Abstract
Loss of basement membrane (BM) integrity is typically associated with cancer. Nidogen-1 is an essential component of the BM. Nidogen-1 is a substrate for cathepsin-S (CatS) which is released into the tumor microenvironment. Measuring nidogen-1 degraded by CatS may therefore have biomarker potential in cancer. The aim of this study was to investigate if CatS-degraded nidogen-1 was detectable in serum and a possible biomarker for cancer, a pathology associated with disruption of the BM. A competitive enzyme-linked immunosorbent assay (NIC) was developed with a monoclonal mouse antibody specific for a CatS cleavage site on human nidogen-1. Dilution and spiking recovery, inter- and intravariation, as well as accuracy were evaluated. Serum levels were evaluated in patients with breast cancer, small cell lung cancer (SCLC), and non-SCLC (NSCLC) and in healthy controls. The results indicated that the NIC assay was specific for nidogen-1 cleaved by CatS. Inter- and intraassay variations were 9% and 14%, respectively. NIC was elevated in NSCLC as compared to healthy controls (P < .001), breast cancer (P < .01), and SCLC (P < .5). The diagnostic power (area under the receiver operating characteristics) of NIC for NSCLC as compared to all other samples combined was 0.83 (95% confidence interval: 0.71-0.95), P < .0001. In conclusion, nidogen-1 degraded by CatS can be quantified in serum by the NIC assay. The current data strongly suggest that cathepsin-S degradation of nidogen-1 is strongly associated with NSCLC, which needs validation in larger clinical cohorts.
Introduction
A major component of the tumor microenvironment is the extracellular matrix (ECM). Disruption of the healthy tissue homeostasis and altered turnover of the ECM are hallmarks of cancer and lead to progression of the disease [1]. The basement membrane (BM) is a specialized layer of ECM that underlies and surrounds epithelial cells, endothelial cells, and mesenchymal cells and hereby is essential for embryonic development as well as for maintaining tissue architecture and cellular polarization [2]. The BM also serves as a barrier for cell invasion, and breaching of the BM and loss of BM integrity have been associated with an invasive phenotype in cancer [3]. Thus, it is important to develop cancer biomarkers associated with BM disruptions.
Nidogens are highly conserved proteins that are found in almost all BMs [4]. Two different nidogens exist: nidogen-1 (entactin) and nidogen-2. Nidogen-1 is the most widely expressed and comprises three globular domains (G1, G2, and G3) connected by two rod-shaped domains. Nidogen-1 serves as the linker between other BM proteins, namely, collagen type IV, perlecan, and laminin [5]. Hereby, nidogen-1 plays a key role in the BM assembly and stabilization [6], as is also evident by the lung and heart abnormalities and perinatal death observed in nidogen-deficient mice as a direct result of BM changes [7].
The interaction between nidogen-1 and other BM components has been studied both in vitro and in vivo [8], [9], [10]. Type IV collagen and perlecan bind to the G2 domain, and laminin binds to the G3 domain [5]. Especially the binding between nidogen-1 and laminin has been extensively studied [11], [12], [13]. Inhibition of this interaction results in defects in BM formation in both organ and co-culture systems [14], [15], [16], [17]. Moreover, it has been shown that mice lacking nidogen-1 have impaired wound healing [18].
Nidogen-1 is a substrate for cathepsin-S (CatS), but not cathepsin B, H, K, and L [19]. Degradation of nidogen-1 by CatS results in impaired binding between nidogen-1 and type IV collagen, laminin, and perlecan and hereby affects the properties of the BM [19]. Cathepsins are thought to play an important role in cancer and have functional roles in growth, angiogenesis, invasion, and metastasis of tumor cells [20]. In general, cathepsins become active at acidic pH such as in lysosomes [21] but are inactive at neutral pH such as in the extracellular environment [22]. CatS, however, retains most of its activity at neutral pH [23].
CatS is produced in both tumor cells and tumor-associated macrophages (TAMs) [24], and CatS has been shown to promote growth, angiogenesis, migration, invasion, and mestatasis in different cancer types [24], [25], [26], [27]. As nidogen-1 is a key protein of the BM and CatS is an important protein for cancer biology, this interaction may provide a biological set-point, with increased accuracy for cancer and biomarker development.
We hypothesized that nidogen-1 degraded by CatS may have biomarker potential in cancer as it may reflect loss of BM integrity, a phenomenon which is typically associated with procancer events and an invasive phenotype. The aim of this study was to enable noninvasive assessment of CatS-degraded nidogen-1 and investigate this as a biomarker for cancer.
Material and Methods
Selection of Peptides
The following three main CatS cleavage-sites (↓) on nidogen-1 were previously identified by N-terminal sequencing and published by Sage et al. [19]: 1) NH3-…LLPL459↓APVG…, located within the central part of the G2 domain; 2) NH3-…HERE693↓QHILG…, located in the thyroglobulin-like (Tg) domain adjacent to the G3 domain; and 3) NH3-…RQDL859↓QGSPE…, located at the N-terminal part of the G3 domain. We chose to target the second cleavage site located outside the globular domains in the direction toward the N-terminus, i.e. NH3-…HERE693↓. We chose to name this target NIC.
To generate an antibody specific for the selected CatS cleavage site on nidogen-1, a sequence of 11 amino acids just adjacent to the cleavage site was chosen as the target: NH3-… VEKTRCQHERE↓. This amino acid sequence was used to design the selection peptide. The sequence was blasted for homology to other human proteins using NPS@: Network Protein Sequence Analysis with the UniprotKB/Swiss-prot database [28] and found to be unique to human nidogen-1.
All synthetic peptides (Table 1) were purchased from the American Peptide Company, Vista, CA. A biotinylated screening peptide (Biotin-VEKTRCQHERE) was included to coat the streptavidin-coated plates. To test for specificity of the antibody, a truncated selection peptide (EVEKTRCQHER) missing the one amino acid closest to the cleavage site and an elongated selection peptide (VEKTRCQHEREH), with an additional amino acid added C-terminally to the cleavage site, as well as a nonsense selection peptide (APVGGIIGWM) and a nonsense biotinylated screening peptide (Biotin-APVGGIIGWM), were included in the assay development. The latter two correspond to the cleavage site located within the central part of the G2 domain (↓APVG…). The immunogenic peptide (KLH-VEKTRCQHERE) was generated by covalently cross-linking the selection peptide to Keyhole Limpet Hemocyanin (KLH) carrier protein using succinimidyl 4-(N-maleimidomethyl)cyclohexane-1-carboxylate (Thermo Scientific, Waltham, MA; cat. #22336).
Table 1.
Amino Acid Sequence of the Synthetic Peptides Used for Antibody Production and Assay Validation
| Selection peptide | VEKTRCQHERE |
|---|---|
| Immunogenic peptide | KLH-VEKTRCQHERE |
| Screening peptide | Biotin-VEKTRCQHERE |
| Truncated selection peptide | EVEKTRCQHER |
| Elongated selection peptide | VEKTRCQHEREH |
| Nonsense selection peptide | APVGGIIGWM |
| Nonsense screening peptide | Biotin-APVGGIIGWM |
Antibody Development: Immunization and Hybridoma Screening
Six-week-old BALB/c mice (n = 6) were immunized subcutaneously with 200 μl of emulsified antigen and 50 μg of the immunogenic peptide (KLH-VEKTRQHERE) using Freund's incomplete adjuvant (Sigma-Aldrich, St. Louis, MO). Immunizations were repeated every second week, and blood samples were collected from the second immunization until stable serum antibody titer levels were reached. The mice with the highest antibody titer rested for a month and were then boosted intravenously with 50 μg immunogenic peptide in 100 μl of 0.9% sodium chloride solution. Three days later, the spleen was isolated for cell fusion. The fusion procedure was performed as described elsewhere [29]. Briefly, mouse spleen cells were fused with SP2/0 myeloma cells to produce hybridoma cells. The hybridoma cells were cloned in culture dishes, and limited dilution was used to secure monoclonal growth. Native reactivity and peptide binding of the monoclonal antibodies were evaluated by displacement using human serum samples and the selection peptide (VEKTRCQHERE) in a preliminary enzyme-linked immunosorbent assay (ELISA) using 10 ng/ml of screening peptide on a streptavidin-coated microtiter plate (Roche, Basel, Switzerland; cat. #11940279) and the supernatant from the growing monoclonal hybridoma cells (containing the antibodies). The clones with best reactivity were purified using protein-G-columns according to the manufacturer's instructions (GE Healthcare Life Sciences, Little Chalfont, UK; cat. #17–0404-01). Two hybridoma clones qualified to a screening for their ability to react competitively only with the selection peptide (VEKTRCQHERE) and not with the truncated (EVEKTRCQHER), elongated (VEKTRCQHEREH), and nonsense (APVGGIIGWM) selection peptides. One hybridoma clone was chosen for further assay development. Optimal incubation buffer, time, and temperature, as well the optimal concentrations of antibody and screening peptide, were determined from checkerboard analysis.
Proteolysis of Nidogen-1 In Vitro
Human recombinant (hr-) nidogen-1 (R&D Systems, Minneapolis, MN; cat. #2570-ND) was dissolved in CatS buffer [100 mM NaH2PO4(H2O)2, 2 mM DTT, 0.01% Brij-35, pH 7.4] or matrix metalloproteinase (MMP) buffer (50 mM Tris, 150 mM NaCl, 10 mM CaCl2, 10 μM ZnCl, pH 7.5) to a final concentration of 125 μg/ml. The nidogen-1 solutions were incubated at 37°C for 1 and 24 hours with or without the addition of hr.-cathepsin S (Calbiochem, Whitehouse Station, NJ; cat. #219343), hr.-MMP-1 (R&D Systems, cat. #901-MP), hr.-MMP-7 (R&D Systems, cat. #907-MP), or hr.-MMP-9 (R&D Systems, cat. #911-MP) in final concentrations of 1.25 μg/ml, resulting in an enzyme-to-protein ratio of 1:100. The chosen MMPs had previously been shown to degrade nidogen-1 [11], [30]. Enzymatic activity tests were performed in parallel with the actual cleavage of nidogen-1. The reaction was stopped by adding E-64 or EDTA (final concentration of 1 μM) to CatS and MMP solutions, respectively. CatS and MMP buffer with relevant proteases were included as controls. The experiment was repeated twice. Samples were stored at −80°C until analysis. The cleavage of nidogen-1 was confirmed by silver staining according to the manufacturer's instructions (SilverXpress, Invitrogen, cat. #LC6100) (Supplementary Figure 1A), and a Western blot was performed to investigate if the CatS-generated fragments were of expected sizes (Supplementary Figure 1B).
NIC Assay Protocol
The competitive ELISA procedure was as follows: a 96-well streptavidin-coated microtiter plate was coated with 2.5 ng/ml of screening peptide (Biotin-VEKTRCQHERE) dissolved in assay buffer (50 mM Tris-BTB, 8 g/l NaCl, pH 8.0). The plate was incubated for 30 minutes at 20°C in darkness and was subsequently washed five times in washing buffer (20 mM Tris, 50 mM NaCl, pH 7.2). Then, 20 μl of selection peptide (VEKTRCQHERE) or sample (e.g., serum) was added to appropriate wells. This was followed by immediately adding 100 μl of monoclonal antibody dissolved in assay buffer to a concentration of 20 ng/ml. The plate was incubated for 3 hours at 20°C followed by ×5 wash in washing buffer. Then, 100 μl of goat anti-mouse POD-conjugated IgG antibody (Thermo Scientific, Waltham, MA; cat. #31437) diluted 1:6000 in assay buffer (final concentration of 130 ng/ml) was added to each well. The plate was incubated for 1 hour at 20°C followed by five washes in washing buffer. Next, 100 μl of tetramethylbenzinidine (Kem-En-Tec, Taastrup, Denmark; cat. #438OH) was added; the plate was incubated for 15 minutes at 20°C, and 100 μl of stopping solution (1% H2SO4) was added. All incubation steps were performed while the plate was shaking with 300 rpm. Finally, the optical density was measured in a VersaMax ELISA microplate reader at 450 nm with 650 nm as reference. A four-parametric mathematical fit model was used to plot a calibration curve. Data were analyzed using the SoftMax Pro v.6.3 software.
Technical Evaluation of the NIC ELISA
The lower limit of detection (LLOD) was determined from 21 measurements of the zero sample (assay buffer) and was calculated as the mean + 3 standard deviations. The upper limit of detection (ULOD) was determined from 10 independent runs of the highest selection peptide concentration employed in the standard curve and calculated as the mean back-calibration concentration + 3 standard deviations. The lower limit of quantification (LLOQ) was calculated as the highest NIC levels quantifiable in serum with a coefficient of variation below 30% reproduced from three independent runs of serum samples diluted stepwise. The intraassay variation was calculated as the mean coefficient of variance (CV %) within plates, and the interassay variation was calculated as the mean CV % between plates. The inter- and intraassay variations were determined by 10 independent runs of 8 quality control samples and 2 internal controls covering the detection range (LLOD-ULOD), with each run consisting of double determinations of the samples. The eight samples included two human serum samples, two animal serum samples spiked with the selection peptide, and four pools of human serum samples spiked with the selection peptide. Two-fold dilutions of human serum and plasma EDTA samples were used to calculate linearity as a percentage of dilution recovery of the undiluted sample. Accuracy was determined by spiking human serum and plasma EDTA samples with two-fold dilutions of the selection peptide and calculating the percentage spiking recovery using the expected concentration (serum and peptide combined) as reference. The effect of repeated freezing and thawing of the samples was determined for three human serum and plasma EDTA samples in four freeze/thaw cycles. The freeze-thaw recovery was calculated with the first cycle as reference. Analyte stability was determined for three human serum and plasma EDTA samples after 24 hours of storage at either 4°C or 20°C. Recovery was calculated with samples stored at −20°C as reference. Interference was determined by adding a low/high content of hemoglobin (0.155/0.310 mM), lipemia/lipids (4.83/10.98 mM), and biotin (30/90 ng/ml) to a serum sample of known concentration. Recovery percentage was calculated with the normal serum sample as reference.
Patient Samples
Serum samples consisted of two cohorts. An overview is given in Table 2. Appropriate Institutional Review Board/Independent Ethical Committee approved sample collection, and all the patients filed informed consent. The first cohort was acquired from the commercial vendor Proteogenex (Culver City, CA). The second cohort was a combination of cancer patient samples acquired from the commercial vendor Asterand (Detroit, MI) and healthy control samples obtained from another study population (reg. no. KA99070gm). According to Danish law, it is not required to get additional ethical approval when measuring biomarkers in previously collected samples. All investigations were carried out in accordance with the Helsinki Declaration.
Table 2.
Patient Characteristics
| Study Cohort | Size | Age | Gender | Tumor Stage |
|||
|---|---|---|---|---|---|---|---|
| #1 | n | Median, Range | Females | I | II | III | IV |
| Controls | 8 | 55, 44-65 | 6 | - | - | - | - |
| Breast cancer | 8 | 55, 34-62 | 8 | - | 3 | 5 | - |
| NSCLC | 8 | 62, 47-77 | 1 | 1 | 2 | 3 | 2 |
| #2 | n | Median, Range | Females | I | II | III | IV |
| Controls | 43 | 72, 60-82 | 43 | - | - | - | - |
| Breast cancer | 13 | 56, 43-69 | 12 | - | 11 | 2 | - |
| NSCLC | 12 | 58, 47-80 | 3 | 5 | 3 | 4 | - |
| SCLC | 8 | 57, 46-82 | 2 | 2 | 1 | 4 | 1 |
Statistical Analysis
Serum levels of NIC were compared by one-way analysis of variance (ANOVA) adjusted for multiple comparisons with Holm-Sidak's test. The area under the receiver operating characteristics (AUROC) curves was used to assess the diagnostics power of NIC. Data were considered statistically significant when P < .05. GraphPad Prism v6.05 and MedCalc Statistical Software v14.8.1 were used to perform statistical analysis.
Results
Specificity of the NIC Assay
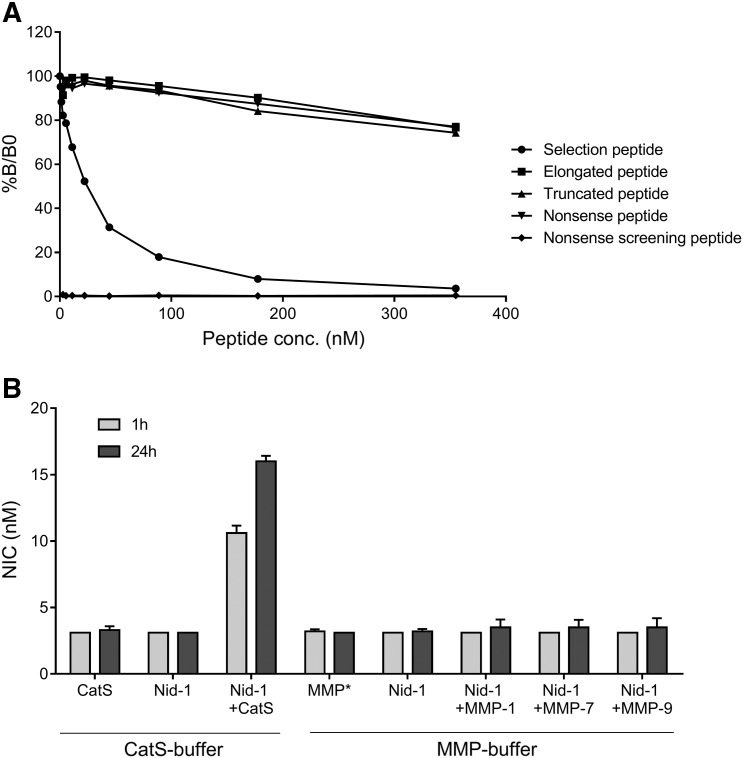
The specificity of the competitive NIC ELISA was evaluated by assessing the percentage of inhibition induced by the selection peptide (VEKTRCQHERE) as compared to an elongated selection peptide (VEKTRCQHEREH), a truncated selection peptide (EVEKTRCQHER), a nonsense selection peptide (APVGGIIGWM), and a nonsense screening peptide (Biotin-APVGGIIGWM) (the latter two correspond to the cleavage site located within the central part of the G2 domain on nidogen-1). Results are shown in Figure 1A. The selection peptide clearly inhibited the signal in a dose-dependent manner, whereas the signal was only slightly inhibited by the truncated, elongated, and nonsense peptides at the highest concentrations. No reactivity was detected toward the nonsense screening peptide. In addition, the levels of NIC generated by incubating nidogen-1 with CatS and various MMPs were investigated. As shown in Figure 1B, CatS generated NIC in a time-dependent manner (10.6 nM and 16.0 nM at 1 and 24 hours of incubation, respectively). Approximately five-fold higher levels of NIC were detected after incubating nidogen-1 with CatS for 24 hours. In contrast, no NIC was generated either without proteases (below LLOD of 3.1 nM)or when incubating with MMP-1, MMP-7, or MMP-9 (ranging from below LLOD of 3.1 nM to 3.5 nM). Altogether, this indicates that the antibody is specific and that the assay accurately measures nidogen-1 cleaved by CatS at amino acid position 693.
Figure 1.
Specificity of the NIC assay. (A) The percentage of inhibition at given concentrations in the competitive NIC-2 ELISA tested with the selection peptide (VEKTRCQHERE), an elongated selection peptide (VEKTRCQHEREH), a truncated selection peptide (EVEKTRCQHER), a nonsense selection peptide (APVGGIIGWM), and a nonsense screening peptide (Biotin-APVGGIIGWM). %B/B0: B equals the OD at × nM peptide and B0 equals the OD at 0 nM peptide. (B) NIC levels measured after cleavage of human recombinant nidogen-1 (Nid-1) with CatS, and MMP-1, -7, and -9. Values below the LLOD were assigned the LLOD. *Representative of MMP-1, -7, and -9.
Technical Evaluation of the NIC Assay
The overall technical performance of the NIC assay is listed in Table 3. The assay had an LLOD of 3.1 nM and a ULOD of 260 nM. The LLOQ was 3.2 nM. Intraassay variation was 9% and the interassay variation was 14%, below the acceptance level of 10% and 15%, respectively. All analyte recoveries were accepted if within 100 ± 20%. The dilution recovery was 104% and 92% for serum and plasma EDTA, respectively, indicating good linearity when diluting the samples. Spiking recovery for the standard peptide was also acceptable in serum (91%) and plasma EDTA (94%), indicating that these sample matrices do not affect assay response. The analyte was recovered in both serum and plasma EDTA after four freeze-thaw cycles with 95% and 85% recoveries, respectively. The analyte was also recovered after storage at 4°C and 20°C for 24 hours, resulting in 88% and 82% recoveries for serum and 90% and 98% recoveries for plasma EDTA at 4°C and 20°C, respectively. Together, this indicates that the analyte is relatively stable. No interference was detected from either low or high contents of biotin, lipids (lipemia), or hemoglobin, with recoveries ranging from 81% to 105%.
Table 3.
Technical Validation of the NIC Assay
| Technical Validation Step | Results |
|---|---|
| Detection range (LLOD-ULOD) | 3.1-260 nM |
| LLOQ | 3.2 nM |
| Intraassay variation | 9% |
| Interassay variation | 14% |
| Dilution recovery in serum⁎ | 104% (80-115%) |
| Dilution recovery in plasma EDTA⁎ | 92% (81-100%) |
| Spiking recovery in serum⁎ | 91% (83-105%) |
| Spiking recovery in plasma EDTA⁎ | 94% (80-102%) |
| Freeze-thaw recovery in serum⁎ | 95% (89-101%) |
| Freeze-thaw recovery in plasma EDTA⁎ | 85% (78-101%) |
| Analyte stability serum 24 h, 4°C/20°C⁎ | 88% (80-94%)/82% (71-87%) |
| Analyte stability plasma EDTA 24 h, 4°C/20°C⁎ | 90% (86-94%)/98% (95-105%) |
| Interference biotin, low/high⁎ | 98%/118% |
| Interference lipemia, low/high⁎ | 93%/81% |
| Interference hemoglobin, low/high⁎ | 105%/101% |
Percentages are reported as mean with range shown in brackets.
Evaluation of NIC Levels in Serum from Patients with Lung Cancer, Patients with Breast Cancer, and Healthy Controls
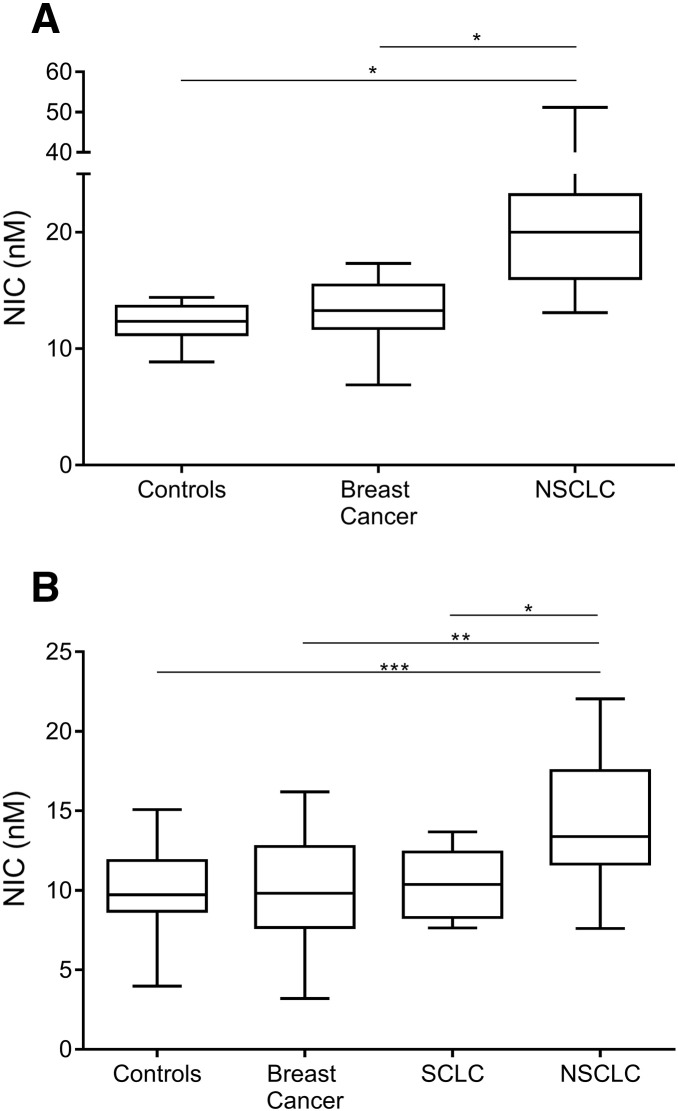
NIC levels were measured in serum from two different patient cohorts. As shown in Figure 2A, NIC was significantly elevated (P > .05) in serum from NSCLC patients as compared to healthy controls and breast cancer patients. Median NIC in the NSCLC patients was 20 nM, ranging from 13.1 to 51.1 nM. Median NIC in the breast cancer patients was 13.3 nM, ranging from 6.9 to 17.3 nM, and median NIC in the healthy controls was 12.4 nM, ranging from 8.9 to 14.4 nM. As shown in Figure 2B, NIC was significantly elevated in serum from NSCLC patients as compared to healthy controls (P < .001), breast cancer patients (P < .01), and patients with SCLC (P < .05). In this cohort, median NIC in the NSCLC patients was 13.4 nM, ranging from 7.6 to 22.1 nM. Median NIC in the SCLC patients was 10.4 nM, ranging from 7.6 to 13.7 nM; median NIC in the breast cancer patients was 9.8 nM, ranging from 3.3 to 16.2 nM; and median NIC in the healthy controls was 9.7 nM, ranging from 4.0 to 15.1 nM.
Figure 2.
Evaluation of NIC in serum from two study cohorts. (A) Proteogenex samples; (B) Asterand samples. Box and whiskers show data from minimum to maximum. Data were compared by one-way ANOVA adjusted for multiple comparisons. Values below the LLOQ were assigned the LLOQ (n = 1, breast cancer, B). *P < .05, **P < .01, ***P < .001.
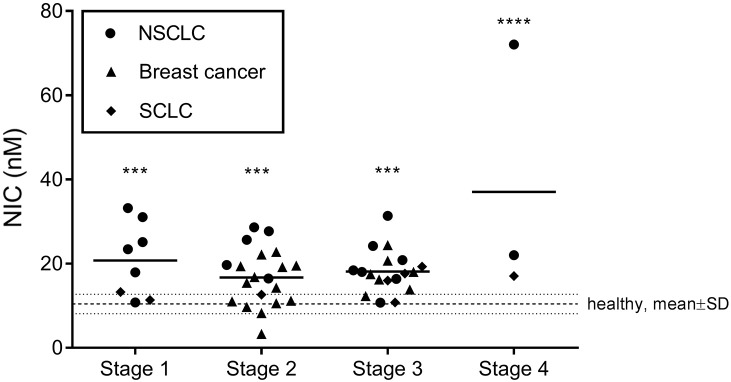
NIC levels were also assessed according to tumor stage, an important clinical parameter in cancer. Results are shown in Figure 3 for all cancers combined. In detail, all stages of disease had elevated levels of NIC as compared to the healthy controls, whereas no difference could be detected between stages. This indicates that NIC is generated independent of tumor stage.
Figure 3.
Evaluation of NIC in serum according to tumor stage. Samples were pooled according to tumor stage. Data are presented as scatter plots with each cancer type identifiable. Data were compared to healthy controls by one-way ANOVA adjusted for multiple comparisons. ***P < .001, ****P < .0001.
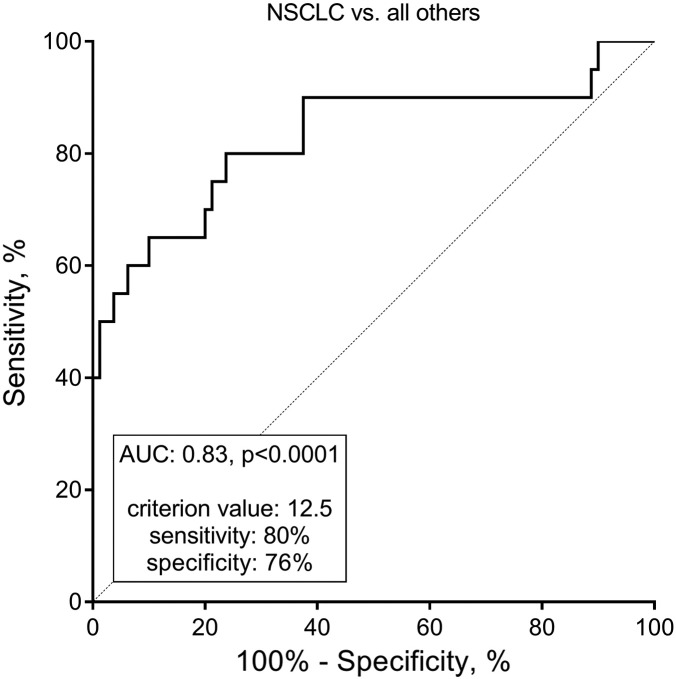
To analyze the diagnostic power of NIC with respect to NSCLC, both cohorts were pooled and grouped in “NSCLC” versus“all others,” and the AUROC was calculated. The AUROC was 0.83 (95% confidence intervals: 0.71-0.95), P < .0001. The ROC curve, as well as the sensitivity and specificity at a preliminary estimated optimal cutoff value, is shown in Figure 4. These findings indicate that NIC is able to separate NSCLC patients from the other subjects combined. Altogether, the present findings suggest that NIC may be highly associated with NSCLC.
Figure 4.
ROC analysis evaluating the ability of NIC to separate NSCLC from all other samples included in this study combined (breast cancer, SCLC, and healthy controls). AUC, area under the ROC curve.
Discussion
In the present study, we describe the development and validation of a robust competitive ELISA that enables noninvasive measurements of fragments of nidogen-1 degraded specifically by CatS (NIC). Moreover, to our knowledge, this is the first study to show that nidogen-1 degraded specifically by CatS may have biomarker potential for NSCLC.
CatS cleavage of nidogen-1 resulted in a specific fragment containing a neo-epitope with the amino acid composition VEKTRCQHERE (the NIC domain). Hereby, we were able to verify the findings by Sage et al. showing that nidogen-1 is cleaved by CatS at amino acid position 693 (NH3-…HERE693) which is located in the thyroglobulin-like (Tg) domain adjacent to the G3 domain [19].
The elevated NIC levels found in serum from patients with NSCLC as compared to patients with breast cancer, patients with SCLC, and healthy controls suggest that NSCLC patients have increased CatS-mediated degradation of nidogen-1. This was further backed by the diagnostic accuracy of NIC (AUROC 0.83) for NSCLC when compared to all other subjects. In detail, for NSCLC, the average NIC levels were slightly lower in cohort 2 compared to cohort 1 (Figure 2). This, together with the relatively large variation within each group of patients, suggests however that NIC may not serve as a diagnostic tool for lung cancer. Instead, NIC levels may be indicative of a specific pathological event in a patient's tumor, and this pathological event seems to be more associated with NSCLC as compared to the other cancer types analyzed. One would expect the amount of BM in the given tumor type to be a limiting factor, and whether the NSCLC cancer subtype has more BM (nidogen-1) compared to the other types of cancer tested here remains to be established.
Intuitively, one would expect NIC, by reflecting altered BM homeostasis, to be highly associated with an invasive phenotype and potential for metastasizing. As NIC levels were elevated in all stages of disease especially in NSCLC, this indicates that CatS degradation of nidogen-1 is ongoing in both early and late stages of disease independent of tumor stage and that NIC may be applied in the early onset of disease. In this context, it is important to get a deeper understanding of the role of CatS in relation to tumor-associated events. CatS has been associated with different cancer types and protumorigenic events [24], [25], [26], [27]. However, the exact mechanisms leading to increased expression of cathepsins in tumors have not been investigated in detail [20].
The NIC domain was specifically generated by CatS and not by MMPs that had previously been shown to degrade nidogen-1 [11], [30]. This is important because different nidogen-1 fragments may reflect different (patho)physiological events, i.e., nidogen-1 degraded by CatS may reflect different disease activity patterns as compared to, for instance, nidogen-1 degraded by an MMP [31]. The two other described CatS cleavage sites, which are located within the G2 and G3 domains on nidogen-1, are essential for the tertiary assembly of the BM [5], [8]. Whether cleavage at the NIC domain is associated with similar or distinct (patho)physiological features remains to be established.
CatS has been shown to be released from TAMs [24], and it has been shown that, when co-culturing lung cancer cell lines with TAMs isolated from NSCLC patients, cell invasion was increased and associated with an upregulation of the expression of CatS in the cancer cells [32]. Moreover, it has been shown that CatS through its ECM-degrading properties affects angiogenesis [33]. Circulating plasma levels of nidogen-1 have been detected in ovarian cancer, especially at the advanced stage of disease [27]. Here, nidogen-1 levels were measured using a sandwich ELISA developed with commercially available antibodies from R&D Systems (cat. #DY2570); however, the exact epitopes were unknown. Thus, in contrast to samples measured with the NIC assay, it remains to be established how nidogen-1 ended up in circulation from these ovarian cancer patients. In fact, a complex network of proteolytic enzymes is involved in tumor progression [34] and should be considered when evaluating the impact of circulating fragments of nidogen-1. Nonetheless, our data indicate that CatS is definitely involved in cleavage of nidogen-1 and may be particularly associated with NSCLC. Still, in addition to the tissue of origin, the degree of angiogenesis and invasion with associated dynamic alterations in BM remodeling from both the desmoplastic reaction and altered proteolytic activity may be highly contributing to the differences observed in the patients.
It has been shown that nidogen-1 can regulate specific gene expression in mammary epithelial cells [35]. When the expression of β-casein was investigated as a function of nidogen quality/composition, a fragment of nidogen containing the laminin-1 binding domain but lacking the type IV collagen binding domain was able to reduce the expression of β-casein. In contrast, a fragment of nidogen containing the type IV collagen binding domain but lacking the laminin-111 binding domain was not. These findings both suggest that nidogen enhances the effect of laminin-111 binding to its receptors, which in turns supports mammary gland differentiation and associated gene expression. This is similar to observations described by Weaver et al. showing that it is possible to repress the tumorigenic phenotype of breast cancer by modulating the ECM and its receptors [36]. Together, these findings suggests that specific (not all) nidogen-1 fragments may affect the cell-BM interaction and hereby possibly contribute to tumorigenesis by affecting cell phenotype. Thus, hypothetically, specific fragment of nidogen-1, such as NIC, may reflect particular protumorigenic events.
Concerning the efficacy of anticancer drugs, ECM-modifying drugs are currently investigated as possible interventions for modulating the tumor microenvironment and obtain tumor shrinkage or more efficient delivery of chemotherapeutic drugs and targeted therapies. Interestingly, it has been shown that antibody-mediated blockage of CatS enhances the efficacy of chemotherapy [37]. Moreover, this antibody has been suggested as a direct strategy for the treatment of solid tumors [38]. Potentially, the NIC assay could be used for predicting which patients are most likely to benefit from such treatment or for monitoring early efficacy of treatment. Further studies are needed to investigate this, and the current study provides the tool for such investigations.
An important parameter of the NIC assay is related to robustness, i.e., technical and analytical evaluation. Technical evaluation of NIC included the establishment of detection range, sensitivity, inter- and intraassay variation, linearity/precision (dilution recovery), analyte stability, and interference, which all were within acceptable limits. Analytical evaluation of the NIC assay refers to the specificity of the assay and to whether the assay detects the analyte it was designed to detect in serum/plasma EDTA. The specificity was evaluated by displacement analysis (Figure 1A) and by testing reactivity toward intact and cleaved nidogen-1 (Figure 1B). Accuracy was evaluated by spiking the analyte into the relevant matrices (serum and plasma EDTA) and calculating the percentage recovery (also within acceptable limits). Thus, the NIC assay was technically and analytically robust, indicating that the assay is specific and able to perform accurately over a period of time.
The present study is limited by relatively small population sizes and the cross-sectional designs when evaluating NIC in relation to cancer. Moreover, clinical information was limited, not allowing for any correlation analysis between the pathophysiology of individual patients, or particular tumor phenotype, to the measured NIC levels. In addition, gender could be a confounding factor, as the controls were primarily female while the vast proportion of the NSCLC samples were from males. However, in opposition to this, a significant difference in NIC levels was also detected between more gender-matched samples of different tumor types (e.g., NSCLC versus SCLC). Larger longitudinal studies are necessary to validate the present findings and to investigate any potential diagnostic and prognostic applicability of NIC. Moreover, to get a better understanding of biological relevance of NIC, it would be highly relevant to correlate to other BM degradation products associated with specific pathological events.
Conclusion
In conclusion, CatS-degraded nidogen-1 can be quantified in serum by the technically robust NIC assay. NIC levels were elevated in NSCLC patients as compared to breast cancer patients, SCLC patients, and healthy controls. If validated in larger clinical studies, the NIC assay may serve as a noninvasive biomarker for cancer and provide important insight into tumor biology.
The following are the supplementary data related to this article.
Declaration of Interests
All authors are employed at Nordic Bioscience A/S involved in biomarker development and research.
Acknowledgement
We acknowledge the Danish Research Foundation for providing funding for this study.
References
- 1.Lu P, Weaver VM, Werb Z. The extracellular matrix: a dynamic niche in cancer progression. J Cell Biol. 2012;196:395–406. doi: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.LeBleu VS, Macdonald B, Kalluri R. Structure and function of basement membranes. Exp Biol Med. 2007;232:1121–1129. doi: 10.3181/0703-MR-72. [DOI] [PubMed] [Google Scholar]
- 3.Glentis A, Gurchenkov V, Vignjevic DM. Assembly, heterogeneity, and breaching of the basement membranes. Cell Adh Migr. 2014;8:236–245. doi: 10.4161/cam.28733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yurchenco PD, Patton BL. Developmental and pathogenic mechanisms of basement membrane assembly. Curr Pharm Des. 2009;15:1277–1294. doi: 10.2174/138161209787846766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ho MSP, Böse K, Mokkapati S, Nischt R, Smyth N. Nidogens-extracellular matrix linker molecules. Microsc Res Tech. 2008;71:387–395. doi: 10.1002/jemt.20567. [DOI] [PubMed] [Google Scholar]
- 6.Chung AE, Durkin ME. Entactin: structure and function. Am J Respir Cell Mol Biol. 1990;3:275–282. doi: 10.1165/ajrcmb/3.4.275. [DOI] [PubMed] [Google Scholar]
- 7.Bader BL, Smyth N, Nedbal S, Miosge N, Baranowsky A, Mokkapati S, Murshed M, Nischt R. Compound genetic ablation of nidogen 1 and 2 causes basement membrane defects and perinatal lethality in mice. Mol Cell Biol. 2005;25:6846–6856. doi: 10.1128/MCB.25.15.6846-6856.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aumailley M, Battaglia C, Mayer U, Reinhardt D, Nischt R, Timpl R, Fox JW. Nidogen mediates the formation of ternary complexes of basement membrane components. Kidney Int. 1993;43:7–12. doi: 10.1038/ki.1993.3. [DOI] [PubMed] [Google Scholar]
- 9.Kohfeldt E, Sasaki T, Göhring W, Timpl R. Nidogen-2: a new basement membrane protein with diverse binding properties. J Mol Biol. 1998;282:99–109. doi: 10.1006/jmbi.1998.2004. [DOI] [PubMed] [Google Scholar]
- 10.Fox JW, Mayer U, Nischt R, Aumailley M, Reinhardt D, Wiedemann H, Mann K, Timpl R, Krieg T, Engel J. Recombinant nidogen consists of three globular domains and mediates binding of laminin to collagen type IV. EMBO J. 1991;10:3137–3146. doi: 10.1002/j.1460-2075.1991.tb04875.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayer U, Mann K, Timpl R, Murphy G. Sites of nidogen cleavage by proteases involved in tissue homeostasis and remodelling. Eur J Biochem. 1993;217:877–884. doi: 10.1111/j.1432-1033.1993.tb18316.x. [DOI] [PubMed] [Google Scholar]
- 12.Pöschl E, Mayer U, Stetefeld J, Baumgartner R, Holak TA, Huber R, Timpl R. Site-directed mutagenesis and structural interpretation of the nidogen binding site of the laminin gamma1 chain. EMBO J. 1996;15:5154–5159. [PMC free article] [PubMed] [Google Scholar]
- 13.Willem M, Miosge N, Halfter W, Smyth N, Jannetti I, Burghart E, Timpl R, Mayer U. Specific ablation of the nidogen-binding site in the laminin gamma1 chain interferes with kidney and lung development. Development. 2002;129:2711–2722. doi: 10.1242/dev.129.11.2711. [DOI] [PubMed] [Google Scholar]
- 14.Breitkreutz D, Mirancea N, Schmidt C, Beck R, Werner U, Stark H-J, Gerl M, Fusenig NE. Inhibition of basement membrane formation by a nidogen-binding laminin gamma1-chain fragment in human skin-organotypic cocultures. J Cell Sci. 2004;117:2611–2622. doi: 10.1242/jcs.01127. [DOI] [PubMed] [Google Scholar]
- 15.Tunggal J, Wartenberg M, Paulsson M, Smyth N. Expression of the nidogen-binding site of the laminin gamma1 chain disturbs basement membrane formation and maintenance in F9 embryoid bodies. J Cell Sci. 2003;116:803–812. doi: 10.1242/jcs.00293. [DOI] [PubMed] [Google Scholar]
- 16.Kadoya Y, Salmivirta K, Talts JF, Kadoya K, Mayer U, Timpl R, Ekblom P. Importance of nidogen binding to laminin gamma1 for branching epithelial morphogenesis of the submandibular gland. Development. 1997;124:683–691. doi: 10.1242/dev.124.3.683. [DOI] [PubMed] [Google Scholar]
- 17.Ekblom P, Ekblom M, Fecker L, Klein G, Zhang HY, Kadoya Y, Chu ML, Mayer U, Timpl R. Role of mesenchymal nidogen for epithelial morphogenesis in vitro. Development. 1994;120:2003–2014. doi: 10.1242/dev.120.7.2003. [DOI] [PubMed] [Google Scholar]
- 18.Baranowsky A, Mokkapati S, Bechtel M, Krügel J, Miosge N, Wickenhauser C, Smyth N, Nischt R. Impaired wound healing in mice lacking the basement membrane protein nidogen 1. Matrix Biol. 2010;29:15–21. doi: 10.1016/j.matbio.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 19.Sage J, Leblanc-Noblesse E, Nizard C, Sasaki T, Schnebert S, Perrier E, Kurfurst R, Brömme D, Lalmanach G, Lecaille F. Cleavage of nidogen-1 by cathepsin S impairs its binding to basement membrane partners. PLoS One. 2012;7:e43494. doi: 10.1371/journal.pone.0043494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohamed MM, Sloane BF. Cysteine cathepsins: multifunctional enzymes in cancer. Nat Rev Cancer. 2006;6:764–775. doi: 10.1038/nrc1949. [DOI] [PubMed] [Google Scholar]
- 21.Turk V, Stoka V, Vasiljeva O, Renko M, Sun T, Turk B, Turk D. Cysteine cathepsins: from structure, function and regulation to new frontiers. Biochim Biophys Acta-Proteins Proteomics. 2012;1824:68–88. doi: 10.1016/j.bbapap.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turk B, Bieth JG, Björk I, Dolenc I, Cimerman N, Kos J, Öolic A, Stoka V, Turk V. Regulation of the activity of lysosomal cysteine proteinases by pH-induced inactivation and/or endogenous protein inhibitors, cystatins. Biol Chem. 1995;376:225–230. doi: 10.1515/bchm3.1995.376.4.225. [DOI] [PubMed] [Google Scholar]
- 23.Kirschke H, Wiederanders B, Brömme D, Rinne A. Cathepsin S from bovine spleen. Purification, distribution, intracellular localization and action on proteins. Biochem J. 1989;264:467–473. doi: 10.1042/bj2640467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sevenich L, Bowman RL, Mason SD, Quail DF, Elie BT, Brogi E, Brastianos PK, Hahn WC, Holsinger J, Massagué J. Analysis of tumor- and stroma-supplied proteolytic networks reveals a brain metastasis-promoting role for cathepsin S. Nat Cell Biol. 2014;16:876–888. doi: 10.1038/ncb3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan Q, Wang X, Zhang H, Li C, Fan J, Xu J. Silencing cathepsin S gene expression inhibits growth, invasion and angiogenesis of human hepatocellular carcinoma in vitro. Biochem Biophys Res Commun. 2012;425:703–710. doi: 10.1016/j.bbrc.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 26.Yixuan Y, Kiat LS, Yee CL, Huiyin L, Yunhao C, Kuan CP, Hassan A, Ting WT, Manuel ST, Guan YK, Pin LY. Cathepsin S mediates gastric cancer cell migration and invasion via a putative network of metastasis-associated proteins. J Proteome Res. 2010;9:4767–4778. doi: 10.1021/pr100492x. [DOI] [PubMed] [Google Scholar]
- 27.Li L, Zhang Y, Li N, Feng L, Yao H, Zhang R. Nidogen-1: a candidate biomarker for ovarian serous cancer. Jpn J Clin Oncol. 2015;45:176–182. doi: 10.1093/jjco/hyu187. [DOI] [PubMed] [Google Scholar]
- 28.Combet C, Blanchet C, Geourjon C, Deléage G. NPS@: network protein sequence analysis. Trends Biochem Sci. 2000;25:147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 29.Gefter ML, Margulies DH, Scharff MD. A simple method for polyethylene glycol-promoted hybridization of mouse myeloma cells. Somat Genet. 1977;3:231–236. doi: 10.1007/BF01551818. [DOI] [PubMed] [Google Scholar]
- 30.Sires UI, Griffin GL, Broekelmann TJ, Mecham RP, Murphy G, Chung AE, Welgus HG, Senior RM. Degradation of entactin by matrix metalloproteinases. J Biol Chem. 1993;268:2069–2074. [PubMed] [Google Scholar]
- 31.Karsdal MA, Delvin E, Christiansen C. Protein fingerprints — relying on and understanding the information of serological protein measurements. Clin Biochem. 2011;44:1278–1279. doi: 10.1016/j.clinbiochem.2011.08.1135. [DOI] [PubMed] [Google Scholar]
- 32.Wang R, Zhang J, Chen S, Lu M, Luo X, Yao S, Liu S, Qin Y, Chen, Haiquan Tumor-associated macrophages provide a suitable microenvironment for non–small lung cancer invasion and progression. Lung Cancer. 2011;74:188–196. doi: 10.1016/j.lungcan.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 33.Shi GP, Sukhova GK, Kuzuya M, Ye Q, Du J, Zhang Y, Pan JH, Lu ML, Cheng XW, Iguchi A. Deficiency of the cysteine protease cathepsin S impairs microvessel growth. Circ Res. 2003;92:493–500. doi: 10.1161/01.RES.0000060485.20318.96. [DOI] [PubMed] [Google Scholar]
- 34.Mason SD, Joyce JA. Proteolytic networks in cancer. Trends Cell Biol. 2011;21:228–237. doi: 10.1016/j.tcb.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pujuguet P, Simian M, Liaw J, Timpl R, Werb Z, Bissell MJ. Nidogen-1 regulates laminin-1-dependent mammary-specific gene expression. J Cell Sci. 2000;113:849–858. doi: 10.1242/jcs.113.5.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weaver VM, Petersen OW, Wang F, Larabell CA, Briand P, Damsky C, Bissell MJ. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J Cell Biol. 1997;137(1):231–245. doi: 10.1083/jcb.137.1.231. [137] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burden RE, Gormley JA, Kuehn D, Ward C, Kwok HF, Gazdoiu M, McClurg A, Jaquin TJ, Johnston JA, Scott CJ, Olwill SA. Inhibition of Cathepsin S by Fsn0503 enhances the efficacy of chemotherapy in colorectal carcinomas. Biochimie. 2012;94:487–493. doi: 10.1016/j.biochi.2011.08.017. [DOI] [PubMed] [Google Scholar]
- 38.Vázquez R, Astorgues-Xerri L, Bekradda M, Gormley J, Buick R, Kerr P, Cvitkovic E, Raymond E, D'Incalci M, Frapolli R, Riveiro ME. Fsn0503h antibody-mediated blockade of cathepsin S as a potential therapeutic strategy for the treatment of solid tumors. Biochimie. 2014;108:101–107. doi: 10.1016/j.biochi.2014.10.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.