Abstract
Targeted cancer therapies require a precise determination of the underlying biological processes driving tumorigenesis within the complex tumor microenvironment. Therefore, new diagnostic tools that capture the molecular activity at the disease site in vivo are needed to better understand tumor behavior and ultimately maximize therapeutic responses. Matrix metalloproteinases (MMPs) drive multiple aspects of tumorigenesis, and their activity can be monitored using engineered peptide substrates as protease-specific probes. To identify tumor specific activity profiles, local sampling of the tumor microenvironment is necessary, such as through remote control of probes, which are only activated at the tumor site. Alternating magnetic fields (AMFs) provide an attractive option to remotely apply local triggering signals, as they penetrate deep into the body and are not likely to interfere with biological processes due to weak magnetic properties of tissue. Here, we report the design and evaluation of a protease-activity nanosensor that can be remotely activated at the site of disease via an AMF at 515 kHz and 15 kA/m. Our nanosensor was comprised of thermosensitive liposomes containing functionalized protease substrates that were unveiled at the target site by remotely triggered heat dissipation of co-encapsulated magnetic nanoparticles (MNPs). This nanosensor was combined with a unique detection assay to quantify the amount of cleaved substrates in the urine. We applied this spatiotemporally controlled system to determine tumor protease activity in vivo and identified differences in substrate cleavage profiles between two mouse models of human colorectal cancer.
Keywords: thermoliposomes, proteases, activity-based biomarkers, nanosensors, magnetic nanoparticles, hysteresis-loss heating
With the advent of targeted therapies, an increasing emphasis has been placed on establishing a system of precision medicine, such that therapies are individually tailored.1 Robust companion diagnostics are needed to stratify patient disease state, and thus inform therapeutic decisions.1–3 For example, molecular imaging can be used to identify a patient’s vascular permeability to nanotherapeutics.1, 4, 5 Alternatively, analysis of samples acquired by invasive biopsies can be performed to identify therapeutic regimens (e.g. by analyzing receptor expression). An emerging area of targeted therapies is the application of protease-activated agents, as proteases are known to play role in almost every hallmark of cancer.6–9 These enzyme-dependent tools are responsive to the local microenvironment, which can improve the therapeutic windows of numerous agents. Protease-activated antibodies, ‘probodies’, being developed are one such example.10 To harness the full potential of enzyme-activated interventions, functional diagnostics that provide information on the activity and function of particular proteases within the disease environment are necessary.1
Protease activity is typically measured by the cleavage of selective functionalized peptide substrates, which results in contrast generation for imaging purposes.11–14 We have previously developed multiplexed nanosensors called ‘synthetic biomarkers’ that generate detection signals after proteolytic cleavage of peptide substrates.15–17 Unlike in situ imaging sensors, the cleavage fragment enters circulation, is concentrated in the urine, and can be detected by a range of analytical techniques (e.g. mass spectrometry or ELISA) enabling non-invasive measurement of in vivo protease activity. However, like other protease sensors, proteolytic cleavage that occurs in circulation or off-target organs affects the specificity of signal.18, 19 In order to reduce off-target activation, we recently developed photoactivatable synthetic biomarkers that incorporate photolabile groups to shield access to the peptide substrates, and which can reduce the degree of non-specific signal generation.20 This technique, however, is challenging to utilize in vivodue to poor penetration of light into most biological tissues.
Here, we report the development of magnetically actuated protease sensors (MAPS) that rely on exposure to local alternating magnetic fields (AMFs) to release peptide substrates from thermosensitive liposomes into the tumor microenvironment, after which they sample and respond to protease activity. In the MAPS platform, peptide substrates are co-encapsulated in thermosensitive liposomes with magnetic nanoparticles (MNPs), which undergo hysteretic heat dissipation in the presence of AMFs, leading to elevated local temperatures and liposome deformation. We characterize temperature sensitivity of MAPS and responsiveness to AMFs in vitroand then demonstrate that this newly formulated sensor can distinguish different tumor types by profiling protease cleavage specificities across two xenograft mouse models of colorectal cancer.
Matrix metalloproteinases (MMPs) are a family of structurally related, zinc-dependent endopeptidases with important roles in development, tissue injury and repair, and many diseases.21, 22 In cancer, MMPs promote invasion and metastasis, and different tumors often exhibit unique MMP expression profiles.22 Thus, we designed a remotely controllable nanosensor to locally assay MMP profiles in tumors.
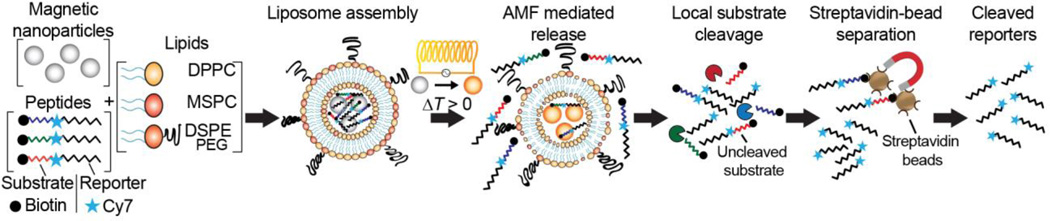
We employed liposomal carriers to entrap peptide substrates and shield them from nonspecific cleavage in the blood stream, and took advantage of the enhanced permeability and retention (EPR) effect to achieve accumulation at tumor sites.23, 24 To enable thermally induced release of protease-cleavable substrates linked to urinary reporters,15 liposomes comprised of thermosensitive bilayers were co-loaded with MNPs that undergo hysteresis and dissipate heat in the presence of AMFs with frequencies in the range of hundreds of kHz (Fig. 1).
Figure 1. Magnetically actuated protease sensors (MAPS).
Thermosensitive liposomes, consisting of the lipids DPPC, MSPC and DPSE-PEG, were encapsulated with magnetic nanoparticles and synthetic peptides. These peptide substrates are each coupled with a near IR dye, Cy7, as a urinary reporter, and an N-terminal biotin enabling streptavidin-bead based separation. Upon exposure to alternating magnetic fields (AMF), heat is dissipated by the co-entrapped MNPs due to hysteresis losses, which in turn melts the thermosensitive bilayer. The permeabilized membrane allows peptides to diffuse to the exterior where they are cleaved by proteases. Cleaved and uncleaved peptides clear into urine, where cleaved reporters are isolated using streptavidin-coated beads.
We chose a clinically approved thermosensitive liposome formulation containing DCCP (1,2-dipalmitoyl-sn-glycero-3-phosphocholine), the most common phosphoglyceride used as a backbone in liposomal bilayer preparations, the lysolipid (MSPC, 1-myristoyl-2-stearoyl-sn-glycero-3-phosphocholine), and DSPE-PEG(2000) (1,2-distearoyl-sn-glycero-3-phosphoetha-nolamine-N-[amino(polyethyleneglycol)-2000]).25 At the phase transition temperature, liposomal bilayers exhibit leaky interfacial regions between solid and melting liquid phases. For our chosen volume ratio of 80:15:5 for DCCP:MSPC:DSPE-PEG, the critical melting temperature was determined as Tm ≈ 41°C,26, 27 and thus, the disruption of our liposomes should require only mild temperature elevation through externally triggered hysteretic heat dissipation.
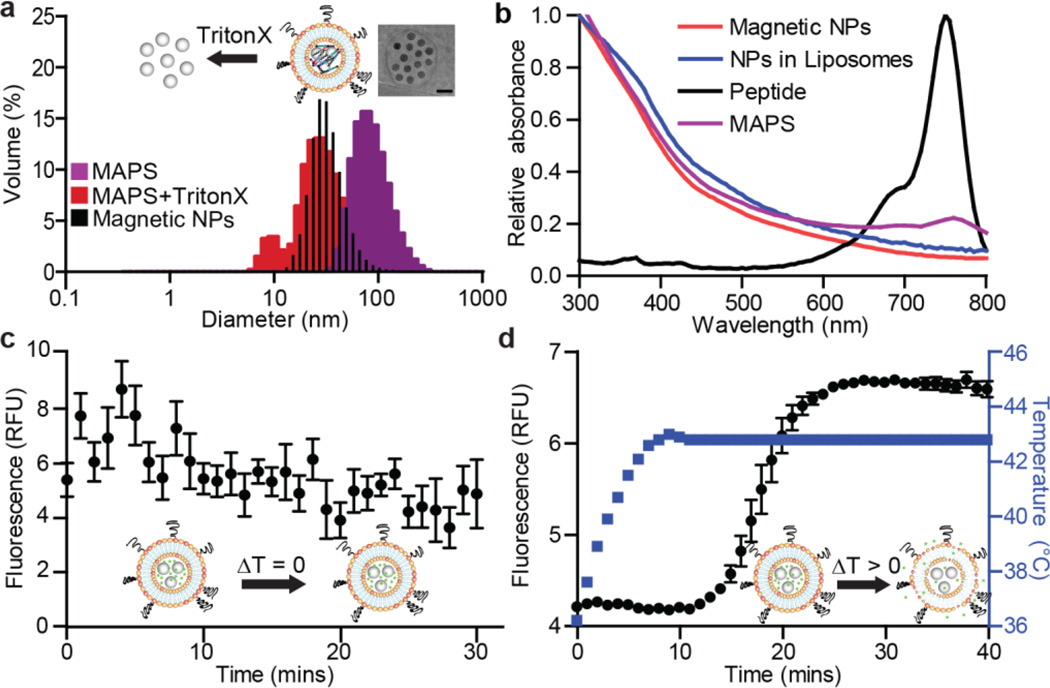
To achieve the required temperature increase, we chose iron oxide MNPs with a diameter of 25 nm (Ocean Nanotech LLC, SHA-25, Fig. 2a). The fully assembled MAPS containing MNPs and peptides exhibited a mean diameter of 100 nm, based on dynamic light scattering (DLS) measurements (Fig. 2a). Successful loading of individual components, i.e. substrates with reporter dye and MNPs, was confirmed by absorbance spectroscopy after filtration (Fig. 2b). To quantify the degree of MNP loading within the MAPS, iron content was assayed by inductively coupled plasma atomic emission spectrometry (ICP-AES) and found to be 1.89 ± 0.15 mg/mL (Fig. S1). Further morphological analysis by transmission electron microscopy (TEM) supported these findings, as MNPs were predominantly entrapped, with only few free MNPs observed (see inset Fig. 2a and Fig. S2).
Figure 2. MAPS characterization: size, composition, magnetic properties, and stability.
(a) Dynamic light scattering (DLS) measurements of MAPS (purple) and disrupted MAPS after addition of 0.1% TritonX, showing the release of coentrapped MNPs (red) (n = 3). DLS measurement of pure MNPs is overlaid in black. Inset: transmission electron microscopy image of an individual MAPS (scale bar: 50 nm). (b) Absorbance spectra of various components of MAPS. (c) Calcein fluorescence release assay at 37°C over 30 min (n = 5, SEM). (d) At higher temperatures, the release of calcein was detected by increases in sample fluorescence (n = 5, SEM).
Prior to applying MAPS to profile MMP activity, we sought to characterize their thermosensitivity and temperature-dependent release profile using a calcein-based assay. Calcein is a membrane impermeable dye that exhibits homoquenching at high concentrations, thus any release of MAPS-loaded calcein will result in a fluorescence increase. We first probed for any change in permeability at 37°C, and found the liposomes to be stable at this temperature, as the fluorescent signal did not increase (Fig. 2c). A substantial release of fluorescence was detected, however, when the temperature of the loaded MAPS samples was raised to 43°C (Fig. 2d). Both the steep slope and magnitude of the response are indicative of a robust and sensitive change in liposome permeability.
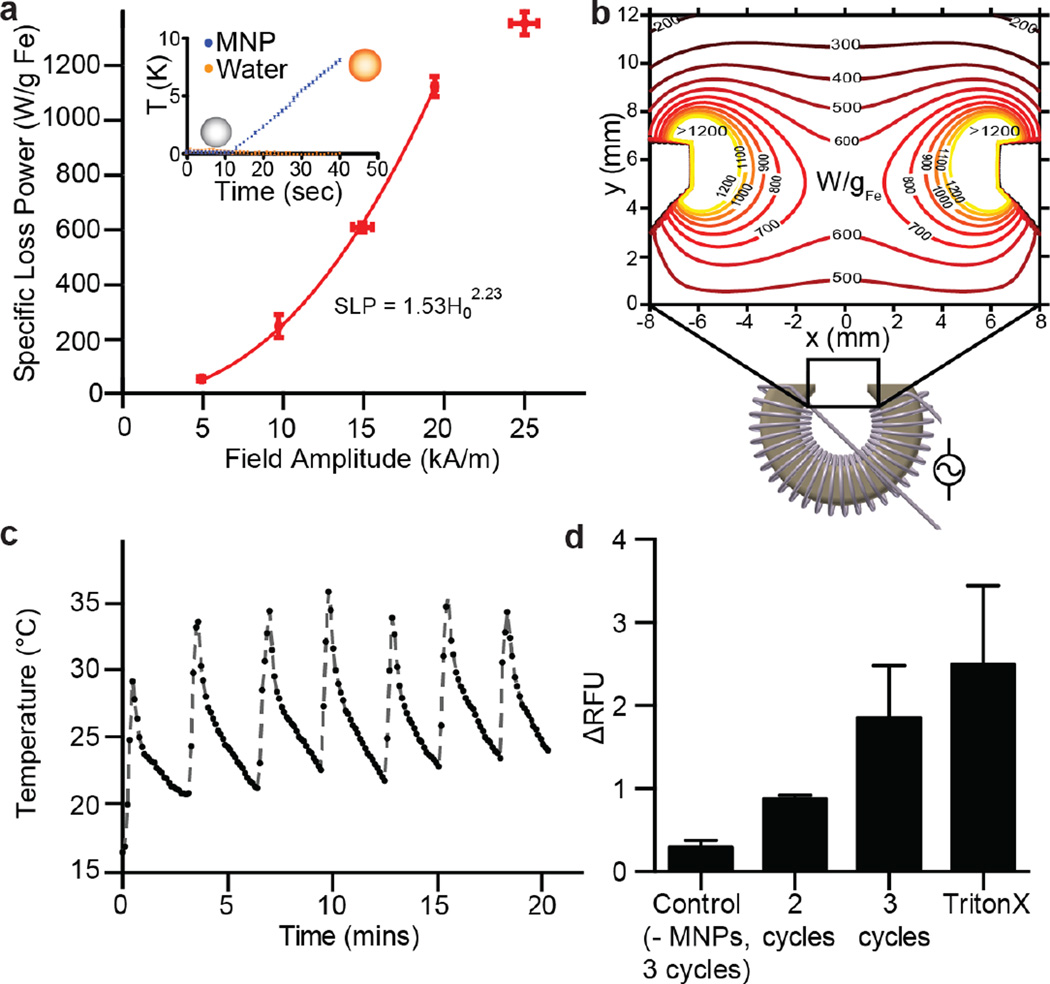
Next, we determined the AMF parameters necessary for sufficient hysteretic heating rates (specific loss power, SLP) in entrapped MNPs to induce magnetothermal melting of the liposomal bilayer. SLP depends on the amplitude and frequency of the externally applied AMF, as well as MNP properties, such as size, shape, and composition.28 For a MNP solution, the SLP is calculated as , where C is the specific heat capacity of water (C = 4.184 J K−1 mL−1), m is the iron concentration (in g(Fe)/mL) and ΔT/Δt is the experimentally measured initial slope of the temperature increase under AMF exposure.
While previous studies have reported liposomes loaded with small iron oxide MNPs in the size range of 5–15 nm23, we instead chose comparatively larger 25 nm MNPs due to their higher SLP. This selection is motivated by dynamic hysteresis modeling,29, 30 combined with a limit on permissible amplitude-frequency products for continuous exposure (Ho×f~5×109 A·m−1·s−1)31. We have previously suggested that maximized heating rates subject to this constraint are achieved by using particles with relaxation times exceeding the timescale set by the AMF frequency, at field magnitudes close to their effective anisotropy field. Additionally, this previous work identified that spherical iron oxide particles with 25 nm diameters exhibit high SLPs, and specifically high individual particle loss powers (IPLPs), at AMF conditions similar to those employed here.29 A detailed analysis of IPLPs for different particle sizes that motivated our particle choice has been published recently.32 Based on calorimetry measurements, an AMF with an amplitude of 15kA/m and frequency of 515 kHz was found to produce an SLP of 610 ± 16 W/g(Fe)(Fig. 3a, and inset). Recent studies have reported a large temperature increase near the surface of nanoparticles,33, 34 and such local heating may play a mechanistic role in release.32 We were further interested in potential particle-particle interactions, given the local MNP accumulation in liposomes that could influence the magnetic response.35 Analysis by vibrating sample magnetometry revealed superparamagnetic behavior with converegnce to nearly the same slope in the limit of a low applied field for both the pure magnetic nanoparticles and suspensions of liposomes with encapsulated MNPs (Fig. S3). However, the encapsulated MNPs approach saturation more gradually than the free particles. Both of these qualtiative features are predicted for noninteracting ensembles of randomly oriented particles with increasing anisotropy.30
Figure 3. Magnetothermal activation: coil design and parameter determination for magnetically induced release.
(a) Optimal AMF parameters were evaluated by calorimetric measurements. Fit line interpolated using a power law valid for field strength magnitudes between 0 and 20 kA/m (n=3, SEM). Inset: fluid temperature increase during 30 s of AMF exposure at 515 kHz and 15 kA/m (n = 5). (b) Technical drawing of coil with ferromagnetic core utilized in studies. Inset shows the distribution of the specific loss power (SLP) for 25 nm particles at 515 kHZ across the 12.5 mm wide gap. SLP values were derived based on the interpolation of the measurements shown in (a) at 515 kHZ and varying field amplitude. (c) Infrared thermal camera measurements of heat dissipation in the coil gap during a duty cycle of 40 s on time and 240 s off. (d) Liposomes prepared with quenched calcein solution and with and without MNPs were exposed to an AMF sequence (515 kHz, 15 kA/m for 40 s). Fluorescence release was quantified. The release profiles were compared to fluorescence signal increase by the addition of TritonX, which destroys the liposomal structure (n ≥ 2, SEM).
To produce the AMF conditions identified above in up to 1 cm large tumors, we designed a coil consisting of litz wire wound around a soft ferromagnetic toroid core with a 12.5 mm gap (Fig. 3b, S4) that delivers the necessary stimulus without significant overheating (Fig. 3c, S5). We determined a duty cycle consisting of 40 sec AMF exposures separated by 240 sec rest, which permitted steady state operation (Fig. S5). Modeling of the expected field amplitude profile was used in combination with interpolated calorimetry data to predict how SLP would vary with position across the exposed tumor volume (Fig. 3b).
To quantify the release dynamics, we exposed samples with and without MNPs to AMF pulses and measured relative fluorescence using the calcein release assay described above. The fluid temperature was monitored with a fiber optic thermometer, ensuring that stray heat from the setup did not exceed 38°C. Exposing liposomes containing MNPs to an increasing number of AMF cycles led to a corresponding increase in fluorescence (Fig. 3d). The relative fluorescence signal did not increase significantly when control samples, which did not contain MNPs, were tested (Fig. 3d). Liposome disruption using the detergent Triton X-100 resulted in fluorescence increase on par with AMF-triggered release (Fig. 3d).
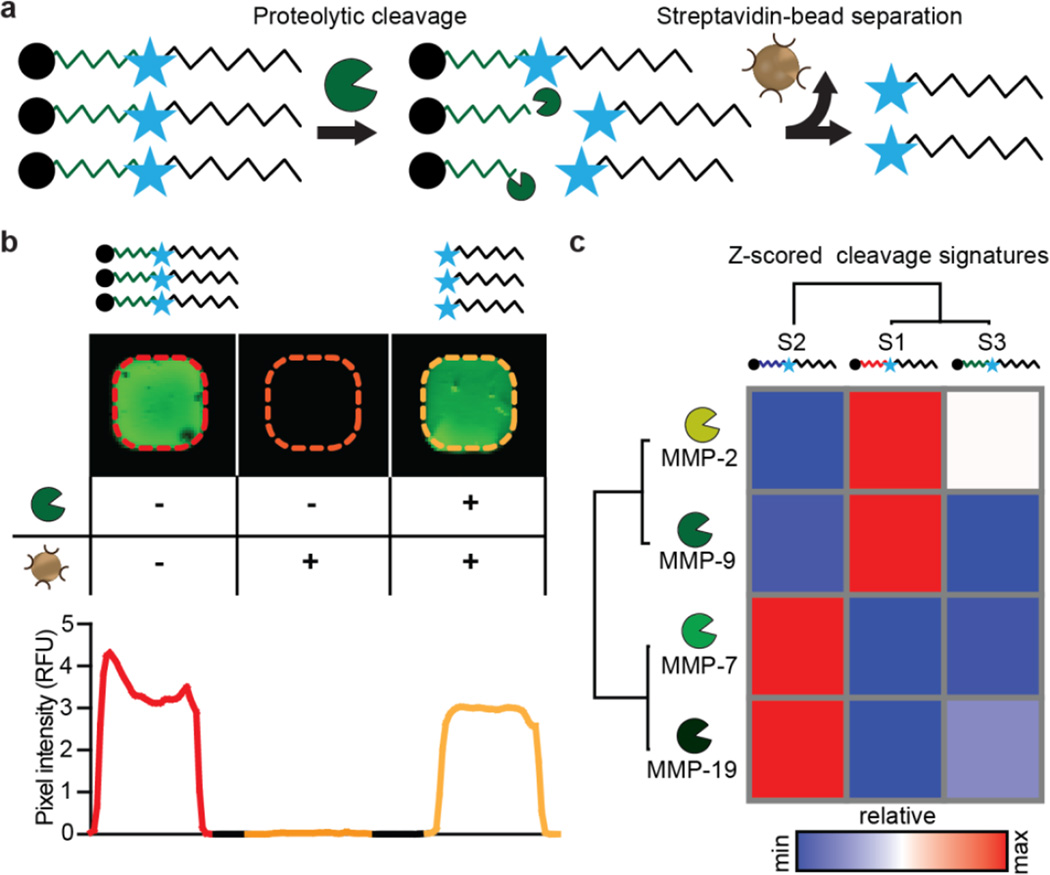
For the purpose of loading the liposomes with candidate tumor responsive nanosensors, we chose three peptide substrates that respond to MMPs, based on our previous work.15 These protease substrates are each linked to a D-stereoisomer of Glutamate Fibrinopeptide coupled with a near infrared (IR) dye, Cy7, as a urinary reporter, similar to our previous synthetic biomarker constructs (Fig. 4a, Table S1). In contrast to our earlier approaches, the peptide-reporter conjugates used here for liposome loading are not tethered directly to nanoparticle surfaces. Since kidney filtration is efficient for molecules less than 5 nm in size, unconjugated peptides could filter through to the urine even in the absence of proteolysis.36 To circumvent this source of false positive signals, we developed an assay to deplete uncleaved substrates. We coupled biotin to the N-terminus of all peptides, which can be efficiently captured using streptavidin beads. Uncleaved substrates will retain the N-terminal biotin and thus be efficiently eliminated (Fig. S6a). We validated this detection method by first confirming the ability to completely remove excess uncleaved substrate by measuring the fluorescence signal of free peptide sequences in phosphate buffered saline (PBS) and 2% urine, pre and post streptavidin bead separation (Fig. S6b, c). Cy7 measurements were robust and could be measured over several log dilutions using an IR fluorescence scanner (Fig. S7). Moreover, we confirmed the shielding mechanism of the liposomal bilayer by incubating MAPS with streptavidin beads and exposure to a permanent magnet (Fig. S8). Using this new detection method, we measured relative proteolysis of the substrates by several MMPs (Fig. 4b, c). By hierarchical clustering, S1 and S3 performed similarly and responded primarily to MMP2 and MMP9. S2 was cleaved efficiently by MMP7 and MMP19 (Fig. 4c).
Figure 4. Characterization of cleavage quantification assay and protease specificities.
(a) Schematic of assay. The N-terminal biotin identifies an uncleaved substrate, which can be depleted using streptavidin beads. Measurements of cleaved reporters are enabled by a Cy7 fluorophore. (b) Left: Cy7 signal of an initial peptide solution. Middle: no fluorescence signal was detected after depletion of uncleaved substrates with streptavidin beads Right: addition of MMP9 and subsequent streptavidin depletion results in similar fluorescent levels as initial peptide solution, showing robust cleavage of the substrate. (c) Recombination assay was performed for three distinct peptides substrates. Data was clustered via hierarchical clustering (one minus Pearson correlation) revealing substrate cleavage patterns.
With all of the components of our MAPS platform validated in vitrowe next sought to assay the performance of our magnetically-actuated, thermosensitive particles bearing MMP substrate reporters in vivo. As a first step, we determined the blood half-life of fluorescently labeled liposomes in healthy Swiss Webster mice to be approximately 1 hour, which we hypothesized to be sufficient to allow for passive accumulation at the tumor (Fig. S9a). To identify the optimal time point for remote AMF triggering of reporter release from nanosensors, we measured accumulation of liposomes in organs and in tumor xenografts of the human colon cancer cell line LS174T, which has been used extensively for in vivo cancer models and secretes active MMPs, including MMP2, 9 (Fig. S9b).37 We determined that the optimal time for tumor activation of the MAPS was 3 hours post-administration, as there should be relatively low blood concentration (due to half-life of approximately one hour) and reasonable tumor accumulation for specific activation.
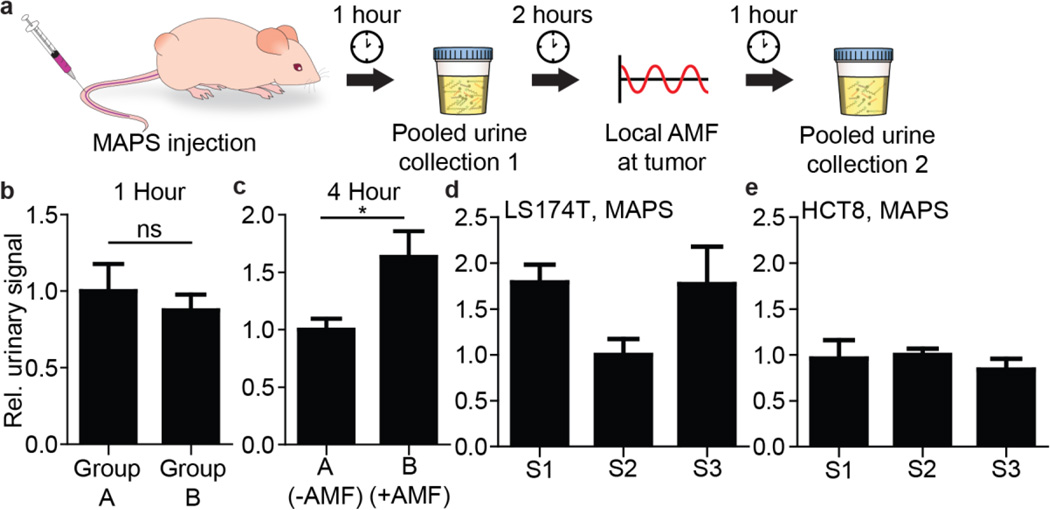
We applied MAPS to profile tumor protease activity in vivo. In order to assay for the specific release of MAPS-delivered reporters, we first intravenously injected MAPS-S3 in two cohorts of LS174T flank tumor-bearing mice. To establish a baseline signal and thus control for any non-specific liposome release, and/or background noise, urine from both cohorts was collected one hour after MAPS administration. Two hours later, mice from one group were exposed to AMF by positioning the flank tumor within the 12.5 mm gap present in the custom-made AMF coil, and a second urine collection was performed an hour after the AMF treatment (Fig. 5a, S10). As expected, at the one-hour time point, prior to AMF exposure, all mice displayed uniform, low-level urine reporter fluorescence (Fig. 5b), likely a result of non-specific leakage from the liposomes and subsequent proteolytic cleavage. In contrast, following the application of two AMF pulses to the tumor site, a statistically significant fluorescence increase was observed in the urine samples, only for the activated group (Fig. 5c). This outcome demonstrates the proof-of-principle, heat-dependent local release of labeled peptide substrates from MAPS, exposing them to proteolytic cleavage.
Figure 5. MAPS enables in vivo profiling of protease activity in tumors.
(a) Schematic of the in vivo profiling assay. One hour after MAPS administration, urine was collected to measure background signal. Three hours post injection, AMF was applied locally at the tumor to one group and urine collected one hour after activation. (b) Prior to activation, two cohorts of mice show similar urine reporter concentrations. (c) After activation, significantly greater urine reporter concentrations can be detected in the group exposed to AMF (n = 5, *P < 0.05 Student’s t-test; data normalized to Group A). (d) MAPS urinary signatures after activation across the three substrates for LS174T and (e) HCT-8 reveal that S1 and S3 are cleaved at greater rate relative to S2 in LS174T tumors (n ≥ 5 per group; data normalized to S2 signal for both LS174T and HCT8).
Having established that our MAPS constructs are sensitive to targeted release via AMF administration, we next aimed to test whether our MAPS(S1-S3) could be used to distinguish between tumor-derived cell lines, established from distinct patients, due to differences in their in vivo MMP profiles. The ability to classify clinical tumors based on MMP profiles would provide valuable input during therapy selection. For comparison, we chose another human colon carcinoma line, HCT-8, which has been validated previously to secrete lower levels of MMP9 than LS174T in vitro.19 Additionally, we measured MMP2 levels in cell culture supernatant by ELISA, and found that HCT-8 cells secrete lower amounts of MMP2 compared to LS174T (Fig. S11a ). We tested for cleavage of our substrates by cell-secreted proteases by employing fluorescently quenched versions and exposing them to conditioned supernatant. From these in vitro cleavage assays, S1 was cleaved most significantly by LS174T proteases, whereas minimal cleavage of S2 and S3 was observed (Fig. S11b). In vitro proteolysis was abrogated in the presence of Marimastat, an MMP inhibitor. In contrast, none of the substrates were efficiently cleaved by HCT-8 secreted proteases (Fig. S11c).
Following the in vitro optimization steps, we generated flank tumors using the two cell lines. Following administration of the pooled sets of MAPS particles, we applied the in vivo activation protocol and collected urine in an effort to detect distinct profiles of protease activity between LS174T and HCT8 tumors. Indeed, in response to the localized, in vivo activation of our magnetothermally-triggered nanosensors, we measured elevations in both MAPS-S1 and MAPS-S3 urinary signals, relative to the levels of the MAPS-S2 cleaved substrates in LS174T tumor-bearing mice (Fig. 5d). In contrast, all three constructs generated similar urine signals with respect to each other when tested in mice with HCT8-derived tumors (Fig. 5e), which is consistent with the relatively low MMP2 and 9 secretion rates, as S1 and S3 respond strongly to MMP2 and 9 (Fig. 4c). This differing pattern of substrate release highlights that MAPS have the capacity to distinguish the protease activities present in the tumors derived from two different colon cancer types. Injection of free peptide into healthy mice showed that S3 had the lowest background cleavage, validating its previous application as a robust protease substrate for profiling protease activity of cancers with high sensitivity and specificity (Fig. S12a).17 In contrast to the MAPS signature of LS174T flank tumor-bearing mice, injection of free peptides (i.e. unencapsulated in liposomes) into these mice, which should primarily sample blood activity (including proteases secreted from the tumor into the blood),19 showed similar S2 and S3 signals and an elevated S1 signal (Fig. S12b). Taken together, these results emphasize the importance of shielding protease access to circulating, activity-based reporters when seeking to sample local protease activities, and highlight the capability of MAPS to distinguish in vivo protease profiles of two human colon cancer lines with differing protease levels (Fig. S11 and S12).
In summary, we report an approach to measure tumor protease activity in vivo with greater specificity using remotely addressable nanosensors. We demonstrate that peptides are shielded in our thermosensitive MNP-containing liposomes and can be locally released in the tumor microenvironment via external application of AMFs. This system is not hindered by optical windows and allows for deep-tissue activation. We demonstrate that this technique is able to identify differences in MMP profiles across two in vivo human colorectal cancer xenograft models. The urinary reporters employed can be readily multiplexed (e.g. by mass encoding15) to enable high-content profiling of tumors. Furthermore, multimodal diagnosis and profiling could be enabled by magnetic resonance imaging for the magnetic nanoparticles within the liposomes. We primarily apply MAPS to profile MMP activity, but this approach is readily applicable to a variety of enzyme systems. MAPS will prove valuable in the development, validation, and application of protease-targeted therapeutics.
Materials and Methods
Synthesis of peptide substrates and liposomes
Peptides were synthesized by CPC Scientific, Inc. For full peptide sequence and description see supplementary information. Briefly, peptide-reporter tandems are comprised of an N-terminal biotin for depletion, followed by protease substrate, and then D-stereoisomer of Glutamate Fibrinopeptide conjugated to Cy7 for urinary measurements. Liposomes were prepared by applying the lipid-film hydration method with subsequent sequential extrusion. A lipid composition of 11.18 mg of dipalmitoylphosphatidyl-choline (DCCP), 1.31 mg monostearoylphosphatidylcholine (MSPC) and 2.51 mg poly(ethylene glycol)-conjugated distearoylphosphatidylethanolamine, DSCP-PEG-2000, was dissolved in 1.5 mL isopropanol, shortly sonicated and 3 aliquots of each 0.5 mL were dried under gentle nitrogen flow. All components were purchased from Avanti Polar lipids. The formed lipid cakes were then kept at least for 12 hours under vacuum. A volume of 300 µl trizma-based hydration media was prepared and mixed with magnetic nanoparticles (Ocean Nanotech, SHA-25) at a final iron concentration of 2.5 mg/mL and DMSO-based peptide solutions at a concentration of 2µM. The solution was pre-warmed to 65°C and added to the liposomal cake which was hydrated for 1 hour at 65°C in a water bath under continuous agitation. For in vitro release studies, 80 mM calcein was added to the trizma solution instead of peptide substrates. At this concentration, the self-quenching properties of calcein in solution were ensured. After hydration, the liposomes were extruded sequentially using 400, 200 and 100 nm large filter membranes to narrow the size distribution. The solutions were then purified from excess particles and free substrates by gravity column filtration. The resulting size was quantified by dynamic light scattering and peptide and iron concentration were measured by absorbance scans. The final solutions for in vivo injection were equally adjusted to 0.5 µM for peptides S1, S2 and S3 by dilution in PBS.
In vitro thermo-release studies
Temperature stability and kinetic release profile were measured in a fluorescence plate reader (Tecan) by suspending MAPS samples of 80 µl in 384 well plates. Temperature was set and kept at 37°C and increased to 43°C for kinetic release measurements when crossing the melting temperature. Calcein release was determined by measuring the increase of the fluorescence signal for an excitation wavelength of λex= 494 nm and emission wavelength λem= 517 nm.
Magnetic activation of thermosensitive liposomes
Magnetic activation of the liposomes was performed using a custom AMF setup. Two coils were fabricated and specifically designed to fit the requirements for in vitro and in vivo experiments. A toroid composed of a soft ferromagnetic material optimized for high frequency power transformers (Ferroxcube 3F3) was used as coil core. A transformer circuit with a resistive ballast in the primary circuit was used to generate high, stable currents in the secondary while simultaneously matching the impedance of the variable frequency 200 W amplifier (1020L, Electronics & Innovation). In the secondary, the coil acted as the resistive and inductive elements of an RLC resonance circuit, with a high voltage series capacitor setting the resonant frequency. The field magnitude was measured by a custom built probe employing a pickup loop and an oscilloscope. A simple cooling system with circulating ice water was coupled to the coil via silicone tubing and an electric fan was positioned in proximity to the coil. For in vitro release studies and calorimetric measurements of the particles, temperature measurements were conducted using an AMF insensitive fiber optic temperature probe and recorded during AMF exposure. SLP measurements were repeated 3 times and control samples with only water were measured after every 4 trials to determine the background heating rate. All samples were 0.1 mL with a MNP concentration of approximately 2 mg/mL. The SLP value measured was normalized to the metal content determined by elemental analysis. In vitro release studies were temperature monitored and fluid temperatures did not exceed T=39°C due to background heating. Measurements were repeated three times and calcein release was evaluated in a multi-well plate fluorescence reader as described above.
In vitro recombinant protease assays
MMPs (~100 nM working concentration, Enzo Life Sciences) were added to substrates in 384-well plates in activity buffer (50 mM Tris, 150 mM NaCl, 5 mM CaCl2, 1 μM ZnCl2) containing 1% BSA. After one hour, uncleaved peptide was extracted using Dynabeads Strepatividin C1 (Life Technologies) as per manufacturer protocols. An excess of Dynabeads was used.
Cell culture and secreted protease activity assay
LS174T and HCT-8 cells were cultured in Eagle’s Minimal Essential Medium (ATCC) supplemented with 10% FBS (Gibco) and 1% penicillin-streptomycin (CellGro). Cells were passaged when confluence reached 80%. To isolate secreted proteases, after cells were plated, cells were washed and replaced in serum-free media. Media was collected and MMP2 was measured in supernatant using a Quantikine MMP-2 kit following manufacturer protocols (R&D Systems). Secretion was normalized to number of cells and days in culture. A similar approach was used when collecting supernatants for measuring proteolysis of S1-3.
Pharmacokinetic studies
Wild-type, female Swiss Webster mice (4-6 wk, Taconic) were infused intravenously via the tail vein with liposomes carrying a near IR dye (VT750, Perkin Elmer). Blood was withdrawn retro-oribtally (~10 μL) and then immediately transferred into 90 μL of PBS with 5 mM EDTA and spun at 1000xg to pellet blood cells. Concentration of liposome was measured using an Odyssey Infrared scanner (Li-Cor Inc.). Nude mice bearing LS174T tumors (see below) were infused with labeled liposomes. Mice were sacrificed at different timepoints, followed by necropsy to remove organs and tumors. Organ accumulation was measured using an Odyssey scanner and quantified using ImageJ (NIH).
In vivo cancer model studies
Female nude mice (4-6 week, Taconic) were inoculated subcutaneously with 3 × 106 LS174T cells and HCT-8 cells on the hind flank and allowed to grow. Two weeks after inoculation, tumor-bearing mice were infused with MAPS. Suspensions were diluted to each 0.5 µM peptide concentration in 200 µl sterile PBS. Immediately after infusion, mice were placed in an in-house devised urine collector with a 96 well plate base. Urine was collected and stored at −80 °C. For analysis, urine was diluted from 25-fold in PBS. Reporter concentration was quantified by Cy7 fluorescence measurements in the Odyseey Scanner and compared to a ladder (SI Fig. 9).
Statistics and data analysis
All statistical analyses were performed in GraphPad (Prism 5.0). Statistical significance and individual tests are described in figure legends. Heatmaps and hierarchical clusters were generated using GENE-E (Broad Institute). Data were clustered by one minus Pearson correlation.
Supplementary Material
Acknowledgments
We thank Dr. H. Fleming (MIT) for critical reading and editing of the manuscript, Dr. A. Warren (MIT) and Dr. E. Kwon (MIT) for technical instruction and valuable insight, and the Koch Biopolymers & Proteomics Core for assistance. This study was supported in part by the Ludwig Center for Molecular Oncology, a Koch Institute Support Grant P30-CA14051 from the National Cancer Institute (Swanson Biotechnology Center), and a Core Center Grant P30-ES002109 from the National Institute of Environmental Health Sciences. This research was conducted with Government support awarded by DoD, Air Force Office of Scientific Research, National Defense Science and Engineering Graduate (NDSEG) Fellowship, 32 CFR 168a, which funds M.G.C. J.S.D. thanks the National Science Foundation Graduate Research Fellowship Program for support. S.S. gratefully acknowledges the support by the Swiss National Science Foundation (SNSF) through the “Early Postdoc Mobility Fellowship” and funding provided by the DAAD, German Academic Exchange Service. P.A. thanks the Defense Advanced Research Project Agency (DARPA) for partial support of this study through Young Faculty Award and ElectRx (HR0011-15-C-0155) programs. S.N.B. is a Howard Hughes Medical Institute Investigator.
Footnotes
Supporting Information Available
Supplementary figures 1-12. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Friedman AA, Letai A, Fisher DE, Flaherty KT. Nat. Rev. Cancer. 2015;15(12):747–756. doi: 10.1038/nrc4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Papadopoulos N, Kinzler KW, Vogelstein B. Nat. Biotechnol. 2006;24(8):985–995. doi: 10.1038/nbt1234. [DOI] [PubMed] [Google Scholar]
- 3.Hori SS, Gambhir SS. Sci. Transl. Med. 2011;3(109):109ra116–109ra116. doi: 10.1126/scitranslmed.3003110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller MA, Gadde S, Pfirschke C, Engblom C, Sprachman MM, Kohler RH, Yang KS, Laughney AM, Wojtkiewicz G, Kamaly N, Bhonagiri S, Pittet MJ, Farokhzad OC, Weissleder R. Sci. Transl. Med. 2015;7(314):314ra183–314ra183. doi: 10.1126/scitranslmed.aac6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gambhir SS. Nat. Rev. Cancer. 2002;2(9):683–693. doi: 10.1038/nrc882. [DOI] [PubMed] [Google Scholar]
- 6.Hanahan D, Weinberg RA. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 7.López-Otín C, Bond JS. J. Biol. Chem. 2008;283(45):30433–30437. doi: 10.1074/jbc.R800035200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi KY, Swierczewska M, Lee S, Chen X. Theranostics. 2012;2(2):156–178. doi: 10.7150/thno.4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fortelny N, Cox JH, Kappelhoff R, Starr AE, Lange PF, Pavlidis P, Overall CM. PLoS Biol. 2014;12(5):e1001869. doi: 10.1371/journal.pbio.1001869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desnoyers LR, Vasiljeva O, Richardson JH, Yang A, Menendez EEM, Liang TW, Wong C, Bessette PH, Kamath K, Moore SJ, Sagert JG, Hostetter DR, Han F, Gee J, Flandez J, Markham K, Nguyen M, Krimm M, Wong KR, Liu S, Daugherty PS, West JW, Lowman HB. Sci. Transl. Med. 2013;5(207):207ra144–207ra144. doi: 10.1126/scitranslmed.3006682. [DOI] [PubMed] [Google Scholar]
- 11.Mahmood U, Weissleder R. Mol. Cancer Ther. 2003;2(5):489–496. [PubMed] [Google Scholar]
- 12.Hilderbrand SA, Weissleder R. Curr. Opin. Chem. Biol. 2010;14(1):71–79. doi: 10.1016/j.cbpa.2009.09.029. [DOI] [PubMed] [Google Scholar]
- 13.Whitney M, Savariar EN, Friedman B, Levin RA, Crisp JL, Glasgow HL, Lefkowitz R, Adams SR, Steinbach P, Nashi N, Nguyen QT, Tsien RY. Angew. Chem. Int. Ed. 2013;52(1):325–330. doi: 10.1002/anie.201205721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whitley MJ, Cardona DM, Lazarides AL, Spasojevic I, Ferrer JM, Cahill J, Lee C-L, Snuderl M, Blazer DG, Hwang ES, Greenup RA, Mosca PJ, Mito JK, Cuneo KC, Larrier NA, O’Reilly EK, Riedel RF, Eward WC, Strasfeld DB, Fukumura D, Jain RK, Lee WD, Griffith LG, Bawendi MG, Kirsch DG, Brigman BE. Sci. Transl. Med. 2016;8(320):320ra4–320ra4. doi: 10.1126/scitranslmed.aad0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwong GA, von Maltzahn G, Murugappan G, Abudayyeh O, Mo S, Papayannopoulos IA, Sverdlov DY, Liu SB, Warren AD, Popov Y, Schuppan D, Bhatia SN. Nat. Biotechnol. 2013;31(1):63–70. doi: 10.1038/nbt.2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin KY, Kwong GA, Warren AD, Wood DK, Bhatia SN. ACS Nano. 2013 doi: 10.1021/nn403550c. 13090923114600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warren AD, Kwong GA, Wood DK, Lin KY, Bhatia SN. Proc. Natl. Acad. Sci. 2014;111(10):3671–3676. doi: 10.1073/pnas.1314651111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller MA, Barkal L, Jeng K, Herrlich A, Moss M, Griffith LG, Lauffenburger DA. Integr. Biol. 2011;3(4):422–438. doi: 10.1039/c0ib00083c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwong GA, Dudani JS, Carrodeguas E, Mazumdar EV, Zekavat SM, Bhatia SN. Proc. Natl. Acad. Sci. 2015 doi: 10.1073/pnas.1506925112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dudani JS, Jain PK, Kwong GA, Stevens KR, Bhatia SN. ACS Nano. 2015;9(12):11708–11717. doi: 10.1021/acsnano.5b05946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kessenbrock K, Plaks V, Werb Z. Cell. 2010;141(1):52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Egeblad M, Werb Z. Nat. Rev. Cancer. 2002;2(3):161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 23.Torchilin VP. Nat. Rev. Drug Discov. 2005;4(2):145–160. doi: 10.1038/nrd1632. [DOI] [PubMed] [Google Scholar]
- 24.Allen TM, Cullis PR. Adv. Drug Deliv. Rev. 2013;65(1):36–48. doi: 10.1016/j.addr.2012.09.037. [DOI] [PubMed] [Google Scholar]
- 25.Needham D, Park J-Y, Wright AM, Tong J. Faraday Discuss. 2013;161:515–534. doi: 10.1039/c2fd20111a. discussion 563-589. [DOI] [PubMed] [Google Scholar]
- 26.Li L, ten Hagen TLM, Schipper D, Wijnberg TM, van Rhoon GC, Eggermont AMM, Lindner LH, Koning GA. J. Control. Release Off. J. Control. Release Soc. 2010;143(2):274–279. doi: 10.1016/j.jconrel.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 27.Li L, ten Hagen TLM, Hossann M, Süss R, van Rhoon GC, Eggermont AMM, Haemmerich D, Koning GA. J. Control. Release Off. J. Control. Release Soc. 2013;168(2):142–150. doi: 10.1016/j.jconrel.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 28.Glöckl G, Hergt R, Zeisberger M, Dutz S, Nagel S, Weitschies W. J. Phys. Condens. Matter. 2006;18(38):S2935. [Google Scholar]
- 29.Christiansen MG, Senko AW, Chen R, Romero G, Anikeeva P. Appl. Phys. Lett. 2014;104(21):213103. [Google Scholar]
- 30.Carrey J, Mehdaoui B, Respaud M. J. Appl. Phys. 2011;109(8):083921. [Google Scholar]
- 31.Hergt R, Dutz S. J. Magn. Magn. Mater. 2007;311(1):187–192. [Google Scholar]
- 32.Romero G, Christiansen MG, Stocche Barbosa L, Garcia F, Anikeeva P. Adv. Funct. Mater. 2016 n/a–n/a. [Google Scholar]
- 33.Dong J, Zink JI. ACS Nano. 2014;8(5):5199–5207. doi: 10.1021/nn501250e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riedinger A, Guardia P, Curcio A, Garcia MA, Cingolani R, Manna L, Pellegrino T. Nano Lett. 2013;13(6):2399–2406. doi: 10.1021/nl400188q. [DOI] [PubMed] [Google Scholar]
- 35.Allia P, Tiberto P, Coisson M, Chiolerio A, Celegato F, Vinai F, Sangermano M, Suber L, Marchegiani G. J. Nanoparticle Res. 2011;13(11):5615–5626. [Google Scholar]
- 36.Soo Choi H, Liu W, Misra P, Tanaka E, Zimmer JP, Itty Ipe B, Bawendi MG, Frangioni JV. Nat. Biotechnol. 2007;25(10):1165–1170. doi: 10.1038/nbt1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brand K, Baker AH, Perez-Cantó A, Possling A, Sacharjat M, Geheeb M, Arnold W. Cancer Res. 2000;60(20):5723–5730. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.