Abstract
Background:
Selection of stably expressing housekeeping genes (HKGs) is a crucial step in gene expression analysis. However, there are no universal HKGs for all experiments, and they should be determined by each biologic condition. The aim of this study was to detect appropriate HKGs for kidney cells cultured in long-term hypoxia.
Materials and Methods:
Based on a screening step using a microarray data available from gene expression omnibus database, a set of candidate HKGs were chosen to be assessed in human kidney cells cultured in hypoxic or normoxic conditions for about 2 weeks in a time course manner. The stability of gene expression was assessed by refFinder, a web-based tool that integrates four computational programs (geNorm, Normfinder, BestKeeper, and the comparative ΔΔCt method).
Results:
GAPDH and ACTB were the most stable genes in hypoxia treated cells whereas, B2M and ACTB were the best HKGs in cells cultured in normoxia. When both hypoxia and normoxia treated cells from all time points were evaluated together, GAPDH and ACTB equally showed the most stability.
Conclusion:
As in relative quantification of real-time polymerase chain reaction data, the same HKGs should be selected for all groups, we believe that GAPDH and ACTB are suitable HKGs for studies on the effect of hypoxia on cultured kidney cells.
Keywords: Gene expression analysis, housekeeping gene, hypoxia, kidney
Introduction
Oxygen is vital for the existence and appropriate function of most living organisms. Tissue hypoxia, a condition in which oxygen consumption exceeds oxygen delivery[1] is a common event in degenerative diseases and malignancies.[2,3] For instance, it is shown that bidirectional exacerbation of hypoxia and tubulointerstitial fibrosis in chronic kidney disease forms a vicious cycle, leading to end-stage renal disease.[4,5]
Our understanding of the hypoxia pathway mainly comes from gene expression analysis experiments with real-time polymerase chain reaction (PCR) being a common method in these studies.[6] Because of some variations in enzyme efficiency, unequal starting materials and the presence of inhibitors, data normalization is a pivotal step in this technique. Assessment of the expression of housekeeping genes (HKGs) is the most common approach for this purpose. HKGs are assumed to have constant expression rates and not to be influenced by different experiment conditions. However, this assumption is not true for frequently used HKGs in all situations. For instance, Schmittgenand Zakrajsek showed a big variation in GAPDH and ACTB, two frequently used HKGs, during stimulation of fibroblasts with serum.[7] de Kok et al. also showed that tissue type is a key parameter in the selection of an appropriate HKG as tissues differ in the most stable HKGs.[8] Therefore, it is generally recommended that for quantitative gene expression analysis in a given biological state, the most stable HKGs be selected among a set of HKGs in the context of a specific cell or tissue type.
Hypoxia affects a broad spectrum of cellular functions with a wide verity of genes being regulated by hypoxia pathway.[9] Therefore, determining genes with a near constant expression rate for data normalization in real-time PCR can be a big challenge in studied on hypoxia. In the current study, we have performed a time-course study on human kidney primary culture cells incubated in hypoxia or normoxia conditions for 2 weeks to determine the most stable HKGs from a set of genes selected from a microarray experiment.
Materials and Methods
Microarray data reanalysis
Microarray data were downloaded from gene expression omnibus (GEO) database at http://www.ncbi.nlm.nih.gov/geo/ and is accessible through GEO series accession number GSE11341 deposited by Costello et al.[10] In this experiment the responses of primary human pulmonary microvascular endothelial cells- (HMVEC-L) and cardiac microvascular endothelium to hypoxia was assessed in a time course manner. We employed GEO2R to re-analyze this data. The gene expression of endothelial cells incubated for 3, 24 and 48 h in hypoxic conditions (1% O2) was compared to normoxia (21% O2). Adjusted P value was calculated by Benjamini–Hochberg method. The relative expressions of six common HKGs, namely GAPDH, TFRC, ACTB, GUSB, TBP, and B2M, in different time points were determined, and coefficient of variation (CV) for each gene was calculated.
Cell culture
Pieces of about 1 cm3 size were extracted from the kidney cortex of a patient subjected to total nephrectomy. The tissue was minced with a surgical blade and incubated in collagenase I (Sigma, St Louis, USA) for 45 min. The cellular content was harvested by centrifugation in 300 g for 5 min and after washing with phosphate buffered saline (PBS) (GIBCO-BRL, Grand Island, USA) was cultured in dulbecco's modified eagle medium (Bioidea, Tehran, Iran) containing 10% fetal bovine serum (GIBCO-BRL) as well as penicillin G (100 U/mL; Sigma), streptomycin (100 mg/mL; Sigma) in 37°C and 5% CO2. After 24 h the flask was washed with PBS twice and fresh medium added. Cells were cultured either in normoxic or hypoxic (1% O2) conditions.
RNA isolation and cDNA synthesis
In days 0, 1, 3, 5, 7, 9, 11 and 13, cells cultured in normoxia or hypoxia were harvested for gene expression analysis. RNA was extracted with RNX-plus (Sinagen, Tehran, Iran) according to the manufacturer protocol. RNA purity and concentration were measured by BioSpectrometer® (Eppendorf, Hamburg, Germany). Random hexamer primed cDNA synthesis was carried out using revert aid first strand cDNA synthesis kit (Thermo Scientific, Rockford, USA).
Primer design
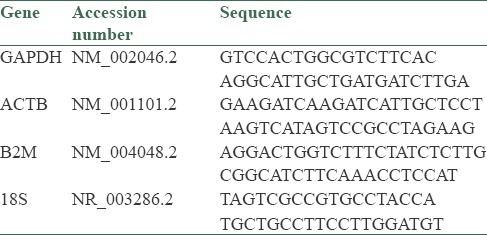
Exon junction primers for human GAPDH, ACTB, B2M and 18s rRNA were designed by AlleleID version 7.6 (Primer Biosoft, Palo Alto, USA). Primer specificity was assessed by NCBI BLAST. Primer sequences are shown in Table 1.
Table 1.
The sequence of primers
Real-time polymerase chain reaction
The expression of GAPDH, ACTB, B2M and 18s rRNA was assessed with Maxima™ SYBR Green qPCR Master Mix (Thermo Scientific) and Rotor Gene 6000 Real-Time PCR Machine (Corbett, Sydney, Australia). The temperature profile was as follows: An initial step of 95°C for 10 min and then 40 cycles of 95°C for 15 s and 60°C for 1 min.
Data analysis
For choosing the most stable HKG, raw cycle threshold values were analyzed with ref Finder at cotton expressed sequence tag database available at http://www.leonxie.com/referencegene.php.
Results
To determine the most stable reference genes in hypoxia, first we re-analyzed the data of a time course study carried out on human pulmonary endothelial cells[11] and evaluated the expression of six common HKGs (GAPDH, TFRC, ACTB, GUSB, TBP, and B2M). Cells in normoxia were assumed as control, and the expression fold changes were calculated for 3, 24 and 48 h incubation in hypoxia [Figure 1]. To determine the stability of expression, CV% of the fold changes in these 3 time points was calculated for each gene. According to this parameter, ACTB, GAPDH, and B2M show the least variations. In addition to these three gene, we also selected 18s rRNA that is previously proposed by Caradec et al. as an appropriate HKG in hypoxia.[11]
Figure 1.
The relative expression of six common housekeeping genes after 3, 24, and 48 h of hypoxic conditioning of lung endothelial cells were assessed in a gene expression omnibus microarray dataset (accession number GSE11341). Coefficient of variation% for the expression fold change in the three time points was calculated for each gene
To further assess the suitability of ACTB, GAPDH, B2M, and 18s rRNA as HKGs in hypoxia, human kidney-derived cells were cultured in hypoxic or normoxic conditions and quantitative gene expression analysis was performed every 2 days for about 2 weeks. Real-time PCR data were analyzed by refFinder, a web-based comprehensive tool that integrates four different computational programs (geNorm, Normfinder, BestKeeper, and the comparative ΔΔCt method) to compare and rank the tested candidate HKGs. According to the ranking of each program, it assigns a weight to an individual gene and then calculates the geometric mean of their weights. According to this parameter, in hypoxia samples, GAPDH and ACTB were the most stable genes and 18s rRNA was the least stable one [Figure 2a]. However, in normoxia, B2M and ACTB had the best score and again 18s rRNA was the worst [Figure 2b]. When samples from both groups were analyzed together, GAPDH and ACTB, with equal scores, showed to be the most suitable HKGs [Figure 2c].
Figure 2.
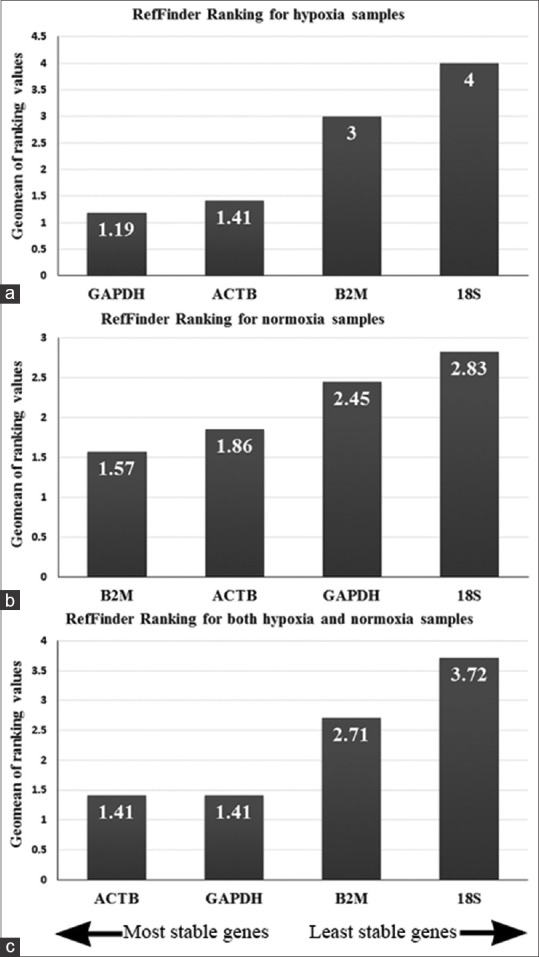
Real-time polymerase chain reaction data were analyzed with ref finder web-based tool to assess the stability of four housekeeping genes in human kidney cells cultured in hypoxia (a) or normoxia (b) when hypoxia and normoxia treated cells were analyzed together, GAPDH and ACTB had the best rank of stability (c)
Discussion
Real-Time PCR has become a pervasive method for accurate quantification of gene expression at the level of mRNA. To rely on data obtained from this method, it is crucial to choose appropriate reference genes for data normalization. However, there are no common HKGs for all experiments as the stability of gene expression is condition-specific. Hence, it is necessary to establish the most stable HKGs in each biological condition. Here, to determine stable reference genes in hypoxia, after a preliminary screening step with microarray data, we cultured human kidney-derived cells for different duration, from 1 day to about 2 weeks, in hypoxic conditions. Although, in most experiment designs, gene expressions are assessed compared to one starting control group (day 0), here we have concurrently assessed both normoxia and hypoxia groups in a time-course manner. Another advantage of this study is the long duration of in vitro hypoxia incubation. To the best of our knowledge, the 13 days culture in hypoxic conditions is the longest reported duration for such intervention.
In spite of lots of studies designed on the effects of hypoxia on the stability of gene expression, the data are yet contradictory. Examining several cell lines, de Kok et al. proposed that HPRT is an appropriate reference gene for experiments on hypoxia.[9] Whereas, Foldager et al. recommend RPL13A, B2M and RPII as suitable HKGs in hypoxic cultured human chondrocytes.[12] In another study, it was shown that RPLP0 and TFRC are suitable HKGs for human umbilical vein endothelial cell cultured in hypoxia.[13] A study by Caradec et al. shed light on this controversial issue; assessing the expression stability of 10 reference genes in four cell lines in different levels of hypoxia, they identified that both the type of cell and oxygen concentration have remarkable effect on the stability rank of the measured genes.[14] In agreement with this finding, in the current study we observed different stable genes for hypoxia and normoxia samples. Indeed, for hypoxia treated samples, GAPDH and ACTB revealed to be the most stable genes and in normoxia, B2M and ACTB were the best. However, as in relative quantification of real-time PCR data, treated samples are compared to controls with same set of HKGs, we believe that for selection of an appropriate HKG, the overall stability in all study groups should be considered. When samples from both groups and all time points were analyzed together, we found that GAPDH and ACTB equally had the best stability score. These data are in agreement with a study by Tan et al. demonstrating the stability of ACTB and GAPDH in hypoxia-preconditioned cardiosphere-derived cells.[15]
Conclusion
Our findings show that ACTB and GAPDH are suitable to be used as HKGs in gene expression analysis experiments within hypoxia treated kidney cells. However, we suggest that systematic selection of appropriate reference genes for each experiment condition be considered as a prerequisite step in gene expression analysis studies.
Financial support and sponsorship
Isfahan University of Medical sciences.
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
This study was supported by grant number 193092 from Isfahan University of medical sciences.
References
- 1.Fraisl P, Aragonés J, Carmeliet P. Inhibition of oxygen sensors as a therapeutic strategy for ischaemic and inflammatory disease. Nat Rev Drug Discov. 2009;8:139–52. doi: 10.1038/nrd2761. [DOI] [PubMed] [Google Scholar]
- 2.Higgins DF, Kimura K, Iwano M, Haase VH. Hypoxia-inducible factor signaling in the development of tissue fibrosis. Cell Cycle. 2008;7:1128–32. doi: 10.4161/cc.7.9.5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fine LG, Bandyopadhay D, Norman JT. Is there a common mechanism for the progression of different types of renal diseases other than proteinuria? Towards the unifying theme of chronic hypoxia. Kidney Int Suppl. 2000;75:S22–6. [PubMed] [Google Scholar]
- 4.Fine LG, Norman JT. Chronic hypoxia as a mechanism of progression of chronic kidney diseases: From hypothesis to novel therapeutics. Kidney Int. 2008;74:867–72. doi: 10.1038/ki.2008.350. [DOI] [PubMed] [Google Scholar]
- 5.Legrand M, Mik EG, Johannes T, Payen D, Ince C. Renal hypoxia and dysoxia after reperfusion of the ischemic kidney. Mol Med. 2008;14:502–16. doi: 10.2119/2008-00006.Legrand. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nat Protoc. 2006;1:1559–82. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- 7.Schmittgen TD, Zakrajsek BA. Effect of experimental treatment on housekeeping gene expression: Validation by real-time, quantitative RT-PCR. J Biochem Biophys Methods. 2000;46:69–81. doi: 10.1016/s0165-022x(00)00129-9. [DOI] [PubMed] [Google Scholar]
- 8.de Kok JB, Roelofs RW, Giesendorf BA, Pennings JL, Waas ET, Feuth T, et al. Normalization of gene expression measurements in tumor tissues: Comparison of 13 endogenous control genes. Lab Invest. 2005;85:154–9. doi: 10.1038/labinvest.3700208. [DOI] [PubMed] [Google Scholar]
- 9.Liu W, Shen SM, Zhao XY, Chen GQ. Targeted genes and interacting proteins of hypoxia inducible factor-1. Int J Biochem Mol Biol. 2012;3:165–78. [PMC free article] [PubMed] [Google Scholar]
- 10.Costello CM, Howell K, Cahill E, McBryan J, Konigshoff M, Eickelberg O, et al. Lung-selective gene responses to alveolar hypoxia: Potential role for the bone morphogenetic antagonist gremlin in pulmonary hypertension. Am J Physiol Lung Cell Mol Physiol. 2008;295:L272–84. doi: 10.1152/ajplung.00358.2007. [DOI] [PubMed] [Google Scholar]
- 11.Nagelkerke A, Mujcic H, Wouters B, Span PN. 18S is an appropriate housekeeping gene for in vitro hypoxia experiments. Br J Cancer. 2010;103:590. doi: 10.1038/sj.bjc.6605754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foldager CB, Munir S, Ulrik-Vinther M, Søballe K, Bünger C, Lind M. Validation of suitable house keeping genes for hypoxia-cultured human chondrocytes. BMC Mol Biol. 2009;10:94. doi: 10.1186/1471-2199-10-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bakhashab S, Lary S, Ahmed F, Schulten HJ, Bashir A, Ahmed FW, et al. Reference genes for expression studies in hypoxia and hyperglycemia models in human umbilical vein endothelial cells. G3 (Bethesda, Md) 2014;4:2159–65. doi: 10.1534/g3.114.013102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caradec J, Sirab N, Keumeugni C, Moutereau S, Chimingqi M, Matar C, et al. ‘Desperate house genes’: The dramatic example of hypoxia. Br J Cancer. 2010;102:1037–43. doi: 10.1038/sj.bjc.6605573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan SC, Carr CA, Yeoh KK, Schofield CJ, Davies KE, Clarke K. Identification of valid housekeeping genes for quantitative RT-PCR analysis of cardiosphere-derived cells preconditioned under hypoxia or with prolyl-4-hydroxylase inhibitors. Mol Biol Rep. 2012;39:4857–67. doi: 10.1007/s11033-011-1281-5. [DOI] [PMC free article] [PubMed] [Google Scholar]



