Abstract
Protein synthesis in all cells begins with recruitment of the small ribosomal subunit to the initiation codon in a messenger RNA. In some eukaryotic viruses, RNA upstream of the coding region forms an internal ribosome entry site (IRES) that directly binds to the 40S ribosomal subunit and enables translation initiation in the absence of many canonical translation initiation factors. The hepatitis C virus (HCV) IRES RNA requires just two initiation factors, eukaryotic initiation factor (eIF) 2 and eIF3, to form preinitiation 48S ribosomal complexes that subsequently assemble into translation-competent ribosomes. Using an RNA-based affinity purification approach, we show here that HCV IRES RNA facilitates eIF2 function through its interactions with eIF3 and the 40S ribosomal subunit. Although the wild-type IRES assembles normally into 48S and 80S ribosomal complexes in human cell extract, mutant IRES RNAs become trapped at the 48S assembly stage. Trapped 48S complexes formed by IRES mutants with reduced eIF3 binding affinity nonetheless contain eIF3, consistent with inherent eIF3–40S subunit affinity. Intriguingly, however, one of these IRES mutants prevents stable association of both eIF3 and eIF2, preventing initiator tRNA deposition and explaining the block in 80S assembly. In contrast, an IRES mutant unable to induce a conformational change in the 40S subunit, as observed previously by single-particle cryoelectron microscopy, blocks 80S formation at a later stage in assembly. These data suggest that the IRES RNA coordinates interactions of eIF3 and eIF2 on the ribosome required to position the initiator tRNA on the mRNA in the ribosomal peptidyl-tRNA site (P site).
Keywords: eukaryotic initiation factor
Initiation of protein synthesis in eukaryotes requires the ordered assembly of ribosomal preinitiation complexes, beginning with the association of the small (40S) ribosomal subunit with an mRNA (reviewed in refs. 1–3). Cap-dependent translation involves initiation factor protein association with the 7-methyl guanosine moiety at the mRNA 5′ end, leading to 40S ribosome binding and scanning to the initiation codon before association with the 60S ribosomal subunit to form an active 80S ribosome (1). An alternate pathway, called internal translation initiation, is a cap-independent mechanism of recruiting, positioning, and activating the eukaryotic protein synthesis machinery, driven by structured RNA sequences called internal ribosome entry sites (IRESs) located in the mRNA 5′ untranslated region (UTR). These sequences have been identified in numerous viral RNAs, and there is evidence suggesting that certain cellular mRNAs may also contain IRES elements (4, 5).
In hepatitis C virus (HCV), a human pathogen and world-wide health threat, the minimal sequence and secondary structure requirements of IRES-driven translation have been defined (6–10), the presence and architecture of an IRES RNA tertiary fold has been described (11, 12), and the identities and binding sites of necessary cofactors have been determined (12, 13). Because the IRES RNA functionally replaces the eukaryotic initiation factor (eIF) 4F protein complex (eIF4E, eIF4G, and eIF4A), translation initiation in HCV requires just two of the canonical initiation factors, eIF2 and eIF3, for correct positioning of the initiator tRNA during cap-independent initiation (13). The IRES RNA, one of the most conserved regions of the entire HCV genome, directly and specifically binds the 40S ribosomal subunit and eIF3 (12, 14, 15). This ternary complex joins with eIF2/GTP/initiator tRNA to form a 48S particle in which the tRNA is positioned in the P site of the 40S subunit, base-paired to the start codon of the mRNA (2). Upon hydrolysis of GTP, eIF2 releases the initiator tRNA and dissociates from the complex. A second GTP hydrolysis step involving initiation factor eIF5B then enables the 60S ribosomal subunit to associate, forming a functional 80S ribosome that initiates viral protein synthesis (13, 16).
The HCV IRES RNA has been shown to fold under physiological salt conditions into a defined three-dimensional structure whose integrity is essential for efficient IRES-mediated translation (11). As revealed by single-particle cryoelectron microscopy, the elongated HCV IRES structure binds the 40S ribosomal subunit and induces a significant conformational change that closes the mRNA binding cleft (17). This result suggested a possible mechanism by which the IRES might position the viral mRNA in the ribosomal P site without the assistance of canonical initiation factors. Although these experiments showed how the IRES actively manipulates the 40S ribosomal subunit, it remained unclear why a direct interaction of the HCV IRES and initiation factor eIF3 is required during translation initiation. eIF3, an ≈700 kDa complex comprising at least 12 subunits in humans, prevents premature association of 40S and 60S ribosomal subunits by a mechanism that has yet to be determined (18, 19). It seems to play a critical role in the assembly of active 80S ribosomes during both cap-dependent and IRES-mediated translation initiation, but the molecular basis for its activity in either case is not known.
To investigate how the HCV IRES assembles functional human 80S ribosomes, we developed an RNA-based affinity purification method suitable for isolating IRES-associated ribosomal complexes from human cell extracts. We show that mutant forms of the IRES form “trapped” 48S preinitiation complexes that are incapable of efficient assembly into 80S ribosomes. Mass spectrometry and Western blotting of the protein components of these complexes revealed two distinct defects in 80S assembly. Surprisingly, one mutant that is unable to bind tightly to eIF3 blocks the stable association of eIF2, and consequently, deposition of the initiator tRNA in the complex. In contrast, a mutant IRES with normal binding affinity for the 40S subunit and eIF3, but incapable of inducing a conformational change in the 40S subunit, is defective at a later stage in the 80S ribosome assembly pathway. These data suggest that the IRES–eIF3 interaction stabilizes the productive association of both eIF3 and eIF2 in the 48S complex, enabling the delivery and positioning of initiator tRNA. The IRES may thus function in place of initiation factors that carry out a similar activity during cap-dependent translation initiation.
Materials and Methods
Plasmid Construction and RNA Transcription. A DNA fragment encoding the HCV IRES sequence with three MS2 recognition hairpin sequences at the 5′ end was generated by two rounds of PCR by using a previous HCV IRES construct (11) as the template and three synthetic oligonucleotides as primers: 5′-CGGA AT TCTA ATACGACTCACTATAGCGTACA-CCATCAGGGTACGAGCTAGCCCATGGCGTACAC-CATCAGGGTACGACTAGC-3′, 5′-ATCAGGGTACGAC-TAGCTAGATCTCGTACACCATCAG GGTACGTCTA-GAGGTACCGATCACTCCCCTGTGAGGA ACTAC-3′, and 5′-CGGGATCCTTTTTCTT TGAGGT TA AGGAT-TTG-3′. The resulting DNA fragment was ligated into the EcoRI and BamHI restriction sites of pUC19 to form the parent plasmid for all subsequent constructs. Derivative plasmids encoding IRES mutants G(266–268)C, IIIa_Comp (nucleotides 162–165, AGUA changed to UCAU), U228C, ΔIIIb (nucleotides 175–224 replaced by GAAA), and domain (Dom) III (nucleotides 40–119 deleted) were generated by using QuikChange mutagenesis (Stratagene) (IRES numbering system as described in ref. 11). All constructs were verified by DNA sequencing. IRES RNA was produced by in vitro transcription by using purified T7 RNA polymerase and purified by denaturing gel electrophoresis as described (11).
Expression and Purification of Chimeric Affinity Tag Recognition Protein. The maltose-binding protein (MBP)-MS2 fusion protein construct was a gift from Josep Vilardell (Centre de Recerca Genòmica, Barcelona). The MBP-MS2 protein was expressed in Escherichia coli strain DH5α grown in LB medium plus 2% glucose at 37°C as described (20, 21). Cells were grown until the OD600 reached 0.5, and then 0.5 mM isopropyl β-d-thiogalactoside (IPTG) was added to the medium to induce protein expression. After 3 h, cells were pelleted, resuspended in buffer A (20 mM Hepes, pH 7.5/200 mM KCl/1 mM EDTA) plus tablets of proteinase inhibitor mixture (Roche) and lysed by sonication on ice. The supernatant was applied to an amylose column, and MBP-MS2 protein was eluted with a buffer containing 20 mM Hepes (pH 7.5), 20 mM KCl, 1 mM EDTA, 10% glycerol, and 5 mM maltose. Contaminating nucleic acid was removed by applying the protein to a Heparin Hi-trap column (Amersham Pharmacia) and eluting with a 100–600 mM KCl gradient. Purified protein was dialyzed into a buffer containing 20 mM Hepes (pH 7.5), 100 mM KCl, and 10% glycerol and stored at a concentration of 4 mg/ml at –80°C.
Assembly and Purification of Protein Synthesis Initiation Complexes. The concentration of HeLa cell lysate capable of binding the IRES RNA (active concentration) was determined by using a stoichiometric filter binding assay (12) as described. Binding of MS2 hairpin-tagged IRES RNA with MBP-MS2 fusion protein was confirmed by using a native gel mobility shift assay. Small-scale assembly of translation initiation complexes was performed by incubating ≈3 μg of 5′ end 32P-labeled IRES RNA with 50 μl of HeLa cell cytoplasmic extract (OD260 of 50 per ml) in 500 μl of buffer B (20 mM Tris, pH 7.5/100 mM KCl/2.5 mM MgCl2/2 mM DTT) for 30 min at 37°C. 48S and 80S complexes were resolved by centrifugation at 274,355 × g for 3.5 h through a 10–50% sucrose gradient; gradient fractions were analyzed by dot-blotting onto nitrocellulose membranes and quantitated by using a PhosphorImager (Molecular Dynamics) and imagequant software. To assemble protein initiation complexes for affinity purification, 4.8 ml of HeLa cell cytoplasmic extract (OD260 of 50 per ml) was diluted into 1 liter of buffer B and incubated with IRES RNA (2 nM final concentration) at 37°C for 30 min. At these concentrations ≈50% of the input IRES RNA was bound to ribosomal complexes, ensuring the greatest sensitivity to changes in IRES binding affinity for 40S and/or eIF3 in the assay. MBP-MS2 protein (0.6 ml of a 4 mg/ml solution) was then added into the solution and incubated at 37°C for 30 min. Subsequently, the initiation complexes were affinity-selected by binding to an amylose column; after washing with 10 column volumes of buffer B, IRES-containing complexes were eluted with 5 mM maltose in buffer B. Mock purification using untagged wild-type IRES RNA yielded undetectable protein or RNA after elution from the amylose column. Each affinity-selected sample was concentrated to 3 ml, layered onto a 30-ml 10–50% sucrose gradient in buffer B, and centrifuged at 23,000 rpm in a Beckman SW28 rotor at 4°C for 15 h. Translation initiation complexes were recovered by fractionating the gradient and detecting the absorbance at 260 nm. SDS/PAGE and mass spectrometric analysis verified the presence of ribosomal and/or eIF3 proteins.
Western Blots. Proteins within purified initiation complexes were separated by SDS/PAGE and transferred to poly(vinylidene difluoride) membranes (Millipore); eIF2α was detected with polyclonal rabbit anti-eIF2 antiserum (1:1,000 dilution, Cell Signaling Technology, Beverly, MA), whereas eIF3 was detected with a monoclonal antibody to the eIF3a subunit (22). Protein bands were visualized by incubation with an appropriate alkaline phosphatase-conjugated secondary antibody (Sigma).
Results and Discussion
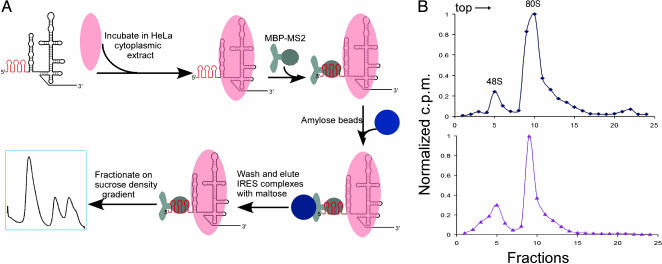
Affinity Purification of IRES-Bound Translation Initiation Complexes. Previously, we observed that mutant forms of the HCV IRES RNA show defects in translation initiation complex assembly in rabbit reticulocyte lysate (23), suggesting that it might be possible to dissect the IRES-mediated assembly pathway. We used an RNA-based affinity tagging method, previously developed for purification of spliceosomes (20, 21), to isolate IRES-containing human translation initiation complexes assembled in HeLa cell cytoplasmic extract. Three tandem RNA hairpin sequences that form binding sites for the bacteriophage MS2 coat protein (24) were introduced at the 5′ end of the wild-type and five mutant forms of the HCV IRES RNA (Fig. 1A). The presence of the 5′ MS2 recognition hairpin sequence does not affect the assembly of wild-type IRES RNA into 48S and 80S translation complexes upon incubation in HeLa cell cytoplasmic extract (Fig. 1B). For affinity purification experiments, tagged IRES RNAs were incubated in HeLa cell extract diluted to a concentration at which ≈50% of the input RNA bound to ribosomal complexes (Fig. 2). Although efficient 80S ribosome formation occurs in the concentrated extract (Fig. 1B), not all 48S complexes proceed to 80S ribosomes under these more dilute conditions (Fig. 2 A). However, dilution of the extract to the approximate Kd concentration for IRES-40S subunit association ensures the greatest sensitivity to changes in IRES binding affinity for 40S and/or eIF3 in the assay. After incubation, IRES RNA was bound to an MBP-MS2 fusion protein and purified by amylose affinity chromatography. IRES-bound translation initiation complexes were separated by sucrose density gradient centrifugation, yielding highly purified samples of 48S and 80S ribosomal fractions. This method ensures that all isolated complexes contain the full-length IRES RNA, because the 5′ hairpins are required for affinity selection and the 3′ region of the IRES is required for ribosome binding.
Fig. 1.
IRES-containing ribosomal complex purification. (A) Strategy for affinity purification of IRES-containing complexes from HeLa cell extract. HCV IRES RNAs containing three MS2 recognition hairpins at the 5′ end (shown in red) were incubated in HeLa cell cytoplasmic extract; IRES-bound complexes were affinity-purified by binding to a chimeric MBP-MS2 fusion protein and isolating by amylose affinity chromatography (see Materials and Methods). Translation initiation complexes were then separated and purified by sucrose density gradient centrifugation. (B) Ribosomal complex assembly in HeLa cell lysate using 32P-end-labeled wild-type IRES (Upper) or 32P-end-labeled MS2-tagged wild-type IRES (Lower). Ribosomal complexes were fractionated by sucrose density gradient centrifugation, and radioactivity in each fraction was determined by PhosphorImager analysis (see Materials and Methods), with values normalized by dividing by the maximum cpm value observed; sedimentation was from left to right.
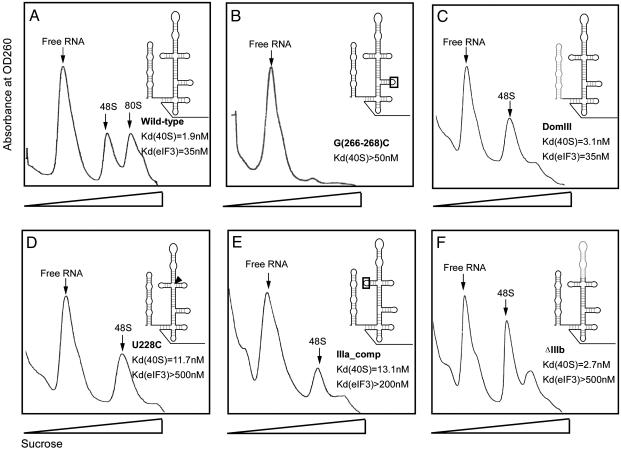
Fig. 2.
Sucrose density gradient analysis of translation complexes bound by wild-type and mutant forms of the HCV IRES. Affinity-tagged IRES RNAs were incubated in HeLa cell extract at a concentration of extract yielding half-maximal binding of wild-type IRES. Plots of absorbance at 260 nm versus sucrose density are shown; peaks corresponding to free RNA, 48S, and 80S are indicated (verified by denaturing PAGE and electrospray mass spectrometry). (A) Wild-type IRES. (B–F) Translation-defective IRES constructs containing the following mutations and translation complex binding defects relative to the wild-type HCV IRES (12, 23). (B) G(266–268)C mutation in the IIId loop, >25-fold-reduced 40S binding affinity. (C) DomIII, no change in 40S or eIF3 binding affinities. (D) U228C, >15-fold-reduced eIF3 binding affinity. (E) IIIa_Comp, >6-fold-reduced eIF3 binding affinity. (F) ΔIIIb, >15-fold-reduced eIF3 binding affinity.
Mutant IRES RNAs Form Trapped 48S Complexes. The wild-type HCV IRES RNA mediates formation of both 48S and 80S ribosomal complexes that have been analyzed in detail by using electrospray ionization mass spectrometry (Y.Y., H.J., J.A.D. and J.L., unpublished results). The identity of 48S and 80S particles was initially confirmed by using denaturing gel electrophoresis to analyze the protein content of each sample, revealing the distinctive protein banding patterns specific to 48S and 80S complexes (data not shown). Five variants of the HCV IRES with mutations in functionally critical regions (12, 25) were tested for 80S ribosome assembly in HeLa cell extract. One of these, an IRES construct with a mutation in the IIId loop [G(266–268)C] that reduces 40S binding affinity by >25-fold (12), was tested and shown to prevent association into 48S complexes (Fig. 2B). A DomIII IRES construct containing a deletion of the DomII region required to induce a conformational change in the 40S ribosomal subunit (17) forms 48S complexes that are incapable of efficient 80S ribosome assembly (Fig. 2C). Two mutants with changes in the IIIabc region that reduce 40S subunit and eIF3 binding affinity by >6-fold [U228C and IIIa_Comp (12)] also form 48S complexes but few or no 80S ribosomes (Fig. 2 D and E). Finally, an IRES mutant with a deletion of the IIIb stem-loop required for eIF3 binding (12, 13, 26) also forms 48S complexes but few or no 80S ribosomes (Fig. 2F). Notably, the G(266–268)C, DomIII, IIIa_Comp, and U228C IRES mutants have all been shown to have significantly reduced translation initiation efficiency in vitro (12, 27–29).
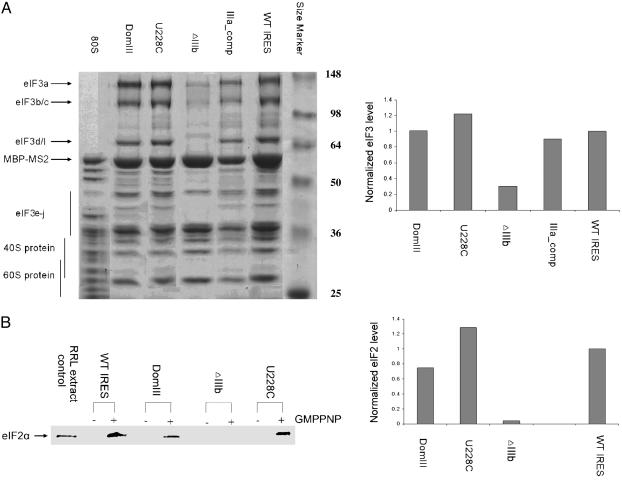
eIF3 Is Present in 48S Complexes Containing Mutant IRES RNAs with Reduced Affinity for eIF3 in Isolation. One possible explanation for the observed block in 80S ribosome formation by mutant forms of the HCV IRES is that initiation factor eIF3 is absent from these complexes. eIF3 has been shown to be essential to the assembly of active ribosomes during both cap-dependent and IRES-mediated translation initiation (18, 30, 31). Although it has been established that HCV IRES binding to eIF3 is necessary for efficient translation initiation (13, 32), the natural affinity of eIF3 for 40S subunits (18) might preclude the need for eIF3 recruitment by the IRES. To investigate the requirement for a high-affinity IRES-eIF3 interaction, 48S complexes containing wild-type IRES RNA or IRES constructs defective in eIF3 binding were affinity purified and fractionated on sucrose density gradients. These samples were then analyzed by both electrospray ionization mass spectrometry and by denaturing PAGE and Western blotting. As expected, eIF3 was present in 48S complexes containing the wild-type IRES. Surprisingly, it was also present in all of the trapped 48S complexes, although the amount of eIF3 in samples containing the ΔIIIb IRES construct was consistently reduced by ≈70%, compared with other IRES constructs (Fig. 3A). These data are consistent with association of eIF3 with the 40S ribosomal subunit independent of binding by the IRES or other mRNAs or initiation factors, although the IIIb region of the IRES clearly stabilizes eIF3 in the complex. This result implies that the requirement for HCV IRES-eIF3 binding is not solely for recruitment of eIF3 into 48S complexes, and suggests a more active role for the IRES in modulating eIF3 function.
Fig. 3.
Analysis of initiation factor presence within translation complexes assembled on wild-type and mutant HCV IRES RNAs. (A Left) Coomassie blue-stained 10% SDS/PAGE gel of affinity-purified 48S and 80S samples; IRES constructs are indicated at the top, protein size markers are labeled at right, and bands corresponding to eIF3, 40S, and 60S proteins are indicated at left. The identities of eIF3 bands were verified by Western blotting (data not shown). (Right) Relative eIF3 levels in wild-type IRES and mutant IRES-bound complexes. For each sample, three eIF3 bands were quantitated by using imagej software, and the summed intensities were divided by the intensity of the MBP-MS2 band as a loading control; all values were normalized to that of the wild-type IRES-containing sample. Each value shown is the average of two independent experiments. (B Left) Western blot analysis of 48S complexes in the absence and presence of the nonhydrolyzable GTP analog GMPPNP using an anti-eIF2α antibody. Equal amounts of sample based on OD260 were applied to each lane. (Right) Relative eIF2 levels in wild-type and mutant IRES-bound complexes. Band intensities were quantitated by using imagej software and divided by the intensity of ribosomal protein bands in each lane to control for sample loading differences; all values were normalized to that of the wild-type IRES-containing sample. Each value shown, except for U228C, is the average of two independent experiments.
Productive IRES–eIF3 Interaction Is Required for eIF2 Association with the 48S Complex. A second possible explanation for the block in 80S ribosome formation by mutant forms of the HCV IRES is that eIF2 function is somehow impaired. eIF2, a three-subunit initiation factor with GTP hydrolyzing activity, brings the initiator tRNA into the 48S particle and positions it at the start codon of the mRNA in the ribosomal P site (33, 34). Upon proper tRNA placement, eIF2 hydrolyzes GTP and releases from the 48S complex, paving the way for subsequent 60S subunit joining to form an active 80S complex. Because there is no scanning involved in HCV IRES-initiated translation, tRNA initiator codon pairing presumably occurs upon 48S complex formation, leading to rapid GTP hydrolysis and eIF2 release. To test the possibility that this step in the 80S assembly pathway might be impaired in mutant IRES-containing complexes, 48S complexes were affinity purified as before except that the HeLa cell extracts were pretreated with 5 mM GMPPNP, a nonhydrolyzable analog of GTP. Under these conditions, the wild-type IRES forms only 48S complexes and eIF2 remains associated with the particle because the essential GTP hydrolysis step cannot occur (2). As shown by Western blotting using antiserum specific for the human eIF2α subunit, eIF2 is detected in the GMPPNP-containing wild-type IRES sample (Fig. 3B). Likewise, eIF2 is present in GMPPNP-blocked 48S complexes containing an IRES point mutant (U228C) with reduced eIF3 binding affinity or an IRES mutant that binds with normal affinity to eIF3 (DomIII). In contrast, eIF2 is not detectable in GMPPNP-blocked 48S complexes containing the ΔIIIb IRES mutant lacking the IIIb region essential for high-affinity eIF3 binding (Fig. 3B). These data show that the IRES stem-loop IIIb interaction with eIF3 affects both eIF3 and eIF2 stability in preinitiation complexes formed during assembly of active ribosomes.
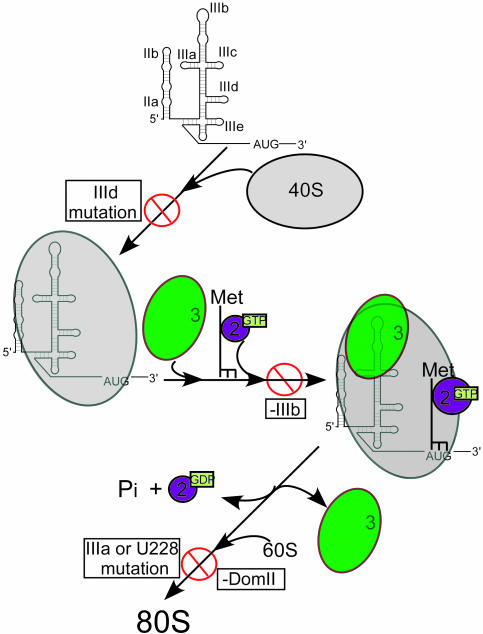
Model for HCV IRES-Mediated Assembly of 80S Ribosomes. By using an affinity purification approach, it has been possible to isolate HCV IRES-containing human initiation complexes and to analyze defects in the 80S ribosome assembly pathway that result from various mutations of the IRES RNA. This method ensures that all purified complexes include the full-length IRES in question, obviating concerns about loss of the IRES element due to mRNA degradation during sample preparation (35). Our results show that mutations in the IRES block formation of active ribosomes at three steps along the assembly pathway (Fig. 4). An IRES mutant [G(266–268)C] with reduced 40S subunit binding affinity in vitro prevents association between the IRES and the 40S subunit, thereby blocking formation of 48S preinitiation complexes. IRES mutants with reduced eIF3 binding affinity in vitro form 48S complexes that are incapable of efficient 80S ribosome formation. An eIF3 recognition-defective IRES mutant (U228C) forms 48S complexes that contain normal levels of eIF3 and eIF2, whereas another (ΔIIIb) assembles into 48S complexes in which both eIF3 and eIF2 are destabilized. These results imply that a productive IRES-eIF3 interaction through the IIIb stem-loop of the IRES is necessary to stabilize the association of both eIF3 and eIF2, possibly affecting the proper positioning of the initiator tRNA on the message through conformational rearrangement of the 48S complex.
Fig. 4.
Model for HCV IRES-coordinated assembly of human translation initiation complexes. 40S subunit binding to IRES RNA with an intact IIId loop is required for IRES-40S association; eIF3 interaction with the IIIb region of the IRES is required for stable association of eIF3 and eIF2 in the 48S complex, whereas eIF3 contacts to the junction of stems IIIa, -b and -c, as well as the 40S conformational change induced by DomII, are necessary for downstream events required for 60S subunit joining.
Finally, a mutant IRES (DomIII) having normal affinity for both the 40S subunit and eIF3 but lacking the DomII element required to produce an observed conformational change in the 40S ribosomal subunit forms trapped 48S complexes with apparently normal eIF3 and eIF2 stability. Both the U228C and DomIII IRES mutants thus result in defects at a later stage in the 80S assembly pathway. These results show that the HCV IRES is exquisitely tuned to provide multiple molecular interactions needed for proper ribosome assembly on viral messages. An interesting possibility is that these functional interactions of the IRES mimic activities carried out by host translation initiation factors during cap-dependent translation initiation. The limited size and modular structure of the HCV IRES lend themselves to further functional dissection of this process that may provide insights into both viral and host cell translation and offer new opportunities for developing anti-HCV therapies.
Acknowledgments
We thank John Hershey, Jamie Cate, Wendy Gilbert, and Ian Macrae for helpful discussions and comments on the manuscript, and Matthias Hentze for critical review of the manuscript. This work was supported by the National Institutes of Health and the Howard Hughes Medical Institute.
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on April 30, 2002.
Abbreviations: MBP, maltose-binding protein; IRES, internal ribosome entry site; HCV, hepatitis C virus; eIF, eukaryotic initiation factor; Dom, domain.
See accompanying Biography on page 16987.
References
- 1.Kozak, M. (1989) J. Cell Biol. 108, 229–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hershey, J. W. B. & Merrick, W. C. (2000) in Translational Control of Gene Expression, eds. Sonenberg, N., Hershey, J. W. B. & Mathews, M. B. (Cold Spring Harbor Lab. Press, Plainview, NY), Vol. 39, pp. 33–88. [Google Scholar]
- 3.Pestova, T. V., Kolupaeva, V. G., Lomakin, I. B., Pilipenko, E. V., Shatsky, I. N., Agol, V. I. & Hellen, C. U. (2001) Proc. Natl. Acad. Sci. USA 98, 7029–7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hellen, C. U. & Sarnow, P. (2001) Genes Dev. 15, 1593–1612. [DOI] [PubMed] [Google Scholar]
- 5.Merrick, W. C. (2004) Gene 332, 1–11. [DOI] [PubMed] [Google Scholar]
- 6.Tsukiyama-Kohara, K., Iizuka, N., Kohara, M. & Nomoto, A. (1992) J. Virol. 66, 1476–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang, C., Sarnow, P. & Siddiqui, A. (1993) J. Virol. 67, 3338–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Honda, M., Ping, L. H., Rijnbrand, R. C., Amphlett, E., Clarke, B., Rowlands, D. & Lemon, S. M. (1996) Virology 222, 31–42. [DOI] [PubMed] [Google Scholar]
- 9.Reynolds, J. E., Kaminski, A., Carroll, A. R., Clarke, B. E., Rowlands, D. J. & Jackson, R. J. (1996) RNA 2, 867–878. [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao, W. D. & Wimmer, E. (2001) J. Virol. 75, 3719–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kieft, J. S., Zhou, K., Jubin, R., Murray, M. G., Lau, J. Y. & Doudna, J. A. (1999) J. Mol. Biol. 292, 513–529. [DOI] [PubMed] [Google Scholar]
- 12.Kieft, J. S., Zhou, K., Jubin, R. & Doudna, J. A. (2001) RNA 7, 194–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pestova, T. V., Shatsky, I. N., Fletcher, S. P., Jackson, R. J. & Hellen, C. U. (1998) Genes Dev. 12, 67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sizova, D. V., Kolupaeva, V. G., Pestova, T. V., Shatsky, I. N. & Hellen, C. U. (1998) J. Virol. 72, 4775–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buratti, E., Tisminetzky, S., Zotti, M. & Baralle, F. E. (1998) Nucleic Acids Res. 26, 3179–3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, J. H., Pestova, T. V., Shin, B. S., Cao, C., Choi, S. K. & Dever, T. E. (2002) Proc. Natl. Acad. Sci. USA 99, 16689–16694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spahn, C. M., Kieft, J. S., Grassucci, R. A., Penczek, P. A., Zhou, K., Doudna, J. A. & Frank, J. (2001) Science 291, 1959–1962. [DOI] [PubMed] [Google Scholar]
- 18.Benne, R. & Hershey, J. W. (1978) J. Biol. Chem. 253, 3078–3087. [PubMed] [Google Scholar]
- 19.Chaudhuri, J., Chowdhury, D. & Maitra, U. (1999) J. Biol. Chem. 274, 17975–17980. [DOI] [PubMed] [Google Scholar]
- 20.Jurica, M. S., Licklider, L. J., Gygi, S. R., Grigorieff, N. & Moore, M. J. (2002) RNA 8, 426–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou, Z., Sim, J., Griffith, J. & Reed, R. (2002) Proc. Natl. Acad. Sci. USA 99, 12203–12207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanner, S. B., Reynolds, A. B., Vines, R. R. & Parsons, J. T. (1990) Proc. Natl. Acad. Sci. USA 87, 3328–3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kieft, J. S., Grech, A., Adams, P. & Doudna, J. A. (2001) Cold Spring Harbor Symp. Quant. Biol. 66, 277–283. [DOI] [PubMed] [Google Scholar]
- 24.LeCuyer, K. A., Behlen, L. S. & Uhlenbeck, O. C. (1995) Biochemistry 34, 10600–10606. [DOI] [PubMed] [Google Scholar]
- 25.Kolupaeva, V. G., Pestova, T. V. & Hellen, C. U. (2000) J. Virol. 74, 6242–6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kieft, J. S., Zhou, K., Grech, A., Jubin, R. & Doudna, J. A. (2002) Nat. Struct. Biol. 9, 370–374. [DOI] [PubMed] [Google Scholar]
- 27.Buratti, E., Gerotto, M., Pontisso, P., Alberti, A., Tisminetzky, S. G. & Baralle, F. E. (1997) FEBS Lett. 411, 275–280. [DOI] [PubMed] [Google Scholar]
- 28.Honda, M., Rijnbrand, R., Abell, G., Kim, D. & Lemon, S. M. (1999) J. Virol. 73, 4941–4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang, S., Collier, A. J. & Elliott, R. M. (1999) J. Virol. 73, 2359–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asano, K., Goss Kinzy, T., Merrick, W. C. & Hershey, J. W. B. (1997) J. Biol. Chem. 272, 1101–1109. [DOI] [PubMed] [Google Scholar]
- 31.Nielsen, K. H., Szamecz, B., Valasek, L., Jivotovskaya, A., Shin, B. S. & Hinnebusch, A. G. (2004) EMBO J. 23, 1166–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pestova, T. V., Hellen, C. U. & Shatsky, I. N. (1996) Mol. Cell. Biol. 16, 6859–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta, N. K., Roy, A. L., Nag, M. K., Kinzy, T. G., MacMillan, S. E., Hileman, R. E., Dever, T. E., Wu, S., Merrick, W. C. & Hershey, J. W. B. (1990) in Transcriptional Control of Gene Expression, eds. McCarthy, J. E. G. & Tuite, M. F. (Springer, Berlin), pp. 521–526.
- 34.Marintchev, A., Kolupaeva, V. G., Pestova, T. V. & Wagner, G. (2003) Proc. Natl. Acad. Sci. USA 100, 1535–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozak, M. (2001) Mol. Cell. Biol. 21, 1899–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]