Abstract
NAD (Nicotinamide Adenine Dinucleotide) biosynthesis is vital for bacterial physiology and plays an important role in cellular metabolism. A naturally occurring vitamin B complex, niacin (nicotinic acid), is a precursor of coenzymes NAD and NADP. Here, we study the impact of niacin on global gene expression of Streptococcus pneumoniae D39 and elucidate the role of NiaR as a transcriptional regulator of niaX, nadC, and pnuC. Transcriptome comparison of the D39 wild-type grown in chemically defined medium (CDM) with 0 to 10 mM niacin revealed elevated expression of various genes, including niaX, nadC, pnuC, fba, rex, gapN, pncB, gap, adhE, and adhB2 that are putatively involved in the transport and utilization of niacin. Niacin-dependent expression of these genes is confirmed by promoter lacZ-fusion studies. Moreover, the role of transcriptional regulator NiaR in the regulation of these genes is explored by DNA microarray analysis. Our transcriptomic comparison of D39 ΔniaR to D39 wild-type revealed that the transcriptional regulator NiaR acts as a transcriptional repressor of niaX, pnuC, and nadC. NiaR-dependent regulation of niaX, nadC, and pnuC is further confirmed by promoter lacZ-fusion studies. The putative operator site of NiaR (5′-TACWRGTGTMTWKACASYTRWAW-3′) in the promoter regions of niaX, nadC, and pnuC is predicted and further confirmed by promoter mutational experiments.
Keywords: niacin, NiaR, Pneumococcus, niaX, nadC, pnuC
Introduction
Bacteria can trigger transcriptional and phenotypic programs to synchronize an adaptive response in reaction to environmental fluctuations or stresses (Edwards et al., 2013). This not only relies on the number of virulence factors it possesses, but also on the proper use of nutrients available in the human niches (Phillips et al., 1990; Titgemeyer and Hillen, 2002). A number of important vitamins and co-factors are required by bacteria to survive and grow successfully. Streptococcus pneumoniae, a major Gram-positive human pathogen and nasopharyngeal colonizer, encounters different environmental factors and has to fine-tune its gene expression accordingly (Bogaert et al., 2004; Kadioglu et al., 2008).
Niacin (nicotinic acid), a naturally occurring vitamin B complex, is a precursor of coenzymes NAD and NADP, and plays an important role in electron transfer during metabolic processes (Wei et al., 2014). Niacin has long been used for the treatment of lipid disorders and cardiovascular disease (Wei et al., 2014). It can regulate the activity of microbial two-component systems and, subsequently, modulate the genes and phenotypes that are controlled by these regulatory proteins (McPheat et al., 1983). Particularly, niacin has been reported to repress the expression of many genes including virulence factors in Bordetella pertussis, such as pertussis toxin, adenylate cyclase toxin, and filamentous hemagglutinin (Schneider and Parker, 1982; McPheat et al., 1983; Cotter and DiRita, 2000; Cummings et al., 2006). Furthermore, the two-component system BvgA/BvgS, which is known to have a role in the regulation of virulence and colonization, becomes inactive in B. pertussis when niacin is present in the medium (Miller et al., 1989). Similarly, the Escherichia coli EvgA/EvgS system that confers multidrug resistance and acid tolerance is regulated by niacin (Masuda and Church, 2002, 2003; Eguchi et al., 2003; Nishino et al., 2003). Both the BvgA/BvgS system of B. pertussis and the EvgA/EvgS system of E. coli are part of a family of proteins that utilize a multistep phosphor-relay to trigger their responsive pathways.
It has been proposed that in S. pneumoniae niacin enters the cell through NiaX and is converted to nicotinate (nicotinic acid)-mononucleotide by PncB (Johnson et al., 2015). Nicotinate mononucleotide is then converted to nicotinic acid adenine dinucleotide by NadD, whereafter NadE converts nicotinic acid adenine dinucleotide to nicotine adenine dinucleotide (NAD) (Johnson et al., 2015). Another important enzyme glyceraldehyde-3-phosphate dehydrogenase (GAP) is a highly conserved and a multifunctional protein with significant activity in several fundamental cell pathways (Sirover, 2011). Usually, the dehydrogenase reactions of metabolic pathways have been deemed the major sources of NADPH. Nevertheless, the importance of transhydrogenases, glucose dehydrogenases, and non-phosphorylating glyceraldehyde 3- phosphate dehydrogenase (GAPN), is becoming eminent, suggesting that the traditional view is over-simplistic (Sauer U. et al., 2004; Matsubara et al., 2011; Bräsen et al., 2014). As NAD is a vital cofactor used by all living organisms, all bacterial species make use of the pathways to reduce NAD+ to NADH (Jurtshuk, 1996). NAD+ is also used by bacteria as a substrate for dehydrogenases involved in breaking down aldehydes and alcohols (Nobelmann and Lengeler, 1996; Kotrbova-Kozak et al., 2007; Luong et al., 2015). Furthermore, several cellular processes in bacterial and mammalian cells also use NAD, for instance DNA ligation and repair, redox recycling in the pyruvate dehydrogenase pathway, and synthesis of acetyl-CoA for the tricarboxylic acid cycle (Ishino et al., 1986; Satoh and Lindahl, 1992; Wilkinson et al., 2001; Chalkiadaki and Guarente, 2012; Chiarugi et al., 2012; Patel et al., 2014).
YrxA (NiaR) was found to be a niacin-responsive repressor of NAD de novo synthesis in Bacillus subtilis and transcriptional regulation of NAD biosynthesis in bacteria having orthologs of B. subtilis yrxA was determined using a comparative genomic approach and expression studies (Rodionov et al., 2008a). NiaR family members are generally conserved in the Bacillus/Clostridium group and in the unrelated Fusobacteria and Thermotogales lineages (Rodionov et al., 2008a). The NiaR regulon is not limited to the transcriptional regulation of the nadABC but in some species it also covers niacin salvage (the pncAB genes) and contains uncharacterized membrane proteins putatively involved in niacin transport (Rodionov et al., 2008a). Moreover, members of the NiaP family (involved in niacin uptake) are not only conserved in bacteria but also in multicellular eukaryotes, including humans, suggesting the putative involvement of NiaP in niacin utilization in these organisms (Rodionov et al., 2008a).
This study explains the transcriptomic response of S. pneumoniae D39 to niacin and regulation of niaX, pnuC, and nadC genes. We established that the transcriptional regulator NiaR acts as a transcriptional repressor for niaX, pnuC, and nadC genes involved in niacin uptake and utilization. The putative operator site (5′-TACWRGTGTMTWKACASYTRWAW-3′ where R = A/G, K = G/T, S = G/C, Y = T/C, W = A/T and M = A/C) of NiaR in the promoter regions of niaX, pnuC, and nadC is predicted, and subsequently confirmed by mutating NiaR operator sites in the respective promoters.
Materials and methods
Bacterial strains and growth conditions
Bacterial strains and plasmids used in this study are listed in Table 1. S. pneumoniae D39 was grown as described previously (Kloosterman et al., 2006; Afzal et al., 2014). For β-galactosidase assays, derivatives of S. pneumoniae D39 were grown in chemically defined medium (CDM) (Neves et al., 2002) with or without 10 mM niacin. CDM was prepared without niacin. For selection on antibiotics, media were supplemented with the following concentrations of antibiotics: 150 μg/ml spectinomycin and 2.5 μg/ml tetracycline for S. pneumoniae, and 100 μg/ml ampicillin for E. coli. All bacterial strains used in this study were stored in 10% (v/v) glycerol at −80°C. For PCR amplification, chromosomal DNA of S. pneumoniae D39 (Lanie et al., 2007) was used as a template. Primers used in this study are based on the sequence of the S. pneumoniae D39 genome and listed in Table 2.
Table 1.
List of strains and plasmids used in this study.
| Strain/plasmid | Description | Source |
|---|---|---|
| S. PNEUMONIAE | ||
| D39 | Serotype 2 strain. cps 2 | Laboratory of P. Hermans. |
| MA1300 | D39 ΔniaR; SpecR | This study |
| MA1301 | D39 ΔbgaA:: PniaX-lacZ; TetR | This study |
| MA1302 | D39 ΔbgaA:: PpnuC-lacZ; TetR | This study |
| MA1303 | D39 ΔbgaA:: PnadC-lacZ; TetR | This study |
| MA1304 | MA1300 ΔbgaA:: PniaX-lacZ; TetR | This study |
| MA1305 | MA1300 ΔbgaA:: PpnuC-lacZ; TetR | This study |
| MA1306 | MA1300 ΔbgaA:: PnadC-lacZ; TetR | This study |
| MA1307 | D39 ΔbgaA:: PniaX-M-lacZ; TetR | This study |
| MA1308 | D39 ΔbgaA:: PnuC-M-lacZ; TetR | This study |
| MA1309 | D39 ΔbgaA:: PnadC-R1-M-lacZ; TetR | This study |
| MA1310 | D39 ΔbgaA:: PnadC-R2-M-lacZ; TetR | This study |
| MA1311 | D39 ΔbgaA:: Pfba-lacZ; TetR | This study |
| MA1312 | D39 ΔbgaA:: Prex-lacZ; TetR | This study |
| MA1313 | D39 ΔbgaA:: PgapN-lacZ; TetR | This study |
| MA1314 | D39 ΔbgaA:: PpncB-lacZ; TetR | This study |
| MA1315 | D39 ΔbgaA:: Pgap-lacZ; TetR | This study |
| MA1316 | D39 ΔbgaA:: PadhE-lacZ; TetR | This study |
| MA1317 | D39 ΔbgaA:: PadhB2-lacZ; TetR | This study |
| E. COLI | ||
| EC1000 | KmR; MC1000 derivative carrying a single copy of the pWV1 repA gene in glgB | Laboratory collection |
| PLASMIDS | ||
| pPP2 | AmpR TetR; promoter-less lacZ. For replacement of bgaA with promoter lacZ fusion. Derivative of pPP1 | Halfmann et al., 2007 |
| pMA1301 | pPP2 PniaX-lacZ | This study |
| pMA1302 | pPP2 PpnuC-lacZ | This study |
| pMA1303 | pPP2 PnadC-lacZ | This study |
| pMA1304 | pPP2 PniaX-M-lacZ | This study |
| pMA1305 | pPP2 PnuC-M-lacZ | This study |
| pMA1306 | pPP2 PnadC-R1-M-lacZ | This study |
| pMA1307 | pPP2 PnadC-R1-M-lacZ | This study |
| pMA1308 | pPP2 Pfba-lacZ | This study |
| pMA1309 | pPP2 Prex-lacZ | This study |
| pMA1310 | pPP2 PgapN-lacZ | This study |
| pMA1311 | pPP2 PpncB-lacZ | This study |
| pMA1312 | pPP2 Pgap-lacZ | This study |
| pMA1313 | pPP2 PadhE-lacZ | This study |
| pMA1314 | pPP2 PadhB2-lacZ | This study |
Table 2.
List of primers used in this study.
| Name | Nucleotide Sequence (5′ → 3′) | Restriction site* |
|---|---|---|
| niaX-F | CATGGAATTCTCAAACCTGAAGGTGGAGAT | EcoRI |
| niaX-R | CATGGGATCCGCATAACAATTGGAATCAAAATCG | BamHI |
| pnuC-F | CATGGAATTCCCATATGATTCTTTCTAATGAGTTG | EcoRI |
| pnuC-R | CATGGGATCCGCAAATAAGTATGCATCATTTCTCC | BamHI |
| nadC-F | CATGGAATTCCCAATGGCTAGAGCAATGGC | EcoRI |
| nadC-R | CATGGGATCCCATCTTCTCGCAAGGCTGC | BamHI |
| niaX-M-R | CATGGGATCCCACAAGAATCTCCTTTTTAACGGCATATGTACTAGTATGG | BamHI |
| pnuC-M-F | CATGGAATTCCATGATTTTCTAAAATTTTACTACAAAGACGGTTGAC | EcoRI |
| nadC-R1-M-F | CATGGAATTCGACTATTATACACAAAAAAAATACAATTACCTTGACCATTGTA | EcoRI |
| nadC-R2-M-F | CATGGAATTCTACACAAAAAAAATACAATTGTCTTGACAATTACATTGACCCTTGTT | EcoRI |
| NiaR-1 | GCCATGTTCTTGTCGCCC | - |
| NiaR-2 | GCATAGGCGCGCCCAAGAGTTGGAGCAGGGC | AscI |
| NiaR-3 | CGATTGCGGCCGCGCCGAAACACAACAAGACC | NotI |
| NiaR-4 | CGCTGGTCTGGTTATGCC | - |
| fba-F | CATGGAATTCCGTCCAAGACTAGGGAGAG | EcoRI |
| fba-R | CATGGGATCCGCATAACCGTTGTCACGGG | BamHI |
| rex-F | CATGGAATTCCCTCATGGATAGCTTGGTAG | EcoRI |
| rex-R | CATGGGATCCGCTGTAGCTTTTGGAATAGC | BamHI |
| gapN-F | CATGGAATTCGGTTTGGCTGTCCCCAACC | EcoRI |
| gapN-R | CATGGGATCCGTCATGGCTGGAACTGTACC | BamHI |
| pncB-F | CATGGAATTCGCTATGGCGAATGGGCTC | EcoRI |
| pncB-R | CATGGGATCCCTGGTACAAGTCCGTGTGC | BamHI |
| gap-F | CATGGAATTCCGTTACGCTATGAATAATAAGGG | EcoRI |
| gap-R | CATGGGATCCCGACCGATACGTCCGAAACC | BamHI |
| adhE-F | CATGGAATTCGCGCTTACCTGTAAATCCC | EcoRI |
| adhE-R | CATGGGATCCGAACCAACTCATCTACGTGC | BamHI |
| adhB2-F | CATGGAATTCGCAACCTACCTAGATGGCG | EcoRI |
| adhB2-R | CATGGGATCCGCACAATAGCGTCTGTTGGC | BamHI |
| NiaR-Conf-1 | GGAGATTCTTGTGAATACACGG | - |
| NiaR-Conf-2 | GATAATATCTCTGGTAGTAAGTCTG | - |
| Spec-R | GCTAAGCGGCCGCACTAAACGAAATAAACGC | NotI |
| Spec-F | GCTATGGCGCGCCCTAATCAAAATAGTGAGGAGG | AscI |
Restriction sites are underlined.
Construction of a niaR mutant
A niaR mutant (MA1300) was constructed in S. pneumoniae D39 by allelic replacement with a spectinomycin-resistance cassette. Primer pairs niaR-1/niaR-2 and niaR-3/niaR-4 were used to generate PCR fragments of the left and right flanking regions of niaR using Phusion® High-Fidelity DNA polymerase. PCR products of left and right flanking regions of niaR contain AscI and NotI sites, respectively. The spectinomycin-resistance marker, which was amplified by primers SpecR/SpecF from pORI38, also contains AscI and NotI sites on its ends. Then, by restriction and ligation, the left and right flanking regions of niaR were fused to the spectinomycin-resistance gene. The resulting ligation products were transformed to S. pneumoniae D39 wild-type and selection of the mutant was done on the appropriate concentration of spectinomycin. Deletion of niaR was further verified by PCR using primer pair NiaR-Conf-1/NiaR-Conf-2 and DNA sequencing.
Construction of promoter lacZ-fusions and their use in β-galactosidase assays
Chromosomal transcriptional lacZ-fusions to niaX, pnuC, and nadC promoters were constructed in pPP2 (Halfmann et al., 2007) with primer pairs mentioned in Table 2, resulting in pMA1301-03, respectively. These constructs were further introduced into D39 wild-type and D39 ΔniaR (MA1300) resulting in strains MA1301-03 and MA1304-06, respectively. The following lacZ-fusions of PniaX, PpnuC, and PnadC with mutations in the NiaR site were made in pPP2 (Halfmann et al., 2007) using the primer pairs mentioned in Table 2: PniaX-M (mutation in the niaR site), PpnuC-M (mutation in the niaR site), PnadC-R1 (mutation in the niaR site 1), and PnadC-R2 (mutation in the niaR site 2), resulting in plasmids pMA1304-07, respectively. These constructs were introduced into the S. pneumoniae D39 wild-type strain, resulting in strains MA1307-1310, respectively. Similarly, chromosomal transcriptional lacZ-fusions to fba, rex, gapN, pncB, gap, adhE, and adhB2 promoters were constructed in pPP2 (Halfmann et al., 2007) with primer pairs mentioned in Table 2, resulting in pMA1308-14, respectively. These constructs were further introduced into D39 wild-type resulting in strains MA1311-17, respectively. All plasmid constructs were further checked for the presence of the right insert by PCR and DNA sequencing.
β-galactosidase assays were performed as described before (Israelsen et al., 1995; Halfmann et al., 2007) using cells that were harvested in the mid-exponential growth phase, and grown in CDM (Neves et al., 2002) with or without niacin as mentioned in the results section.
Microarray analysis
Microarray analysis was performed as described before (Afzal et al., 2015; Shafeeq et al., 2015). For DNA microarray analysis of S. pneumoniae in the presence of niacin, the transcriptome of S. pneumoniae D39 wild-type grown in replicates in CDM with 10 mM niacin was compared to that grown in CDM with 0 mM niacin and harvested at their respective mid-exponential growth phases.
For DNA microarray analysis of D39 ΔniaR, the transcriptome of S. pneumoniae D39 ΔniaR was compared to S. pneumoniae D39 wild-type grown in replicates in complete CDM and harvested at respective mid-exponential growth phases. Complete CDM contains 8 μM of niacin. The procedures for DNA microarray analysis were performed as described previously (Afzal et al., 2015; Shafeeq et al., 2015). For the identification of differentially expressed genes, a Bayesian p < 0.001 and a fold-change cut-off > 1.5 was applied. Microarray data have been submitted to GEO (Gene Expression Omnibus) under accession numbers GSE94511 and GSE94513.
Results
Niacin-dependent gene regulation in S. pneumoniae D39
Microarray comparison of S. pneumoniae D39 grown in CDM with 0 mM to same strain grown in CDM with 10 mM niacin was performed to explore the impact of niacin on the transcriptome of S. pneumoniae D39 wild-type. CDM was prepared without niacin. A number of genes/operons were differentially expressed under our tested conditions (Table 3). A particular gene cluster (spd-0093-0095) was significantly upregulated in the absence of niacin. This gene cluster codes for three hypothetical proteins, which are putative membrane proteins. Another gene cluster (spd-1798-1802) was significantly upregulated in the absence of niacin. This gene cluster consists of a DNA-binding response regulator (encoded by spd-1798), a sensor histidine kinase (encoded by spd-1799), two hypothetical proteins (encoded by spd-1800 and spd-1802) and an ABC transporter (encoded by spd-1801). Some genes that appear to be a part of a gene cluster were also downregulated under our tested conditions (spd-0113-15 and spd-0122-24). All of these genes code for hypothetical proteins and the role of these genes warrants further investigation.
Table 3.
Summary of the transcriptome comparison of S. pneumoniae D39 wild-type grown in CDM with 0 mM niacin to grown in CDM with 10 mM niacin.
| D39 taga | Functionb | Ratioc |
|---|---|---|
| UPREGULATED GENES | ||
| spd_0093 | Hypothetical protein | 3.1 |
| spd_0094 | Hypothetical protein | 2.8 |
| spd_0095 | Hypothetical protein | 2.4 |
| spd_0474 | Hypothetical protein | 4.6 |
| spd_0475 | CAAX amino terminal protease family protein | 3.5 |
| spd_0526 | Fructose-1,6-bisphosphate aldolase, class II, Fba | 1.5 |
| spd_0976 | Redox-sensitive transcriptional regulator Rex | 1.5 |
| spd_1004 | Glyceraldehyde-3-phosphate dehydrogenase, NADP-dependent, GapN | 3.5 |
| spd_1091 | Substrate-specific component predicted niacin ECF transporter, NiaX | 1.8 |
| spd_1250 | NAD+ synthetase, NadE | 1.5 |
| spd_1251 | Nicotinate phosphoribosyltransferase, putative, PncB | 1.9 |
| spd_1640 | Ribosyl nicotinamide transporter, PnuC-like, PnuC | 4.2 |
| spd_1798 | DNA-binding response regulator | 2.1 |
| spd_1799 | Sensor histidine kinase, putative | 2.0 |
| spd_1800 | Hypothetical protein | 2.4 |
| spd_1801 | ABC transporter, ATP-binding protein | 2.0 |
| spd_1802 | Hypothetical protein | 2.2 |
| spd_1823 | Glyceraldehyde-3-phosphate dehydrogenase, type I, Gap | 1.7 |
| spd_1824 | Hypothetical protein | 2.2 |
| spd_1826 | Nicotinate-nucleotide pyrophosphorylase, NadC | 4.4 |
| spd_1827 | Hypothetical protein | 3.1 |
| spd_1833 | PTS system, IIA component | 1.7 |
| spd_1834 | Alcohol dehydrogenase, iron-containing, AdhE | 5.8 |
| spd_1865 | Alcohol dehydrogenase, zinc-containing, AdhB2 | 1.7 |
| spd_1874 | LysM domain protein | 3.7 |
| DOWNREGULATED GENES | ||
| spd_0113 | Hypothetical protein | −2.9 |
| spd_0114 | Hypothetical protein | −3.1 |
| spd_0115 | Hypothetical protein | −2.7 |
| spd_0122 | Hypothetical protein | −2.2 |
| spd_0123 | Hypothetical protein | −2.4 |
| spd_0124 | Hypothetical protein | −2.0 |
Gene numbers refer to D39 locus tags.
D39 annotation/TIGR4 annotation (Lanie et al., 2007).
Ratio represents the fold increase/decrease in the expression of genes in CDM with 0 mM Niacin to CDM with 10 mM Niacin. Errors in the ratios never exceeded 10% of the given values.
Putative niacin biosynthesis pathway genes were significantly upregulated in the absence of niacin (fba, rex, gapN, niaX, pncB-nadE, pnuC, gap, spd-1824, nadC, adhE, and adhB2). fba codes for a fructose-bisphosphate aldolase, whereas rex encodes a redox-sensitive transcriptional regulator. Similarly, gapN encodes a glyceraldehyde-3-phosphate dehydrogenase that is involved in generation of NADPH from NADH. pncB encodes a nicotinate phosphoribosyltransferase that converts nicotinate into nicotinate D-ribonucleotide and vice versa, whereas nadE encodes a NAD+ synthetase that converts deamino-NAD+ to NAD+ and adhE codes for an alcohol dehydrogenase. gap encodes another glyceraldehyde-3-phosphate dehydrogenase and adhE codes for an iron-containing alcohol dehydrogenase, whereas adhB2 encodes a zinc-containing alcohol dehydrogenase. NiaX (encoded by niaX) is a substrate-specific component predicted niacin ECF transporter, whereas PnuC (encoded by pnuC) is a ribosyl nicotinamide transporter. NadC (encoded by nadC) is a nicotinate-nucleotide pyrophosphorylase and has been proposed to convert quinolinate formed from alanine, aspartate, and glutamate, and tryptophan metabolism into nicotinate D-ribonucleotide (Kanehisa et al., 2014).
Niacin-dependent expression of fba, rex, gapN, niaX, pncB, pnuC, gap, spd-1824, nadC, adhE, and adhB2
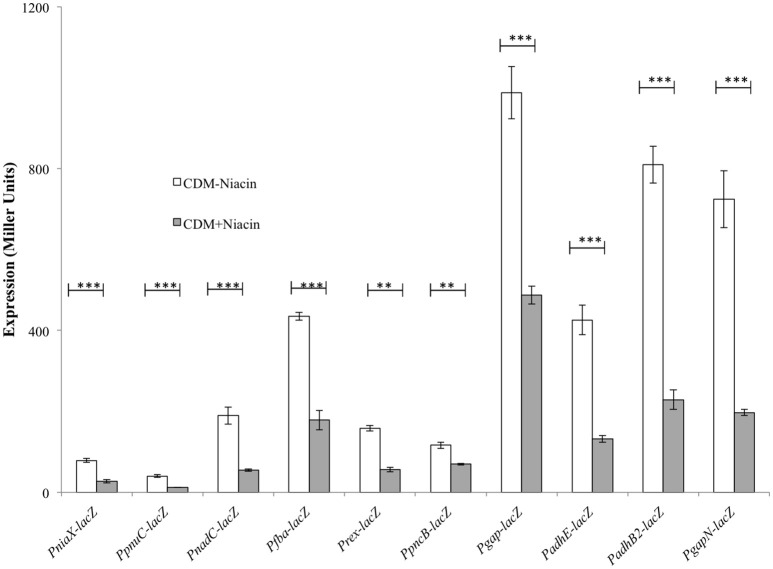
Our niacin-dependent microarray data mentioned above indicated the role of niacin in the regulation of fba, rex, gapN, niaX, pncB, pnuC, gap, spd-1824, nadC, adhE, and adhB2. To confirm our microarray results and further study the effect of niacin on the expression of fba, rex, gapN, niaX, pncB, pnuC, gap, spd-1824, nadC, adhE, and adhB2, we performed β-galactosidase assays with promoter lacZ-fusions of these genes constructed in S. pneumoniae D39 wild-type. Our β-galactosidase data demonstrated that the expression of Pfba-lacZ, Prex-lacZ, PgapN-lacZ, PniaX-lacZ, PpncB-lacZ, PpnuC-lacZ, Pgap-lacZ, PnadC-lacZ, PadhE-lacZ, and PadhB2-lacZ increased significantly in the absence of niacin in the medium (Figure 1). These data further confirm our microarray data described above and suggest the role of niacin in the regulation of these genes.
Figure 1.
Expression levels (in Miller units) of PniaX-lacZ, PpnuC-lacZ, PnadC-lacZ, Pfba-lacZ, Prex-lacZ, PpncB-lacZ, Pgap-lacZ, PadhE-lacZ, PadhB2-lacZ, and PgapN-lacZ in CDM with 0 and 10 mM niacin in S. pneumoniae D39 wild-type. Standard deviations of three independent experiments are indicated in bars. Statistical significance of the differences in the expression levels was determined by one-way ANOVA (NS, not significant, **P < 0.001, and ***P < 0.0001).
Microarray analysis of D39 ΔniaR
Niacin genes are mostly regulated by a transcriptional regulator NiaR in different bacteria (Novichkov et al., 2010). In Firmicutes and Thermotogales, transcriptional regulator NiaR regulates the NAD biosynthesis and salvage of niacin (Rodionov et al., 2008a). NiaR was first studied in B. subtilis as a niacin-responsive transcriptional repressor that binds to its DNA targets in the presence of niacin (Rossolillo et al., 2005). NiaR belongs to a unique protein family, which possesses an N-terminal HTH (Helix-Turn-Helix) DNA binding domain (PF08279) and a C-terminal effector binding domain, called the 3H domain (PF02829). S. pneumoniae also possesses a NiaR transcriptional regulator, which might be involved in the regulation of the niacin-regulated genes described above. Therefore, we decided to further study the role of NiaR in the regulation of these genes.
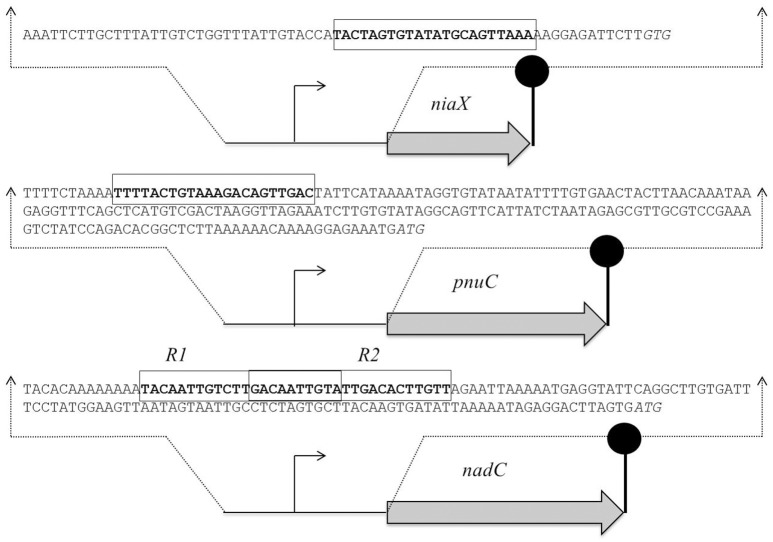
A deletion mutant of the niaR gene was constructed and microarray comparison of S. pneumoniae D39 ΔniaR to D39 wild-type grown in complete CDM was performed to investigate the role of NiaR in S. pneumoniae D39. Complete CDM contains 8 μM of niacin. Table 4 summarizes the transcriptome changes induced by the deletion of niaR in S. pneumoniae D39. Expression of niaR was downregulated about 3-fold confirming the niaR deletion in D39 ΔniaR. Expression of nadC, niaX, and pnuC was upregulated significantly in D39 ΔniaR, suggesting the role of NiaR as a transcriptional repressor of niaX, nadC, and pnuC in S. pneumoniae D39. Expression of spd-1824 and spd-1827 (coding for hypothetical proteins) was also upregulated. Spd-1827 is localized adjacent to nadC (spd-1826), but transcribed in opposite direction.
Table 4.
Summary of transcriptome comparison of S. pneumoniae D39 ΔniaR compared to the D39 wild-type grown in complete CDM.
| D39 taga | Functionb | Ratioc |
|---|---|---|
| spd_1091 | Substrate-specific component predicted niacin ECF transporter, NiaX | 2.1 |
| spd_1093 | Transcriptional regulator, biotin repressor family protein, NiaR | −2.7 |
| spd_1640 | Ribosyl nicotinamide transporter, PnuC-like, PnuC | 1.5 |
| spd_1824 | Hypothetical protein | 3.5 |
| spd_1826 | Nicotinate-nucleotide pyrophosphorylase, NadC | 7.2 |
| spd_1827 | Hypothetical protein | 3.1 |
Complete CDM contains 8 μM of niacin.
Gene numbers refer to D39 locus tags.
D39 annotation/TIGR4 annotation (Lanie et al., 2007).
Ratio represents the fold increase/decrease in the expression of genes in D39 ΔniaR compared to the D39 wild-type in complete CDM. Errors in the ratios never exceeded 10% of the given values.
Role of NiaR as a transcriptional repressor of niaX, nadC, and pnuC
To further investigate the role of NiaR in the regulation of niaX, nadC, and pnuC, we transformed the lacZ-fusions of the promoter regions of niaX, nadC, and pnuC into D39 ΔniaR and performed β-galactosidase assays in complete CDM (Figure 2). The results of the β-galactosidase assays showed that the activity of all these promoters increased significantly in D39 ΔniaR compared to the D39 wild-type, confirming the role of NiaR as a transcriptional repressor of niaX, nadC, and pnuC.
Figure 2.
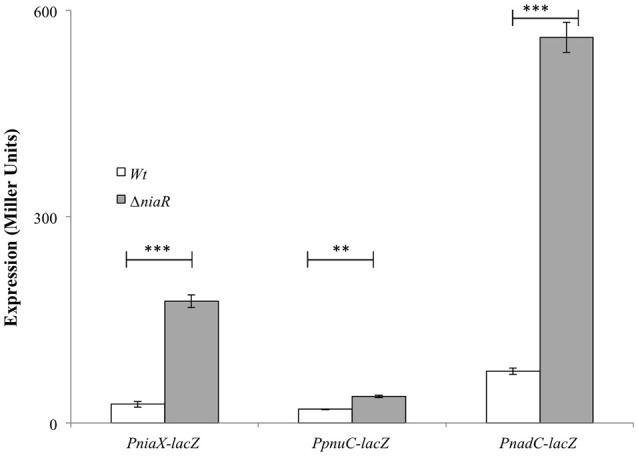
Expression levels (in Miller units) of PniaX-lacZ, PpnuC-lacZ, and PnadC-lacZ in complete CDM in S. pneumoniae D39 wild-type and D39 ΔniaR. Standard deviations of three independent experiments are indicated in bars. Statistical significance of the differences in the expression levels was determined by one-way ANOVA (NS, not significant, **P < 0.001, and ***P < 0.0001).
Prediction and confirmation of the NiaR site in PniaX, PnadC, and PpnuC
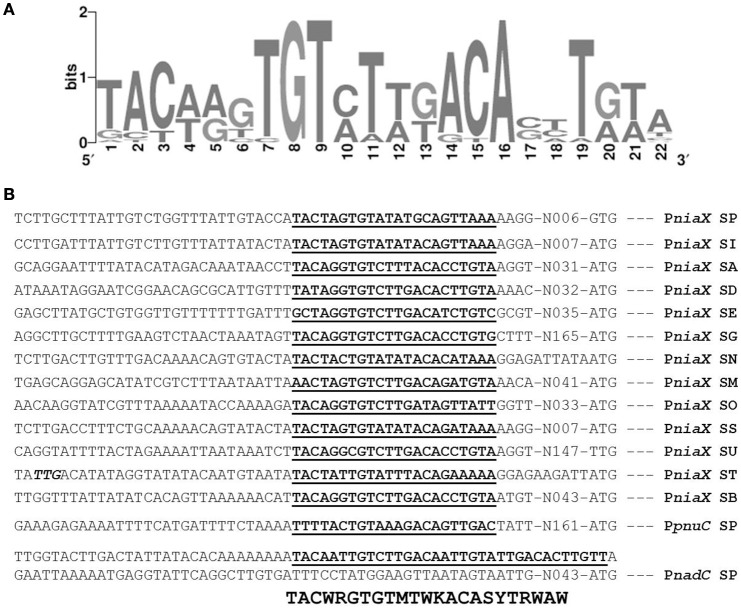
The promoter regions of all of the upregulated genes, including spd_1824 and spd_1827, were analyzed by Genome2D software (Baerends et al., 2004) and a MEME motif sampler search (Bailey and Elkan, 1994). A 22-bp palindromic-like sequence was found in the promoter regions of niaX, nadC, and pnuC (Figure 3). This DNA sequence might serve as the NiaR operator site in S. pneumoniae. PniaX from different streptococci was also analyzed for the presence of NaiR site. The NiaR site present in the promoter region of niaX of different streptococci is shown in Figure 4. Weight matrix based on these putative NiaR sites (5′- TACWRGTGTMTWKACASYTRWAW -3′) was constructed (Figure 4).
Figure 3.
Organization of the NiaR-regulated genes in S. pneumoniae D39. Putative NiaR operator sequences are rectangle and translational initiation sites are italicized, whereas the lollipop structures represent the putative transcriptional terminators. See text for further details.
Figure 4.
Identification of the NiaR operator site. (A) Weight matrix of the identified NiaR operator site in the promoter regions of niaX, nadC, and pnuC. (B) Position of the NiaR operator site in the promoter region of niaX, nadC, and pnuC in different streptococci. Putative NiaR operator sites are bold and underlined. SP, S. pneumoniae; SI, Streptococcus mitis; SA, Streptococcus agalactiae; SD, Streptococcus dysgalactiae; SE, Streptococcus equi; SG, Streptococcus gallolyticus; SN, Streptococcus gordonii; SM, Streptococcus mutans; SO, Streptococcus pyogenes; SS, Streptococcus sanguinis; SU, Streptococcus suis; ST, Streptococcus thermophiles; and SB, Streptococcus uberis.
The predicted NiaR operator site present in the promoter regions of niaX, nadC, and pnuC was further verified by promoter mutational experiment. For this purpose, we made transcriptional lacZ-fusions of PniaX, PpnuC, and PnadC, where conserved bases in the putative NiaR sites were mutated in PniaX (5′- TACTAGTGTATATGCAGTTAAA-3′ to 5′- TACTAGTACATATGCCGTTAAA -3′), PpnuC (5′- TTTTACTGTAAAGACAGTTGAC -3′ to 5′- TTTTACTACAAAGACGGTTGAC -3′), PnadC-R1 (5′- TACAATTGTCTTGACAATTGTA -3′ to 5′- TACAATTACCTTGACCATTGTA -3′), and PnadC-R2 (5′- GACAATTGTATTGACACTTGTT -3′ to 5′- GACAATTACATTGACCCTTGTT -3′). β-galactosidase assays were performed on cells grown in complete CDM. Complete CDM contains 8 μM of niacin. The expression of PniaX and PpnuC with mutated conserved bases of NiaR operator sites increased significantly in S. pneumoniae D39 wild-type, confirming that the predicted NiaR sites present in the promoter regions of niaX and pnuC are active and intact in S. pneumoniae (Figure 5). Two putative operator sites for NiaR are present in PnadC (R1 and R2). We mutated both sites individually and performed β-galactosidase assays. We could only observe derepression (caused by NiaR) in the activity of PnadC when NiaR operator site 2 (R2) was mutated and did not observe any change in the activity of PnadC due to mutation in NiaR operator site 1 (R1) (Figure 5). These data suggest that operator site 2 (R2) is the functional operator site in PnadC.
Figure 5.
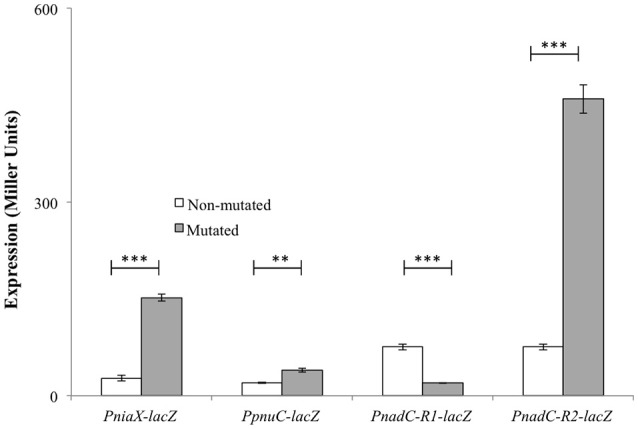
Expression levels (in Miller units) of PniaX-lacZ, PpnuC-lacZ, and PnadC-lacZ with mutated and non-mutated NiaR operator sites in S. pneumoniae D39 wild-type grown in complete CDM. Standard deviations of three independent experiments are indicated in bars. Statistical significance of the differences in the expression levels was determined by one-way ANOVA (NS, not significant, **P < 0.001, and ***P < 0.0001).
Discussion
NAD is an essential cofactor used by all living organisms. NAD synthesis is a tightly regulated intracellular process in bacteria (Huang et al., 2009). Bacteria acquire NAD in two main ways: through de novo synthesis and through the salvage pathway. Some bacteria do not have the ability to de novo synthesize NAD and must make use of the salvage pathway to import niacin or nicotinamide riboside through the substrate importers NiaX and PnuC, respectively. The de novo pathway synthesizes NAD from aspartic acid, whereas the salvage pathway brings intermediates many steps downstream into the NAD de novo synthesis pathway (Rodionov et al., 2008b). NiaX and PnuC are the two major importers in the NAD salvage pathway, where NiaX is responsible for niacin uptake, and PnuC transports nicotinamide riboside (Herbert et al., 2003; Sauer E. et al., 2004; Rodionov et al., 2008a, 2009). Our current study demonstrates the transcriptomic response of S. pneumoniae to niacin and reveals that a number of genes including pnuC, pncB, and nadC are differentially expressed under the tested conditions. We further demonstrate that a transcriptional regulator NiaR acts as a transcriptional repressor of niaX, pnuC, and nadC in the presence of niacin.
An extracellular protein capable of modifying nicotinamide mononucleotide to an importable form appears to help NiaX and PnuC for importing nicotinamide mononucleotide or there may be another import system in S. pneumoniae (Johnson et al., 2015). There is significant variability between PnuC homologs (Jaehme et al., 2014), and the PnuC homologs from Haemophilus influenzae and Salmonella typhimurium do not import nicotinamide mononucleotide, but can transform it to an importable form for PnuC (nicotinamide riboside) through NadN or AphA, respectively (Kemmer et al., 2001; Grose et al., 2005). The PnuC proteins from H. influenzae, S. typhimurium, and S. pneumoniae all possess the motif for nicotinamide mononucleotide binding. Nevertheless, PnuC homologs from many other organisms lack the consensus binding residues (Kemmer et al., 2001; Sauer E. et al., 2004; Grose et al., 2005). These observations indicate that different groups of NAD salvage substrate importers (annotated as PnuC) import nicotinamide riboside and/or nicotinamide mononucleotide, and that NiaX imports niacin and/or nicotinamide mononucleotide as preferred substrates. Moreover, the amino acids in Salmonella PnuC curtailing import of nicotinamide mononucleotide are not conserved in the pneumococci, suggesting that the pneumococcal PnuC may permit this substrate along with nicotinamide riboside. Although, both PnuC and NiaX in S. pneumoniae may have acquired the ability to import nicotinamide mononucleotide, an extra importer (that is yet to be characterized) may also be present (Johnson et al., 2015). The role of PnuC in pneumococcal pathogenesis has been studied and PnuC could be a potential viable small molecule therapeutic target to halt disease progression in the host (Johnson et al., 2015). The proposed NAD pathway in S. pneumoniae states that niacin and nicotinamide enter the cells through NiaX, and PnuC transports nicotinamide riboside to the inside of the cell, whereas the transporter for nicotinamide mononucleotide is unknown (Johnson et al., 2015). spd-1411 encodes a nicotinamidase (PncA) that converts nicotinamide into niacin, which is further converted into nicotinate mononucleotide by a nicotinic acid phosphoribosyltransferase (PncB) (Johnson et al., 2015). The nicotinate mononucleotide is then converted into NAD by NadD and NadE. Moreover, NadD (nicotinate/nicotinamide nucleotide adenylyltransferase) converts nicotinamide riboside and nicotinamide mononucleotide into NAD (Johnson et al., 2015). Nicotinamide riboside augmentation has been attributed to several advantageous functions in the host, including shielding against mitochondrial myopathy (Khan et al., 2014), hearing loss (Brown et al., 2014) and obesity (Cantó et al., 2012). These functions may not be due to increasing NAD synthesis (Frederick et al., 2015), but may be due to overall bioavailability. While nicotinamide riboside is required for pathogen and host, luckily pneumococcal PnuC and its homologous in other bacteria do not have sequence homology to any proteins in the animal kingdom. Hence, PnuC could be a potential therapeutic target in bacterial species shielding this pathway without mammalian significance as has been effectively shown with H. influenzae (Sauer E. et al., 2004).
NiaR orthologs have been found in 30 out of 45 species from the Bacillus/Clostridium group (Firmicutes), in addition to the diverged groups of the Fusobacteria and Thermotogales and for the Bacillus/Clostrida group another DNA binding site was proposed (Rodionov et al., 2008a). There are two different types of DNA-binding sites of NiaR i.e., type I operator found in Firmicutes and Fusobacteria, and type II in the Thermotogales. The niacin-responsive transcription factor NiaR (known as YrxA in B. subtilis) was first recognized as a nicotinic acid-responsive repressor of the de novo NAD biosynthesis operon (nadABC) in B. subtilis (Rossolillo et al., 2005). NiaR regulation of the niacin salvage genes pncB (in Lactobacillus plantarum), pncA (in Streptococcus pyogenes, Streptococcus equi, and Clostridium tetani), and/or the RNam salvage transporter pnuC (in S. pneumoniae and Streptococcus mutans) (Rodionov et al., 2008a) is less common. Moreover, the NiaR regulon contains membrane proteins that putatively have a role in niacin uptake. The most abundant NiaP family is found in ten NiaR-containing species (Bacilli, Lactobacilli and Thermotogales) in addition to several species that do not have the NiaR regulator (Rodionov et al., 2008a). Among Streptococci and Clostridia, NiaX is found in twelve genomes, and NiaY is found in five genomes (Bacilli and Clostridia). Several lines of genomic evidence support the putative involvement of these gene families in niacin uptake including the predicted co-regulation with NAD biosynthesis and niacin salvage genes, and co-occurrence with the niacin salvage genes pncB-pncA (Rodionov et al., 2008a). Our study demonstrates that niaX, pnuC, and nadC are the genes that have a putative NiaR operator site in their promoter regions and are repressed by NiaR in the presence of niacin. We have further confirmed the NiaR operator sites in the promoter regions of niaX, pnuC, and nadC by mutagenesis studies. There are some other genes that are differentially expressed under our tested conditions (fba, rex, gapN, pncB-nadE, gap, spd-1824, spd-1827, adhE, and adhB2). The change in the expression of these genes suggests that these genes may have a role in the transport and biosynthesis of niacin or they may be upregulated due to some indirect effect of niacin genes. These genes do not have a putative NiaR operator site in their promoter regions suggesting the role of another transcriptional regulator in the regulation of fba, rex, gapN, pncB-nadE, gap, spd-1824, spd-1827, adhE, and adhB2. Therefore, we propose that the study of the regulatory mode of the above-mentioned genes would shed light on this possibility.
Author contributions
Substantial contributions to the conception or design of the work; or the acquisition, analysis, or interpretation of data for the work: MA, SS, and OK. Drafting the work or revising it critically for important intellectual content: MA, SS, and OK. Final approval of the version to be published: MA, SS, and OK. Agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved: MA, SS, and OK.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
MA is supported by the Government College University, Faisalabad, Pakistan under the faculty development program of HEC Pakistan.
References
- Afzal M., Manzoor I., Kuipers O. P. (2015). A fast and reliable pipeline for bacterial transcriptome analysis case study: serine-dependent gene regulation in Streptococcus pneumoniae. J. Vis. Exp. 98:e52649 10.3791/52649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afzal M., Shafeeq S., Kuipers O. P. (2014). LacR is a repressor of lacABCD and LacT is an activator of lacTFEG, constituting the lac gene cluster in Streptococcus pneumoniae. Appl. Environ. Microbiol. 80, 5349–5358. 10.1128/aem.01370-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerends R. J. S., Smits W. K., de Jong A., Hamoen L. W., Kok J., Kuipers O. P. (2004). Genome2D: a visualization tool for the rapid analysis of bacterial transcriptome data. Genome Biol. 5:R37. 10.1186/gb-2004-5-5-r37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T. L., Elkan C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. Biol. 2, 28–36. [PubMed] [Google Scholar]
- Bogaert D., De Groot G. R., Hermans P. W. (2004). Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect. Dis. 4, 144–154. 10.1016/S1473-3099(04)00938-7 [DOI] [PubMed] [Google Scholar]
- Bräsen C., Esser D., Rauch B., Siebers B. (2014). Carbohydrate metabolism in Archaea: current insights into unusual enzymes and pathways and their regulation. Microbiol. Mol. Biol. Rev. 78, 89–175. 10.1128/MMBR.00041-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K. D., Maqsood S., Huang J.-Y., Pan Y., Harkcom W., Li W., et al. (2014). Activation of SIRT3 by the NAD+ precursor nicotinamide riboside protects from noise-induced hearing loss. Cell Metab. 20, 1059–1068. 10.1016/j.cmet.2014.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantó C., Houtkooper R. H., Pirinen E., Youn D. Y., Oosterveer M. H., Cen Y., et al. (2012). The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab. 15, 838–847. 10.1016/j.cmet.2012.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalkiadaki A., Guarente L. (2012). Sirtuins mediate mammalian metabolic responses to nutrient availability. Nat. Rev. Endocrinol. 8, 287–296. 10.1038/nrendo.2011.225 [DOI] [PubMed] [Google Scholar]
- Chiarugi A., Dölle C., Felici R., Ziegler M. (2012). The NAD metabolome–a key determinant of cancer cell biology. Nat. Rev. Cancer 12, 741–752. 10.1038/nrc3340 [DOI] [PubMed] [Google Scholar]
- Cotter P. A., DiRita V. J. (2000). Bacterial virulence gene regulation: an evolutionary perspective. Annu. Rev. Microbiol. 54, 519–565. 10.1146/annurev.micro.54.1.519 [DOI] [PubMed] [Google Scholar]
- Cummings C. A., Bootsma H. J., Relman D. A., Miller J. F. (2006). Species- and strain-specific control of a complex, flexible regulon by Bordetella BvgAS. J. Bacteriol. 188, 1775–1785. 10.1128/JB.188.5.1775-1785.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards R. L., Bryan A., Jules M., Harada K., Buchrieser C., Swanson M. S. (2013). Nicotinic acid modulates Legionella pneumophila gene expression and induces virulence traits. Infect. Immun. 81, 945–955. 10.1128/IAI.00999-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi Y., Oshima T., Mori H., Aono R., Yamamoto K., Ishihama A., et al. (2003). Transcriptional regulation of drug efflux genes by EvgAS, a two-component system in Escherichia coli. Microbiology 149, 2819–2828. 10.1099/mic.0.26460-0 [DOI] [PubMed] [Google Scholar]
- Frederick D. W., Davis J. G., Dávila A., Agarwal B., Michan S., Puchowicz M. A., et al. (2015). Increasing NAD synthesis in muscle via nicotinamide phosphoribosyltransferase is not sufficient to promote oxidative metabolism. J. Biol. Chem. 290, 1546–1558. 10.1074/jbc.M114.579565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grose J. H., Bergthorsson U., Xu Y., Sterneckert J., Khodaverdian B., Roth J. R. (2005). Assimilation of nicotinamide mononucleotide requires periplasmic AphA phosphatase in Salmonella enterica. J. Bacteriol. 187, 4521–4530. 10.1128/JB.187.13.4521-4530.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann A., Hakenbeck R., Bruckner R. (2007). A new integrative reporter plasmid for Streptococcus pneumoniae. FEMS Microbiol. Lett. 268, 217–224. 10.1111/j.1574-6968.2006.00584.x [DOI] [PubMed] [Google Scholar]
- Herbert M., Sauer E., Smethurst G., Kraiss A., Hilpert A.-K., Reidl J. (2003). Nicotinamide ribosyl uptake mutants in Haemophilus influenzae. Infect. Immun. 71, 5398–5401. 10.1128/IAI.71.9.5398-5401.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N., De Ingeniis J., Galeazzi L., Mancini C., Korostelev Y. D., Rakhmaninova A. B., et al. (2009). Structure and function of an ADP-ribose-dependent transcriptional regulator of NAD metabolism. Structure 17, 939–951. 10.1016/j.str.2009.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino Y., Shinagawa H., Makino K., Tsunasawa S., Sakiyama F., Nakata A. (1986). Nucleotide sequence of the lig gene and primary structure of DNA ligase of Escherichia coli. Mol. Gen. Genet. 204, 1–7. 10.1007/BF00330179 [DOI] [PubMed] [Google Scholar]
- Israelsen H., Madsen S. M., Vrang A., Hansen E. B., Johansen E. (1995). Cloning and partial characterization of regulated promoters from Lactococcus lactis Tn917-lacZ integrants with the new promoter probe vector, pAK80. Appl. Environ. Microbiol. 61, 2540–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaehme M., Guskov A., Slotboom D. J. (2014). Crystal structure of the vitamin B3 transporter PnuC, a full-length SWEET homolog. Nat. Struct. Mol. Biol. 21, 1013–1015. 10.1038/nsmb.2909 [DOI] [PubMed] [Google Scholar]
- Johnson M. D. L., Echlin H., Dao T. H., Rosch J. W. (2015). Characterization of NAD salvage pathways and their role in virulence in Streptococcus pneumoniae. Microbiology 161, 2127–2136. 10.1099/mic.0.000164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurtshuk P. (1996). Bacterial Metabolism, in Medical Microbiology, ed Baron S. (Galveston, TX: University of Texas Medical Branch at Galveston; ). [PubMed] [Google Scholar]
- Kadioglu A., Weiser J. N., Paton J. C., Andrew P. W. (2008). The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat. Rev. Microbiol. 6, 288–301. 10.1038/nrmicro1871 [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Sato Y., Kawashima M., Furumichi M., Tanabe M. (2014). Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 42, D199–D205. 10.1093/nar/gkt1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemmer G., Reilly T. J., Schmidt-Brauns J., Zlotnik G. W., Green B. A., Fiske M. J., et al. (2001). NadN and e (P4) are essential for utilization of NAD and nicotinamide mononucleotide but not nicotinamide riboside in Haemophilus influenzae. J. Bacteriol. 183, 3974–3981. 10.1128/JB.183.13.3974-3981.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan N. A., Auranen M., Paetau I., Pirinen E., Euro L., Forsström S., et al. (2014). Effective treatment of mitochondrial myopathy by nicotinamide riboside, a vitamin B3. EMBO Mol. Med. 6, 721–731. 10.1002/emmm.201403943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman T. G., Bijlsma J. J. E., Kok J., Kuipers O. P. (2006). To have neighbour's fare: extending the molecular toolbox for Streptococcus pneumoniae. Microbiology 152, 351–359. 10.1099/mic.0.28521-0 [DOI] [PubMed] [Google Scholar]
- Kotrbova-Kozak A., Kotrba P., Inui M., Sajdok J., Yukawa H. (2007). Transcriptionally regulated adhA gene encodes alcohol dehydrogenase required for ethanol and n-propanol utilization in Corynebacterium glutamicum R. Appl. Microbiol. Biotechnol. 76, 1347–1356. 10.1007/s00253-007-1094-6 [DOI] [PubMed] [Google Scholar]
- Lanie J. A., Ng W. L., Kazmierczak K. M., Andrzejewski T. M., Davidsen T. M., Wayne K. J., et al. (2007). Genome sequence of Avery's virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J. Bacteriol. 189, 38–51. 10.1128/JB.01148-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luong T. T., Kim E.-H., Bak J. P., Nguyen C. T., Choi S., Briles D. E., et al. (2015). Ethanol-induced alcohol dehydrogenase E (AdhE) potentiates pneumolysin in Streptococcus pneumoniae. Infect. Immun. 83, 108–119. 10.1128/IAI.02434-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda N., Church G. M. (2002). Escherichia coli gene expression responsive to levels of the response regulator EvgA. J. Bacteriol. 184, 6225–6234. 10.1128/JB.184.22.6225-6234.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda N., Church G. M. (2003). Regulatory network of acid resistance genes in Escherichia coli. Mol. Microbiol. 48, 699–712. 10.1046/j.1365-2958.2003.03477.x [DOI] [PubMed] [Google Scholar]
- Matsubara K., Yokooji Y., Atomi H., Imanaka T. (2011). Biochemical and genetic characterization of the three metabolic routes in Thermococcus kodakarensis linking glyceraldehyde 3-phosphate and 3-phosphoglycerate. Mol. Microbiol. 81, 1300–1312. 10.1111/j.1365-2958.2011.07762.x [DOI] [PubMed] [Google Scholar]
- McPheat W. L., Wardlaw A. C., Novotny P. (1983). Modulation of Bordetella pertussis by nicotinic acid. Infect. Immun. 41, 516–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. F., Roy C. R., Falkow S. (1989). Analysis of Bordetella pertussis virulence gene regulation by use of transcriptional fusions in Escherichia coli. J. Bacteriol. 171, 6345–6348. 10.1128/jb.171.11.6345-6348.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves A. R., Ventura R., Mansour N., Shearman C., Gasson M. J., Maycock C., et al. (2002). Is the glycolytic flux in Lactococcus lactis primarily controlled by the redox charge? Kinetics of NAD(+) and NADH pools determined in vivo by 13C NMR. J. Biol. Chem. 277, 28088–28098. 10.1074/jbc.M202573200 [DOI] [PubMed] [Google Scholar]
- Nishino K., Inazumi Y., Yamaguchi A. (2003). Global analysis of genes regulated by EvgA of the two-component regulatory system in Escherichia coli. J. Bacteriol. 185, 2667–2672. 10.1128/JB.185.8.2667-2672.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobelmann B., Lengeler J. W. (1996). Molecular analysis of the gat genes from Escherichia coli and of their roles in galactitol transport and metabolism. J. Bacteriol. 178, 6790–6795. 10.1128/jb.178.23.6790-6795.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novichkov P. S., Laikova O. N., Novichkova E. S., Gelfand M. S., Arkin A. P., Dubchak I., et al. (2010). RegPrecise: a database of curated genomic inferences of transcriptional regulatory interactions in prokaryotes. Nucleic Acids Res. 38, D111–D118. 10.1093/nar/gkp894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M. S., Nemeria N. S., Furey W., Jordan F. (2014). The pyruvate dehydrogenase complexes: structure-based function and regulation. J. Biol. Chem. 289, 16615–16623. 10.1074/jbc.R114.563148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips N. J., John C. M., Reinders L. G., Gibson B. W., Apicella M. A., Griffiss J. M. (1990). Structural models for the cell surface lipooligosaccharides of Neisseria gonorrhoeae and Haemophilus influenzae. Biomed. Environ. Mass Spectrom. 19, 731–745. 10.1002/bms.1200191112 [DOI] [PubMed] [Google Scholar]
- Rodionov D. A., De Ingeniis J., Mancini C., Cimadamore F., Zhang H., Osterman A. L., et al. (2008b). Transcriptional regulation of NAD metabolism in bacteria: NrtR family of Nudix-related regulators. Nucleic Acids Res. 36, 2047–2059. 10.1093/nar/gkn047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov D. A., Hebbeln P., Eudes A., ter Beek J., Rodionova I. A., Erkens G. B., et al. (2009). A novel class of modular transporters for vitamins in prokaryotes. J. Bacteriol. 191, 42–51. 10.1128/JB.01208-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov D. A., Li X., Rodionova I. A., Yang C., Sorci L., Dervyn E., et al. (2008a). Transcriptional regulation of NAD metabolism in bacteria: genomic reconstruction of NiaR (YrxA) regulon. Nucleic Acids Res. 36, 2032–2046. 10.1093/nar/gkn046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossolillo P., Marinoni I., Galli E., Colosimo A., Albertini A. M. (2005). YrxA is the transcriptional regulator that represses de novo NAD biosynthesis in Bacillus subtilis. J. Bacteriol. 187, 7155–7160. 10.1128/JB.187.20.7155-7160.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh M. S., Lindahl T. (1992). Role of poly(ADP-ribose) formation in DNA repair. Nature 356, 356–358. 10.1038/356356a0 [DOI] [PubMed] [Google Scholar]
- Sauer E., Merdanovic M., Mortimer A. P., Bringmann G., Reidl J. (2004). PnuC and the utilization of the nicotinamide riboside analog 3-aminopyridine in Haemophilus influenzae. Antimicrob. Agents Chemother. 48, 4532–4541. 10.1128/AAC.48.12.4532-4541.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer U., Canonaco F., Heri S., Perrenoud A., Fischer E. (2004). The soluble and membrane-bound transhydrogenases UdhA and PntAB have divergent functions in NADPH metabolism of Escherichia coli. J. Biol. Chem. 279, 6613–6619. 10.1074/jbc.M311657200 [DOI] [PubMed] [Google Scholar]
- Schneider D. R., Parker C. D. (1982). Effect of pyridines on phenotypic properties of Bordetella pertussis. Infect. Immun. 38, 548–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafeeq S., Afzal M., Henriques-Normark B., Kuipers O. P. (2015). Transcriptional profiling of UlaR-regulated genes in Streptococcus pneumoniae. Genomics Data 4, 57–59. 10.1016/j.gdata.2015.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirover M. A. (2011). On the functional diversity of glyceraldehyde-3-phosphate dehydrogenase: biochemical mechanisms and regulatory control. Biochim. Biophys. Acta 1810, 741–751. 10.1016/j.bbagen.2011.05.010 [DOI] [PubMed] [Google Scholar]
- Titgemeyer F., Hillen W. (2002). Global control of sugar metabolism: a gram-positive solution. Antonie Van Leeuwenhoek 82, 59–71. 10.1023/A:1020628909429 [DOI] [PubMed] [Google Scholar]
- Wei Z., Fu Y., Zhou E., Tian Y., Yao M., Li Y., et al. (2014). Effects of niacin on Staphylococcus aureus internalization into bovine mammary epithelial cells by modulating NF-κB activation. Microb. Pathog. 71–72, 62–67. 10.1016/j.micpath.2014.03.005 [DOI] [PubMed] [Google Scholar]
- Wilkinson A., Day J., Bowater R. (2001). Bacterial DNA ligases. Mol. Microbiol. 40, 1241–1248. 10.1046/j.1365-2958.2001.02479.x [DOI] [PubMed] [Google Scholar]