Abstract
The aryl hydrocarbon receptor (AHR) was for many years of interest only to pharmacologists and toxicologists. However, this protein has fundamental roles in biology that are being revealed through studies in diverse animal species. The AHR is an ancient protein. AHR homologs exist in most major groups of modern bilaterian animals, including deuterostomes (chordates, hemichordates, echinoderms) and the two major clades of protostome invertebrates [ecdysozoans (e.g. arthropods and nematodes) and lophotrochozoans (e.g. molluscs and annelids)]. AHR homologs also have been identified in cnidarians such as the sea anemone Nematostella and in the genome of Trichoplax, a placozoan. Bilaterians, cnidarians, and placozoans form the clade Eumetazoa, whose last common ancestor lived approximately 600 million years ago (MYA). The presence of AHR homologs in modern representatives of all these groups indicates that the original eumetazoan animal possessed an AHR homolog. Studies in invertebrates and vertebrates reveal parallel functions of AHR in the development and function of sensory neural systems, suggesting that these may be ancestral roles. Vertebrate animals are characterized by the expansion and diversification of AHRs, via gene and genome duplications, from the ancestral protoAHR into at least five classes of AHR-like proteins: AHR, AHR1, AHR2, AHR3, and AHRR. The evolution of multiple AHRs in vertebrates coincided with the acquisition of high-affinity binding of halogenated and polynuclear aromatic hydrocarbons and the emergence of adaptive functions involving regulation of xenobiotic-metabolizing enzymes and roles in adaptive immunity. The existence of multiple AHRs may have facilitated subfunction partitioning and specialization of specific AHR types in some taxa. Additional research in diverse model and non-model species will continue to enrich our understanding of AHR and its pleiotropic roles in biology and toxicology.
Keywords: Ah receptor, aryl hydrocarbon receptor, bHLH-PAS, dioxin, evolution, development, metazoan, vertebrate, fish, genome duplication, gene expression
1. Introduction
The aryl hydrocarbon receptor (AHR) was initially identified because of its role in regulating the induction of drug-metabolizing enzymes and in mediating the extreme toxic potency of chlorinated dibenzo-p-dioxins and related compounds [1,2]. For two decades after its discovery, this protein was of interest only to pharmacologists and toxicologists [3–5]. However, early on it was recognized by some investigators that study of this receptor and its high-affinity ligand 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) offered potential insights into fundamental biological processes, in the same way that other potent toxins and toxicants have been used to understand cellular functions [6]. Now, it is widely acknowledged that the AHR has multiple functions beyond toxicology [7–10], and the past decade has ushered in a new and exciting era in AHR biology as experts from a variety of fields in biomedicine have turned their attention to elucidate the roles of this pleiotropic protein in cell and developmental biology, immunology, and human disease.
Despite the emerging understanding of the AHR, fundamental questions remain concerning its molecular mechanisms of action (including possible non-genomic mechanisms), target genes beyond the well-known genes encoding biotransformation enzymes, and networks of interactions with other signaling pathways. Similarly, the precise mechanisms underlying most AHR-dependent toxic effects of AHR ligands are not yet known. Complementing the studies of biomedical scientists, research by biologists in other fields has provided a comparative perspective that has yielded insights into the variety of biological functions carried out by AHRs in diverse species. The identification and characterization of AHRs in powerful model species such as Mus musculus [11,12], Caenorhabditis elegans [13], Drosophila melanogaster [14], Danio rerio [15], and Nematostella vectensis [16] has been especially valuable because the tools available for those species have facilitated manipulative experiments to assess AHR functions. Understanding the variety of AHR functions in biology may enable a better understanding of the mechanisms by which exposure to AHR ligands leads to toxicity.
Beyond the established model systems, a broader elucidation of the evolutionary history of the AHR, and of the bHLH-PAS (basic helix-loop-helix Per-Arnt-Sim) family to which it belongs, can provide a foundation for understanding shared and novel features of AHR biology, foster insight into toxic mechanisms, and support extrapolation and prediction of responses to chemicals among species [17,18]. To understand AHR evolution, we look at modern representatives of early-diverging groups. In what organisms did AHR first appear? What were the original functions? Are there fundamental features of AHR action that are conserved throughout its evolutionary history? What new features and roles have evolved and how do they vary among taxa?
2. AHR origins: The AHR is an ancient protein
What were the first organisms to have an AHR? Early studies using radioligand binding assays with [3H]TCDD or 2-azido-3-[125I]iodo-7,8-dibromodibenzo-p-dioxin suggested that AHR was a vertebrate protein [19,20]. Subsequently, the identification of AHR homologs in C. elegans [13,21] and D. melanogaster [14] revealed that AHR was more broadly distributed in multiple animal phyla. We now know that AHR is present throughout metazoans (Fig. 1). When did it first emerge? We cannot know for sure, but we can obtain clues by looking at genomes of modern descendants of some of the earliest diverging metazoans and their relatives.
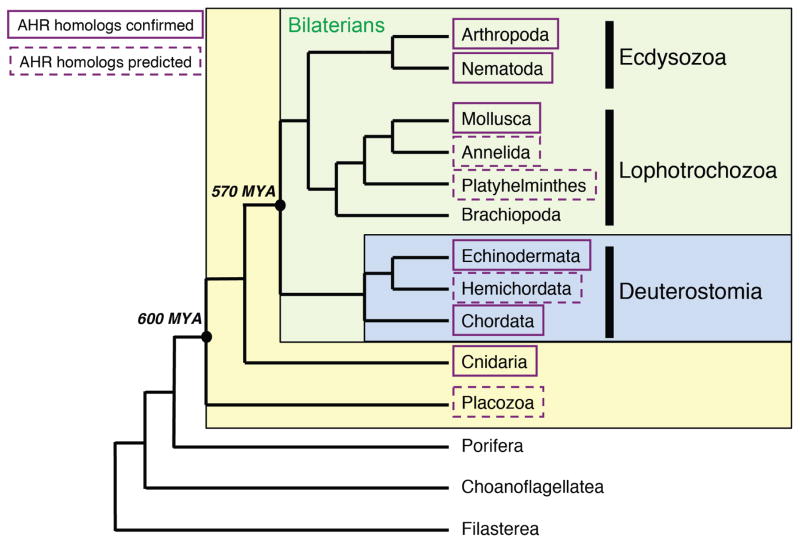
Figure 1. Presence of AHR homologs in holozoans.
The tree shows the relationships of selected metazoan (animal) taxa and related unicellular eukaryotes and whether they possess AHR homologs. Ecdysozoa and Lophotrochozoa together comprise the protostomes. Protostomes and deuterostomes are bilaterian animals (green-shaded box). Solid boxes represent groups containing species from which AHR homologs have been confirmed by cloning. Dashed boxes occur around groups with AHR homologs predicted from sequenced genomes. The large yellow-shaded box encompasses the eumetazoans, a group that includes all the taxa in which AHR has been identified to date. See text for additional information. Phylogenetic relationships of choanoflagellates and filastereans after Torruella [149].
Filastereans (represented by the genus Capsaspora) are not metazoans, but they are the closest unicellular relatives of metazoans and possess many of the transcription factors that are important for metazoan development [22,23]. Capsaspora has four bHLH-PAS proteins but no recognizable AHR homolog [22].
Choanoflagellates such as Monosiga and Salpingoeca also are single-celled but at certain stages can form aggregates held together by cell adhesion proteins, considered a primitive type of multicellularity [24–26]. Choanoflagellates have a few bHLH-PAS proteins but no clear AHR homolog [22].
Sponges (phylum Porifera; e.g. genus Amphimedon), which exhibit embryonic and larval stages and possess a large set of metazoan-specific developmental transcription factors, are considered the oldest extant metazoan lineage [27]. The Amphimedon genome encodes three bHLH-PAS proteins that resemble the ARNT/BMAL, HIF/SIM/TRH, and CLOCK proteins of more recently diverging animals, but again no AHR [28].
The most ancient metazoan lineage with a clearly recognizable AHR in its genome is the placozoan Trichoplax [29,30]. Placozoans have three cell layers and a variety of transcription factors involved in metazoan development and cell fate specification, but no recognizable specialized sensory or nerve cells. The three bHLH-PAS proteins in Trichoplax include an AHR homolog that shows high amino acid sequence similarity to human AHR in its bHLH domain (84%), and substantial but lower similarity in its PAS-A (43%) and PAS-B (51%) domains. Nothing is known about the function of the placozoan AHR.
An AHR homolog is also found in a cnidarian, the starlet sea anemone Nematostella vectensis. This species, which is often studied because of its phylogenetic position basal to the bilaterian metazoans, has a nervous system, sensory organs, and a full toolkit of metazoan developmental regulatory proteins, including many shown to interact with AHR signaling in mammals (e.g. notch, hes, wnt, fgf). The N. vectensis genome encodes seven bHLH-PAS proteins, including homologs of HIF, SIM/TRH, ARNT, BMAL, and CLOCK in addition to the AHR and a second AHR-like protein [16,31]. Functional characterization of the AHR suggests differences compared to AHRs of vertebrate animals. For example, when expressed in vitro the N. vectensis AHR protein does not exhibit specific binding of [3H]TCDD or [3H]beta-naphthoflavone ([3H]BNF), prototypical ligands for vertebrate AHRs [16]. In addition, unlike vertebrate AHRs the N. vectensis AHR does not interact with ARNT or BMAL in vitro, suggesting that it may act independent of ARNT. In situ hybridization shows that AHR is expressed during larval development at the base of the apical tuft (a sensory structure) and later in the developing tentacles [16]. The expression patterns of AHR and ARNT are non-overlapping at most of these stages, providing additional evidence for ARNT-independent function of the N. vectensis AHR.
The results in N. vectensis strongly suggest that the common ancestor of cnidarians and bilaterian animals already possessed the modern animal set of familiar bHLH-PAS proteins, including AHR. The functions of this early AHR are unknown, but studies in modern bilaterians (protostomes and deuterostomes) have provided some clues.
3. AHRs in protostomes: Key roles in development of sensory systems
The protostomes include most of the major invertebrate phyla and two key model species that have provided important insights into possible ancestral functions of AHR [32,33].
An AHR homolog (called AHR-1) in the nematode C. elegans [13,21] resembles the cnidarian AHR in its inability to bind typical AHR ligands [13,34]. AHR-1 is expressed during embryonic and larval development and primarily in developing neurons, including touch receptor neurons, GABAergic motor neurons, interneurons, and sensory neurons that contact the pseudocoelomic fluid [35]. Loss of AHR-1 function results in defective neuronal migration and axonal pathfinding, altered touch neuron fate, and changes in locomotor and social feeding behaviors [35–37]. The role of AHR-1 in neuronal development requires ARNT (AHA-1) [35,36] and may involve regulation of wnt signaling [38]. In addition to its roles in development, AHR-1 appears to have an ongoing role in regulating the expression of oxygen-sensing guanylate cyclases involved in the control of feeding behavior [39,40]. Together, these results support a role for AHR-1 in neuronal differentiation, migration, and cell fate determination [32] as well as post-embryonic neuronal functions.
Another powerful model, the fruit fly D. melanogaster, has also provided important insights into the pleiotropic developmental roles of AHR. The fly AHR homolog, the product of the spineless (ss) locus, is expressed in larval eye-antennal imaginal discs, the regions destined to becoming adult eyes and antennae [14]. Loss-of-function mutations demonstrate that ss specifies the identity of the distal segments of antennae (multi-sensory structures) and legs and the formation of mechanosensory bristles (the loss of which is reflected in the name “spineless”) [14]; in the antennae ss appears to have a specific role in development of olfactory sensillae [41]. The action of ss in controlling development of antennae and distal leg require the fly ARNT homolog tango (tgo) [42] and may in part involve the repression of gene expression by ss/tgo complexes [43].
Later in development, ss has a role in specifying photoreceptor cell fate in ommatidia of the developing compound eye. Stochastic expression of ss in specific photoreceptors determines the type of rhodopsin that is expressed, thus controlling color sensitivity in D. melanogaster [44,45] and other insects [46]. As seen for antenna and leg development, the role of ss in controlling photoreceptor cell fate requires tgo (ARNT) [47]. The expression of ss is maintained in these ss-specified photoreceptor subtypes in adults [45] and thus may be necessary to maintain the pattern of rhodopsin expression.
Yet another developmental role of the D. melanogaster AHR homolog is in controlling dendrite morphology on dendritic arborization (da) sensory neurons in the fly peripheral nervous system [48,49]. The effect of ss varies in different types of da neurons, with the end result of diversifying dendrite morphology. Although this role was originally suggested to be independent of tgo/ARNT [48], more recent results indicate that co-expression of tgo is in fact necessary [47]. Because the C. elegans AHR has a similar role in controlling dendritic branching complexity, this has been suggested as an ancestral role of AHR [37].
The common theme of the research in C. elegans and D. melanogaster, with circumstantial support from studies in the cnidarian N. vectensis, is one of pleiotropic roles of AHR in controlling the development (cell fate and differentiation) and function of sensory structures and neural systems [18,32,33,49]. These functions appear to involve both activation and repression of gene expression by AHR [37,43].
What is the role of ligands in the functions of protostome and cnidarian AHR homologs? Although these AHRs do not appear to bind typical (i.e. vertebrate) AHR ligands [13,16,34] and there is some evidence for constitutive, ligand-independent activity [32,42,50], it is also possible that there are endogenous ligands or other regulatory mechanisms involved [51–53]. Nevertheless, we refer to these proteins as “protoAHRs” (Table 1) to highlight the apparently substantial differences in ligand specificity between these proteins and their vertebrate homologs, which function (at least in part) as true “aryl hydrocarbon receptors.” It is important to note, however, that all of the evidence currently available is consistent with the idea that protoAHRs and vertebrate AHRs are true orthologs (i.e. descended from the same gene in the most recent common bilaterian ancestor [54]).
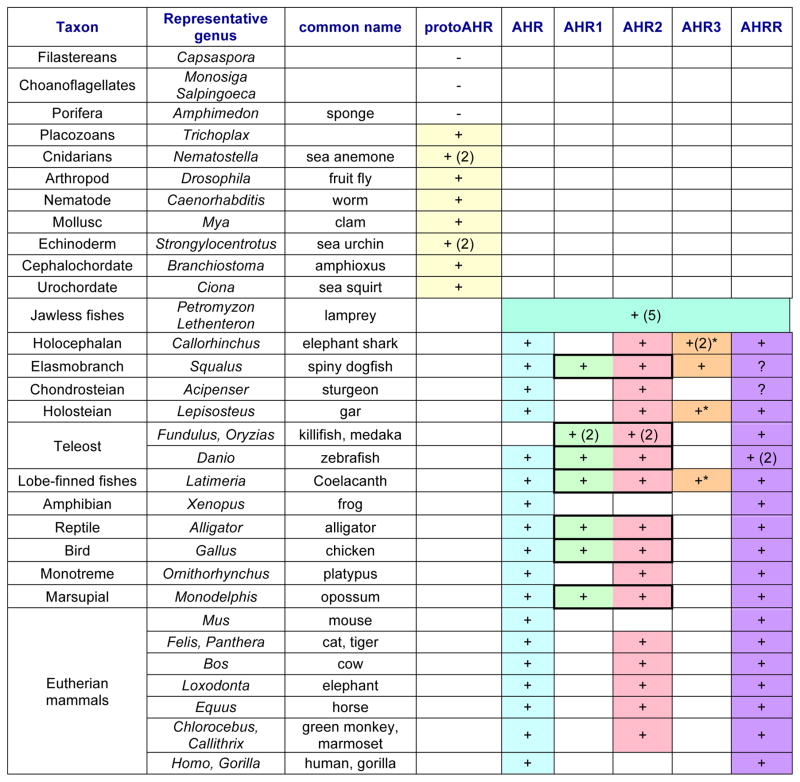
Table 1.
Phylogenetic distribution of multiple AHR forms in metazoans.
This table presents a summary of our current thinking regarding AHR multiplicity and the phylogenetic distribution of various AHR forms. Results are derived from published papers and from our analyses of numerous phylogenetic trees and syntenic relationships (shared synteny). The conclusions here should be considered tentative and will be subject to revision as additional sequences and species are examined. See sections 3 and 4 of text for details regarding the different types of AHRs.
Note: bold boxes indicate a demonstrated tandem arrangement of AHR1 and AHR2 genes.
The classification of these as AHR3 is uncertain. Resolution will require additional sequences and synteny information.
4. AHR in deuterostomes: Expansion through gene and genome duplications
The other major group of bilaterian animals, the deuterostomes, includes echinoderms, hemichordates, and chordates (Fig. 1). Predicted AHR homologs are found in genomes of the echinoderm Strongylocentrotus (sea urchin) [55,56], hemichordate Saccoglossus (acorn worm) [57], and invertebrate chordates such as the cephalochordate Branchiostoma (amphioxus) [58,59] and urochordate Ciona (sea squirt) [60]. The Ciona AHR, like other invertebrate AHRs, does not bind TCDD (unpublished data), but nothing is known about the function of the other invertebrate deuterostome AHRs.
In contrast to the echinoderms, hemichordates, and invertebrate chordates, the vertebrate chordate lineage is notable for the diversification of AHRs (and other bHLH-PAS proteins [61]) (Table 1), a result of vertebrate- and teleost-specific whole genome duplications as well as an early tandem duplication of AHR [61]. It is in the vertebrates where we first see AHRs that exhibit high-affinity binding of TCDD [18], AHR-dependent regulation of genes encoding xenobiotic-metabolizing CYP1 enzymes [62,63], and high sensitivity to toxic effects of dioxin-like compounds [64].
In the oldest extant vertebrate group, Agnatha (jawless fishes), represented by the lampreys Petromyzon marinus [65] and Lethenteron japonicum [66], we see a remarkable expansion, with five predicted AHR genes in each species ([67] and S. Karchner unpublished; Table 1). This increase in AHR genes was likely a result of the two whole genome duplications that occurred early in vertebrate evolution, more than 450 million years ago; current evidence suggests that both of these preceded the divergence of agnathan cyclostomes (jawless vertebrates) and gnathostomes (jawed vertebrates) [65,68] (but see also [69]).
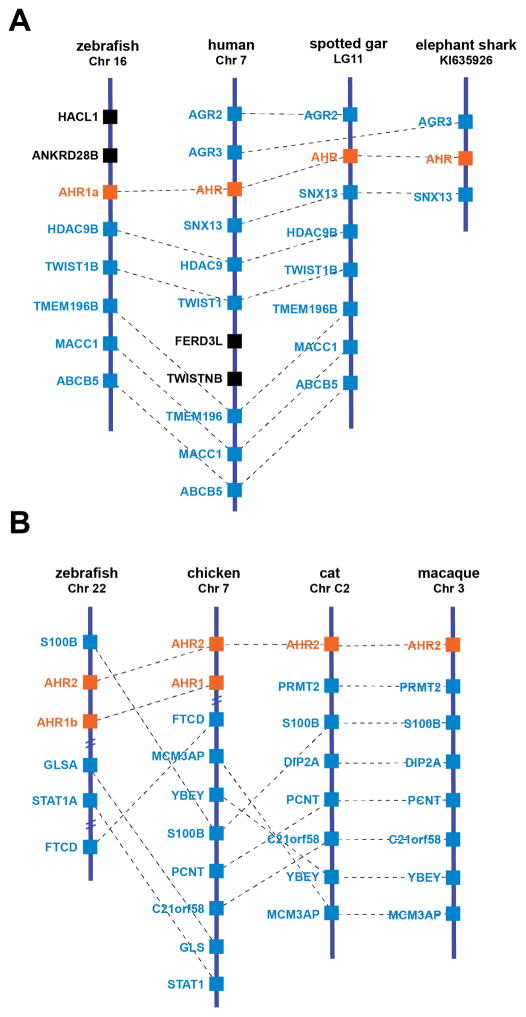
In the jawed vertebrates (Chondrichthyes [cartilaginous fishes] and Osteichthyes [bony fishes] and their descendants, including tetrapods), our current understanding—supported by phylogenetic analyses and information from shared synteny—is that there are at least five classes of AHR-related genes (Table 1). We use AHR to refer to vertebrate orthologs of the AHR originally identified in mammals [11,12]. AHR genes are found in nearly all vertebrates, including sharks, gar, and sturgeon. However, teleosts (the largest group of ray-finned fishes) are notable in that most of those studied to date lack this AHR. The exception is zebrafish, where the enigmatic “AHR1a” [70–72] appears to be an AHR based on our recent analysis of shared synteny with human AHR and other AHR genes (Fig. 2A).
Figure 2. Analysis of shared synteny supports AHR classification.
(A) Zebrafish AHR1a and related AHR genes in earlier-diverging fishes may be orthologous to human AHR and related AHR genes. (B) Predicted AHR2 genes found in several mammals exhibit shared synteny with AHR2 genes from fish and birds. Analysis of syntenic relationships was performed using Genomicus [150] and manual scanning of sequenced genomes.
AHR1 and AHR2 are paralogs derived from a tandem gene duplication that occurred prior to the divergence of cartilaginous and bony fish lineages [21,61,71,73]. Although AHR1 was originally thought to be orthologous to mammalian AHR [73], more recent analysis of additional AHR sequences reveals that AHR and AHR1 represent distinct lineages (Table 1). AHR1 and AHR2 orthologs are found in tandem in cartilaginous fishes [74], bony fishes (which usually have duplicated AHR1-AHR2 pairs; see below) [61,71], coelacanth (a lobe-finned fish) [75], birds [76,77], reptiles [78], and an early diverging marsupial mammal, the opossum Monodelphis ([79]; C. Panti, S. Karchner, M. Hahn, unpublished) (Table 1). Based on Xenopus genomes and other analyses, the AHR1/AHR2 pair has been lost from at least some amphibians [80] (Table 1). Although orthologs of AHR1 and AHR2 are not found in rodents or humans, there are predicted AHR2 genes in several genomes in the mammalian orders Carnivora, Cetartiodactyla, and Primates (both old world and new world monkeys, but not great apes; S. Karchner, R. Merson, and M. Hahn, unpublished) (Table 1). These occur without an adjacent AHR1, but the identity as AHR2 is supported by phylogenetic analyses (not shown) as well as by shared synteny with species possessing tandem AHR1-AHR2 pairs (Fig. 2B).
AHR1 and AHR2 genes are often found as duplicated pairs in teleosts. Thus, most teleost genomes include both an AHR1a-AHR2a tandem pair and an AHR1b-AHR2b tandem pair [61,71,81]; these duplicated pairs are thought to have arisen as part of the teleost-specific whole-genome duplication [82,83]. A prominent exception is the zebrafish (D. rerio), which has only one AHR1-AHR2 pair (orthologous to AHR1b-AHR2b in other teleosts) [61,71] and a separate AHR (currently designated AHR1a, but likely an “AHR”, as noted above) [71,72]. Interestingly, the recently sequenced genome of gar, representing the holosteian lineage, which diverged prior to the teleost-specific genome duplication [84], does not contain an AHR1-AHR2 pair. However, it has—in addition to an AHR—an AHR2 that is orthologous to other fish AHR2 genes, based on phylogenetic analysis and shared synteny; thus, the tandem AHR1 appears to have been lost in this species. Similarly, sturgeon (a chondrosteian) has an AHR and an AHR2, but no AHR1 [85].
AHR3 is a novel AHR found originally in elasmobranchs (a subclass of cartilaginous fishes encompassing true sharks, skates, and rays; R. Merson & M. Hahn, unpublished; see also [61]) (Table 1). In the shark Squalus acanthias both AHR2 and AHR3 (but not AHR1) bind TCDD and are transcriptionally active in heterologous expression systems (Merson et al., in preparation). The genome of the elephant shark [86], a representative of the cartilaginous fish subclass Holocephali (chimaeras), contains two possible AHR3-like genes. AHR3-like genes are also found in some early diverging fishes such as lamprey, gar, and coelacanth (Table 1) but they form a distinct clade in phylogenetic analyses. The resolution of AHR3 relationships will require analysis of additional species and the completion of genome assemblies to assess the genomic context of this locus. AHR3 and AHR3-like genes do not appear elsewhere in the vertebrates.
AHRR, first identified in mouse [87], is distinct from AHR, AHR1, AHR2, and AHR3 and acts via multiple mechanisms to repress signaling through AHR and some other pathways [88]. The PAS-B region of AHRR, which in other AHR-related proteins forms the ligand-binding domain, is missing or highly divergent in AHRR [87]; consistent with that, AHRRs do not bind [3H]TCDD or [3H]BNF [89]. AHRR has been retained in nearly all vertebrate groups (Table 1), suggesting that it has an important physiological function. Recent findings regarding the possible roles of AHRR in the immune system [90,91], reproduction [92,93], and carcinogenesis [94,95] support that notion.
5. Functional divergence of a pleiotropic protein: Shared and divergent roles of metazoan AHRs
What are the ancestral roles of AHRs and how have they changed during metazoan evolution? We look for evidence in functions that may be shared among modern animals whose most recent common ancestor lived long ago, e.g. protostomes and deuterostomes, but these roles can be difficult to identify given the substantial developmental and physiological differences among long-diverged lineages.
Studies in protostomes provide evidence for pleiotropic roles of AHR in controlling the development and function of sensory structures and neural systems and these have been suggested as ancestral roles of AHR [18,32,33,37,49]. Possible roles of AHR in sensory/neural systems are less well understood in vertebrate animals, but results from AHR loss-of-function studies and effects of TCDD on neural development in fish and mammals [96–103] suggest that this is an area worth further exploration. For example, roles for AHR in developing GABAergic systems and in controlling dendrite growth may be shared by vertebrate and invertebrate species [36,37,96,99,102,104].
Other developmental and physiological roles of AHR have been identified in vertebrates, including some involving vascular development, reproductive function, immunological development, and stem cell biology [7,8,33,105–107]. Some of these developmental roles may explain the special sensitivity of vertebrate early life stages to disruption by AHR ligands [108,109]. However, the possible connection of these roles to those of invertebrate AHRs is not obvious. Conceivably, shared features of AHR function may be more readily identified at the level of molecular interactions such as those involving gene regulation or protein-protein interactions. For example, interaction with wnt signaling appears to be a shared, and thus possibly evolutionarily conserved, feature of vertebrate and invertebrate AHRs [38,110–113]. A comparative analysis of gene regulatory networks involving AHR in a variety of vertebrate and invertebrate model systems could illuminate additional ancient molecular interactions [40,114–116].
What is the role of ARNT and AHR-ARNT interactions with AHR response elements (AHREs, also called DREs and XREs) in ancestral and modern functions of AHRs? At least some of the toxic effects of AHR ligands in mammals require both ARNT dimerization [117] and DNA binding [118]. Similarly, some of the developmental roles in invertebrate species are ARNT-dependent and/or involve interactions with AHRE sequences similar to those found in vertebrates [35,36,42,47]. Yet there also is evidence for ARNT-independent functions in Nematostella [16] and, increasingly, evidence for ARNT-independent or AHRE-independent roles of AHR in vertebrates [119–122].
The increased AHR diversity in vertebrate animals appears to have been accompanied by (and perhaps enabled) the emergence of new AHR adaptive functions, including regulation of the inducible expression of genes encoding xenobiotic-metabolizing enzymes such as cytochrome P450s (CYPs) in response to chemicals. Although CYPs and other biotransformation enzymes are inducible in C. elegans, the nematode AHR does not appear to be involved in this response [123]. In insects, there is evidence that AHR and ARNT homologs regulate the basal expression, but not the xenobiotic-inducible expression, of CYP6B1, which is involved in detoxification of dietary phytotoxins [124]. The first clear evidence for AHR-dependent regulation of inducible CYP1 genes is in jawed vertebrates [62,64]. How this association between AHR and CYP regulation evolved remains a mystery. However, one clue may be the intriguing tandem arrangement of AHR and CYP1-like genes in the urochordate Ciona [63], suggesting a possible mechanism whereby auto-regulation of AHR might have become co-opted by CYP1 through physical proximity on the chromosome.
As we have noted earlier, the evolution of the ability of AHR to engage in high-affinity ligand binding associated with ligand-dependent adaptive functions was a vertebrate innovation [18], perhaps driven by a need to detoxify halogenated marine natural products [125–128]. Ironically, this new ligand-dependence of AHR also introduced a mechanism by which some persistent halogenated aromatic hydrocarbons could cause toxicity through high-affinity AHR binding and sustained AHR activation. Invertebrate animals, which have AHRs that lack the ability to bind dioxin-like compounds, are generally insensitive to the toxicity of these chemicals [18,64].
Additional AHR-mediated adaptive functions that may have first emerged in vertebrate animals are those involving the regulation of innate and adaptive immunity [7,129]. Some of these immunological roles of AHR appear to involve endogenous and microbiota-derived ligands [130–134], although it is not yet known whether AHR affinity for some of these ligands (many of them indole derivatives) evolved in parallel with these immune functions.
In addition to novel functions, the AHR expansion in vertebrates may have enabled AHR isoform specialization, through subfunction partitioning and subsequent functional refinement. AHRR may be one example of that, whereby loss of ligand-binding through degeneration of the PAS-B ligand-binding domain [89] led to a specialization for repression of gene expression.
Other examples of AHR specialization involve the apparent partitioning of tissue expression patterns, ligand specificity, and target gene specificity among multiple AHR paralogs in some non-mammalian vertebrates. Whereas most mammals have a single pleiotropic AHR, through which various classes of ligands must all act, there is evidence that the multiple AHRs of fishes, birds, reptiles, and amphibians have partitioned some of these functions. For example, the set of three AHR genes in zebrafish have evolved very different functional properties and expression patterns involving partitioning of ligand specificity and developmental versus adaptive roles [70–72,135–137]. Whereas zebrafish AHR2 appears to mediate most of the gene induction and developmental toxicity of TCDD, PCB-126, and some polycyclic aromatic hydrocarbons [138–143], AHR1a is preferentially involved in the response to some non-halogenated compounds such as leflunomide, pyrene, and oxygenated PAHs [70,136,144] and AHR1b may have a tissue-specific developmental role [137]. Similarly, AHR paralog-specific differences in ligand structure-activity relationships (e.g. for halogenated vs. non-halogenated ligands) or target gene specificity have been observed in chicken [76,145], alligator [78], and frog [146].
6. Conclusions
From the information summarized above (and other data that could not be covered in a brief review such as this) we offer some conclusions, some of which must necessarily be considered tentative.
Clearly, the AHR is an ancient protein, which has existed for more than 600 million years of animal evolution and should be considered part of the fundamental metazoan toolkit. In modern (living) invertebrates, AHR has roles in the development of sensory structures, including sensory neural systems; these may be some of the most ancient roles of metazoan AHRs. AHRs have undergone substantial diversification in the vertebrate chordates; this diversification was likely facilitated by the gene and genome duplications occurring prior to and after the vertebrate radiation. AHRs are pleiotropic, with multiple functions that vary by cell type and developmental stage within single species as well as among animal taxa. In some cases, those multiple functions are partitioned among AHR isoforms within a species. The emergence of the adaptive functions of AHR in the vertebrates is associated with the acquisition of high-affinity binding of planar aromatic hydrocarbons, which appears to be a vertebrate innovation. This broadened the capacity for inducible detoxification of xenobiotics but also introduced a mechanism by which some persistent, high-affinity ligands could cause toxicity.
It is worth noting that all of the information we have about AHR functions—including ligand-binding, protein-protein interactions, DNA binding, gene regulation, and developmental roles—is from studies in modern animals. All of these species—from Trichoplax to humans—can be considered “advanced” in that they are the result of a long evolutionary process; i.e. they are the “survivors.” These modern animals are the descendants of earlier species in which resided the ancestral functions of AHR that we seek to understand. Although we cannot turn the clock back to study these ancestors, it is now possible, through ancestral sequence reconstruction, to resurrect and study ancestral proteins [147,148]. The evolution of AHR ligand-dependence and ligand specificity, in particular, may be revealed by reconstruction and analysis of the ancestral AHR proteins that existed at key points in metazoan evolution, such as the emergence of bilaterians or the base of the vertebrate radiation.
There is an intuitive appeal to the hypothesis that the AHR had ancestral roles in the development of sensory structures and neurons that were later co-opted for novel roles in chemical sensing and adaptive responses. Studies of AHRs in new model and non-model systems will help to illuminate or refute that idea. One thing is indisputable: the AHR will continue to intrigue and surprise us over the next decade as its manifold roles are revealed.
Highlights.
The AHR is an ancient protein, part of the fundamental eumetazoan genetic tool kit.
Ancestral roles of AHR likely included control of sensory neural development.
AHR has undergone expansion in vertebrates, generating at least 5 AHR types.
Adaptive roles of AHR in xenobiotic sensing may have emerged in vertebrates.
Modern AHRs exhibit both ancestral and recently evolved roles in cell biology.
Acknowledgments
This paper is based in part on an invited presentation at the AHR 2016 Symposium, “The Aryl Hydrocarbon Receptor as a Central Mediator of Health and Disease,” held at the University of Rochester Medical Center from August 3-6, 2016. The authors thank Drs. Ann Tarrant and Wade Powell for comments on an earlier version of the manuscript, and Diana Franks for assistance with AHR assays in a variety of species over many years. M.E.H. and S.I.K are grateful for the long-term support of our AHR research from the National Institute of Environmental Health Sciences (NIEHS) through grants R01ES006272 and P42ES007381 (Superfund Research Program at Boston University). We also acknowledge support from a WHOI Independent Study Award funded by the Andrew W. Mellon Foundation Endowed Fund for Innovative Research. R.R.M. acknowledges support from the NIH National Center for Research Resources RI-INBRE (P20RR016457), National Science Foundation EPSCoR Cooperative Agreement #EPS-1004057, a MDIBL New Investigator Award funded by ME-INBRE (P20RR016463), and NIEHS grant P30ES003828. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies. The funding agencies had no role in the preparation of the report or the decision to submit the manuscript for publication.
Abbreviations
- AHR
aryl hydrocarbon receptor
- AHRR
AHR Repressor
- AHRE
AHR response element
- bHLH-PAS
basic helix-loop-helix Per-Arnt-Sim
- BNF
beta-naphthoflavone
- CYP
cytochrome P450
- MYA
million years ago
- da
dendritic arborization
- TCDD
2,3,7,8-tetrachlorodibenzo-p-dioxin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References cited
Papers of particular interest are noted:
• of special interest
•• of outstanding interest
- 1.Poland A, Glover E, Kende AS. Stereospecific, high-affinity binding of 2,3,7,8-tetrachlorodibenzo-p-dioxin by hepatic cytosol. J Biol Chem. 1976;251:4936–4946. [PubMed] [Google Scholar]
- 2.Okey AB. An aryl hydrocarbon receptor odyssey to the shores of toxicology: the Deichmann Lecture, International Congress of Toxicology-XI. Toxicol Sci. 2007;98:5–38. doi: 10.1093/toxsci/kfm096. [DOI] [PubMed] [Google Scholar]
- 3.Poland A, Knutson JC. 2,3,7,8-Tetrachlorodibenzo-p-dioxin and related halogenated aromatic hydrocarbons: examination of the mechanism of toxicity. Annual Reviews of Pharmacology and Toxicology. 1982;22:517–554. doi: 10.1146/annurev.pa.22.040182.002505. [DOI] [PubMed] [Google Scholar]
- 4.Hankinson O. The aryl hydrocarbon receptor complex. Annual Review of Pharmacology and Toxicology. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- 5.Whitlock JP. Genetic and molecular aspects of 2,3,7,8-tetrachlorodibenzo-p-dioxin action. Annual Reviews of Pharmacology and Toxicology. 1990;30:251–277. doi: 10.1146/annurev.pa.30.040190.001343. [DOI] [PubMed] [Google Scholar]
- 6•.Poland A, Kende A. 2,3,7,8-Tetrachlorodibenzo-p-dioxin: Environmental contaminant and molecular probe. Federation Proceedings. 1976;35:2404–2411. This insightful early paper drew parallels between TCDD and other highly toxic chemicals that have been used to probe biological systems. It predicted that understanding the mechanism of TCDD action through the AHR would lead to fundamental understanding in cell biology. [PubMed] [Google Scholar]
- 7.Esser C, Rannug A. The aryl hydrocarbon receptor in barrier organ physiology, immunology, and toxicology. Pharmacol Rev. 2015;67:259–279. doi: 10.1124/pr.114.009001. [DOI] [PubMed] [Google Scholar]
- 8.Stockinger B, Di Meglio P, Gialitakis M, Duarte JH. The aryl hydrocarbon receptor: multitasking in the immune system. Annu Rev Immunol. 2014;32:403–432. doi: 10.1146/annurev-immunol-032713-120245. [DOI] [PubMed] [Google Scholar]
- 9.Bock KW. From dioxin toxicity to putative physiologic functions of the human Ah receptor in homeostasis of stem/progenitor cells. Biochem Pharmacol. 2016 doi: 10.1016/j.bcp.2016.06.015. [DOI] [PubMed] [Google Scholar]
- 10.Mulero-Navarro S, Fernandez-Salguero PM. New trends in aryl hydrocarbon receptor biology. Front Cell Dev Biol. 2016;4:45. doi: 10.3389/fcell.2016.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burbach KM, Poland A, Bradfield CA. Cloning of the Ah receptor cDNA reveals a distinctive ligand-activated transcription factor. Proceedings of the National Academy of Sciences US A. 1992;89:8185–8189. doi: 10.1073/pnas.89.17.8185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ema M, Sogawa K, Watanabe N, Chujoh Y, Matsushita N, Gotoh O, Funae Y, Fujii-Kuriyama Y. cDNA cloning and structure of mouse putative Ah receptor. Biochemical and Biophysical Research Communications. 1992;184:246–253. doi: 10.1016/0006-291x(92)91185-s. [DOI] [PubMed] [Google Scholar]
- 13.Powell-Coffman JA, Bradfield CA, Wood WB. Caenorhabditis elegans orthologs of the aryl hydrocarbon receptor and its heterodimerization partner the aryl hydrocarbon receptor nuclear translocator. Proceedings of the National Academy of Sciences US A. 1998;95:2844–2849. doi: 10.1073/pnas.95.6.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duncan DM, Burgess EA, Duncan I. Control of distal antennal identity and tarsal development in Drosophila by spineless-aristapedia, a homolog of the mammalian dioxin receptor. Genes and Development. 1998;12:1290–1303. doi: 10.1101/gad.12.9.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carney SA, Prasch AL, Heideman W, Peterson RE. Understanding dioxin developmental toxicity using the zebrafish model. Birth Defects Res A Clin Mol Teratol. 2006;76:7–18. doi: 10.1002/bdra.20216. [DOI] [PubMed] [Google Scholar]
- 16•.Reitzel AM, Passamaneck YJ, Karchner SI, Franks DG, Martindale MQ, Tarrant AM, Hahn ME. Aryl hydrocarbon receptor (AHR) in the cnidarian Nematostella vectensis: comparative expression, protein interactions, and ligand binding. Development Genes and Evolution. 2014;224:13–24. doi: 10.1007/s00427-013-0458-4. This paper characterizes the AHR in an early-diverging metazoan, providing insight into possible ancestral roles, including possible ARNT-independent functions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hahn ME, Karchner SI. Chapter 27. Structural and functional diversification of AHRs during metazoan evolution. In: Pohjanvirta R, editor. The Ah receptor in Biology and Toxicology. John Wiley & Sons, Inc; 2012. pp. 389–403. [Google Scholar]
- 18.Hahn ME. Aryl hydrocarbon receptors: Diversity and evolution. Chemico-Biological Interactions. 2002;141:131–160. doi: 10.1016/s0009-2797(02)00070-4. [DOI] [PubMed] [Google Scholar]
- 19.Denison MS, Hamilton JW, Wilkinson CF. Comparative studies of aryl hydrocarbon hydroxylase and the Ah receptor in nonmammalian species. Comp Biochem Physiol. 1985;80C:319–324. doi: 10.1016/0742-8413(85)90063-5. [DOI] [PubMed] [Google Scholar]
- 20.Hahn ME, Poland A, Glover E, Stegeman JJ. Photoaffinity labeling of the Ah receptor: Phylogenetic survey of diverse vertebrate and invertebrate species. Arch Biochem Biophys. 1994;310:218–228. doi: 10.1006/abbi.1994.1160. [DOI] [PubMed] [Google Scholar]
- 21•.Hahn ME, Karchner SI, Shapiro MA, Perera SA. Molecular evolution of two vertebrate aryl hydrocarbon (dioxin) receptors (AHR1 and AHR2) and the PAS family. Proc Nat’l Acad Sci US A. 1997;94:13743–13748. doi: 10.1073/pnas.94.25.13743. This paper was the first report of multiple AHR genes in vertebrate animals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sebe-Pedros A, de Mendoza A, Lang BF, Degnan BM, Ruiz-Trillo I. Unexpected repertoire of metazoan transcription factors in the unicellular holozoan Capsaspora owczarzaki. Mol Biol Evol. 2011;28:1241–1254. doi: 10.1093/molbev/msq309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suga H, Chen Z, de Mendoza A, Sebe-Pedros A, Brown MW, Kramer E, Carr M, Kerner P, Vervoort M, Sanchez-Pons N, et al. The Capsaspora genome reveals a complex unicellular prehistory of animals. Nat Commun. 2013;4:2325. doi: 10.1038/ncomms3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fairclough SR, Chen Z, Kramer E, Zeng Q, Young S, Robertson HM, Begovic E, Richter DJ, Russ C, Westbrook MJ, et al. Premetazoan genome evolution and the regulation of cell differentiation in the choanoflagellate. Salpingoeca rosetta Genome Biol. 2013;14:R15. doi: 10.1186/gb-2013-14-2-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I, et al. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richter DJ, King N. The genomic and cellular foundations of animal origins. Annu Rev Genet. 2013;47:509–537. doi: 10.1146/annurev-genet-111212-133456. [DOI] [PubMed] [Google Scholar]
- 27.Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier ME, Mitros T, Richards GS, Conaco C, Dacre M, Hellsten U, et al. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature. 2010;466:720–726. doi: 10.1038/nature09201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simionato E, Ledent V, Richards G, Thomas-Chollier M, Kerner P, Coornaert D, Degnan BM, Vervoort M. Origin and diversification of the basic helix-loop-helix gene family in metazoans: insights from comparative genomics. BMC Evol Biol. 2007;7:33. doi: 10.1186/1471-2148-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Srivastava M, Begovic E, Chapman J, Putnam NH, Hellsten U, Kawashima T, Kuo A, Mitros T, Salamov A, Carpenter ML, et al. The Trichoplax genome and the nature of placozoans. Nature. 2008;454:955–960. doi: 10.1038/nature07191. [DOI] [PubMed] [Google Scholar]
- 30.Gyoja F. A genome-wide survey of bHLH transcription factors in the Placozoan Trichoplax adhaerens reveals the ancient repertoire of this gene family in metazoan. Gene. 2014;542:29–37. doi: 10.1016/j.gene.2014.03.024. [DOI] [PubMed] [Google Scholar]
- 31.Reitzel AM, Behrendt L, Tarrant AM. Light entrained rhythmic gene expression in the sea anemone Nematostella vectensis: the evolution of the animal circadian clock. PLoS One. 2010;5:e12805. doi: 10.1371/journal.pone.0012805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Powell-Coffman JA, Qin H. Chapter 28. Invertebrate AHR homologs: Ancestral functions in sensory systems. In: Pohjanvirta R, editor. The Ah receptor in Biology and Toxicology. John Wiley & Sons, Inc; 2012. pp. 405–411. [Google Scholar]
- 33•.McMillan BJ, Bradfield CA. The aryl hydrocarbon receptor sans xenobiotics: endogenous function in genetic model systems. Mol Pharmacol. 2007;72:487–498. doi: 10.1124/mol.107.037259. This is an outstanding review of developmental roles of AHR in diverse animal models. [DOI] [PubMed] [Google Scholar]
- 34•.Butler RB, Kelley ML, Powell WH, Hahn ME, Van Beneden RJ. An aryl hydrocarbon receptor homologue from the soft-shell clam, Mya arenaria: Evidence that invertebrate AHR homologues lack TCDD and BNF binding. Gene. 2001;278:223–234. doi: 10.1016/s0378-1119(01)00724-7. This paper demonstrated the inability of invertebrate AHRs (arthropods, nematodes, and molluscs) to bind typical AHR ligands TCDD and β-naphthoflavone. [DOI] [PubMed] [Google Scholar]
- 35••.Qin H, Powell-Coffman JA. The Caenorhabditis elegans aryl hydrocarbon receptor, AHR-1, regulates neuronal development. Dev Biol. 2004;270:64–75. doi: 10.1016/j.ydbio.2004.02.004. This is one of two elegant papers that first identified the neurodevelopmental roles of AHR in C. elegans. [DOI] [PubMed] [Google Scholar]
- 36••.Huang X, Powell-Coffman JA, Jin Y. The AHR-1 aryl hydrocarbon receptor and its cofactor the AHA-1 aryl hydrocarbon receptor nuclear translocator specify GABAergic neuron cell fate in C. elegans. Development. 2004;131:819–828. doi: 10.1242/dev.00959. This is one of two elegant papers that first identified the neurodevelopmental roles of AHR in C. elegans. [DOI] [PubMed] [Google Scholar]
- 37•.Smith CJ, O’Brien T, Chatzigeorgiou M, Spencer WC, Feingold-Link E, Husson SJ, Hori S, Mitani S, Gottschalk A, Schafer WR, et al. Sensory neuron fates are distinguished by a transcriptional switch that regulates dendrite branch stabilization. Neuron. 2013;79:266–280. doi: 10.1016/j.neuron.2013.05.009. This paper describes a role for AHR in specifying the cell fate of sensory neurons and identifies control of dendritic branching complexity as an evolutionarily conserved AHR function. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J, Li X, Jevince AR, Guan L, Wang J, Hall DH, Huang X, Ding M. Neuronal target identification requires AHA-1-mediated fine-tuning of Wnt signaling in C. elegans. PLoS Genet. 2013;9:e1003618. doi: 10.1371/journal.pgen.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qin H, Zhai Z, Powell-Coffman JA. The Caenorhabditis elegans AHR-1 transcription complex controls expression of soluble guanylate cyclase genes in the URX neurons and regulates aggregation behavior. Dev Biol. 2006;298:606–615. doi: 10.1016/j.ydbio.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 40.Aarnio V, Heikkinen L, Peltonen J, Goldsteins G, Lakso M, Wong G. Transcriptional profiling reveals differential expression of a neuropeptide-like protein and pseudogenes in aryl hydrocarbon receptor-1 mutant Caenorhabditis elegans. Comp Biochem Physiol Part D Genomics Proteomics. 2014;9:40–48. doi: 10.1016/j.cbd.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 41.Dong PD, Dicks JS, Panganiban G. Distal-less and homothorax regulate multiple targets to pattern the Drosophila antenna. Development. 2002;129:1967–1974. doi: 10.1242/dev.129.8.1967. [DOI] [PubMed] [Google Scholar]
- 42•.Emmons RB, Duncan D, Estes PA, Kiefel P, Mosher JT, Sonnenfeld M, Ward MP, Duncan I, Crews ST. The spineless-aristapedia and tango bHLH-PAS proteins interact to control antennal and tarsal development in Drosophila. Development. 1999;126:3937–3945. doi: 10.1242/dev.126.17.3937. This paper shows that Drosophila AHR function requires ARNT and that Drosophila AHR-ARNT complexes can recognize mammalian AHR response elements. They also present evidence that AHR function in Drosophila is ligand-independent. [DOI] [PubMed] [Google Scholar]
- 43.Kozu S, Tajiri R, Tsuji T, Michiue T, Saigo K, Kojima T. Temporal regulation of late expression of Bar homeobox genes during Drosophila leg development by Spineless, a homolog of the mammalian dioxin receptor. Dev Biol. 2006;294:497–508. doi: 10.1016/j.ydbio.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 44••.Wernet MF, Mazzoni EO, Celik A, Duncan DM, Duncan I, Desplan C. Stochastic spineless expression creates the retinal mosaic for colour vision. Nature. 2006;440:174–180. doi: 10.1038/nature04615. This paper shows that Drosophila AHR acts as a switch to control the cell fate of photoreceptors, determining the type of rhodopsins expressed and thus regulating the development of color vision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnston RJ, Jr, Desplan C. Interchromosomal communication coordinates intrinsically stochastic expression between alleles. Science. 2014;343:661–665. doi: 10.1126/science.1243039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perry M, Kinoshita M, Saldi G, Huo L, Arikawa K, Desplan C. Molecular logic behind the three-way stochastic choices that expand butterfly colour vision. Nature. 2016;535:280–284. doi: 10.1038/nature18616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thanawala SU, Rister J, Goldberg GW, Zuskov A, Olesnicky EC, Flowers JM, Jukam D, Purugganan MD, Gavis ER, Desplan C, et al. Regional modulation of a stochastically expressed factor determines photoreceptor subtypes in the Drosophila retina. Dev Cell. 2013;25:93–105. doi: 10.1016/j.devcel.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim MD, Jan LY, Jan YN. The bHLH-PAS protein Spineless is necessary for the diversification of dendrite morphology of Drosophila dendritic arborization neurons. Genes Dev. 2006;20:2806–2819. doi: 10.1101/gad.1459706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crews ST, Brenman JE. Spineless provides a little backbone for dendritic morphogenesis. Genes Dev. 2006;20:2773–2778. doi: 10.1101/gad.1487706. [DOI] [PubMed] [Google Scholar]
- 50.Kudo K, Takeuchi T, Murakami Y, Ebina M, Kikuchi H. Characterization of the region of the aryl hydrocarbon receptor required for ligand dependency of transactivation using chimeric receptor between Drosophila and Mus musculus. Biochim Biophys Acta. 2009;1789:477–486. doi: 10.1016/j.bbagrm.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 51.Nguyen LP, Bradfield CA. The search for endogenous activators of the aryl hydrocarbon receptor. Chem Res Toxicol. 2008;21:102–116. doi: 10.1021/tx7001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mackowiak B, Wang H. Mechanisms of xenobiotic receptor activation: Direct vs. indirect. Biochim Biophys Acta. 2016;1859:1130–1140. doi: 10.1016/j.bbagrm.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Denison MS, Soshilov AA, He G, DeGroot DE, Zhao B. Exactly the same but different: promiscuity and diversity in the molecular mechanisms of action of the aryl hydrocarbon (dioxin) receptor. Toxicological sciences. 2011;124:1–22. doi: 10.1093/toxsci/kfr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fitch WM. Distinguishing homologous from analogous proteins. Syst Zool. 1970;19:99–113. [PubMed] [Google Scholar]
- 55.Goldstone JV, Hamdoun A, Cole BJ, Howard-Ashby M, Nebert DW, Scally M, Dean M, Epel D, Hahn ME, Stegeman JJ. The chemical defensome: environmental sensing and response genes in the Strongylocentrotus purpuratus genome. Dev Biol. 2006;300:366–384. doi: 10.1016/j.ydbio.2006.08.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sodergren E, Weinstock GM, Davidson EH, Cameron RA, Gibbs RA, Angerer RC, Angerer LM, Arnone MI, Burgess DR, Burke RD, et al. The genome of the sea urchin Strongylocentrotus purpuratus. Science. 2006;314:941–952. doi: 10.1126/science.1133609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simakov O, Kawashima T, Marletaz F, Jenkins J, Koyanagi R, Mitros T, Hisata K, Bredeson J, Shoguchi E, Gyoja F, et al. Hemichordate genomes and deuterostome origins. Nature. 2015;527:459–465. doi: 10.1038/nature16150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li KL, Lu TM, Yu JK. Genome-wide survey and expression analysis of the bHLH-PAS genes in the amphioxus Branchiostoma floridae reveal both conserved and diverged expression patterns between cephalochordates and vertebrates. Evodevo. 2014;5:20. doi: 10.1186/2041-9139-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Putnam NH, Butts T, Ferrier DE, Furlong RF, Hellsten U, Kawashima T, Robinson-Rechavi M, Shoguchi E, Terry A, Yu JK, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- 60.Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, De Tomaso A, Davidson B, Di Gregorio A, Gelpke M, Goodstein DM, et al. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- 61.Hahn ME, Karchner SI, Evans BR, Franks DG, Merson RR, Lapseritis JM. Unexpected diversity of aryl hydrocarbon receptors in non-mammalian vertebrates: Insights from comparative genomics. Journal of Experimental Zoology. 2006;305A:693–706. doi: 10.1002/jez.a.323. [DOI] [PubMed] [Google Scholar]
- 62.Hahn ME, Woodin BR, Stegeman JJ, Tillitt DE. Aryl hydrocarbon receptor function in early vertebrates: Inducibility of cytochrome P4501A in agnathan and elasmobranch fish. Comparative Biochemistry and Physiology. 1998;120C:67–75. doi: 10.1016/s0742-8413(98)00007-3. [DOI] [PubMed] [Google Scholar]
- 63•.Goldstone JV, Goldstone HM, Morrison AM, Tarrant A, Kern SE, Woodin BR, Stegeman JJ. Cytochrome P450 1 genes in early deuterostomes (tunicates and sea urchins) and vertebrates (chicken and frog): origin and diversification of the CYP1 gene family. Mol Biol Evol. 2007;24:2619–2631. doi: 10.1093/molbev/msm200. This paper presents a detailed phylogenetic analysis of the CYP1 gene family and discusses the evolutionary history of AHR-dependent regulation of CYP1 expression. [DOI] [PubMed] [Google Scholar]
- 64.Hahn ME. The aryl hydrocarbon receptor: A comparative perspective. Comparative Biochemistry and Physiology. 1998;121C:23–53. doi: 10.1016/s0742-8413(98)10028-2. [DOI] [PubMed] [Google Scholar]
- 65.Smith JJ, Kuraku S, Holt C, Sauka-Spengler T, Jiang N, Campbell MS, Yandell MD, Manousaki T, Meyer A, Bloom OE, et al. Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat Genet. 2013;45:415–421. 421e411–412. doi: 10.1038/ng.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mehta TK, Ravi V, Yamasaki S, Lee AP, Lian MM, Tay BH, Tohari S, Yanai S, Tay A, Brenner S, et al. Evidence for at least six Hox clusters in the Japanese lamprey (Lethenteron japonicum) Proc Natl Acad Sci U S A. 2013;110:16044–16049. doi: 10.1073/pnas.1315760110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hahn ME, Sakai JA, Greninger D, Franks DG, Merson RR, Karchner SI. Structural and functional characterization of the aryl hydrocarbon receptor in an early diverging vertebrate, the lamprey Petromyzon marinus. Marine Environmental Research. 2004;58:137–138. [Google Scholar]
- 68.Canestro C, Albalat R, Irimia M, Garcia-Fernandez J. Impact of gene gains, losses and duplication modes on the origin and diversification of vertebrates. Semin Cell Dev Biol. 2013;24:83–94. doi: 10.1016/j.semcdb.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 69.Smith JJ, Keinath MC. The sea lamprey meiotic map improves resolution of ancient vertebrate genome duplications. Genome Res. 2015;25:1081–1090. doi: 10.1101/gr.184135.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70•.Goodale BC, La Du JK, Bisson WH, Janszen DB, Waters KM, Tanguay RL. AHR2 mutant reveals functional diversity of aryl hydrocarbon receptors in zebrafish. PLoS ONE. 2012;7:e29346. doi: 10.1371/journal.pone.0029346. This paper describes the phenotype of AHR2-mutant zebrafish and provides new insight into the ligand-specific and regulatory diversity of the three zebrafish AHRs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Karchner SI, Franks DG, Hahn ME. AHR1B, a new functional aryl hydrocarbon receptor in zebrafish: tandem arrangement of ahr1b and ahr2 genes. Biochemical Journal. 2005;392:153–161. doi: 10.1042/BJ20050713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Andreasen EA, Hahn ME, Heideman W, Peterson RE, Tanguay RL. The zebrafish (Danio rerio) aryl hydrocarbon receptor type 1 (zfAHR1) is a novel vertebrate receptor. Molecular Pharmacology. 2002;62:234–249. doi: 10.1124/mol.62.2.234. [DOI] [PubMed] [Google Scholar]
- 73.Karchner SI, Powell WH, Hahn ME. Identification and functional characterization of two highly divergent aryl hydrocarbon receptors (AHR1 and AHR2) in the teleost Fundulus heteroclitus Evidence for a novel subfamily of ligand-binding basic helix-loop-helix Per-ARNT-Sim (bHLH-PAS) factors. J Biol Chem. 1999;274:33814–33824. doi: 10.1074/jbc.274.47.33814. [DOI] [PubMed] [Google Scholar]
- 74.Merson RR, Mattingly CJ, Planchart AJ. Tandem duplication of aryl hydrocarbon receptor (AHR) genes in the genome of the spiny dogfish shark (Squalus acanthias) Bulletin of the Mount Desert Island Biological Laboratory. 2009;48:43–44. [Google Scholar]
- 75.Amemiya CT, Alfoldi J, Lee AP, Fan S, Philippe H, Maccallum I, Braasch I, Manousaki T, Schneider I, Rohner N, et al. The African coelacanth genome provides insights into tetrapod evolution. Nature. 2013;496:311–316. doi: 10.1038/nature12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yasui T, Kim EY, Iwata H, Franks DG, Karchner SI, Hahn ME, Tanabe S. Functional characterization and evolutionary history of two aryl hydrocarbon receptor isoforms (AhR1 and AhR2) from avian species. Toxicol Sci. 2007;99:101–117. doi: 10.1093/toxsci/kfm139. [DOI] [PubMed] [Google Scholar]
- 77.Lee JS, Iwabuchi K, Nomaru K, Nagahama N, Kim EY, Iwata H. Molecular and functional characterization of a novel aryl hydrocarbon receptor isoform, AHR1beta, in the chicken (Gallus gallus) Toxicol Sci. 2013;136:450–466. doi: 10.1093/toxsci/kft192. [DOI] [PubMed] [Google Scholar]
- 78.Oka K, Kohno S, Ohta Y, Guillette LJ, Jr, Iguchi T, Katsu Y. Molecular cloning and characterization of the aryl hydrocarbon receptors and aryl hydrocarbon receptor nuclear translocators in the American alligator. Gen Comp Endocrinol. 2016;238:13–22. doi: 10.1016/j.ygcen.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 79.Mikkelsen TS, Wakefield MJ, Aken B, Amemiya CT, Chang JL, Duke S, Garber M, Gentles AJ, Goodstadt L, Heger A, et al. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature. 2007;447:167–177. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- 80.Lavine JA, Rowatt AJ, Klimova T, Whitington AJ, Dengler E, Beck C, Powell WH. Aryl Hydrocarbon Receptors in the Frog Xenopus laevis: Two AhR1 Paralogs Exhibit Low Affinity for 2,3,7,8-Tetrachlorodibenzo-p-Dioxin (TCDD) Toxicol Sci. 2005;88:60–72. doi: 10.1093/toxsci/kfi228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Reitzel AM, Karchner SI, Franks DG, Evans BR, Nacci DE, Champlin D, Vieira VM, Hahn ME. Genetic variation at aryl hydrocarbon receptor (AHR) loci in populations of Atlantic Killifish (Fundulus heteroclitus) inhabiting polluted and reference habitats. BMC Evolutionary Biology. 2014;14:6. doi: 10.1186/1471-2148-14-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Glasauer SM, Neuhauss SC. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol Genet Genomics. 2014;289:1045–1060. doi: 10.1007/s00438-014-0889-2. [DOI] [PubMed] [Google Scholar]
- 83.Postlethwait J, Amores A, Cresko W, Singer A, Yan YL. Subfunction partitioning, the teleost radiation and the annotation of the human genome. Trends Genet. 2004;20:481–490. doi: 10.1016/j.tig.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 84.Braasch I, Gehrke AR, Smith JJ, Kawasaki K, Manousaki T, Pasquier J, Amores A, Desvignes T, Batzel P, Catchen J, et al. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat Genet. 2016;48:427–437. doi: 10.1038/ng.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Doering JA, Wiseman S, Beitel SC, Giesy JP, Hecker M. Identification and expression of aryl hydrocarbon receptors (AhR1 and AhR2) provide insight in an evolutionary context regarding sensitivity of white sturgeon (Acipenser transmontanus) to dioxin-like compounds. Aquat Toxicol. 2014;150:27–35. doi: 10.1016/j.aquatox.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 86.Venkatesh B, Lee AP, Ravi V, Maurya AK, Lian MM, Swann JB, Ohta Y, Flajnik MF, Sutoh Y, Kasahara M, et al. Elephant shark genome provides unique insights into gnathostome evolution. Nature. 2014;505:174–179. doi: 10.1038/nature12826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87•.Mimura J, Ema M, Sogawa K, Fujii-Kuriyama Y. Identification of a novel mechanism of regulation of Ah (dioxin) receptor function. Genes and Development. 1999;13:20–25. doi: 10.1101/gad.13.1.20. This paper was the first to identify AHRR as an AHR-related repressor of AHR signaling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hahn ME, Allan LL, Sherr DH. Regulation of constitutive and inducible AHR signaling. Complex interactions Involving the AHR repressor. Biochem Pharmacol. 2009;77:485–497. doi: 10.1016/j.bcp.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Karchner SI, Franks DG, Powell WH, Hahn ME. Regulatory interactions among three members of the vertebrate aryl hydrocarbon receptor family: AHR repressor, AHR1, and AHR2. J Biol Chem. 2002;277:6949–6959. doi: 10.1074/jbc.M110779200. [DOI] [PubMed] [Google Scholar]
- 90.Vogel CF, Chang WL, Kado S, McCulloh K, Vogel H, Wu D, Haarmann-Stemmann T, Yang G, Leung PS, Matsumura F, et al. Transgenic overexpression of aryl hydrocarbon receptor repressor (AhRR) and AhR-mediated induction of CYP1A1, cytokines, and acute toxicity. Environ Health Perspect. 2016;124:1071–1083. doi: 10.1289/ehp.1510194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Brandstatter O, Schanz O, Vorac J, Konig J, Mori T, Maruyama T, Korkowski M, Haarmann-Stemmann T, von Smolinski D, Schultze JL, et al. Balancing intestinal and systemic inflammation through cell type-specific expression of the aryl hydrocarbon receptor repressor. Sci Rep. 2016;6:26091. doi: 10.1038/srep26091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Burris HH, Baccarelli AA, Byun HM, Cantoral A, Just AC, Pantic I, Solano-Gonzalez M, Svensson K, Tamayo y Ortiz M, Zhao Y, et al. Offspring DNA methylation of the aryl-hydrocarbon receptor repressor gene is associated with maternal BMI, gestational age, and birth weight. Epigenetics. 2015;10:913–921. doi: 10.1080/15592294.2015.1078963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Watanabe M, Sueoka K, Sasagawa I, Nakabayashi A, Yoshimura Y, Ogata T. Association of male infertility with Pro185Ala polymorphism in the aryl hydrocarbon receptor repressor gene: Implication for the susceptibility to dioxins. Fertil Steril. 2004;82(Suppl 3):1067–1071. doi: 10.1016/j.fertnstert.2004.06.027. [DOI] [PubMed] [Google Scholar]
- 94.Zudaire E, Cuesta N, Murty V, Woodson K, Adams L, Gonzalez N, Martinez A, Narayan G, Kirsch I, Franklin W, et al. The aryl hydrocarbon receptor repressor is a putative tumor suppressor gene in multiple human cancers. J Clin Invest. 2008;118:640–650. doi: 10.1172/JCI30024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hosoya T, Harada N, Mimura J, Motohashi H, Takahashi S, Nakajima O, Morita M, Kawauchi S, Yamamoto M, Fujii-Kuriyama Y. Inducibility of cytochrome P450 1A1 and chemical carcinogenesis by benzo[a]pyrene in AhR repressor-deficient mice. Biochem Biophys Res Commun. 2008;365:562–567. doi: 10.1016/j.bbrc.2007.11.016. [DOI] [PubMed] [Google Scholar]
- 96.Dever DP, Adham ZO, Thompson B, Genestine M, Cherry J, Olschowka JA, DiCicco-Bloom E, Opanashuk LA. Aryl hydrocarbon receptor deletion in cerebellar granule neuron precursors impairs neurogenesis. Dev Neurobiol. 2016;76:533–550. doi: 10.1002/dneu.22330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Latchney SE, Hein AM, O’Banion MK, DiCicco-Bloom E, Opanashuk LA. Deletion or activation of the aryl hydrocarbon receptor alters adult hippocampal neurogenesis and contextual fear memory. J Neurochem. 2013;125:430–445. doi: 10.1111/jnc.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Collins LL, Williamson MA, Thompson BD, Dever DP, Gasiewicz TA, Opanashuk LA. 2,3,7,8-Tetracholorodibenzo-p-dioxin exposure disrupts granule neuron precursor maturation in the developing mouse cerebellum. Toxicol Sci. 2008;103:125–136. doi: 10.1093/toxsci/kfn017. [DOI] [PubMed] [Google Scholar]
- 99.Kimura E, Ding Y, Tohyama C. AhR signaling activation disrupts migration and dendritic growth of olfactory interneurons in the developing mouse. Sci Rep. 2016;6:26386. doi: 10.1038/srep26386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kimura E, Kubo K, Matsuyoshi C, Benner S, Hosokawa M, Endo T, Ling W, Kohda M, Yokoyama K, Nakajima K, et al. Developmental origin of abnormal dendritic growth in the mouse brain induced by in utero disruption of aryl hydrocarbon receptor signaling. Neurotoxicol Teratol. 2015;52:42–50. doi: 10.1016/j.ntt.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 101.Del Pino Sans J, Clements KJ, Suvorov A, Krishnan S, Adams HL, Petersen SL. Developmental exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin may alter LH release patterns by abolishing sex differences in GABA/glutamate cell number and modifying the transcriptome of the male anteroventral periventricular nucleus. Neuroscience. 2016;329:239–253. doi: 10.1016/j.neuroscience.2016.04.051. [DOI] [PubMed] [Google Scholar]
- 102.Hays LE, Carpenter CD, Petersen SL. Evidence that GABAergic neurons in the preoptic area of the rat brain are targets of 2,3,7,8-tetrachlorodibenzo-p-dioxin during development. Environ Health Perspect. 2002;110(Suppl 3):369–376. doi: 10.1289/ehp.02110s3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Petersen SL, Curran MA, Marconi SA, Carpenter CD, Lubbers LS, McAbee MD. Distribution of mRNAs encoding the arylhydrocarbon receptor, arylhydrocarbon receptor nuclear translocator, and arylhydrocarbon receptor nuclear translocator-2 in the rat brain and brainstem. J Comp Neurol. 2000;427:428–439. doi: 10.1002/1096-9861(20001120)427:3<428::aid-cne9>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 104.Puga A, Tomlinson CR, Xia Y. Ah receptor signals cross-talk with multiple developmental pathways. Biochem Pharmacol. 2005;69:199–207. doi: 10.1016/j.bcp.2004.06.043. [DOI] [PubMed] [Google Scholar]
- 105.Boitano AE, Wang J, Romeo R, Bouchez LC, Parker AE, Sutton SE, Walker JR, Flaveny CA, Perdew GH, Denison MS, et al. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science. 2010;329:1345–1348. doi: 10.1126/science.1191536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Smith BW, Rozelle SS, Leung A, Ubellacker J, Parks A, Nah SK, French D, Gadue P, Monti S, Chui DH, et al. The aryl hydrocarbon receptor directs hematopoietic progenitor cell expansion and differentiation. Blood. 2013;122:376–385. doi: 10.1182/blood-2012-11-466722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gasiewicz TA, Singh KP, Bennett JA. The Ah receptor in stem cell cycling, regulation, and quiescence. Ann N Y Acad Sci. 2014;1310:44–50. doi: 10.1111/nyas.12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Peterson RE, Theobald HM, Kimmel GL. Developmental and Reproductive Toxicity of Dioxins and Related Compounds - Cross-Species Comparisons. CRC Critical Reviews in Toxicology. 1993;23:283–335. doi: 10.3109/10408449309105013. [DOI] [PubMed] [Google Scholar]
- 109.King-Heiden TC, Mehta V, Xiong KM, Lanham KA, Antkiewicz DS, Ganser A, Heideman W, Peterson RE. Reproductive and developmental toxicity of dioxin in fish. Mol Cell Endocrinol. 2012;354:121–138. doi: 10.1016/j.mce.2011.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schneider AJ, Branam AM, Peterson RE. Intersection of AHR and Wnt signaling in development, health, and disease. Int J Mol Sci. 2014;15:17852–17885. doi: 10.3390/ijms151017852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yoshioka W, Peterson RE, Tohyama C. Molecular targets that link dioxin exposure to toxicity phenotypes. J Steroid Biochem Mol Biol. 2011;127:96–101. doi: 10.1016/j.jsbmb.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Vezina CM, Lin TM, Peterson RE. AHR signaling in prostate growth, morphogenesis, and disease. Biochem Pharmacol. 2009;77:566–576. doi: 10.1016/j.bcp.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mathew LK, Simonich MT, Tanguay RL. AHR-dependent misregulation of Wnt signaling disrupts tissue regeneration. Biochem Pharmacol. 2009;77:498–507. doi: 10.1016/j.bcp.2008.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114•.Tijet N, Boutros PC, Moffat ID, Okey AB, Tuomisto J, Pohjanvirta R. Aryl hydrocarbon receptor regulates distinct dioxin-dependent and dioxin-independent gene batteries. Mol Pharmacol. 2006;69:140–153. doi: 10.1124/mol.105.018705. This paper is the first to systematically determine genes regulated by AHR in the presence and absence of a xenobiotic ligand. [DOI] [PubMed] [Google Scholar]
- 115.Hayes KR, Zastrow GM, Nukaya M, Pande K, Glover E, Maufort JP, Liss AL, Liu Y, Moran SM, Vollrath AL, et al. Hepatic transcriptional networks induced by exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Chem Res Toxicol. 2007;20:1573–1581. doi: 10.1021/tx7003294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sun RX, Chong LC, Simmons TT, Houlahan KE, Prokopec SD, Watson JD, Moffat ID, Lensu S, Linden J, P’ng C, et al. Cross-species transcriptomic analysis elucidates constitutive aryl hydrocarbon receptor activity. BMC Genomics. 2014;15:1053. doi: 10.1186/1471-2164-15-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nukaya M, Walisser JA, Moran SM, Kennedy GD, Bradfield CA. Aryl hydrocarbon receptor nuclear translocator in hepatocytes is required for aryl hydrocarbon receptor-mediated adaptive and toxic responses in liver. Toxicol Sci. 2010;118:554–563. doi: 10.1093/toxsci/kfq305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bunger MK, Glover E, Moran SM, Walisser JA, Lahvis GP, Hsu EL, Bradfield CA. Abnormal liver development and resistance to 2,3,7,8-tetrachlorodibenzo-p-dioxin toxicity in mice carrying a mutation in the DNA-binding domain of the aryl hydrocarbon receptor. Toxicol Sci. 2008;106:83–92. doi: 10.1093/toxsci/kfn149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tanos R, Patel RD, Murray IA, Smith PB, Patterson AD, Perdew GH. Ah receptor regulates the cholesterol biosynthetic pathway in a dioxin response element-independent manner. Hepatology. 2012;55:1994–2004. doi: 10.1002/hep.25571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Patel RD, Murray IA, Flaveny CA, Kusnadi A, Perdew GH. Ah receptor represses acute-phase response gene expression without binding to its cognate response element. Lab Invest. 2009;89:695–707. doi: 10.1038/labinvest.2009.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Vogel CF, Matsumura F. A new cross-talk between the aryl hydrocarbon receptor and RelB, a member of the NF-kappaB family. Biochem Pharmacol. 2009;77:734–745. doi: 10.1016/j.bcp.2008.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jackson DP, Li H, Mitchell KA, Joshi AD, Elferink CJ. Ah receptor-mediated suppression of liver regeneration through NC-XRE-driven p21Cip1 expression. Mol Pharmacol. 2014;85:533–541. doi: 10.1124/mol.113.089730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jones LM, Rayson SJ, Flemming AJ, Urwin PE. Adaptive and specialised transcriptional responses to xenobiotic stress in Caenorhabditis elegans are regulated by nuclear hormone receptors. PLoS One. 2013;8:e69956. doi: 10.1371/journal.pone.0069956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Brown RP, McDonnell CM, Berenbaum MR, Schuler MA. Regulation of an insect cytochrome P450 monooxygenase gene (CYP6B1) by aryl hydrocarbon and xanthotoxin response cascades. Gene. 2005;358:39–52. doi: 10.1016/j.gene.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 125.Stegeman JJ, Hahn ME. Biochemistry and molecular biology of monooxygenases: Current perspectives on forms, functions, and regulation of cytochrome P450 in aquatic species. In: Malins DC, Ostrander GK, editors. Aquatic Toxicology: Molecular, Biochemical and Cellular Perspectives. CRC/Lewis; 1994. pp. 87–206. [Google Scholar]
- 126.DeGroot DE, Franks DG, Higa T, Tanaka J, Hahn ME, Denison MS. Naturally-Occurring Marine Brominated Indoles are Aryl Hydrocarbon Receptor Ligands/Agonists. Chemical Research in Toxicology. 2015;28:1176–1185. doi: 10.1021/acs.chemrestox.5b00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Vetter W, Hahn ME, Tomy G, Ruppe S, Vatter S, Chahbane N, Lenoir D, Schramm K-W, Scherer G. Biological activity and physico-chemical parameters of the marine halogenated natural products 2,3,3′,4,4′,5,5′-heptachloro-2′-methyl-1,2′-bipyrrole (Q1) and 2,4,6-tribromoanisole (TBA) Archives of Environmental Contamination and Toxicology. 2005;48:1–9. doi: 10.1007/s00244-004-0049-5. [DOI] [PubMed] [Google Scholar]
- 128.Tittlemier SA, Kennedy SW, Hahn ME, Reddy CM, Norstrom RJ. Naturally-produced halogenated dimethyl bipyrroles bind to the Ah receptor and induce cytochrome P4501A and porphyrin accumulation in chicken embryo hepatocytes. Environmental Toxicology and Chemistry. 2003;22:1497–1506. [PubMed] [Google Scholar]
- 129.Quintana FJ, Sherr DH. Aryl hydrocarbon receptor control of adaptive immunity. Pharmacol Rev. 2013;65:1148–1161. doi: 10.1124/pr.113.007823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Moura-Alves P, Fae K, Houthuys E, Dorhoi A, Kreuchwig A, Furkert J, Barison N, Diehl A, Munder A, Constant P, et al. AhR sensing of bacterial pigments regulates antibacterial defence. Nature. 2014;512:387–392. doi: 10.1038/nature13684. [DOI] [PubMed] [Google Scholar]
- 131.Bessede A, Gargaro M, Pallotta MT, Matino D, Servillo G, Brunacci C, Bicciato S, Mazza EM, Macchiarulo A, Vacca C, et al. Aryl hydrocarbon receptor control of a disease tolerance defence pathway. Nature. 2014;511:184–190. doi: 10.1038/nature13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, Zecchi R, D’Angelo C, Massi-Benedetti C, Fallarino F, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 133.Li Y, Innocentin S, Withers DR, Roberts NA, Gallagher AR, Grigorieva EF, Wilhelm C, Veldhoen M. Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell. 2011;147:629–640. doi: 10.1016/j.cell.2011.09.025. [DOI] [PubMed] [Google Scholar]
- 134.Hubbard TD, Murray IA, Bisson WH, Lahoti TS, Gowda K, Amin SG, Patterson AD, Perdew GH. Adaptation of the human aryl hydrocarbon receptor to sense microbiota-derived indoles. Sci Rep. 2015;5:12689. doi: 10.1038/srep12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fraccalvieri D, Soshilov AA, Karchner SI, Franks DG, Pandini A, Bonati L, Hahn ME, Denison MS. Comparative analysis of homology models of the Ah receptor ligand binding domain: verification of structure-function predictions by site-directed mutagenesis of a non-functional receptor. Biochemistry. 2013;52:714–725. doi: 10.1021/bi301457f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Incardona JP, Day HL, Collier TK, Scholz NL. Developmental toxicity of 4-ring polycyclic aromatic hydrocarbons in zebrafish is differentially dependent on AH receptor isoforms and hepatic cytochrome P4501A metabolism. Toxicol Appl Pharmacol. 2006;217:308–321. doi: 10.1016/j.taap.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 137.Karchner SI, Jenny MJ, Aluru N, Franks DG, Laub LB, Linney E, Williams LM, Teraoka H, Hahn ME. Evidence for developmental versus toxicological roles for zebrafish AHR1b. Toxicological Sciences (The Toxicologist Supplement) 2017 Abstract #1165. [Google Scholar]
- 138.Prasch AL, Teraoka H, Carney SA, Dong W, Hiraga T, Stegeman JJ, Heideman W, Peterson RE. Aryl Hydrocarbon Receptor 2 Mediates 2,3,7,8-Tetrachlorodibenzo-p-dioxin Developmental Toxicity in Zebrafish. Toxicological Sciences. 2003;76:138–150. doi: 10.1093/toxsci/kfg202. [DOI] [PubMed] [Google Scholar]
- 139.Jönsson ME, Jenny MJ, Woodin BR, Hahn ME, Stegeman JJ. Role of AHR2 in the expression of novel cytochrome P450 1 family genes, cell cycle genes, and morphological defects in developing zebra fish exposed to 3,3′,4,4′,5-pentachlorobiphenyl or 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol Sci. 2007;100:180–193. doi: 10.1093/toxsci/kfm207. [DOI] [PubMed] [Google Scholar]
- 140.Billiard SM, Timme-Laragy AR, Wassenberg DM, Cockman C, Di Giulio RT. The role of the aryl hydrocarbon receptor pathway in mediating synergistic developmental toxicity of polycyclic aromatic hydrocarbons to zebrafish. Toxicol Sci. 2006;92:526–536. doi: 10.1093/toxsci/kfl011. [DOI] [PubMed] [Google Scholar]
- 141.Incardona JP, Linbo TL, Scholz NL. Cardiac toxicity of 5-ring polycyclic aromatic hydrocarbons is differentially dependent on the aryl hydrocarbon receptor 2 isoform during zebrafish development. Toxicology and applied pharmacology. 2011;257:242–249. doi: 10.1016/j.taap.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 142.Van Tiem LA, Di Giulio RT. AHR2 knockdown prevents PAH-mediated cardiac toxicity and XRE- and ARE-associated gene induction in zebrafish (Danio rerio) Toxicol Appl Pharmacol. 2011;254:280–287. doi: 10.1016/j.taap.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Scott JA, Incardona JP, Pelkki K, Shepardson S, Hodson PV. AhR2-mediated, CYP1A-independent cardiovascular toxicity in zebrafish (Danio rerio) embryos exposed to retene. Aquat Toxicol. 2011;101:165–174. doi: 10.1016/j.aquatox.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 144.Knecht AL, Goodale BC, Truong L, Simonich MT, Swanson AJ, Matzke MM, Anderson KA, Waters KM, Tanguay RL. Comparative developmental toxicity of environmentally relevant oxygenated PAHs. Toxicology and applied pharmacology. 2013;271:266–275. doi: 10.1016/j.taap.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lee JS, Kim EY, Iwata H. Dioxin activation of CYP1A5 promoter/enhancer regions from two avian species, common cormorant (Phalacrocorax carbo) and chicken (Gallus gallus): association with aryl hydrocarbon receptor 1 and 2 isoforms. Toxicol Appl Pharmacol. 2009;234:1–13. doi: 10.1016/j.taap.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 146.Freeburg SH, Engelbrecht E, Powell WH. Subfunctionalization of Paralogous Aryl Hydrocarbon Receptors from the Frog Xenopus Laevis: Distinct Target Genes and Differential Responses to Specific Agonists in a Single Cell Type. Toxicol Sci. 2017;155:337–347. doi: 10.1093/toxsci/kfw212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Harms MJ, Thornton JW. Evolutionary biochemistry: revealing the historical and physical causes of protein properties. Nat Rev Genet. 2013;14:559–571. doi: 10.1038/nrg3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Harms MJ, Thornton JW. Analyzing protein structure and function using ancestral gene reconstruction. Curr Opin Struct Biol. 2010;20:360–366. doi: 10.1016/j.sbi.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Torruella G, Derelle R, Paps J, Lang BF, Roger AJ, Shalchian-Tabrizi K, Ruiz-Trillo I. Phylogenetic relationships within the Opisthokonta based on phylogenomic analyses of conserved single-copy protein domains. Mol Biol Evol. 2012;29:531–544. doi: 10.1093/molbev/msr185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Muffato M, Louis A, Poisnel CE, Roest Crollius H. Genomicus: a database and a browser to study gene synteny in modern and ancestral genomes. Bioinformatics. 2010;26:1119–1121. doi: 10.1093/bioinformatics/btq079. [DOI] [PMC free article] [PubMed] [Google Scholar]