Abstract
The secretion of carbohydrate-degrading enzymes by a bacterium sourced from a softwood forest environment has been investigated by mass spectrometry. The findings are discussed in full in the research article “Proteomic insights into mannan degradation and protein secretion by the forest floor bacterium Chitinophaga pinensis” in Journal of Proteomics by Larsbrink et al. ([1], doi: 10.1016/j.jprot.2017.01.003). The bacterium was grown on three carbon sources (glucose, glucomannan, and galactomannan) which are likely to be nutrient sources or carbohydrate degradation products found in its natural habitat. The bacterium was grown on solid agarose plates to mimic the natural behaviour of growth on a solid surface. Secreted proteins were collected from the agarose following trypsin-mediated hydrolysis to peptides. The different carbon sources led to the secretion of different numbers and types of proteins. Most carbohydrate-degrading enzymes were found in the glucomannan-induced cultures. Several of these enzymes may have biotechnological potential in plant cell wall deconstruction for biofuel or biomaterial production, and several may have novel activities. A subset of carbohydrate-active enzymes (CAZymes) with predicted activities not obviously related to the growth substrates were also found in samples grown on each of the three carbohydrates. The full dataset is accessible at the PRIDE partner repository (ProteomeXchange Consortium) with the identifier PXD004305, and the full list of proteins detected is given in the supplementary material attached to this report.
Keywords: Bacterium, Carbohydrate-active enzymes, Mass spectrometry, Plant biomass deconstruction, Protein secretion
Specifications Table
| Subject area | Microbiology, Biochemistry |
| More specific subject area | Bacterial protein secretion and carbohydrate deconstruction |
| Type of data | Tables and figures |
| How data was acquired | The analysis utilised a nanoHPLC-MS/MS system consisting of a Dionex Ultimate 3000 RSLCnano (Thermo Scientific, Bremen, Germany) connected to a Q-Exactive hybrid quadrupole-orbitrap mass spectrometer (Thermo Scientific, Bremen, Germany) with a nano-electrospray ion source. |
| Data format | Raw, analysed |
| Experimental factors | The bacterium was grown on agarose plates containing one of three tested substrates (glucose, glucomannan and galactomannan). Secreted proteins were collected from the solid medium following trypsin hydrolysis performed within the agarose medium. Samples were collected at three time-points during growth. |
| Experimental features | Three biological replicates were collected for each time-point sample. Proteins were hydrolysed by trypsin within the solid medium. Released peptides were then prepared for analysis by mass spectrometry. Two technical replicate experiments were performed for each sample. Raw data was normalised and analysed using the Max Quant programme, with quantification performed using the MaxLFQ algorithm. |
| Data source location | Proteomic data were collected in-house at the Norwegian University of Life Sciences, Ås, Norway |
| Data accessibility | Data is with the article and at PRIDE: PXD004305. |
Value of the data
-
•
The method of collecting proteins from solid medium gave a strong enrichment of secreted proteins. The method of protein preparation is simple, and can be utilised in future work on both bacteria and fungi.
-
•
The data reveal that the secretion of several CAZymes is induced by the polysaccharide glucomannan, while a larger number of CAZymes appear to be constitutively produced. Sequence and domain analysis of the induced CAZymes suggests that many may have novel mannan-related activities.
-
•
Several of the identified CAZymes have no or weak similarity to enzymes of known function, suggesting the possibility of novel activities. We recommend that these enzymes are biochemically characterised in future experiments.
-
•
Several proteins of unknown function were co-upregulated together with relevant CAZymes, and these may also represent novel activities for biomass deconstruction.
-
•
Enzymes identified in this study may prove to be useful new tools in the deconstruction of plant biomass for a biorefinery or similar biotechnological applications.
1. Data
After an initial growth trial in liquid cultures (Fig. 1), C. pinensis was grown on agarose plates containing 0.5% carbon source and quartz filters [2], to minimise the common issues of cell lysis and exo-polysaccharide contamination, as well as to better mimic natural solid state-like conditions. Samples for proteomic analyses were collected in an early-, mid-, and late-stage of growth (time-points t1, t2 and t3): for KGM and glucose plates, sampling was performed on days 2, 4, and 5, and for CGM plates on days 6, 9, and 15. In the final proteomic analysis (summarised in Fig. 2), a protein was counted as ‘present’ in a sample if detected and quantifiable in at least two biological replicates; technical replicates of each sample were merged in MaxQuant to improve quantification. All identified proteins are described in Supplementary Tables 1–3. As the main focus of this work was the discovery of new CAZymes with potential application in the deconstruction of plant biomass, Fig. 3 refers to only the CAZymes found in each sample. A full discussion of this dataset can be found in Larsbrink et al. [1].
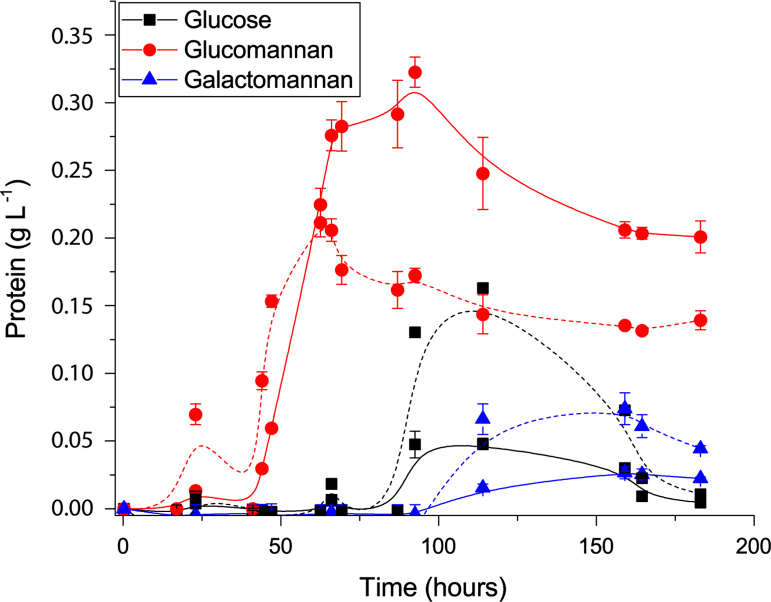
Fig. 1.
Protein secretion by C. pinensis grown in liquid culture containing three different carbon sources. Media contains glucose, konjac glucomannan or carob galactomannan as sole carbon course at 0.5% final concentration. Protein secretion was measured using the Bradford assay to determine protein concentration in the media at various time-points throughout growth. Solid lines: protein concentration in growth medium (g L−1). Dashed lines: protein concentrations normalised for cell density by dividing by A600. Error bars represent one standard deviation from the mean. Compared to glucose and CGM, growth on KGM reached the highest final OD, and the growth curve adhered most closely to the classical three-stage growth profile of a bacterial culture. The glucomannan liquid cultures also showed the highest concentration of protein in the growth medium, as measured by the Bradford assay [3].
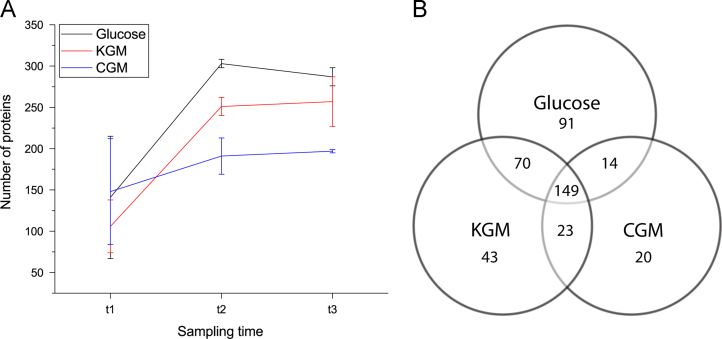
Fig. 2.
Numeric overview of proteins detected after growth on three different carbohydrates. A: The number of proteins detected in the samples at three sampling points during growth. Error bars represent one standard deviation from the mean. The sampling time-points t1, t2 and t3 denote the three stages of growth (early, mid and late) on which sampling was performed. For all substrates, the number of proteins increased between t1 and t2, and then remained relatively stable between t2 and t3. While the total number of detected proteins did not significantly differ at t1, differences emerged at the later time-points. B: Venn diagram showing the similarity and differences between proteins identified for the three different growth conditions. The numbers refer to the total number of identified unique proteins for each substrate.
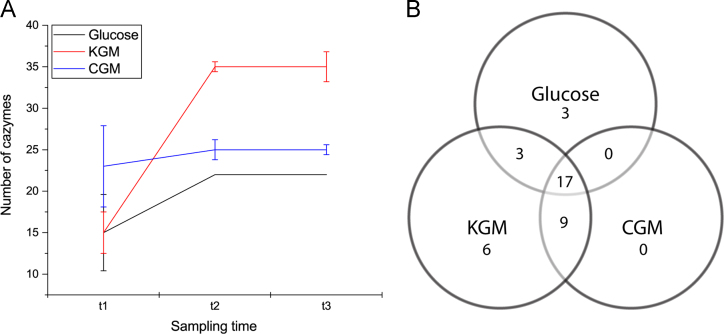
Fig. 3.
Numeric overview of CAZymes detected after growth on three different carbohydrates. A: The number of CAZymes at three time-points during growth. A protein was counted as ‘present’ if detected in at least two replicates for a given substrate. Error bars represent one standard deviation from the mean. B: Venn diagram showing similarities and differences of all 35 CAZymes identified for the three growth conditions at t2.
2. Experimental design, materials and methods
2.1. Carbohydrates
Glucose was obtained from Sigma Aldrich (Stockholm, Sweden). The polysaccharides KGM and CGM were purchased from Megazyme (Wicklow, Ireland).
2.2. Strain growth
All reagents used for bacterial growth were purchased from Sigma-Aldrich, unless otherwise stated, and were of microbiological grade. Chitinophaga pinensis strain UQM 2034 T was propagated at 30 °C on LB agar plates supplemented with kanamycin at 50 µg mL−1, to which the bacterium has innate resistance. To obtain proteins for proteomic analysis, C. pinensis was grown on agarose plates (50 mm diameter). The solid medium contained agarose (1%), M9 medium (prepared according to Miller [4] but lacking any carbohydrate), 50 µg mL−1 kanamycin, and 0.4% (w/v) of either glucose, KGM or CGM. Each plate was cast with a 0.2 µm Pall supor 200 sterile filter (47 mm diameter) laid between two 5 mL beds of medium (total volume 10 mL medium) as described by Bengtsson et al. [2]. Prior to inoculation, C. pinensis was grown in 5 mL LB medium at 30 °C overnight. The cells were harvested by centrifugation for 10 min at 5000 g, washed in 10 ml carbohydrate-free M9 medium, and harvested again by centrifugation. The supernatant fluid was discarded, and the cells were resuspended in carbohydrate-free M9 medium to an OD600 value of 0.5, of which 50 µl was used to inoculate the agarose plates. The plates were incubated at 22 °C until an early, mid or late stage of growth, as estimated from prior visual observations. For KGM and glucose plates, this was days 2, 4, and 5, and for CGM plates this was days 6, 9, and 15. Three biological replicates of each sample were produced. Only two biological replicates were produced for the KGM time-point t1 sample.
2.3. Mass spectrometric analysis of secreted proteins
The process of protein collection, protein hydrolysis, and peptide analysis by mass spectrometry, proceeded essentially as described by Bengtsson et al. [2], and are described below.
2.3.1. Preparation of secreted proteins for MS analysis
Proteins secreted during growth on agarose plates were collected essentially as described by Bengtsson et al. [2]. Proteins were collected from plates at early, mid, and late points during growth, as described above. These three time-points are hereafter referred to as t1, t2 and t3, respectively. The solid medium of a plate was removed from the Petri dish and inverted onto a clean surface. The agarose from directly beneath the filter was stamped out and collected into a pre-weighed 50 mL Falcon tube. The wet mass of the sample was obtained by weighing the tube again. To each gram of sample was added 4 µmol of dithiothreitol. The sample was then heated until the agarose was melted, and vortexed vigorously. The liquefied agarose, containing secreted proteins, was boiled for 30 min, then transferred into a syringe and cooled to room temperature. After solidification, the agarose was extruded, crushing the material. 1 mL of a 100 mM solution of NH4HCO3 was added per gram of sample, giving a final concentration of 50 mM NH4HCO3. To this 2 µg of porcine trypsin (Promega) was added per sample, followed by overnight incubation at 37 °C. The sample was frozen and thawed and then briefly centrifuged. The supernatant liquid contained the extracted trypsin-digested proteins. This supernatant liquid was collected into a 2 mL LoBind tube (Eppendorf) and centrifuged at 16 000g for 10 min to remove any remaining solids. The resulting supernatant liquid was filtered (0.22 µm) into a new eppendorf tube. For mass spectrometric analysis, trifluoroacetic acid (TFA) was added from a 10% (v/v) stock solution to a final concentration of 0.1% (v/v) in the sample. The peptides in this mixture were subsequently purified using a C-18 column (Strata C-18E, Phenomenex, California, USA), and eluted with 80% (v/v) acetonitrile/ 0.1% (v/v) TFA. The eluate containing peptides was vacuum dried, then resuspended in 10 µL 2% (v/v) acetonitrile and 0.1% (v/v) TFA. A subsequent peptide purification step using carboxylate modified magnet beads (Thermo Scientific, USA) was performed as described by Hughes et al. [5], before peptide analysis by LC-MS/MS.
2.3.2. Identification of proteins by mass spectrometry
For peptide analysis by mass spectrometry, a nanoHPLC-MS/MS system consisting of a Dionex Ultimate 3000 RSLCnano (Thermo Scientific, Bremen, Germany) connected to a Q-Exactive hybrid quadrupole-orbitrap mass spectrometer (Thermo Scientific, Bremen, Germany) with a nano-electrospray ion source was used. Samples were loaded onto a trap column (Acclaim PepMap100, C18, 5 µm, 100 Å, 300 µm i.d.×5 mm, Thermo Scientific) and back-flushed onto a 50 cm analytical column (Acclaim PepMap RSLC C18, 2 µm, 100 Å, 75 µm i.d., Thermo Scientific). Equal volumes of all samples were loaded (2×4 µL). Columns were pre-equilibrated in 96% solution A (0.1% (v/v) formic acid), and 4% solution B (80% (v/v) ACN, 0.1% (v/v) formic acid). Peptides were eluted with a 70 min gradient from 4% to 13% (v/v) solution B in 2 min, 13% to 45% B (v/v) in 43 min and finally to 55% B (v/v) in 3 min, followed by a wash phase at 90% B. The flow rate was set to 0.3 µL min−1. By operating the Q-Exactive in data-dependent mode, switching automatically between orbitrap-MS and higher-energy collisional dissociation (HCD) orbitrap-MS/MS acquisition, isolation and fragmentation of the 10 most intense peptide precursor ions at any given time throughout the chromatographic elution was ensured. The selected precursor ions were then excluded for repeated fragmentation for 20 s. The resolution was set to R=70,000 for MS and R=35,000 for MS/MS. Automatic gain control target values were set to 1,000,000 charges and a maximum injection time of 128 ms. Two technical replicates were analysed for each sample. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium [6] via the PRIDE partner repository with the dataset identifier PXD004305.
2.3.3. Bioinformatics and statistical validation
The mass spectrometry data were analysed using MaxQuant [7], [8] version 1.4.1.2. Identification and quantification of proteins were performed using the MaxLFQ algorithm [9], searching against a database containing the full predicted proteome of C. pinensis, generated from the Uniprot database (7179 sequences in total) [10]. The MaxLFQ algorithm uses a non-linear optimisation model to normalise the peptide intensities. Technical replicates were combined in MaxQuant to obtain more reliable quantification values. The database was supplemented with common contaminants such as keratins, trypsin and bovine serum albumin. For estimation of false discovery rates, reversed sequences of all protein entries were concatenated to the database. As variable modifications in the MaxQuant analysis we used protein N-terminal acetylation, oxidation of methionine, conversion of glutamine to pyro-glutamic acid, and deamidation of asparagine and glutamine. Trypsin was used as proteolytic enzyme and two missed cleavages were allowed. The ‘match between runs’ feature of MaxQuant was enabled with default parameters, in order to increase the number of identified peptides and transfer identifications between samples based on accurate mass and retention time [11]. The settings were such that transfer of peptide identifications was only allowed between samples from the same carbon source. All identifications were filtered in order to achieve a protein false discovery rate (FDR) of 1%.
The protein group file from MaxQuant was loaded into Perseus (version 1.5.1.6). The matrix was reduced following a standard MaxQuant procedure by removing proteins categorised as only identified by site, reverse, or as a contaminant, in order to remove false hits from the MaxQuant data files. For a quantification to be considered valid, we used both unique and razor peptides for quantification and required at least two ratio counts. Furthermore, for a protein to be considered as present we required its quantification in at least two of the three biological replicates in at least one time-point (or at least one substrate for comparative analysis). In Perseus the label free quantification (LFQ) intensities were log10 transformed and missing values (proteins not quantified in a given sample) were replaced with a value of zero.
Acknowledgements
This research was supported by the Knut & Alice Wallenberg Foundation via the Wallenberg Wood Science Center, the Swedish Research Council Formas via CarboMat, and the European Research Council through Grant 336355 (“MicroDE”). The authors are grateful to Morten Skaugen and Magnus Ø. Arntzen of NMBU for helpful discussions on troubleshooting and sample clean-up prior to proteomic analysis.
Footnotes
Transparency data associated with this article can be found in the online version at doi:10.1016/j.dib.2017.02.032.
Supplementary data associated with this article can be found in the online version at 10.1016/j.dib.2017.02.032.
Transparency document. Supplementary material
Supplementary material
.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Larsbrink J., Tuveng T.R., Pope P.B., Bulone V., Eijsink V.G., Brumer H., McKee L.S. Proteomic insights into mannan degradation and protein secretion by the forest floor bacterium Chitinophaga pinensis. J. Proteom. 2016 doi: 10.1016/j.jprot.2017.01.003. Under review. [DOI] [PubMed] [Google Scholar]
- 2.Bengtsson O., Arntzen M.O., Mathiesen G., Skaugen M., Eijsink V.G. A novel proteomics sample preparation method for secretome analysis of Hypocrea jecorina growing on insoluble substrates. J. Proteom. 2015 doi: 10.1016/j.jprot.2015.10.017. [DOI] [PubMed] [Google Scholar]
- 3.Bradford M.M. Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 4.Miller J.H. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, USA; 1972. Experiments in Molecular Genetics. [Google Scholar]
- 5.Hughes C.S., Foehr S., Garfield D.A., Furlong E.E., Steinmetz L.M., Krijgsveld J. Ultrasensitive proteome analysis using paramagnetic bead technology. Mol. Syst. Biol. 2014;10:757. doi: 10.15252/msb.20145625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vizcaino J.A., Deutsch E.W., Wang R., Csordas A., Reisinger F., Rios D., Dianes J.A., Sun Z., Farrah T., Bandeira N., Binz P.-A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R.J., Kraus H.-J., Albar J.P., Martinez-Bartolome S., Apweiler R., Omenn G.S., Martens L., Jones A.R., Hermjakob H. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotech. 2014;32(3):223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotech. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 8.Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011;10(4):1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 9.Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell Proteom. 2014;13(9):2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.UniProt-Consortium, UniProt: a hub for protein information, Nucleic Acids Res 43 (Database issue) (2015) D204-D212. [DOI] [PMC free article] [PubMed]
- 11.Nahnsen S., Bielow C., Reinert K., Kohlbacher O. Tools for label-free peptide quantification. Mol. Cell Proteom. 2013;12(3):549–556. doi: 10.1074/mcp.R112.025163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material