Abstract
The mechanosensitive channels of large conductance (MscL) are bacterial membrane proteins that serve as last resort emergency release valves in case of severe osmotic downshock. Sensing bilayer tension, MscL channels are sensitive to changes in the bilayer environment and are, therefore, an ideal test case for exploring membrane protein coupling. Here, we use high-throughput coarse-grained molecular dynamics simulations to characterize MscL gating kinetics in different bilayer environments under the influence of alcohols. We performed over five hundred simulations to obtain sufficient statistics to reveal the subtle effects of changes in the membrane environment on MscL gating. MscL opening times were found to increase with the addition of the straight-chain alcohols ethanol, octanol, and to some extent dodecanol but not with hexadecanol. Increasing concentration of octanol increased the impeding effect, but only up to 10–20 mol %. Our in silico predictions were experimentally confirmed using reconstituted MscL in a liposomal fluorescent efflux assay. Our combined data reveal that the effect of alcohols on MscL gating arises not through specific binding sites but through a combination of the alcohol-induced changes to a number of bilayer properties and their alteration of the MscL–bilayer interface. Our work provides a key example of how extensive molecular simulations can be used to predict the functional modification of membrane proteins by subtle changes in their bilayer environment.
Introduction
Membrane proteins are embedded in a lipid bilayer, and changes in the bilayer environment can affect their function. Hydrophobic bilayer–membrane protein interactions cause the lipid bilayer to adjust locally to embedded proteins,1−3 and vice versa, coupling protein conformation to bilayer properties. To increase their ability to signal, many membrane proteins are posed in a lipid environment close to their conformational transition (e.g., opening/closing), rendering them sensitive to even modest changes in bilayer properties. Thus, small changes in lipid composition or the addition of membrane absorbed drugs or small molecules can lead to changes in membrane protein function without specific binding, by either changing bilayer properties and/or altering the protein–bilayer interface.4−9 However, it remains a challenge to determine which bilayer property changes are the most relevant8 and what the mechanism is at the molecular level.
The mechanosensitive channel of large conductance (MscL) is a quintessential choice for exploring membrane channel functional modification through changes in bilayer environment, as its function is to sense changes in bilayer tension. MscL protects bacteria from lysis upon acute osmotic downshock by opening a large mostly unselective pore, releasing ions and small solutes, and thereby relieving the cytoplasm of osmotic stress.10,11 The crystal structures of Mycobacterium tuberculosis MscL in its closed states have been solved,12,13 revealing a homopentamer with each subunit consisting of two transmembrane helices (TM1 and TM2) connected by a periplasmic loop, a short membrane bound N-terminus, and a C-terminus which forms a water exposed cytosolic helical bundle. MscL has been extensively studied both experimentally and computationally. For reviews on MscL structure and function, see refs (14−17). The sensitivity of MscL gating to changes in the bilayer environment has also been well established. For example, MscL’s gating threshold is affected by changes in bilayer thickness,18 the insertion of amphiphiles,18−20 and other membrane-active molecules such as phytochemicals.21
Computer simulations are a powerful tool to study membrane–protein interactions at the molecular level and, in principle, should be able to predict the effects of changes in the membrane environment on protein conformations. However, two major challenges have proven problematic: first, the difficulty to reach time scales long enough to actually observe the conformational changes; second, membrane-mediated effects can be rather subtle and require simulation of many independent events to obtain statistical relevance. These limitations have prevented the use of atomically detailed models, where MscL gating requires the help of biasing potentials (e.g., see refs (22−28)). However, recent advances in available computational resources and speed up from coarse-grained (CG) methods have paved the way for high-throughput simulation of lipid–protein interaction at near atomistic resolution.29−33 Indeed, using the CG Martini model,34 we have previously shown that MscL can be gated by membrane tension alone, i.e. without the need for biasing potentials.35,36
Here, we further assert the suitability of the Martini model by showing that it can also capture the reclosing of MscL when membrane tension is removed. We then proceed to implement a Martini-based high-throughput protocol that enables us for the first time to explore the effect of bilayer environment on the gating kinetics of a membrane channel. We tested the gating propensity of MscL upon the addition of straight chain alcohols of different lengths to the membrane. Alcohols are known to modify lipid bilayer properties and have been shown to affect the function of a range of membrane proteins (e.g., see Ingolfsson and Andersen37 and references within). Based on over 500 independent CG MD simulations totaling more than 10 ms of effective sampling time, we reveal that short chain alcohols have an impeding effect on the opening threshold of MscL. These in silico predictions were verified using a calcein efflux assay. Further simulations and analysis indicate that the alcohol effect on MscL arises through a combination of different changes in bilayer bulk properties and changes in the interfacial interaction between the bilayer and MscL. Our study opens the way to systematic exploration of the subtle interplay between membrane protein function and membrane properties, providing understanding at a molecular level of detail.
Methods
Simulations and Analysis
All simulations were performed using the Martini coarse-grain (CG) model38−40 and the GROMACS 4.6 simulation package.41 The MscL simulations were done using similar parameters as described previously.35,36 The CG representation of MscL was derived from the crystal structure of the closed state Tb-MscL (PDB ID 2OAR)12,13 using Martini v2.139,40 without an added elastic network. The channel was solvated in a bilayer composed of 500–600 DOPC (dipalmitoyl-phosphatidylcholine, di(18:1)-PC) lipids and around 25k CG water beads (corresponding to about 100k water molecules) using the bilayer builder insane.42 The alcohols—ethanol (eth), octanol (oct), dodecanol (dodec), and hexadecanol (hexdec)—were added directly to the bilayer by lipid replacement. A reference system has the MscL channel embedded in a bilayer composed of the longer tail lipid DEPC (dierucoyl-phosphatidylcholine, di(22:1)-PC). The temperature and pressure were controlled using the Berendsen thermostat (298 K) and barostat.43 The systems were equilibrated for 4–5 μs with 1 bar semi-isotropic pressure coupling. Similar to what was described previously,35 imposed membrane tension (MT) was used to promote channel gating; MT was incrementally applied in seven short (3 ns) simulations to a value of 65–70 mN/m and then kept for 6 μs production runs (also see the Supporting Information, Methods section). Note that to gate the channel in a small bilayer patch on a computationally tractable time scale, higher tensions are required compared to tensions used in experimental assays (∼10 mN/m); see also ref (35). For selected simulations, the bilayer MT was released, using a reverse protocol, and simulated further for 6 μs. In order to measure MscL time to opening, multiple repeated simulations are needed; we performed 30–100 replica simulations for each bilayer environment/condition; see Table S1 for a summary of all simulations. The analysis of simulation trajectories involved the calculation of water fluxes, channel profiles, lipid and alcohol distributions, and several membrane properties. These were carried out partly using the tools provided with the GROMACS package, and partly by custom scripts mainly written in Python and using the MDAnalysis package.44,45 Taken together the simulations total over 2700 μs of simulation time or, accounting for the ∼4-fold faster diffusion at the CG Martini level,39 over 10 ms of molecular dynamics. For further details on the simulation setup and details on all analysis methods, see the Supporting Information, Methods section.
MscL Fluorescence Assay
The effect of alcohols on MscL function was measured using a calcein fluorescence assay.46 Liposomes were prepared from azolectin (a phospholipid mixture extracted from soybeans) with encapsulated calcein fluorophore. When entrapped in vesicles at high concentration calcein self-quenches, such that the fluorescence increases when calcein is released from the vesicles. Calcein-loaded azolectin large unilamellar vesicles (LUVs) were made by extrusion, E. coli G22C MscL mutant at 1:50 protein to lipid (w/w) ratio was incorporated, and external calcein was removed using a Sephadex G50 size-exclusion column. The shorter alcohols, eth and oct, were added to the same vesicle suspension (volume % added to the aqueous phase) and incubated for 3–5 min. The longer alcohols, dodec and hexdec, are, however, insoluble in water, and vesicles were therefore made with 10 mol % of these alcohols already in the azolectin lipid phase before preparation. Note that the variation between different vesicle preparations is much greater than within the same vesicle batch so the dodec and hexdec experiments should only be considered as indicative of their effect.
To activate the G22C mutant MscL 1 mM final concentration of [2-(trimethylammonium)ethyl] methanethiosulfonate bromide (MTSET) was added and the calcein release from the vesicles monitored. Calcein fluorescence was monitored using a Varian Cary Eclipse Fluorimeter. The samples were excited at 495 nm, and emission was recorded at 515 nm. At the end of the experiment, 0.5% (v/v) final concentration of Triton X-100 was added to dissolve all vesicles and measure the maximal fluorescence. All data were normalized using initial fluorescence as 0% calcein release and fluorescence after Triton X-100 addition as 100% release. Maximum release was recorded right before Triton X-100 addition. Control vesicles without MscL were prepared, and only nominal calcein release was observed (at the relevant time scales) both with and without added alcohols.
Results and Discussion
Reversible Gating of MscL in Silico
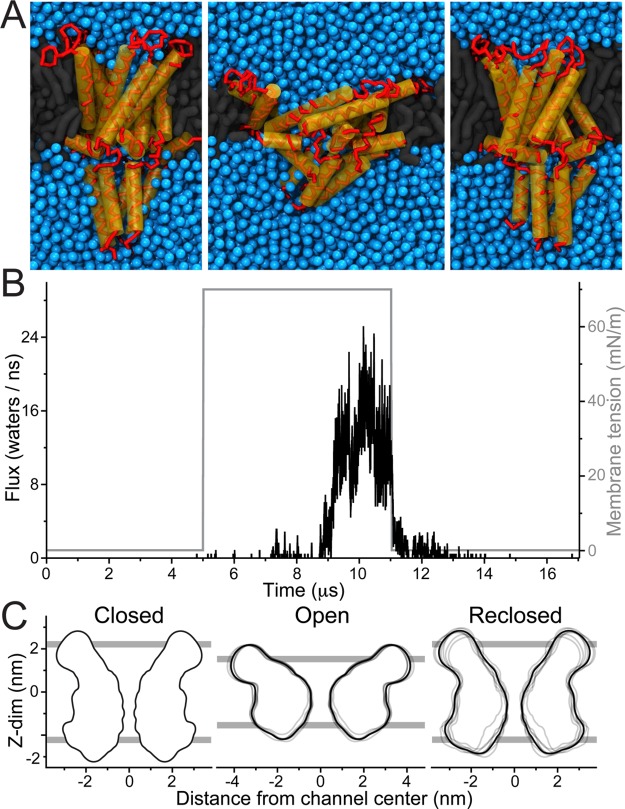
As shown previously, MscL can be gated using CG Martini simulations by applying tension to the membrane.35,36 To illustrate our current simulation protocol, a typical MscL gating event is shown in Figure 1. A single MscL protein is embedded in a DOPC bilayer and equilibrated for 5 μs in the absence of membrane tension (MT) (Figure 1A,B left). During this equilibration phase, the channel remains shut. Then, MT is applied to the bilayer, in a number of incremental steps reaching 70 mN/m. With applied tension the bilayer becomes thinner, and the channel leans down, adjusting to the bilayer thickness. At the same time, the extracellular cavity of the channel expands. Initially, the gate stays closed, with only occasional spurious waters slipping through the channel. This is followed by a flickering behavior (partial opening and reclosing) before eventually the channel fully opens (Figure 1A,B middle). When MT is removed the progress is reversed, i.e. after a gradual channel rearrangement the channel recloses and water flux is blocked (Figure 1A,B right). The channel’s progression can also be followed by looking at the area of pore cross sections at different depths (see Figure S1).
Figure 1.
MscL reversible gating. MscL was embedded in a DOPC bilayer and equilibrated for 5 μs, and membrane tension (MT) was applied and simulated for 6 μs. The MT was then removed and further simulated for 6 μs. (A) Representative snapshots of MscL: without MT (closed, left), with applied MT (open state, middle), and after removing MT (reclosed, right). (B) The water flux across the channel (black line) and applied MT (gray line) is shown for a continuous set of equilibration, applied tension, and after removing tension simulations. Here the water flux is reported as the average number of water molecules (4 × number of CG beads) traversing the channel per ns. (C) Channel profile of each of eight selected open and reclosing cycles, collected over the last 1 μs of each segment (gray lines; average of all eight profiles in black). The closed (pre-MT) profile was obtained from the single equilibration simulation. For clarity the cytoplasmic C-terminus was excluded from the profiles. The average lipid phosphate group position is shown as gray bars.
To characterize the closing of MscL in more detail, in Figure 1C we overlay the profiles of eight independently opened channels in the open and reclosed states, together with their averaged structure; see Supporting Information, Methods section for details on profile calculation. Some structural divergence is visible in the open replicates (Figure 1C middle), and even more so in the reclosed replicates (Figure 1C right). The reclosed profiles (especially their average) are nonetheless quite similar, functionally and structurally, to the crystal equilibration profile (see Figure 1C left, and Figure S2 for details on a particularly divergent case). This infers that the ensemble of closed structures has indeed physiological relevance and that the Martini model is suitable for studying the full gating cycle. The ensembles of open and reclosed structures also parallel the different paths the system can take from the closed to the open state, and from the open to the reclosed state. These are visible from the flux/pore-radius plots in Figure S2, which show that the MscL opening and reclosing proceeds, for the different replicates, through diverse pore constriction states that do not always correlate equally with flux.
High-Throughput Simulations Reveal MscL Gating Kinetics
Channel opening and reclosing are intrinsically stochastic in nature, and multiple repeats are therefore needed for their characterization. To capture the opening kinetics multiple simulations with MT were performed, n = 30–100, for each bilayer condition tested, see Table S1 for a list of all simulations. For each simulation, the time-to-channel-opening was defined by a minimum threshold of water flux through the channel; see Supporting Information, Methods section for details. The channel time-to-opening was evaluated by analyzing the decay of the closed channel fraction across all repeats for a given condition. Figure 2A shows Kaplan–Meier survival plots of the closed channel fraction as a function of simulation time for DOPC (n = 100) and DOPC with 10 mol % octanol in the bilayer (n = 50). At time zero, just after MT has been applied, all channels are closed. After an initial lag phase, during which the channels adjust to the decrease in bilayer thickness, channels start to open. The number of closed channels follows approximately a single exponential decay. All the simulations were stopped at 6 μs resulting in a number of simulations ending with the channel still closed. The same type of closed channel fraction survival plots is shown in Figure S3 for all bilayer conditions tested, and all, to the first approximation, follow a single exponential decay with a delay (see Figure S4 and Table S2). It is worth noting, at least in the case of DOPC with the highest number of replica simulations (n = 100), that the closed channel decay appears multiexponential and can be better fit using a stretched exponential47 with a delay; see Figure S5.
Figure 2.
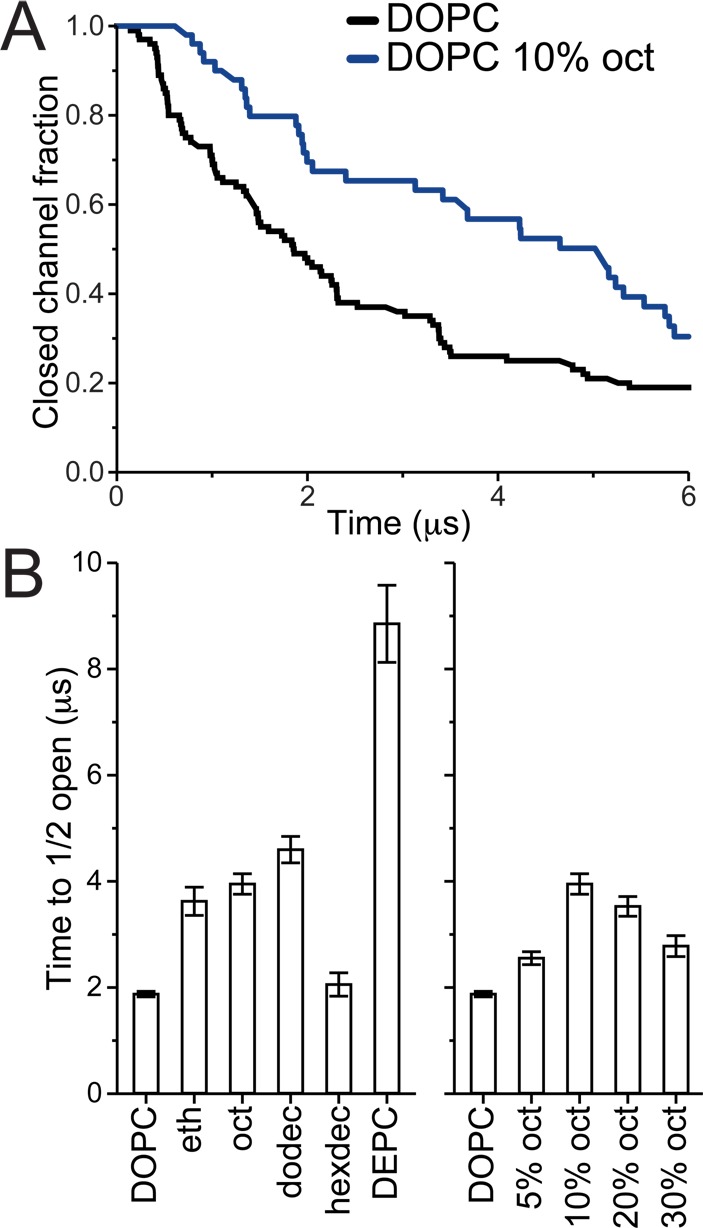
MscL opening time. MscL channels were equilibrating for 4–5 μs in different bilayer environments, MT was applied, and the time to channel opening was evaluated over multiple simulations. (A) Kaplan–Meier plots showing the closed channel fraction decay with simulation time for DOPC (n = 100) and DOPC with 10% octanol (n = 50). All tested bilayer conditions are shown in Figure S3. (B) The time for half the channel population to be opened (t1/2) was estimated. For each bilayer environment, the closed channel fraction was fit using a single exponential decay with a delay (see Figure S4 and Table S2); then t1/2 and t1/2 error were estimated from the fits (see the Supporting Information, Methods section for details).
Prediction of Increased Gating Threshold in the Presence of Alcohols
When comparing channel opening in DOPC and DOPC with 10 mol % octanol in the bilayer (Figure 2A) it is clear that, in silico, octanol makes opening of MscL more difficult. To compare all different conditions we estimated the time it takes for half of the channel population to open (t1/2). For each bilayer condition the closed channel fraction was fit using a single exponential decay with a delay (see Figure S4 for fits and Table S2 for fit parameters and calculated t1/2). In Figure 2B we compare t1/2 for systems containing 10 mol % of the straight-chain alcohols ethanol (eth), octanol (oct), dodecanol (dodec), and hexadecanol (hexdec). The shorter alcohols (eth, oct, and dodec) all show similar behavior, increasing t1/2 by a factor of 2–2.5 compared to pure DOPC. The longer alcohol, hexdec, has nearly no effect. This size cutoff effect of straight-chain alcohols is consistent with what has been observed with straight-chain alcohols on many other channels, e.g., refs (37 and 48−50). The effect of oct was explored in greater detail (Figure 2B, right); DOPC bilayers with 5, 10, 20, and 30 mol % oct were compared. Increasing the oct concentration makes it harder to open MscL until a point where adding more has less effect. This point is reached between 10 and 20 mol %. As a control for testing the effect of bilayer environment on MscL gating characteristics, we also compared DOPC to a much thicker bilayer composed of DEPC lipids (Figure 2B). In DEPC, the MscL time to opening is much slower, around 5-fold, consistent with established literature.18 Together, our results show that the addition of short chain alcohols increases the opening time of MscL, comparably to an increase in gating threshold: the gating behavior of MscL in DOPC with 10 mol % oct at a MT of 70 mN/m is roughly the same as in pure DOPC at a lower MT of 65 mN/m (see Figure S3C and Table S2).
Fluorescence Dequenching Assay Confirms in Silico Results
The effects of alcohols on MscL gating were verified in lipid vesicles using a calcein fluorescence dequenching assay.46 The G22C MscL mutant was incorporated in fluorophore-filled vesicles, and the channels were activated by adding MTSET (a positively charged sulfhydryl reagent). MTSET reacts with Cys22 and weakens the MscL gate allowing the channel to activate without applied tension.51 Channel activation was monitored as the increase in fluorescence when calcein exited LUVs through open MscL channels. A scheme of the assay is shown in Figure 3A. Figure 3B shows representative calcein release curves for DOPC only (black line) and DOPC with octanol added to the aqueous phase (blue lines). We quantified the changes in MscL activity with the addition of alcohols as the change in slope of the calcein release compared to DOPC control (k/kcont.) (see Figure 3C). Overall, these results confirm our in silico findings. The shorter alcohols (eth, oct, and probably also dodec) reduce MscL activity, whereas the longer hexdec has no (or even reverse) effect. In addition, for octanol the same reversal of potency with increasing concentration is observed as in the simulations.
Figure 3.
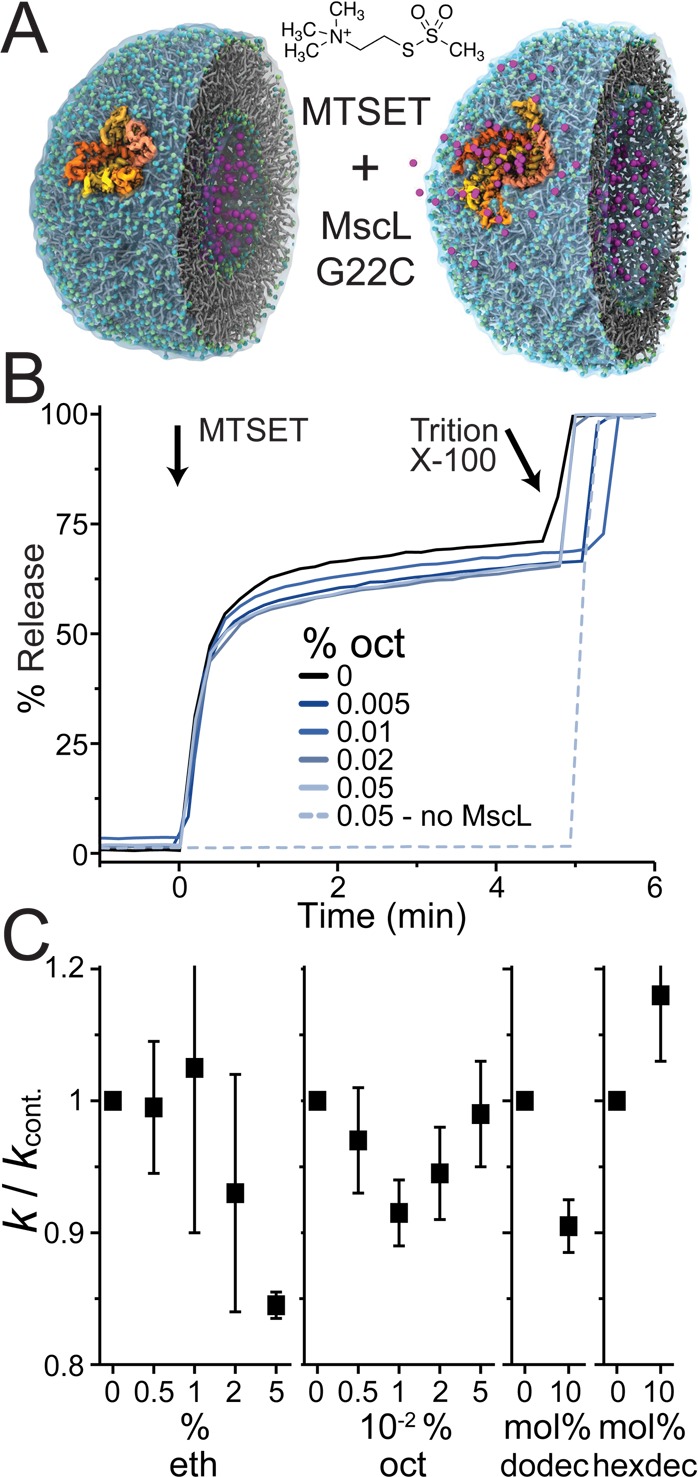
Alcohols affect MscL channels function. MscL channels were reconstituted into calcein-loaded vesicles. Channel activation was initiated by exposure to MTSET, and the release of calcein through open MscL channels monitored as an increase in fluorescence. (A) A cartoon illustration of the fluorescence assay (the structure of the vesicle with embedded MscL used to create this cartoon is from Louhivuori et al.36). (B) Representative calcein release curves are shown with DOPC only (black line) and % added octanol to the aqueous phase (blue lines). A control without MscL channels to test for octanol effects on vesicle integrity is also shown (dotted blue line). (C) Changes in MscL activity with the addition of alcohols were quantified as the change in slope of the calcein release compared to DOPC-only control (k/kcont.). For ethanol (eth) and octanol (oct) the same vesicle preparation was used for each experiment and aliquots with different amounts of added alcohol tested (volume % added to the aqueous phase), avg ± se, n = 3. Dodecanol (dodec) and hexadecanol (hexdec) are insoluble in water, and therefore, vesicles were made with 10 mol % of these alcohols already in the lipid phase. The average k from vesicles made with and without the alcohols were compared (avg ± se, n = 4). Because the variation between different vesicle preparations is much greater than within the same vesicle batch, the dodec and hexdec experiments should only be considered as indicative.
Mechanism of Alcohol Influence
We have demonstrated that short straight-chain alcohols affect MscL gating, increasing the opening threshold. It is well-known that alcohols change bilayer properties (e.g., refs (48, 52−59)) and that MscL is sensitive to changes in the bilayer environment. Therefore, it might be possible to directly correlate specific changes in bilayer properties to changes in MscL function. In order to explore this possibility we measured the area per lipid, bilayer thickness, area compressibility, lipid diffusion, and average tail order parameter for all the bilayer environments tested, in the absence of MscL, both with and without MT (see Figure 4 and Table S3). In the case of DEPC, the increase in membrane thickness alone provides a clear rationale for the observed increase in gating threshold, as the hydrophobic thickness of MscL is much larger, and better-matching, in the closed than in the open state (cf. Figure 1A). The effects of alcohols on membrane properties, however, were only modest. Interestingly, the short chain alcohols caused a slight reduction in bilayer thickness, consistent with previous studies.53,54,58−60 Such a thinning should increase the propensity of MscL toward the hydrophobically thinner open state, with a consequently lower gating threshold. Alcohol-induced changes in bilayer thickness, therefore, cannot explain the increased gating threshold. Alcohols have also been reported to reduce bilayer bending and area compressibility modulus;54,59 this softening could reduce the effectiveness of the applied MT and increase MscL opening times. Although possible, it is hard to imagine a biphasic change in elasticity with oct concentration as observed with their effect on MscL gating. Besides, in our simulations, the alcohols show little effect on area compressibility (see Table S3). Looked at together, there is no simple correlation of the measured changes in bilayer properties to the alcohols effect on MscL opening times. This could indicate that a complex combination of changes in bilayer properties is involved, further complicated by our observation that the effect of alcohols on the bilayer with and without MT is not the same (Table S3).
Figure 4.
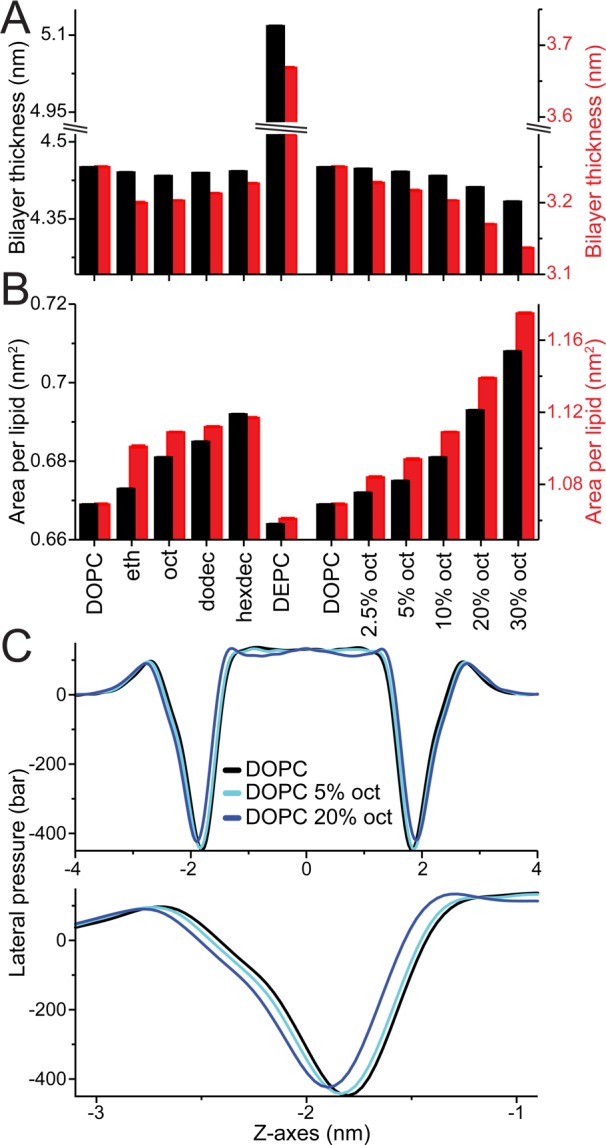
Changes in bilayer properties. Bilayer properties were calculated for all considered bilayer environments both without (black) and with applied membrane tension (red, 65 mN/m). Bilayer thickness (A) and area per lipid (B) are shown; Table S3 lists these properties as well as area compressibility, average tail order parameter, and lipid diffusion. Standard errors (se) are shown but hardly visible, as all values are <0.001. Note, the axes are highly zoomed and set so the DOPC values with and without MT are aligned; this is arbitrary and done to highlight their differences and similar trends, respectively. (C) LPPs for DOPC only (black line) and DOPC with 5 and 20% oct (light and dark blue lines, respectively) centered at the bilayer middle; for the whole bilayer (top) and zoomed in at the bilayer water interface (bottom).
A compound property that can capture these effects is the bilayer lateral pressure profile (LPP). The LPP is a measure of the local stress across the membrane and can affect membrane protein function.61 In fact, a significant part of the free energy of channel opening can come from work done against the pressure profile, rendering MscL sensitive to changes in LPP (e.g., refs (62−66)). Simulation studies of straight-chain alcohols in bilayers have shown that alcohols can change the LPP.57,67 We calculated the LPP for all the bilayer environments tested. Figure 4C show the LPPs for DOPC only (black line) and DOPC with 5% and 20% octanol (light and dark blue lines, respectively); LPPs for all the tested environments are shown in Figure S6. We see that short chain alcohols affect the LPP and, in particular, shift and reduce the negative peak in the LPP arising from the interfacial membrane/water tension. This finding is consistent with previous simulations.57,67 The change of the LPP in the interfacial region is important, as the area expansion of MscL during gating occurs mainly in the interfacial region64 (see also Figure 1A). An expansion in the interfacial region is favorable due to the large negative peak of the LPP in this region and contributes significantly to the overall free energy of channel opening.64 The contribution of the LPP to the free energy change ΔΔG of channel opening can be estimated from ΔΔG = ΔγΔa, where Δγ denotes the change in membrane/water interfacial tension upon the addition of alcohol and Δa the difference in interfacial area of MscL between the open and closed channel states. For an estimated area expansion of MscL of 5 nm2 (see ref (64)), a change in interfacial tension of only 2 mN/m (40 bar over 0.5 nm, i.e., roughly the effect of adding 20% octanol; cf. Figure 4C) would result in a ΔΔG ≈ 2 kT. A reduction of the interfacial tension by alcohols would therefore stabilize the closed state, in line with our computational and experimental data. However, we note that the above line of reasoning does also not account for the full story: the alcohols modify the LPP at more than just the interface (see Figures 4C, S6), and the more subtle effects on MscL opening times are not easily correlated to the changes in LPP.
Instead of modifying bulk membrane properties, the alcohols might also influence MscL directly and/or alter the interface between MscL and the bilayer. To explore the latter possibility, additional simulations with a modified oct were performed. In the modified octanol model (oct Excl) all interactions with the MscL channel were weakened, implying a depletion of oct in the vicinity of the channel. Using the same method as for the other alcohols, MscL’s time to opening was determined for 10% oct Excl (see Figure 5A). Similar to regular oct, the octanol that does not interact with MscL (oct Excl) also increases MscL time to opening, a clear sign of a membrane-mediated mechanism. The change in t1/2, however, is less than half that of regular oct (1.4- compared to 2.1-fold longer), indicating the presence of additional effects. To locate direct interactions between alcohol and MscL, in Figure 5B we show the radially averaged density of oct (top) and oct Excl (bottom) around the MscL channel. Octanols localize primarily at the water–lipid interface, slightly below the lipid phosphate group. In the case of the modified oct, a clear deprivation of the alcohol in the vicinity of the channel is noticeable. In the case of regular oct, oct molecules do approach the MscL channel, but not to the extent exceeding the bulk concentration. In addition, no evidence of specific binding sites is found (see Figure S7). Due to the water solubility of oct and its smaller size compared to the lipids, some density is also observed in places which lipids cannot access such as the water exposed pore and C-terminus, as well as penetrating more deeply between the MscL TM helixes.
Figure 5.
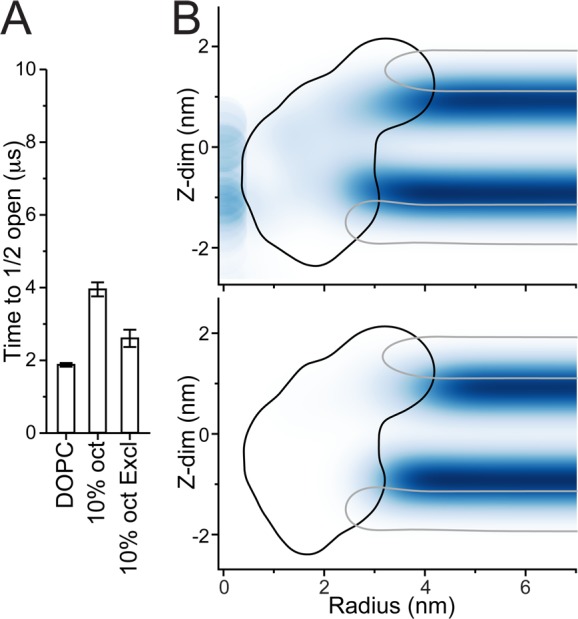
Indirect vs direct effects of octanol. (A) The time for when half the population of channels has opened (t1/2) is shown for octanol that does not interact with the MscL (10% oct Excl). The same DOPC and 10% oct values as in Figure 2B are shown for reference. (B) Octanol density, radially averaged from the channel center, is shown for regular octanol (oct, top) and octanol that is excluded from the channel (oct Excl, bottom); see the Supporting Information, Methods section for details. The outlines of the MscL (black) and DOPC’s PO4 beads (gray) are shown for reference. The apparent significant density of oct in the channel pore is exacerbated due to the small cylindrical volume close to zero distance away from the channel center.
Together, our data indicate that the alcohol-induced stabilization of the closed state—or destabilization of the open state—arises from a combination of changes in a number of bilayer properties and interfacial/solvation effects, but not from specific alcohol-MscL interactions.
Conclusions
We characterized MscL gating and membrane dependence using extensive coarse-grained molecular dynamics simulations. We showed that not only can MscL be opened but also reclosed in silico by applying and removing membrane tension. From numerous repeated simulations MscL opening time could be estimated in different bilayer environments. The effects of increasing bilayer thickness and adding straight-chain alcohols were evaluated, revealing the channel’s complex dependence on the membrane environment. Our simulations showed an increase in MscL opening time with the addition of straight-chain alcohols, an effect which saturates at longer alcohol chain lengths and higher alcohol concentration; these in silico predictions were then verified using a fluorescence assay. A more thorough examination of the effect of octanol on MscL revealed no specific binding sites, but rather that the change in MscL function resulted from a combination of octanol’s changes to different bilayer properties as well as changes in the MscL–bilayer interface.
We shed new light on the mechanism of MscL gating and its membrane dependence, while at the same time illustrating the complexity of membrane protein function modulation by small molecules, which precludes a simple mechanistic interpretation. Our work further demonstrates how extensive molecular simulations can be used to determine the functional dependence of membrane proteins on their bilayer environment, with a level of detail nearly impossible to achieve experimentally.
Acknowledgments
This work was supported by grants from The Netherlands Organization for Scientific Research (NWO): M.N.M. was supported by a Veni grant (No. 722.013.010), S.J.M. was supported by a TOP grant, and H.I.I. was supported by a Rubicon grant. A.K. was supported by a European Research Council Starting Grant (No. 208814). Computer access was granted from the Dutch National Computing Facilities Foundation (NCF) through NWO. Part of this work was performed under the auspices of the U.S. Department of Energy by Lawrence Livermore National Laboratory under Contract DE-AC52-07NA27344. Release number: LLNL-JRNL-715277.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.6b11091.
Supplementary methods, providing details on the simulation set up, force field and the various analysis methods; additional figures showing the MscL pore profile and gating characteristics, channel survival graphs and fits, detailed analysis of channel opening, changes in bilayer LPPs, and octanol occupancy; tables listing all simulations, estimates of channel opening times, and changed in bilayer properties (PDF)
Simulation summary table (XLSX)
The authors declare no competing financial interest.
Supplementary Material
References
- Israelachvili J. N. Biochim. Biophys. Acta, Biomembr. 1977, 469, 221. 10.1016/0005-2736(77)90185-7. [DOI] [PubMed] [Google Scholar]
- Mouritsen O. G.; Bloom M. Biophys. J. 1984, 46, 141. 10.1016/S0006-3495(84)84007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killian J. A. Biochim. Biophys. Acta, Rev. Biomembr. 1998, 1376, 401. 10.1016/S0304-4157(98)00017-3. [DOI] [PubMed] [Google Scholar]
- Huang H. W. Biophys. J. 1986, 50, 1061. 10.1016/S0006-3495(86)83550-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen O. S.; Koeppe R. E. II Annu. Rev. Biophys. Biomol. Struct. 2007, 36, 107. 10.1146/annurev.biophys.36.040306.132643. [DOI] [PubMed] [Google Scholar]
- Andersen O. S. J. Gen. Physiol. 2008, 131, 395. 10.1085/jgp.200810012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh D. Biochim. Biophys. Acta, Biomembr. 2008, 1778, 1545. 10.1016/j.bbamem.2008.01.015. [DOI] [PubMed] [Google Scholar]
- Lundbæk J. A.; Collingwood S. A.; Ingólfsson H. I.; Kapoor R.; Andersen O. S. J. R. Soc., Interface 2010, 7, 373. 10.1098/rsif.2009.0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundbæk J. A.; Koeppe R. E. II; Andersen O. S. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 15427. 10.1073/pnas.1007455107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinac B.; Buechner M.; Delcour A. H.; Adler J.; Kung C. Proc. Natl. Acad. Sci. U. S. A. 1987, 84, 2297. 10.1073/pnas.84.8.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukharev S. I.; Blount P.; Martinac B.; Blattner F. R.; Kung C. Nature 1994, 368, 265. 10.1038/368265a0. [DOI] [PubMed] [Google Scholar]
- Chang G.; Spencer R. H.; Lee A. T.; Barclay M. T.; Rees D. C. Science 1998, 282, 2220. 10.1126/science.282.5397.2220. [DOI] [PubMed] [Google Scholar]
- Steinbacher S.; Bass R.; Strop P.; Rees D. C. In Mechanosensitive Ion Channels, Part A; Current Topics in Membranes; Elsevier: 2007; Vol. 58, pp 1–24. [Google Scholar]
- Hamill O. P.; Martinac B. Physiol. Rev. 2001, 81, 685. [DOI] [PubMed] [Google Scholar]
- Booth I. R.; Edwards M. D.; Black S.; Schumann U.; Miller S. Nat. Rev. Microbiol. 2007, 5, 431. 10.1038/nrmicro1659. [DOI] [PubMed] [Google Scholar]
- Haswell E. S.; Phillips R.; Rees D. C. Structure 2011, 19, 1356. 10.1016/j.str.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koçer A. Curr. Opin. Chem. Biol. 2015, 29, 120. 10.1016/j.cbpa.2015.10.006. [DOI] [PubMed] [Google Scholar]
- Perozo E.; Kloda A.; Cortes D. M.; Martinac B. Nat. Struct. Biol. 2002, 9, 696. 10.1038/nsb827. [DOI] [PubMed] [Google Scholar]
- Martinac B.; Adler J.; Kung C. Nature 1990, 348, 261. 10.1038/348261a0. [DOI] [PubMed] [Google Scholar]
- Mukherjee N.; Jose M. D.; Birkner J. P.; Walko M.; Ingólfsson H. I.; Dimitrova A.; Arnarez C.; Marrink S. J.; Koçer A. FASEB J. 2014, 28, 4292. 10.1096/fj.14-251579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingólfsson H. I.; Thakur P.; Herold K. F.; Hobart E. A.; Ramsey N. B.; Periole X.; de Jong D. H.; Zwama M.; Yilmaz D.; Hall K.; Maretzky T.; Hemmings H. C.; Blobel C.; Marrink S. J.; Koçer A.; Sack J. T.; Andersen O. S. ACS Chem. Biol. 2014, 9, 1788. 10.1021/cb500086e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullingsrud J.; Kosztin D.; Schulten K. Biophys. J. 2001, 80, 2074. 10.1016/S0006-3495(01)76181-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullingsrud J.; Schulten K. Biophys. J. 2003, 85, 2087. 10.1016/S0006-3495(03)74637-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo G.; Marrink S. J.; Mark A. E. Biophys. J. 2003, 84, 2331. 10.1016/S0006-3495(03)75038-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore D. E.; Dougherty D. A. Biophys. J. 2001, 81, 1345. 10.1016/S0006-3495(01)75791-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J.; Voth G. A. Biophys. J. 2008, 94, 3497. 10.1529/biophysj.107.109850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui H.; Kumar R.; Im W. Biophys. J. 2011, 101, 671. 10.1016/j.bpj.2011.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanegas J. M.; Arroyo M. PLoS One 2014, 9, e113947. 10.1371/journal.pone.0113947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom M. S. P.; Scott K. A.; Bond P. J. Biochem. Soc. Trans. 2008, 36, 27. 10.1042/BST0360027. [DOI] [PubMed] [Google Scholar]
- Hall B. A.; Halim K. B. A.; Buyan A.; Emmanouil B.; Sansom M. S. P. J. Chem. Theory Comput. 2014, 10, 2165. 10.1021/ct500003g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingólfsson H. I.; Lopez C. A.; Uusitalo J. J.; de Jong D. H.; Gopal S. M.; Periole X.; Marrink S. J. WIREs Comput. Mol. Sci. 2014, 4, 225. 10.1002/wcms.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenaar T. A.; Pluhackova K.; Moussatova A.; Sengupta D.; Marrink S. J.; Tieleman D. P.; Bockmann R. A. J. Chem. Theory Comput. 2015, 11, 2278. 10.1021/ct5010092. [DOI] [PubMed] [Google Scholar]
- Ingólfsson H. I.; Arnarez C.; Periole X.; Marrink S. J. J. Cell Sci. 2016, 129, 257. 10.1242/jcs.176040. [DOI] [PubMed] [Google Scholar]
- Marrink S. J.; Tieleman D. P. Chem. Soc. Rev. 2013, 42, 6801. 10.1039/c3cs60093a. [DOI] [PubMed] [Google Scholar]
- Yefimov S.; van der Giessen E.; Onck P. R.; Marrink S. J. Biophys. J. 2008, 94, 2994. 10.1529/biophysj.107.119966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louhivuori M.; Risselada H. J.; van der Giessen E.; Marrink S. J. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 19856. 10.1073/pnas.1001316107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingólfsson H. I.; Andersen O. S. Biophys. J. 2011, 101, 847. 10.1016/j.bpj.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrink S. J.; De Vries A. H.; Mark A. E. J. Phys. Chem. B 2004, 108, 750. 10.1021/jp036508g. [DOI] [Google Scholar]
- Marrink S. J.; Risselada H. J.; Yefimov S.; Tieleman D. P.; De Vries A. H. J. Phys. Chem. B 2007, 111, 7812. 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- Monticelli L.; Kandasamy S. K.; Periole X.; Larson R. G.; Tieleman D. P.; Marrink S. J. J. Chem. Theory Comput. 2008, 4, 819. 10.1021/ct700324x. [DOI] [PubMed] [Google Scholar]
- Hess B.; Kutzner C.; van der Spoel D.; Lindahl E. J. Chem. Theory Comput. 2008, 4, 435. 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- Wassenaar T. A.; Ingólfsson H. I.; Böckmann R. A.; Tieleman D. P.; Marrink S. J. J. Chem. Theory Comput. 2015, 11, 2144. 10.1021/acs.jctc.5b00209. [DOI] [PubMed] [Google Scholar]
- Berendsen H. J. C.; Postma J. P. M. J. Chem. Phys. 1984, 81, 3684. 10.1063/1.448118. [DOI] [Google Scholar]
- Michaud-Agrawal N.; Denning E. J.; Woolf T. B.; Beckstein O. J. Comput. Chem. 2011, 32, 2319. 10.1002/jcc.21787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowers R. J.; Linke M.; Barnoud J.; Reddy T. J. E.; Melo M. N.; Seyler S. L.; Domański J.; Dotson D. L.; Buchoux S.; Kenney I. M.; Beckstein O.. MDAnalysis: a Python package for the rapid analysis of molecular dynamics simulations, Proceedings of the 15th Python in Science Conference, Austin, TX, 2016; Benthall S., Rostrup S., Eds.; Scipy: 2016; pp 98–105. [Google Scholar]
- Koçer A.; Walko M.; Feringa B. L. Nat. Protoc. 2007, 2, 1426. 10.1038/nprot.2007.196. [DOI] [PubMed] [Google Scholar]
- Berberan-Santos M. N.; Bodunov E. N.; Valeur B. Chem. Phys. 2005, 315, 171. 10.1016/j.chemphys.2005.04.006. [DOI] [Google Scholar]
- Mitchell D. C.; Lawrence J. T.; Litman B. J. J. Biol. Chem. 1996, 271, 19033. 10.1074/jbc.271.32.19033. [DOI] [PubMed] [Google Scholar]
- Wick M. J.; Mihic S. J.; Ueno S.; Mascia M. P.; Trudell J. R.; Brozowski S. J.; Ye Q.; Harrison N. L.; Harris R. A. Proc. Natl. Acad. Sci. U. S. A. 1998, 95, 6504. 10.1073/pnas.95.11.6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godden E. L.; Harris R. A.; Dunwiddie T. V. J. Pharmacol. Exp. Ther. 2001, 296, 716. [PubMed] [Google Scholar]
- Yoshimura K.; Batiza A.; Kung C. Biophys. J. 2001, 80, 2198. 10.1016/S0006-3495(01)76192-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon R. C.; McComb J. A.; Schreurs J.; Goldstein D. B. J. Pharmacol. Exp. Ther. 1981, 218, 669. [PubMed] [Google Scholar]
- Elliott J. R.; Haydon D. A. Biochim. Biophys. Acta, Biomembr. 1984, 773, 165. 10.1016/0005-2736(84)90561-3. [DOI] [PubMed] [Google Scholar]
- Ly H. V.; Longo M. L. Biophys. J. 2004, 87, 1013. 10.1529/biophysj.103.034280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aagaard T. H.; Kristensen M. N.; Westh P. Biophys. Chem. 2006, 119, 61. 10.1016/j.bpc.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Dickey A. N.; Faller R. Biophys. J. 2007, 92, 2366. 10.1529/biophysj.106.097022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terama E.; Ollila O. H.; Salonen E.; Rowat A. C.; Trandum C.; Westh P.; Patra M.; Karttunen M.; Vattulainen I. J. Phys. Chem. B 2008, 112, 4131. 10.1021/jp0750811. [DOI] [PubMed] [Google Scholar]
- Finol-Urdaneta R. K.; McArthur J. R.; Juranka P. F.; French R. J.; Morris C. E. Biophys. J. 2010, 98, 762. 10.1016/j.bpj.2009.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetter F. W. S.; Hugel T. Biophys. J. 2013, 104, 1049. 10.1016/j.bpj.2013.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klacsová M.; Bulacu M.; Kucerka N.; Uhrikova D.; Teixeira J.; Marrink S. J.; Balgavy P. Biochim. Biophys. Acta, Biomembr. 2011, 1808, 2136. 10.1016/j.bbamem.2011.04.010. [DOI] [PubMed] [Google Scholar]
- Cantor R. S. J. Phys. Chem. B 1997, 101, 1723. 10.1021/jp963911x. [DOI] [Google Scholar]
- Gullingsrud J.; Schulten K. Biophys. J. 2004, 86, 3496. 10.1529/biophysj.103.034322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moe P.; Blount P. Biochemistry 2005, 44, 12239. 10.1021/bi0509649. [DOI] [PubMed] [Google Scholar]
- Ollila O. H. S.; Louhivuori M.; Marrink S. J.; Vattulainen I. Biophys. J. 2011, 100, 1651. 10.1016/j.bpj.2011.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T.; Cranfield C. G.; Deplazes E.; Owen D. M.; Macmillan A.; Battle A. R.; Constantine M.; Sokabe M.; Martinac B. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 8770. 10.1073/pnas.1200051109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iscla I.; Blount P. Biophys. J. 2012, 103, 169. 10.1016/j.bpj.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griepernau B.; Böckmann R. A. Biophys. J. 2008, 95, 5766. 10.1529/biophysj.108.142125. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.