB ECFC-derived cells (normalized to 1). For hsa-miR-21 and hsa-let-7c, CB ECFC-derived cells showed increased expression of these miRNAs (2.06 ± 0.61; 5.24 ± 1.01 fold respectively; p < 0.05, one-way ANOVA) compared to PB ECFC-derived cells (normalized to 1).
Figure 1. Differentially expressed microRNAs between CB and PB ECFC-derived cells.
(A) A heat map of two-way clustering of miRNAs and six samples (CB ECFC-derived cells (n = 3) and PB ECFC-derived cells (n = 3)). The clustering was performed on all samples and on all microRNAs on log2 (Hy3/Hy5) ratios, which passed the filtering criteria on variation across samples. Each row represents one microRNA and each column represents one sample. The colour scale shown at the top illustrates the relative expression level of a microRNA across all samples: red colour represents an expression level above mean, blue colour represents expression level lower than the mean. (B) A heat map of 25 differentially regulated miRNAs between six different batches of CB ECFC-derived cells (n = 3) and PB ECFC-derived cells (n = 3). These miRNAs were identified based on two-tailed t-test calculated between the two sample groups with p-values < 0.01 in all donor samples. (C) Top 25 differentially regulated miRNAs plotted against fold expression difference values calculated for each group versus the pool of samples. MicroRNA’s miR-1908, miR-21 and let-7c were upregulated in CB ECFC-derived cells whilst miR-382, miR-221, miR-100, miR-365, miR-411*, miR-222, miR-29b, miR-146b-5p, miR-409-3p, miR-146a, miR-654-3p, miR-381, miR-337-5p, miR-34a, miR-377, miR-193a-3p, miR-31, miR-139-5p, miR-31*, miR-376c, miR-376a and miR-376b were upregulated in PB ECFC-derived cells. (D) q-RT-PCR validation of three miRNAs upregulated in PB ECFC-derived cells (hsa-miR-193a-3p, hsa-miR-34a and has-miR-376a) and three miRNAs upregulated in CB ECFC-derived cells (hsa-miR-21, hsa-let-7c and hsa-miR-1908) normalised to the RNU44 control (*p < 0.05; one-way ANOVA, Sidak’s multiple comparison).
miR193a-3p limits CB ECFC-derived cell proliferation and cell cycle progression
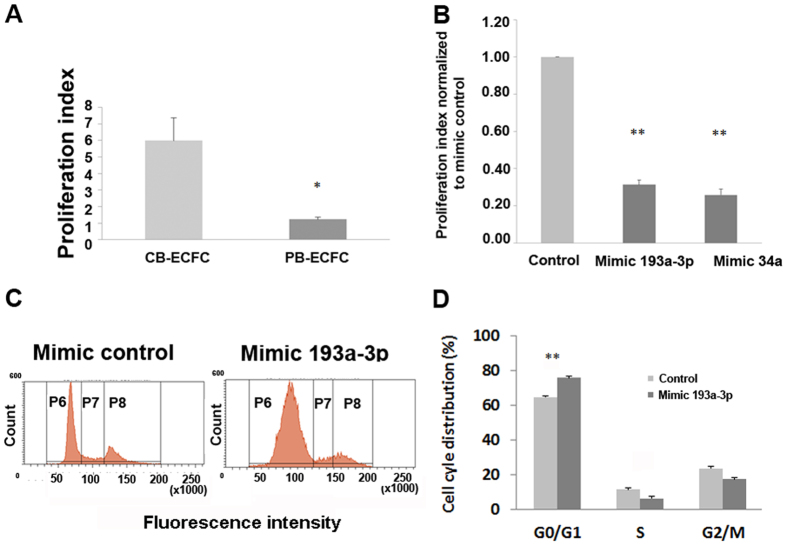
We have previously isolated and demonstrated the difference in functional capacity of adult PB and CB ECFC-derived cells5. These cells have similar cell surface phenotypes as determined by the FACS analyses and by their morphologies (Supplementary Fig. S1); however they differ in their proliferative ability. Notably, PB ECFC-derived cells were on average about 6 times less proliferative in our studies compared to CB ECFC-derived cells as measured by the CyQuant assay (p < 0.05; Student’s t-test; Fig. 2A).
Figure 2. Comparison of CB and PB ECFC-derived cell proliferation.
(A) CB ECFC-derived cells can proliferate approximately 5–6 times more than PB ECFC-derived cells. Proliferation was measured by the CyQuant assay in which the proliferation index was calculated based on fluorescence values at 72 hr normalised to those obtained for each sample on the day of transfection (0 hr). (B) Screen of 2 microRNAs chosen from the microRNA array where CB ECFC-derived cell proliferation was reduced when transfected with 10 nM of miR-193a-3p or miR-34a mimics. Proliferation was measured by the CyQuant proliferation assay at 72 hr post-transfection and data presented as proliferation index normalised to mimic control. (C) Effect of miR-193a-3p and control mimics on CB ECFC-derived cell cycle were assayed using propidium iodide by flow cytometry. CB ECFC-derived cells were transfected with miR-193a-3p or control mimics for 48 hr, fixed in 70% ethanol and stained with propidium iodide (Cell cycle stages; p6 = G0/G1; p7 = S and p8 = G2/M). (D) Cell percentages from flow cytometry represented in bar chart where overexpression of miR-193a-3p resulted in G1/S cell cycle arrest. The values presented are the mean ± S.E.M of three independent experiments (*p < 0.05; **p < 0.01; one-way ANOVA, Sidak’s multiple comparison).
Next, we determined which microRNAs of those differentially regulated in the miRNA arrays for CB compared to PB ECFC-derived cells affected cell proliferation. Initially, we screened 2 different batches of CB ECFC-derived cells transfected with 23 mimics/inhibitors based on those identified in Fig. 1C but excluding hsa-miR411* and hsa-miR-31*, then rescreened 8 of these mimics on CB ECFC-derived cells from 5 different donors and measured their proliferation rate 72 hr post-transfection (data not shown). From these screens, we found that, compared to the control mimic, miR-193a-3p and miR-34a mimics consistently reduced the proliferation of CB ECFC-derived cells (proliferation index compared to the normalized control mimic = 0.31 ± 0.02 and 0.26 ± 0.03 respectively; p < 0.01; one-way ANOVA; Fig. 2B). Transfection efficiency was confirmed using fluorescein tagged mimics (see Supplementary Fig. S2).
Concentrating on miR-193a-3p, and to gain insight into the mechanisms underlying the inhibition of proliferation by miR-193a-3p on human CB ECFC-derived cells, cell cycle analysis was performed (Fig. 2C and D). After transfection of the miR-193a-3p mimic into CB ECFC-derived cells, G1/S arrest was observed. miR-193a-3p overexpression significantly increased the proportion of cells in the G0/G1 phase (75.87% ± 2.92%; p < 0.01; one-way ANOVA) compared with negative control mimic transduced cells (G0/G1: 64.60% ± 2.49%).
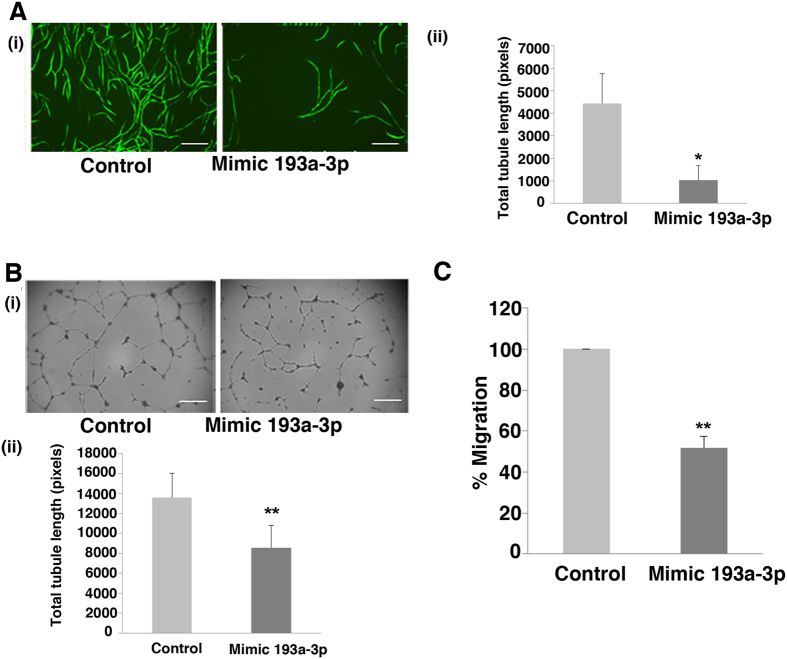
miR-193a-3p reduces CB ECFC-derived cell vascular tubule formation and cell migration
Next, we set out to determine if miR-193a-3p had an effect on ECFC-derived cell migration and vascular tubule formation. We treated 3 batches of CB ECFC-derived cells with control or miR-193a-3p mimic and confirmed that the latter reduced cell proliferation (Fig. 2B; p < 0.05; Student’s t-test). To assess the effect of this miR-193a-3p mimic on vascular tubule formation, we used an in vitro sandwich model, which involved co-culture of hBM MSC and CB ECFC-derived cells (Fig. 3A), and a matrigel assay with CB ECFC-derived cells alone (Fig. 3B). CB ECFC-derived cells transfected with miR-193a-3p mimic demonstrated a significant reduction in total tubule length in the co-culture (Fig. 3A; p < 0.05; Student’s t-test) and matrigel assays (Fig. 3B; p < 0.01; Student’s t-test) compared to the negative control mimic transfected cells. Statistically significant reductions in the numbers of junctions and tubules were also observed with the miR-193a-3p mimic when compared to the negative control mimic. In the vascular co-culture assay (Fig. 3A), the respective number of junctions and number of tubules were 86.1 ± 19.6 vs 8.1 ± 13.2 and 204.2 ± 51.9 vs 34.3 ± 19.8 for the negative control mimic vs miR-193a-3p mimic treatments (p < 0.05 for both; Student’s t-test). In the Matrigel CB-ECFC derived cell assay (Fig. 3B), the respective number of junctions and number of tubules were 83.7 ± 25.5 vs 34.8 ± 17.2 and 175.7 ± 49.7 vs 94.2 ± 31.6 for the negative control mimic vs miR-193a-3p mimic treatments (p < 0.05 for both; Student’s t-test). Furthermore, we observed around a 50% reduced migratory ability in the transwell assay (Fig. 3C) when miR193a-3p mimic was compared to CB ECFC-derived cells transfected with the negative control mimic (p < 0.01; Student’s t-test). These observations indicate that miR-193a-3p is not only an anti-proliferative miRNA, but also an anti-angiogenic/vasculogenic miRNA.
Figure 3. mir-193a-3p effect on CB ECFC-derived cell angiogenic functions.
(A) Transfected CB ECFC-derived cells were plated on hBM MSCs in a 96 well plate and images at x10 magnification were taken at Day 12 (endpoint). Effect of miR-193a-3p mimic on tubule formation of CB ECFC-derived cells were quantified using Angiosys software (n = 3, *p < 0.05). Scale bar, 100 μm. (B) Transfected CB ECFC-derived cells were plated onto growth factor reduced matrigel and allowed to migrate for 18 hr before images at x4 magnification were taken and quantified using Angiosys software (n = 3, p < 0.01). (C) CB ECFC-derived cells transfected with 10 nM miR-193a-3p mimic for 48 hr were suspended in 0.5% FBS in EBM-2 and placed in 8 μm transwells and allowed to migrate towards 10% FBS in EGM-2 growth media (**p < 0.01). The values presented are the mean ± S.E.M of three independent experiments (*p < 0.05; **p < 0.01; Student’s t-test).
miR-193a-3p inhibitor improves PB ECFC-derived cell proliferative, migratory and vascular tubule formation ability in vitro
Based on the anti-angiogenic/vasculogenic action of miR-193a-3p mimic on CB ECFC-derived cells, the effect of a miR193a-3p inhibitor on the relatively less-angiogenic and less proliferative PB ECFC-derived cells was explored. PB ECFC-derived cell numbers increased by 40% (140 ± 9% compared to the control normalized to 100%) over 72 hr post-transfection with 50 nM miR-193a-3p inhibitor, indicating that an inhibition of miR-193a-3p enhances the proliferative potential of adult PB ECFC-derived cells (Fig. 4A; p < 0.05; Student’s t-test). Similarly, miR-193a-3p inhibitor significantly improved tubule formation and cell migration by 41% and 43% respectively (141 ± 7%; 143 ± 10%, respectively) compared to inhibitor control transfected cells (Fig. 4B and C; p < 0.05 for both; Student’s t-test). These results illustrate the potential of the miR-193a-3p inhibitor to not only expand PB ECFC-derived cells in culture but to improve their migratory and vascular tubule formation capabilities.
Figure 4. mir-193a-3p inhibitor increases PB ECFC-derived cell proliferation and related angiogenic functions.
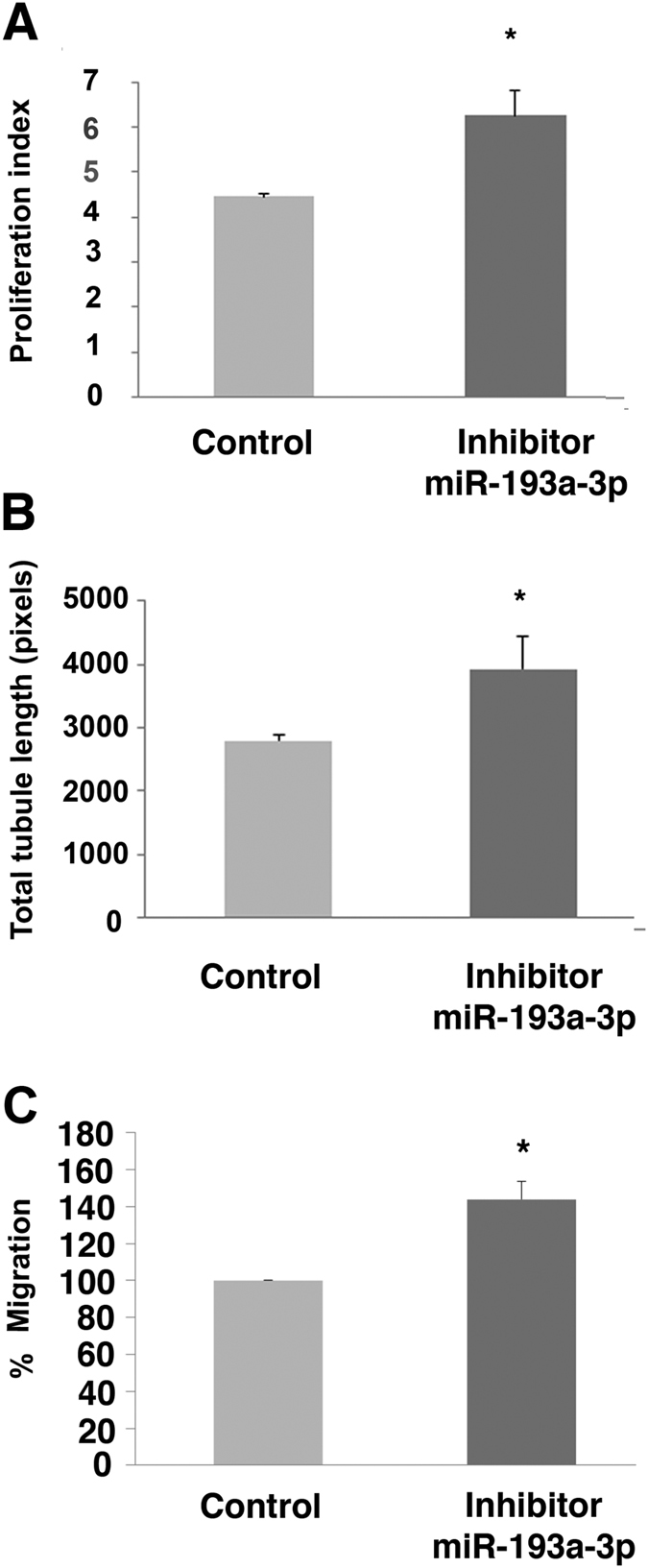
(A) PB ECFC-derived cells transfected with 50 nM miR-193a-3p inhibitor significantly increased cell proliferation compared to cells transfected with inhibitor control (CyQuant assay, n = 3). (B) PB ECFC-derived cells transfected with 50 nM miR-193a-3p inhibitor improved tubule formation in the co-culture assay. Effect of miRNA inhibitors on tubule formation of PB ECFC-derived cells was quantified using Angiosys software. (C) PB ECtFC-derived cells transfected with 50 nM miR-193a-3p inhibitor improved cell migration towards 10% FBS in EGM-2 growth media. The values presented are the mean ± S.E.M of three independent experiments (*p < 0.05; Student’s t-test).
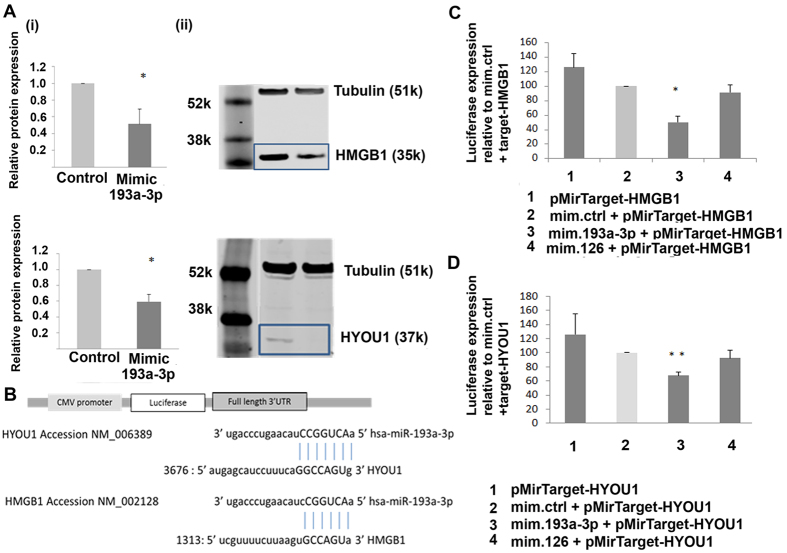
HYOU1 and HMGB1 are direct targets of miR-193a-3p
In order to understand the mechanism of action of miR-193a-3p on ECFC-derived cell function, mass spectrometry was performed on proteins extracted from non-targeting control and miR-193a-3p mimic treated CB ECFC-derived cells to identify potential targets of miR-193a-3p. Fifty one proteins were discovered to be significantly reduced in all 3 samples of miR-193a-3p mimic treated cells compared to the negative control (Table 1). These proteins were then cross checked using 5 target prediction sites to identify potential binding targets for miR-193a-3p, where 12 were predicted targets of miR-193a-3p (see Supplementary Table S1). HYOU1 and HMGB1 proteins with a fold change of 1.9 and 2.6 respectively (Table 1) were chosen for further investigation based on their known role in proliferation in other cell types, and as they have not been confirmed experimentally to be miR193a-3p targets. qPCR studies demonstrated statistically significant 1.88 and 2.33 fold higher levels of HYOU1 and HMGB1 respectively in CB ECFC-derived cells when compared to PB ECFC-derived cells (Supplementary Fig. S3). To validate the CB proteome array results, CB ECFC-derived cells were treated with miR-193a-3p mimic, and then protein lysates collected for Western blot analysis after 48 hr incubation. The results show that HMGB1 and HYOU1 protein levels were reduced approximately 2 fold in cells transfected with miR-193a-3p mimic compared to the control mimic treated cells (Fig. 5A).
Table 1. Differentially regulated proteins between mimic control and miR-193a-3p mimic CB ECFC-derived transfected cells.
| Accession number (UniProt) | Gene symbol | Peptide count | Anova (p) | Fold change | miR-ctrl abundance | miR-193 abundance |
|---|---|---|---|---|---|---|
| Q13418 | ILK | 1 | 2.45E-07 | 66.52 | 459.04 | 6.90 |
| P10155 | TROVE2 | 2 | 3.07E-05 | 18.14 | 823.07 | 45.37 |
| P10768 | ESD | 2 | 0.000177465 | 11.93 | 2060.45 | 172.78 |
| O00154 | ACOT7 | 1 | 9.15E-05 | 10.34 | 1220.57 | 118.02 |
| P35637 | FUS | 7 | 7.70E-11 | 10.11 | 47744.45 | 4721.85 |
| P17612 | PRKACA | 1 | 9.97E-05 | 9.87 | 412.52 | 41.81 |
| P62304 | SNRPE | 1 | 5.34E-06 | 6.75 | 6191.37 | 917.68 |
| P28289 | TMOD1 | 1 | 0.000119239 | 5.64 | 36804.73 | 6522.19 |
| Q9H6R0 | DHX33 | 1 | 8.58E-05 | 5.55 | 2326.65 | 419.27 |
| Q9UHY1 | NRBP1 | 1 | 8.81E-06 | 5.35 | 726.59 | 135.78 |
| Q8IVL6 | LEPREL2 | 2 | 6.52E-10 | 5.23 | 1394.90 | 266.83 |
| Q8WUM0 | NUP133 | 1 | 0.003479238 | 5.20 | 779.99 | 149.88 |
| Q9NQW7 | XPNPEP1 | 2 | 4.97E-06 | 4.86 | 2464.51 | 506.68 |
| Q9H814 | PHAX | 2 | 1.29E-05 | 4.58 | 354.99 | 77.47 |
| P21589 | NT5E | 2 | 9.55E-06 | 4.06 | 3832.37 | 944.69 |
| Q96HP0 | DOCK6 | 2 | 2.60E-07 | 3.87 | 891.69 | 230.35 |
| Q16630 | CPSF6 | 2 | 0.000146707 | 3.79 | 2407.28 | 635.46 |
| Q13283 | G3BP1 | 7 | 1.23E-07 | 3.08 | 22884.07 | 7419.24 |
| P31943 | HNRNPH1 | 8 | 3.61E-06 | 3.08 | 43520.52 | 14120.52 |
| Q13144 | EIF2B5 | 1 | 0.000122997 | 3.03 | 396.01 | 130.75 |
| Q9NZ08 | ERAP1 | 1 | 9.11E-07 | 3.03 | 720.36 | 237.90 |
| P56134 | ATP5J2 | 3 | 1.83E-07 | 2.90 | 9372.84 | 3236.49 |
| Q13617 | CUL2 | 3 | 5.02E-05 | 2.66 | 1224.81 | 459.72 |
| P09429;B2RPK0 | HMGB1 | 9 | 9.58E-12 | 2.61 | 259753.91 | 99477.90 |
| P0C0S5 | H2AFZ | 4 | 5.12E-09 | 2.57 | 106311.36 | 41414.44 |
| P34897;P34896 | SHMT1 | 8 | 3.80E-12 | 2.48 | 39153.06 | 15776.54 |
| P26583 | HMGB2 | 4 | 1.08E-07 | 2.47 | 6248.59 | 2526.31 |
| O00629 | KPNA4 | 1 | 3.88E-05 | 2.45 | 1161.75 | 474.34 |
| Q92900 | UPF1 | 1 | 6.31E-06 | 2.17 | 1053.91 | 485.88 |
| Q7L2H7 | EIF3M | 5 | 7.47E-06 | 2.09 | 5984.93 | 2862.97 |
| P78344 | EIF4G2 | 3 | 2.72E-07 | 2.09 | 2745.60 | 1316.24 |
| O76099 | OR7C1 | 1 | 1.39E-05 | 2.07 | 14322.84 | 6931.60 |
| Q9HB71 | CACYBP | 6 | 5.84E-05 | 2.06 | 27717.15 | 13463.32 |
| P16949 | STMN1 | 3 | 6.43E-08 | 2.04 | 9322.38 | 4565.74 |
| P33991 | MCM4 | 1 | 6.60E-09 | 2.03 | 3383.42 | 1663.71 |
| P63167;Q96FJ2 | DYNLL1 | 2 | 6.02E-07 | 1.98 | 16194.26 | 8159.44 |
| O14980 | XPO1 | 5 | 2.23E-06 | 1.93 | 9380.00 | 4853.49 |
| P09211 | GSTP1 | 14 | 1.33E-06 | 1.91 | 180260.98 | 94165.74 |
| Q9Y4L1 | HYOU1 | 16 | 4.67E-10 | 1.91 | 103604.44 | 54211.42 |
| P12004 | PCNA | 9 | 3.50E-08 | 1.87 | 63904.00 | 34098.36 |
| O95782;O94973 | AP2A1 | 2 | 9.14E-10 | 1.83 | 2036.05 | 1110.96 |
| Q6PIU2 | NCEH1 | 2 | 2.55E-09 | 1.83 | 5022.86 | 2747.12 |
| P40227 | CCT6A | 9 | 1.86E-06 | 1.81 | 26643.10 | 14703.73 |
| Q15365 | PCBP1 | 6 | 6.60E-12 | 1.80 | 96494.89 | 53709.70 |
| Q92973;O14787 | TNPO1 | 9 | 1.57E-05 | 1.74 | 38450.21 | 22089.48 |
| P46778 | RPL21 | 6 | 2.38E-08 | 1.72 | 87866.75 | 51097.70 |
| P18669;P15259;Q8N0Y7 | PGAM1 | 10 | 8.96E-07 | 1.71 | 153025.36 | 89602.32 |
| Q1KMD3;Q6ISU1 | HNRNPUL2 | 5 | 5.88E-08 | 1.65 | 11874.20 | 7177.37 |
| Q06323 | PSME1 | 3 | 3.69E-07 | 1.62 | 11671.43 | 7218.58 |
| O15145 | ARPC3 | 3 | 1.72E-06 | 1.59 | 22377.31 | 14076.88 |
| P53801 | PTTG1IP | 1 | 2.39E-07 | 1.56 | 12013.45 | 7717.98 |
Comparison of proteomic profile of mimic control and miR-193a-3p mimic CB ECFC-derived transfected cells. Fifty one differentially regulated proteins between the two conditions were identified and presented based on highest to lowest fold change between mimic control and miR-193a-3p treated samples.
Figure 5. HYOU1 and HMGB1 are direct targets of miR-193a-3p.
(A) Validation of protein expression of HMGB1 and HYOU1 in control and mimic treated CB ECFC-derived cells using Western blot. The values presented are the mean ± S.E.M of three independent experiments (*p < 0.05; Student’s t-test). Cropped blots were used here and uncropped images of blots are shown in Supplementary Fig. S6. (B) Luciferase constructs were used to test whether miR-193a-3p binds to HMGB1 and HYOU1. The 3′ UTRs of HMGB1 and HYOU1 were subcloned into the CMV-driven pMir-target luciferase vector (Origene). (C) Luciferase activity of plasmid containing firefly luciferase associated with 3′ region of HMGB1 co-transfected separately with miR-193a-3p mimic (10 nM) was reduced. Co-transfection of HMGB1 with a non-targeted miR-126 did not cause a significant decrease in firefly luciferase activity. Data were normalised to the activity of RFP expressed by the HMGB1 plasmid. (D) Luciferase activity of plasmid containing firefly luciferase associated with 3′ region of HYOU1 co-transfected separately with miR-193a-3p mimic (10 nM) was reduced. Co-transfection of HYOU1 with a non-targeted miR-126 did not cause a significant decrease in firefly luciferase activity. Data were normalised to the activity of RFP expressed by the HYOU1 plasmid. The values presented are the mean ± S.E.M of three independent experiments (*p < 0.05; **p < 0.01; one-way ANOVA, Dunnett’s multiple comparison). (Abbreviations: RPP, red fluorescent protein).
To demonstrate the direct interactions of miR-193a-3p with HMGB1 and HYOU1 3′UTRs harbouring the potential binding site to miR-193a-3p, constructs of these regions were cloned into the pMir-Target plasmid downstream of a luciferase reporter gene (Fig. 5B). A significant decrease of 50% and 32% (to 50 ± 8% and 68 ± 5% respectively of the normalized mimic control) in luciferase activity was observed in cells transfected with pMirTarget-HMGB1 and pMirTarget-HYOU1 constructs respectively when treated with 10 nM miR-193a-3p mimic, relative to the same concentration of a control mimic (Fig. 5C and D; p < 0.05 and p < 0.01 respectively; one-way ANOVA). As an additional control, there was no significant difference between cells co-transfected with the target constructs and miR-126, a negative control miRNA validated to not target either HMGB1 or HYOU1 (Fig. 5C and D; 92 ± 11%; p = 0.28 and 92 ± 9%; p = 0.11; one-way ANOVA). Empty vector controls are shown in Supplementary Fig. S4. This provides strong evidence of a direct molecular binding interaction between miR-193a-3p and the 3′UTRs of HMGB1 and HYOU1, indicating that these genes are novel direct targets of miR-193a-3p.
HMGB1 siRNA suppresses CB ECFC-derived cell proliferation and migration
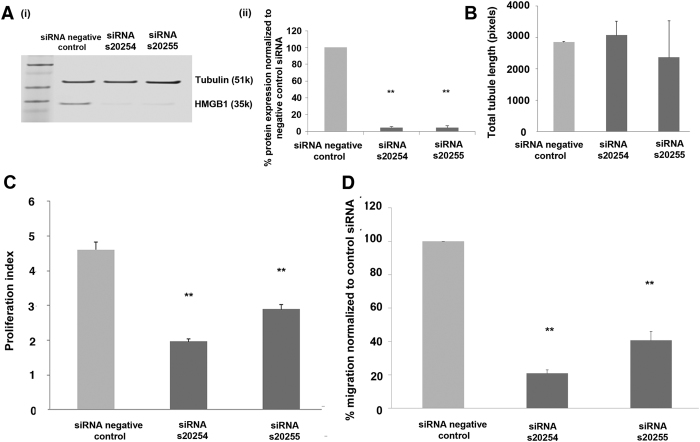
Following the demonstration that HYOU1 and HMGB1 are direct targets of miR-193a-3p, siRNA experiments were carried out to determine whether these targets affect CB ECFC-derived cell angiogenic/vasculogenic functions. CB ECFC-derived cells were transfected with HMGB1 and HYOU1 specific siRNAs. Protein knockdown of approximately 90% was achieved for HMGB1 s20254 and s20255 siRNAs (Fig. 6A; p < 0.01; one-way ANOVA) but only approximately 20–30% for siRNA HYOU1 s20632 and s20634 siRNAs (s20632; p < 0.05; s20634; p < 0.05; one-way ANOVA) (see Supplementary Fig. S5). Since HMGB1 protein was efficiently suppressed using these siRNAs, the functional characteristics of CB ECFC-derived cells upon knockdown of this novel target were examined. HMGB1 siRNAs s20254 and s20255 significantly affected the number of CB ECFC-derived cells generated over 72 hr (Fig. 6C; p < 0.01; one-way ANOVA). Similar results were observed where HMGB1 siRNAs s20254 and s20255 affected CB ECFC-derived cell migration towards 10% EGM-2 growth media containing factors such as VEGF and bFGF (Fig. 6D; p < 0.01; one-way ANOVA). However, HMGB1 siRNAs s20254 and s20255 did not significantly affect tubule formation in the matrigel assay when compared to the control (Fig. 6B; p = 0.87; one-way ANOVA). This indicates that knocking down HMGB1 affects endothelial cell migration towards specific factors and also proliferation but is not necessary for their ability to form vascular tubules in matrigel.
Figure 6. HMGB1 knockdown affects CB ECFC-derived cell proliferation and migratory abilities.
(A) (i) Western blot of HMGB1 protein in CB ECFC-derived cells following transfection of 2 different siRNAs for HMGB1, each at a final concentration of 5 nM. (ii) Western blot quantification of HMGB1 protein levels where the target protein expression was normalised to tubulin expression in each sample. Results show target gene protein levels as a percentage of the negative control siRNA-transfected sample. Cropped blots were used here and uncropped images of blots are shown in Supplementary Fig. S7. (B) No significant effect of HMGB1 siRNA knockdown on the tubule formatting ability of CB ECFC-derived cells. Cells treated with a negative control siRNA, HMGB1 s20254 siRNA, or HMGB1 s20255 siRNA at a final concentration of 5 nM were subjected to a matrigel assay for 18 hr, 48 hr post-transfection. (C) The effect of HMGB1 siRNA knockdown on the proliferative and/or survival ability of CB ECFC-derived cells. Cells treated with either a negative control siRNA, HMGB1 s20254 siRNA, or HMGB1 s20255 siRNA at a final concentration of 5 nM were subjected to a CyQuant proliferation assay at 72 hr to measure proliferation. Fluorescence values were normalised to those obtained on the day of transfection for each sample to obtain a proliferation index for each condition. (D) The effect of HMGB1 siRNA knockdown on the migration ability of CB ECFC-derived cells. Cells treated with a negative control siRNA, HMGB1 s20254 siRNA, or HMGB1 s20255 siRNA at a final concentration of 5 nM were subjected to a transwell assay for 5 hr towards 10% FBS in EGM-2 growth media following 48 hr post-transfection. The values presented are the mean ± S.E.M of three independent experiments (**p < 0.01; one-way ANOVA, Dunnett’s multiple comparison).
Unlike HMGB1 siRNAs, HYOU1 siRNAs did not affect CB ECFC-derived cell proliferation under the conditions used for cell culture (s20632; p = 0.914; s20634; p = 0.813; one-way ANOVA) nor migration (s20632; p = 0.528; s20634; p = 0.343; one-way ANOVA; Supplementary Fig. S5). These siRNA results demonstrate that HMGB1 plays a role in both CB ECFC-derived cell migratory and proliferative ability, which mimics the same effects of miR-193a-3p on CB ECFC-derived cells shown in Fig. 3.
Discussion
A number of microRNAs have already been shown to regulate endothelial cell functions, such as cell proliferation, senescence, migration, differentiation and vascular tubule formation33,34,37,38,41. However, in the past there has been considerable controversy in defining true circulating endothelial lineage progenitor cells. It is now well established that cells which promote revascularization can be identified in the circulation as proangiogenic myeloid/hematopoietic cells (formerly termed early EPCs) and endothelial colony forming cells or ECFCs (formerly termed late EPCs) based on whether they belong to the hematopoietic or endothelial lineages1,42. Additionally, microRNA analyses comparing circulating peripheral blood proangiogenic cells with umbilical cord ECFC-derived cells (HUVEC) have demonstrated mutually exclusive miRNA profiles43,44.
We and others have observed that circulating CB and PB ECFC-derived cells exhibit differences in cell proliferation and vascular tubule formation3,5,6,11. Therefore, in this study, we investigated differences in the post-transcriptional regulation of circulating CB and PB ECFC-derived cells by comparing their microRNA profiles. We identified 25 microRNAs differentially regulated between CB and PB ECFC-derived cells. Within the 22 miRNAs upregulated in PB ECFC-derived cells, we detected miRNAs with both known anti-angiogenic properties (e.g. miR-221, miR-222 and miR-34a)33,34,37,45 or with undefined properties. Of the latter, the miR-193a-3p mimic significantly reduced the proliferative and vasculogenic/angiogenic functions of CB ECFC-derived cells and this miRNA was chosen for further study. Subsequently, miR-193a-3p inhibition in PB ECFC-derived cells resulted in a statistically significant increase in the proliferation rate of these cells. Other studies have examined the role of miR193a (-3p) in tumor cells. For example, in agreement with the effects we observed on cell proliferation, Nakano et al.46 identified miR-193a as the strongest candidate affecting cell proliferation and death in A2780 ovarian cancer cells after performing a gain-of-function miRNA screen. Similarly, miR-193a(-3p) has been implicated in regulating proliferation of different tumor cell types, for example, by targeting c-kit in acute myeloid leukemia cells47, CCND1, ERBB4, Mcl-1, STMN1 and KRAS in ovarian cancer cells46 and ERBB4 in lung cancer cells48.
To understand the potential mechanisms by which miR-193a-3p exerts its anti-proliferative and anti-angiogenic effect on ECFC-derived cells, we performed proteomic analysis on CB ECFC-derived cells that were treated with and without miR-193a-3p mimics and validated our proteome results by using target prediction analyses and luciferase reporter assays. Consequently, we identified HMGB1 and HYOU1 as potential targets of miR-193a-3p in ECFC-derived cells. More importantly, our siRNA knockdown studies resulted in significant effects on HMGB1 related vasculogenic/angiogenic functions. High-mobility group box-1 (HMGB1) protein is a transcription factor that has a dual role as a mobile chromatin protein and as a cytokine49. Amongst its functions, HMGB1 has been implicated in tissue repair and regeneration by promoting chemotaxis of stem/progenitor cells, stimulating the proliferation of tissue resident stem/progenitor cells including endothelial cells and vascular smooth muscle cells, acting as a proangiogenic cytokine and promoting secretion of proangiogenic cytokines such as VEGF and CXCL850,51,52,53,54. Interestingly, HMGB1 has been reported to promote endothelial recruitment via integrin activation50 and endothelial proliferation, migration and angiogenesis via RAGE and/or TLR4 signalling pathways, thus effecting tissue repair55,56,57,58,59,60,61,62,63. Additionally, recent studies using nuclear magnetic resonance and surface plasmon resonance analyses have demonstrated that upon inflammation HMGB1 can form a heterocomplex with CXCL12 which will then signal via CXCR458. HMGB1 binding to CXCL12 induces changes in residues 3–12 of the latter, which are fundamental for the triggering of its receptor, CXCR458. During the initial phase of tissue injury, the CXCL12–HMGB1 complex has been reported to mediate the recruitment of mononuclear cells, thereby cooperating in promoting cell migration in vitro and in vivo58. We have shown previously that CXCR4-CXCL12 interactions in ECFC-derived cell and mesenchymal stromal cell co-cultures are important in vascular tubule formation11, both for endothelial tip-like cell migration and the integration of ECFC-derived cells into developing vascular networks.
Our results have demonstrated for the first time that miR-193a-3p directly suppresses HMGB1 in CB ECFC-derived cells. When HMGB1 is silenced, this affects both vasculogenesis/angiogenesis as shown in both our and other studies50,55,56,57,59,60,61,62,63,64. Given this, other molecules identified in our proteomic screen and/or as potential targets for miR193a-3p may also play a role in modulating angiogenesis. Exemplars include firstly ILK (integrin-linked kinase), which has been reported to promote angiogenesis and to be involved in endothelial cell migration, tubule formation and proliferation in vitro65,66,67,68. Secondly, STMN1 (stathmin) regulates the proliferation, migration, and network formation of cultured endothelial cells as well as angiogenesis in vivo69. Thirdly, CUL2 (cullin 2) is a scaffold protein critical for the assembly of the ubiquitin ligase system and therefore for Hif-alpha stabilization70. Hif-alpha proteins are known to have a role in promoting angiogenesis71. Finally, CD73 (NT5E) has been reported to be involved in tumour angiogenesis72,73,74. These molecules warrant further investigation, which is beyond the scope of this current manuscript, as targets for miR193a-3p.
Conclusion
We have demonstrated that miR-193a-3p regulates the proliferation and the vasculogenic/angiogenic properties of ECFC-derived cells, at least in part, by direct suppression of HMGB1. Therefore, targeting miR-193a-3p in ECFC-derived cells may both enhance the numbers of these cells and improve their vasculo-/angio-genic function.
Data Access
MicroRNA array files and analysis has been have been deposited in NCBI’s Gene Expression Omnibus75 and are accessible through GEO Series accession number GSE81574 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE81574).
Additional Information
How to cite this article: Khoo, C. P. et al. miR-193a-3p interaction with HMGB1 downregulates human endothelial cell proliferation and migration. Sci. Rep. 7, 44137; doi: 10.1038/srep44137 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
The authors are grateful for the support offered by the Cancer Research UK, Breast Cancer Research Foundation and Oxford NHS Biomedical Research Centre to A.L.H. The authors would like to thank Mrs Sandy Britt and Jan Walton for the cord blood collection. These studies have received grant support from the National Institute of Health Research under its Programme Grants Scheme (RP-PG-0310-1003 and RP-PG-0310-1001), Restore: burns and wound healing trust, BBSRC (BRIC grant), the NHSBT Trust Fund and NHS Blood and Transplant. The views expressed in this publication are those of the authors and not necessarily those of NHS, NHS Blood and Transplant, NIHR or the Department of Health.
Footnotes
The authors declare no competing financial interests.
Author Contributions C.P.K. performed all experiments and analyzed the data. C.P.K., M.G.R., G.T., B.K., A.L.H. and S.M.W. conceived the study and revised the manuscript. M.G.R., J.B.S. and R.K. performed experiments, collected and assembled data, performed data analysis and interpreted data. A.L.H. and S.M.W. provided financial support. All authors approved the final manuscript.
References
- Basile D. P. & Yoder M. C. Circulating and tissue resident endothelial progenitor cells. J Cell Physiol 229, 10–16 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram D. A. et al. Vessel wall-derived endothelial cells rapidly proliferate because they contain a complete hierarchy of endothelial progenitor cells. Blood 105, 2783–2786 (2005). [DOI] [PubMed] [Google Scholar]
- Ingram D. A. et al. Identification of a novel hierarchy of endothelial progenitor cells using human peripheral and umbilical cord blood. Blood 104, 2752–2760 (2004). [DOI] [PubMed] [Google Scholar]
- Solomon I. et al. Functional Differences Between Placental Micro- and Macrovascular Endothelial Colony-Forming Cells. Stem Cells Transl Med (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanassopoulos A. et al. Microvessel networks [corrected] pre-formed in artificial clinical grade dermal substitutes in vitro using cells from haematopoietic tissues. Burns 38, 691–701 (2012). [DOI] [PubMed] [Google Scholar]
- Coldwell K. E. et al. Effects of obstetric factors and storage temperatures on the yield of endothelial colony forming cells from umbilical cord blood. Angiogenesis 14, 381–392 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong H. T. et al. Pulmonary artery endothelium resident endothelial colony-forming cells in pulmonary arterial hypertension. Pulm Circ 1, 475–486 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H., Lee S. H., Choi S. H., Asahara T. & Kwon S. M. The sulfated polysaccharide fucoidan rescues senescence of endothelial colony-forming cells for ischemic repair. Stem Cells 33, 1939–1951 (2015). [DOI] [PubMed] [Google Scholar]
- Lin R. Z. et al. Human endothelial colony-forming cells serve as trophic mediators for mesenchymal stem cell engraftment via paracrine signaling. Proc Natl Acad Sci USA 111, 10137–10142 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melero-Martin J. M. et al. Engineering robust and functional vascular networks in vivo with human adult and cord blood-derived progenitor cells. Circ Res 103, 194–202 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newey S. E. et al. The hematopoietic chemokine CXCL12 promotes integration of human endothelial colony forming cell-derived cells into immature vessel networks. Stem Cells Dev 23, 2730–2743 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijmeh H. et al. High proliferative potential endothelial colony-forming cells contribute to hypoxia-induced pulmonary artery vasa vasorum neovascularization. Am J Physiol Lung Cell Mol Physiol 306, L661–671 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel J. et al. Prospective surface marker-based isolation and expansion of fetal endothelial colony-forming cells from human term placenta. Stem Cells Transl Med 2, 839–847 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel J. et al. Self-renewal and high proliferative colony forming capacity of late-outgrowth endothelial progenitors is regulated by cyclin-dependent kinase inhibitors driven by notch signaling. Stem Cells 34, 902–912 (2016). [DOI] [PubMed] [Google Scholar]
- Roubelakis M. G., Tsaknakis G., Pappa K. I., Anagnou N. P. & Watt S. M. Spindle shaped human mesenchymal stem/stromal cells from amniotic fluid promote neovascularization. PLoS One 8, e54747 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker N. G. et al. A chemically defined carrier for the delivery of human mesenchymal stem/stromal cells to skin wounds. Tissue Eng Part C Methods 18, 143–155 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J. A. et al. Distinct contribution of human cord blood-derived endothelial colony forming cells to liver and gut in a fetal sheep model. Hepatology 56, 1086–1096 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. et al. The impact of proliferative potential of umbilical cord-derived endothelial progenitor cells and hypoxia on vascular tubule formation in vitro. Stem Cells Dev 18, 359–375 (2009). [DOI] [PubMed] [Google Scholar]
- Zhou B. et al. A novel function for the haemopoietic supportive murine bone marrow MS-5 mesenchymal stromal cell line in promoting human vasculogenesis and angiogenesis. Br J Haematol 157, 299–311 (2012). [DOI] [PubMed] [Google Scholar]
- Alphonse R. S. et al. Existence, functional impairment, and lung repair potential of endothelial colony-forming cells in oxygen-induced arrested alveolar growth. Circulation 129, 2144–2157 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto K. et al. Intravenous administration of endothelial colony-forming cells overexpressing integrin beta1 augments angiogenesis in ischemic legs. Stem Cells Transl Med 5, 218–226 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo S. C. et al. WKYMVm-induced activation of formyl peptide receptor 2 stimulates ischemic neovasculogenesis by promoting homing of endothelial colony-forming cells. Stem Cells 32, 779–790 (2013). [DOI] [PubMed] [Google Scholar]
- Huang X. T. et al. Intracerebroventricular transplantation of ex vivo expanded endothelial colony-forming cells restores blood-brain barrier integrity and promotes angiogenesis of mice with traumatic brain injury. J Neurotrauma 30, 2080–2088 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang K. T., Coggins M., Xiao C., Rosenzweig A. & Bischoff J. Human vasculogenic cells form functional blood vessels and mitigate adverse remodeling after ischemia reperfusion injury in rats. Angiogenesis 16, 773–784 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo C. P., Pozzilli P. & Alison M. R. Endothelial progenitor cells and their potential therapeutic applications. Regen Med 3, 863–876 (2008). [DOI] [PubMed] [Google Scholar]
- Lee S. H. et al. Genistein promotes endothelial colony-forming cell (ECFC) bioactivities and cardiac regeneration in myocardial infarction. PLoS One 9, e96155 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. H. et al. Selective interference targeting of Lnk in umbilical cord-derived late endothelial progenitor cells improves vascular repair, following hind limb ischemic injury, via regulation of JAK2/STAT3 signaling. Stem Cells 33, 1490–1500 (2015). [DOI] [PubMed] [Google Scholar]
- Moubarik C. et al. Transplanted late outgrowth endothelial progenitor cells as cell therapy product for stroke. Stem Cell Rev 7, 208–220 (2011). [DOI] [PubMed] [Google Scholar]
- Schwarz T. M. et al. Vascular incorporation of endothelial colony-forming cells is essential for functional recovery of murine ischemic tissue following cell therapy. Arterioscler Thromb Vasc Biol 32, e13–21 (2012). [DOI] [PubMed] [Google Scholar]
- Stroncek J. D., Ren L. C., Klitzman B. & Reichert W. M. Patient-derived endothelial progenitor cells improve vascular graft patency in a rodent model. Acta Biomater 8, 201–208 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt S. M. et al. The angiogenic properties of mesenchymal stem/stromal cells and their therapeutic potential. Br Med Bull 108, 25–53 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelley W. C. et al. Changes in the frequency and in vivo vessel-forming ability of rhesus monkey circulating endothelial colony-forming cells across the lifespan (birth to aged). Pediatric Res 71, 156–161, doi: 10.1038/pr.2011.22 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu K. et al. MicroRNAs: Key regulators of endothelial progenitor cell functions. Clin Chim Acta 448, 65–73 (2015). [DOI] [PubMed] [Google Scholar]
- Anand S. A brief primer on microRNAs and their roles in angiogenesis. Vasc Cell 5, 2 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder M. C. Human endothelial progenitor cells. Cold Spring Harb Perspect Med 2, a006692 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebele C. et al. Members of the microRNA-17-92 cluster exhibit a cell-intrinsic antiangiogenic function in endothelial cells. Blood 115, 4944–4950 (2010). [DOI] [PubMed] [Google Scholar]
- Ito T., Yagi S. & Yamakuchi M. MicroRNA-34a regulation of endothelial senescence. Biochem Biophys Res Commun 398, 735–740 (2010). [DOI] [PubMed] [Google Scholar]
- Santulli G. microRNAs and Endothelial (Dys) Function. J Cell Physiol 231, 1638–1644 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welten S. M. et al. Inhibition of 14q32 MicroRNAs miR-329, miR-487b, miR-494, and miR-495 increases neovascularization and blood flow recovery after ischemia. Circ Res 115, 696–708 (2014). [DOI] [PubMed] [Google Scholar]
- Khoo C. P., Micklem K. & Watt S. M. A comparison of methods for quantifying angiogenesis in the Matrigel assay in vitro. Tissue Eng Part C Methods 17, 895–906 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder M. C. Judging a proangiogenic cell by its cover. Blood 114, 756–757 (2009). [DOI] [PubMed] [Google Scholar]
- Goretti E. et al. MicroRNA-16 affects key functions of human endothelial progenitor cells. J Leuk Biol 93, 645–655 (2013). [DOI] [PubMed] [Google Scholar]
- Yoder M. C. Editorial: Early and late endothelial progenitor cells are miR-tually exclusive. J Leuk Biol 93, 639–641 (2013). [DOI] [PubMed] [Google Scholar]
- Zhang Q., Kandic I. & Kutryk M. J. Dysregulation of angiogenesis-related microRNAs in endothelial progenitor cells from patients with coronary artery disease. Biochem Biophys Res Commun 405, 42–46 (2011). [DOI] [PubMed] [Google Scholar]
- Nakano H., Yamada Y., Miyazawa T. & Yoshida T. Gain-of-function microRNA screens identify miR-193a regulating proliferation and apoptosis in epithelial ovarian cancer cells. Int J Oncol 42, 1875–1882 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H. et al. miR-193a-3p functions as a tumor suppressor in lung cancer by down-regulating ERBB4. J Biol Chem 290, 926–940 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams M. et al. miR-193a-3p is a potential tumor suppressor in malignant pleural mesothelioma. Oncotarget 6, 23480–23495 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandolfi F., Altamura S., Frosali S. & Conti P. Key role of DAMP in inflammation, cancer, and tissue repair. Clin Ther. 38, 1017–1028 (2016). [DOI] [PubMed] [Google Scholar]
- Chavakis E. et al. High-mobility group box 1 activates integrin-dependent homing of endothelial progenitor cells. Circ Res 100, 204–212 (2007). [DOI] [PubMed] [Google Scholar]
- Degryse B. et al. The high mobility group (HMG) boxes of the nuclear protein HMG1 induce chemotaxis and cytoskeleton reorganization in rat smooth muscle cells. J Cell Biol 152, 1197–1206 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitola S. et al. Cutting edge: extracellular high mobility group box-1 protein is a proangiogenic cytokine. J Immunol 176, 12–15 (2006). [DOI] [PubMed] [Google Scholar]
- Palumbo R. et al. Extracellular HMGB1, a signal of tissue damage, induces mesoangioblast migration and proliferation. J Cell Biol 164, 441–449 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., O’Hearn S., Kavalukas S. L. & Barbul A. Role of high mobility group box 1 (HMGB1) in wound healing. J Surg Res 176, 343–347 (2012). [DOI] [PubMed] [Google Scholar]
- Lei C. et al. HMGB1 may act via RAGE to promote angiogenesis in the later phase after intracerebral hemorrhage. Neuroscience 295, 39–47 (2015). [DOI] [PubMed] [Google Scholar]
- Lin Q. et al. High-mobility group box-1 mediates toll-like receptor 4-dependent angiogenesis. Arterioscler Thromb Vasc Biol 31, 1024–1032 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdev U. et al. High mobility group box 1 promotes endothelial cell angiogenic behavior in vitro and improves muscle perfusion in vivo in response to ischemic injury. Journal of vascular surgery 55, 180–191 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiraldi M. et al. HMGB1 promotes recruitment of inflammatory cells to damaged tissues by forming a complex with CXCL12 and signaling via CXCR4. J Exp Med 209, 551–563 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlueter C. et al. Angiogenetic signaling through hypoxia: HMGB1: an angiogenetic switch molecule. Am J Pathol 166, 1259–1263 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straino S. et al. High-mobility group box 1 protein in human and murine skin: involvement in wound healing. J Invest Dermatol 128, 1545–1553 (2008). [DOI] [PubMed] [Google Scholar]
- Cottone L. et al. Leukocytes recruited by tumor-derived HMGB1 sustain peritoneal carcinomatosis. Oncoimmunol. 5, e1122860 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Beijnum J. R. et al. Tumor angiogenesis is enforced by autocrine regulation of high-mobility group box 1. Oncogene 32, 363–374 (2013). [DOI] [PubMed] [Google Scholar]
- Wang W. et al. Overexpression of high mobility group box 1 and 2 is associated with the progression and angiogenesis of human bladder carcinoma. Oncol Lett. 5, 884–888 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giffin L., Yan F., Ben Major M. & Damania B. Modulation of Kaposi’s sarcoma-associated herpesvirus interleukin-6 function by hypoxia-upregulated protein 1. J Virol 88, 9429–9441 (2014). Erratum in: J Virol. 88, 12932 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C. et al. Regulation of tumor angiogenesis by integrin-linked kinase (ILK). Cancer Cell; 5, 79–90 (2004). [DOI] [PubMed] [Google Scholar]
- Xie W. et al. Targeting of integrin-linked kinase with small interfering RNA inhibits VEGF-induced angiogenesis in retinal endothelial cells. Ophthalmic Res 49, 139–149 (2013). [DOI] [PubMed] [Google Scholar]
- Guo L., Yu W., Li X., Zhao G. & He P. Targeting of integrin-linked kinase with a small interfering RNA inhibits endothelial cell migration, proliferation and tube formation in vitro. Ophthalmic Res 42, 213–220 (2009). [DOI] [PubMed] [Google Scholar]
- Li Y. et al. Integrin-Linked Kinase Senses Hypoxia During Scar Angiogenesis. Int J Low Extrem Wounds. [Epub ahead of print] (2016). [DOI] [PubMed]
- Miyashita H., Kanemura M., Yamazaki T., Abe M. & Sato Y. Vascular endothelial zinc finger 1 is involved in the regulation of angiogenesis: possible contribution of stathmin/OP18 as a downstream target gene. Arterioscler Thromb Vasc Biol 24, 878–884 (2004). [DOI] [PubMed] [Google Scholar]
- Ghosh G. et al. Hypoxia-induced microRNA-424 expression in human endothelial cells regulates HIF-α isoforms and promotes angiogenesis. J Clin Invest. 120, 4141–4154 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaGory E. L. & Giaccia A. J. The ever-expanding role of HIF in tumour and stromal biology. Nat Cell Biol 8, 356–365 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. et al. Ecto-5′-nucleotidase (CD73) promotes tumor angiogenesis. Clin Exp Metastasis 30, 671–680 (2013). [DOI] [PubMed] [Google Scholar]
- Koszałka P. et al. Specific activation of A3, A2A and A1 adenosine receptors in CD73-knockout mice affects B16F10 melanoma growth, neovascularization, angiogenesis and macrophage infiltration. PLoS One 11, e0151420 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszałka P. et al. Inhibition of CD73 stimulates the migration and invasion of B16F10 melanoma cells in vitro, but results in impaired angiogenesis and reduced melanoma growth in vivo. Oncol Rep. 31, 819–827 (2014). [DOI] [PubMed] [Google Scholar]
- Edgar R., Domrachev M. & Lash A. E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucl Acids Res 30, 207–210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.