Abstract
The A Disintegrin and Metalloproteinase (ADAM) family of endopeptidases plays a role in many solid cancers and includes promising targets for anticancer therapies. Deregulation of ADAM15 has been linked to tumor aggressiveness and cell line studies suggest that ADAM15 overexpression may also be implicated in prostate cancer. To evaluate the impact of ADAM15 expression and its relationship with key genomic alterations, a tissue microarray containing 12,427 prostate cancers was analyzed by immunohistochemistry. ADAM15 expression was compared to phenotype, prognosis and molecular features including TMPRSS2:ERG fusion and frequent deletions involving PTEN, 3p, 5q and 6q. Normal prostate epithelium did not show ADAM15 staining. In prostate cancers, negative, weak, moderate, and strong ADAM15 staining was found in 87.7%, 3.7%, 5.6%, and 3.0% of 9826 interpretable tumors. Strong ADAM15 staining was linked to high Gleason grade, advanced pathological tumor stage, positive nodal stage and resection margin. ADAM15 overexpression was also associated with TMPRSS2:ERG fusions and PTEN deletions (P < .0001) but unrelated to deletions of 3p, 5q and 6q. In univariate analysis, high ADAM15 expression was strongly linked to PSA recurrence (P < .0001). However, in multivariate analyses this association was only maintained if the analysis was limited to preoperatively available parameters in ERG-negative cancers. The results of our study demonstrate that ADAM15 is strongly up regulated in a small but highly aggressive fraction of prostate cancers. In these tumors, ADAM15 may represent a suitable drug target. In a preoperative scenario, ADAM15 expression measurement may assist prognosis assessment, either alone or in combination with other markers.
Abbreviations: ADAM, a disintegrin and metalloproteinase; CHD1, chromodomain-helicase-DNA-binding protein 1; ERG, erythroblast transformation-specific (ETS) related gene; ETS, erythroblast transformation-specific; FISH, fluorescence in situ hybridization; FOXP1, forkhead box protein P1; MAP3K7, mitogen-activated protein kinase kinase kinase 7; MMP, matrix metalloproteinase; PSA, prostate specific antigen; PTEN, phosphatase and tensin homolog; TMA, tissue microarray; TMPRSS2, transmembrane protease, serine 2
Introduction
Prostate cancer is the most prevalent cancer in men in Western societies [1]. Although the majority of prostate cancers behave in an indolent manner, a small subset is highly aggressive and requires extensive treatment [2], [3]. Established preoperative prognostic parameters are limited to Gleason grade and tumor extent on biopsies, prostate-specific antigen (PSA), and clinical stage. These data are statistically powerful, but often insufficient for optimal individual treatment decisions. It is thus hoped that a better understanding of disease biology will eventually lead to the identification of clinically applicable molecular markers that enable a more reliable prediction of prostate cancer aggressiveness.
The human A Disintegrin and Metalloproteinase 15 (ADAM15) is one of more than 20 members of the ADAM family of type I multi-domain transmembrane glycoproteins that function as zinc-dependent endopeptidases (reviewed in [4], [5]). Activated ADAM15 is involved in proteolytic processing of cytokines, growth factors and adhesions molecules [5]. ADAM15 promotes cancer progression in gastric [6], [7], lung [8], [9] and colon cancers [10] as well as melanomas [11]. It is believed that the tumor promoting action of ADAM15 results from disruption of cell–cell [4], [12] and cell-matrix [12] adhesion and from release of membrane-bound growth factors (reviewed in [13], [14]). Accordingly, ADAM15 deregulation has been linked to poor patient outcome in lung and colon cancers [9], [10]. There is accumulating evidence that ADAM15 may also plays a role for prostate cancer biology. Functional studies in PC-3 prostate cancer cells suggest a role of ADAM15 for metastasis, as the capability to migrate through vascular endothelial cells depends on ADAM15 expression [14]. Moreover, a study on 167 clinical prostate cancer specimens suggested a link between ADAM15 overexpression, metastatic phenotype and poor patient prognosis [15].
These promising findings encouraged us to study the putative prognostic value of ADAM15 expression in a large cohort including more than 12,000 prostate cancers that have been assembled in a tissue microarray (TMA) format and for which clinical follow-up data are available.
Materials and Methods
Patients
Radical prostatectomy specimens were available from 12,427 patients, undergoing surgery between 1992 and 2012 at the Department of Urology and the Martini Clinics at the University Medical Center Hamburg-Eppendorf. Histo-pathological data was retrieved from the patient files, including tumor stage, Gleason grade, nodal stage and stage of the resection margin. In addition to the classical Gleason categories, “quantitative” Gleason grading was performed as described before [16]. In brief, for every prostatectomy specimen, the percentages of Gleason 3, 4, and 5 patterns were estimated in cancerous tissues during the regular process of Gleason grading. Gleason 3 + 4 and 4 + 3 cancers were subdivided according to their percentage of Gleason 4. For practical use, we subdivided the 3 + 4 and 4 + 3 cancers in 8 subgroups: 3 + 4 ≤ 5% Gleason 4, 3 + 4 6–10%, 3 + 4 11–20%, 3 + 4 21–30%, 3 + 4 31–49%, 4 + 3 50–60%, 4 + 3 61–80% and 4 + 3 > 80% Gleason 4. In addition, separate groups were defined by the presence of a tertiary Gleason 5 pattern, including 3 + 4 Tert.5 and 4 + 3 Tert. 5. Follow-up data were available for a total of 12,344 patients with a median follow-up of 36 months (range: 1 to 241 months; Table 1). Prostate specific antigen (PSA) values were measured following surgery and PSA recurrence was defined as a postoperative PSA of 0.2 ng/ml and increasing at first of appearance. All prostate specimens were analyzed according to a standard procedure, including a complete embedding of the entire prostate for histological analysis [17]. The TMA manufacturing process was described earlier in detail [18]. In short, one 0.6 mm core was taken from a representative tissue block from each patient. The tissues were distributed among 27 TMA blocks, each containing 144 to 522 tumor samples. For internal controls, each TMA block also contained various control tissues, including normal prostate tissue. The molecular database attached to this TMA contained results on ERG expression in 10,678 [19], ERG break apart FISH analysis in 7099 (expanded from [20]) and deletion status of 5q21 (CHD1) in 7932 (expanded from [21]), 6q15 (MAP3K7) in 6069 (expanded from [22]), PTEN (10q23) in 6704 (expanded from [23]) and 3p13 (FOXP1) in 7081 (expanded from [24]) cancers. The usage of archived diagnostic left-over tissues for manufacturing of tissue microarrays and their analysis for research purposes as well as patient data analysis has been approved by local laws (HmbKHG, §12,1) and by the local ethics committee (Ethics commission Hamburg, WF-049/09 and PV3652). All work has been carried out in compliance with the Helsinki Declaration.
Table 1.
Pathological and Clinical Data of the Arrayed Prostate Cancers
| No. of patients (%) |
||
|---|---|---|
| Study cohort on TMA (N = 12,427) | Biochemical relapse among categories | |
| Follow-up (mo) | ||
| n | 11,665 (93.9%) | 2769 (23.7%) |
| Mean | 48.9 | - |
| Median | 36.4 | - |
| Age (y) | ||
| ≤50 | 334 (2.7%) | 81 (24.3%) |
| 51–59 | 3061 (24.8%) | 705 (23%) |
| 60–69 | 7188 (58.2%) | 1610 (22.4%) |
| ≥70 | 1761 (14.3%) | 370 (21%) |
| Pretreatment PSA (ng/ml) | ||
| <4 | 1585 (12.9%) | 242 (15.3%) |
| 4–10 | 7480 (60.9%) | 1355 (18.1%) |
| 10–20 | 2412 (19.6%) | 737 (30.6%) |
| >20 | 812 (6.6%) | 397 (48.9%) |
| pT stage (AJCC 2002) | ||
| pT2 | 8187 (66.2%) | 1095 (13.4%) |
| pT3a | 2660 (21.5%) | 817 (30.7%) |
| pT3b | 1465 (11.8%) | 796 (54.3%) |
| pT4 | 63 (0.5%) | 51 (81%) |
| Gleason grade | ||
| ≤3 + 3 | 2848 (22.9%) | 234 (8.2%) |
| 3 + 4 | 6679 (53.8%) | 1240 (18.6%) |
| 3 + 4 Tertiary 5 | 433 (3.5%) | 115 (26.6%) |
| 4 + 3 | 1210 (9.7%) | 576 (47.6%) |
| 4 + 3 Tertiary 5 | 646 (5.2%) | 317 (49.1%) |
| ≥4 + 4 | 596 (4.8%) | 348 (58.4%) |
| pN stage | ||
| pN0 | 6970 (91%) | 1636 (23.5%) |
| pN+ | 693 (9%) | 393 (56.7%) |
| Surgical margin | ||
| Negative | 9990 (81.9%) | 1848 (18.5%) |
| Positive | 2211 (18.1%) | 853 (38.6%) |
Percentage in the column “Study cohort on TMA” refers to the fraction of samples across each category. Percentage in column “Biochemical relapse among categories” refers to the fraction of samples with biochemical relapse within each parameter in the different categories. NOTE: Numbers do not always add up to 12,427 in the different categories because of cases with missing data. Abbreviation: AJCC, American Joint Committee on Cancer.
Immunohistochemistry
Freshly cut TMA sections were immunostained on 1 day and in one experiment. Slides were deparaffinized and exposed to heat-induced antigen retrieval for 5 minutes in an autoclave at 121 °C in pH 7,8 Tris-EDTA-Citrate buffer. Primary antibody specific for ADAM15 (rabbit polyclonal antibody, Chemicon (Merck Chemicals GmbH), Schwalbach, Germany; AB19035; dilution 1:150) was applied at 37 °C for 60 minutes. Bound antibody was then visualized using the EnVision Kit (Dako, Glostrup, Denmark) according to the manufacturer's directions. A final score was built from these two parameters according to the following criteria as previously described [20]: Tumors with complete absence of staining were scored as “negative”. Tumors with a score “weak” had a staining intensity of 1+ in ≤70% of tumor cells or 2+ in ≤30% of tumor cells. A score “moderate” was given to cancers with a staining intensity of 1+ in >70% of tumor cells, or 2+ in >30% and ≤70% of tumor cells, or 3+ in ≤30% of tumor cells. The score was “strong” if staining intensity was 2+ in >70% of tumor cells or 3+ in >30% of tumor cells.
Statistics
For statistical analysis, the JMP 10.0.2 software (SAS Institute Inc., Cary, NC, USA) was used. Contingency tables were calculated to study associations between ADAM15 expression and clinicopathological variable, and the Chi-square (Likelihood) test was used to find significant relationships. Kaplan Meier curves were generated for PSA recurrence free survival. The log-Rank test was applied to test the significance of differences between stratified survival functions. Cox proportional hazards regression analysis was performed to test the statistical independence and significance between pathological, molecular, and clinical variables.
Results
Technical Issues
A total of 9826 (79%) tumor samples were interpretable in our TMA analysis. Reasons for non-informative cases (2601; 21%) included lack of tissue samples or absence of unequivocal cancer tissue in the TMA spot.
ADAM15 Expression in Normal and Cancerous Prostate Tissues
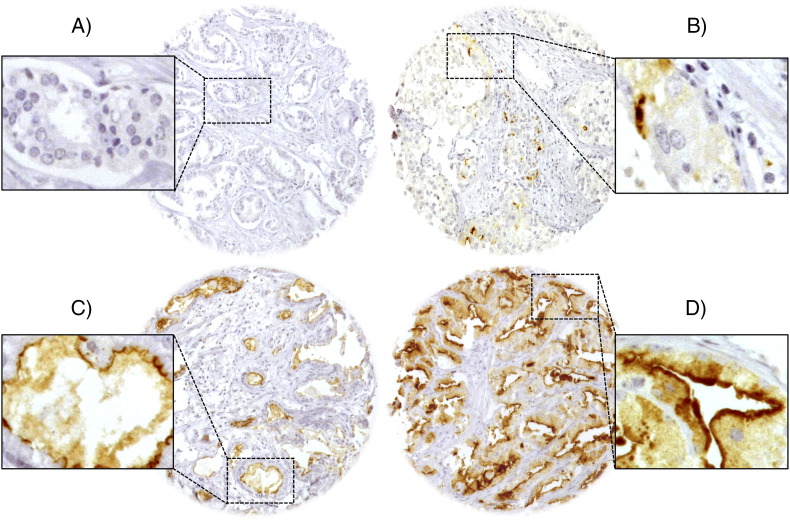
Normal prostate tissue did not show ADAM15 staining. In cancers, ADAM15 immunostaining was typically localized to the apical cell membrane. ADAM15 immunostaining was seen in 12.3% of our 9826 interpretable prostate cancers and was considered weak in 3.7%, moderate in 5.6% and strong in 3.0% of cases. Representative images of ADAM15 immunostainings are given in Figure 1. Elevated ADAM15 expression was significantly linked to advanced pathological tumor stage, Gleason grade, lymph node metastasis (P < .0001, each) and surgical margin (P = .0007). All data are summarized in Table 2. Subgroup analyses revealed that all significant associations to tumor phenotype found in all cancers held also true in the subsets of ERG-negative and ERG-positive cancers (data not shown).
Figure 1.
Representative pictures of ADAM15 immunostaining in prostate cancer with (A) negative, (B) weak, (C) moderate and (D) strong staining. Apical cell membrane staining is seen at 400× magnification in insert 1D).
Table 2.
Association between ADAM15 Immunostaining Results and Prostate Cancer Phenotype in All Cancers
| ADAM15 |
||||||
|---|---|---|---|---|---|---|
| Parameter | N evaluable | Negative (%) | Weak (%) | Moderate (%) | Strong (%) | P value |
| All cancers | 9485 | 88.2 | 3.8 | 4.9 | 3.0 | |
| Tumor stage | <0.0001 | |||||
| pT2 | 6082 | 90.7 | 3.4 | 4.2 | 1.7 | |
| pT3a | 2155 | 84.7 | 4.8 | 5.6 | 4.8 | |
| pT3b-pT4 | 1210 | 82.0 | 4.2 | 7.4 | 6.4 | |
| Gleason grade | <0.0001 | |||||
| ≤3 + 3 | 2261 | 91.9 | 2.7 | 3.5 | 1.9 | |
| 3 + 4 | 5278 | 88.5 | 4.2 | 4.6 | 2.7 | |
| 4 + 3 | 1448 | 84.2 | 4.6 | 6.3 | 4.9 | |
| ≥4 + 4 | 451 | 79.4 | 2.2 | 11.3 | 7.1 | |
| Lymph node metastasis | <0.0001 | |||||
| N0 | 5366 | 87.3 | 4.4 | 4.8 | 3.5 | |
| N+ | 544 | 81.4 | 2.6 | 9.0 | 7.0 | |
| Preoperative PSA level (ng/ml) | 0.4243 | |||||
| <4 | 1154 | 86.9 | 4.2 | 5.0 | 3.9 | |
| 4–10 | 5662 | 88.1 | 4.0 | 5.1 | 2.8 | |
| 10–20 | 1900 | 89.1 | 3.4 | 4.4 | 3.1 | |
| >20 | 664 | 88.9 | 3.3 | 4.4 | 3.5 | |
| Surgical margin | 0.0004 | |||||
| negative | 7506 | 88.8 | 3.7 | 4.7 | 2.7 | |
| positive | 1795 | 85.7 | 4.0 | 6.0 | 4.3 | |
Association with TMPRSS2:ERG Fusion Status and ERG Protein Expression
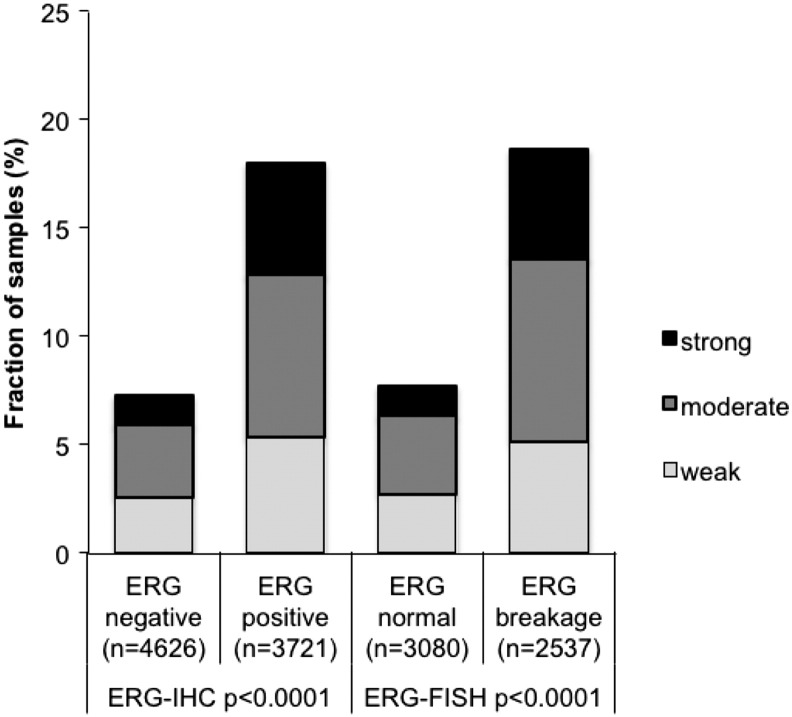
To evaluate whether ADAM15 expression is associated with ERG status in prostate cancers, we used data from previous studies (expanded from [19], [25]). Data on TMPRSS2:ERG fusion status obtained by FISH were available from 5617 and by immunohistochemistry from 8347 tumors with evaluable ADAM15 immunostaining. Data on both ERG FISH and IHC were available from 5401 cancers, and an identical result (ERG IHC positive and break by FISH or ERG IHC negative and missing break by FISH) was found in 5159 of 5617 (91.8%) cancers. ADAM15 staining was more frequent in TMPRSS2:ERG rearranged and ERG-positive prostate cancer. Positive ADAM15 immunostaining was seen in 18.7% (ERG IHC) and 19.7% of ERG FISH positive cancers, but only in 7.8% and 7.9% of cancers without ERG staining and ERG rearrangement (P < .0001, each; Figure 2).
Figure 2.
Association between increasing ADAM15 immunostaining and ERG status (IHC/FISH) in all cancers.
Association to Other Key Genomic Deletions
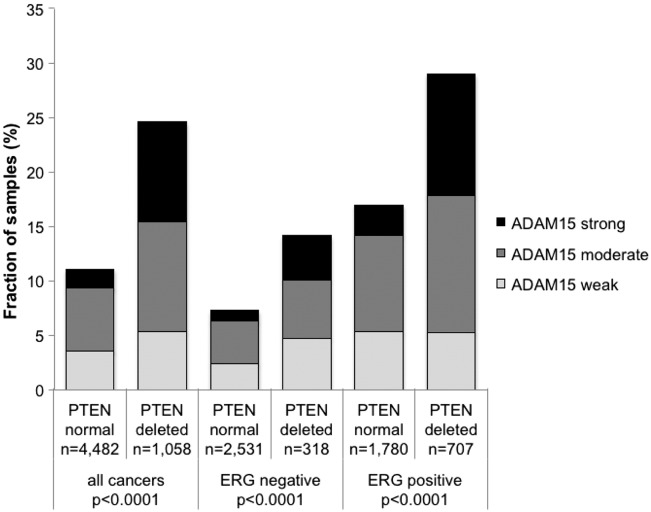
Earlier studies had provided evidence for distinct molecular subgroups of prostate cancers defined by TMPRSS2:ERG fusions and several genomic deletions. We and others had previously described a strong link of PTEN and 3p13 deletions to ERG positivity and of 5q21 and 6q15 deletions to ERG negativity [21], [22], [23], [24]. To examine, whether ADAM15 expression might be particularly associated with one of these genomic deletions, ADAM15 data were compared to the preexisting findings on PTEN (10q23), FOXP1 (3p13), MAP3K7 (6q15) and CHD1 (5q21). Elevated ADAM15 expression levels was strongly linked to PTEN deletions in both ERG-negative and ERG-positive cancers (P < .0001 each, Figure 3). ADAM15 was unrelated to the other genomic deletions (data not shown).
Figure 3.
Association between ADAM15 localization and 10q23 (PTEN) deletion in all cancers, ERG-negative and ERG-positive cancers.
Association with PSA Recurrence
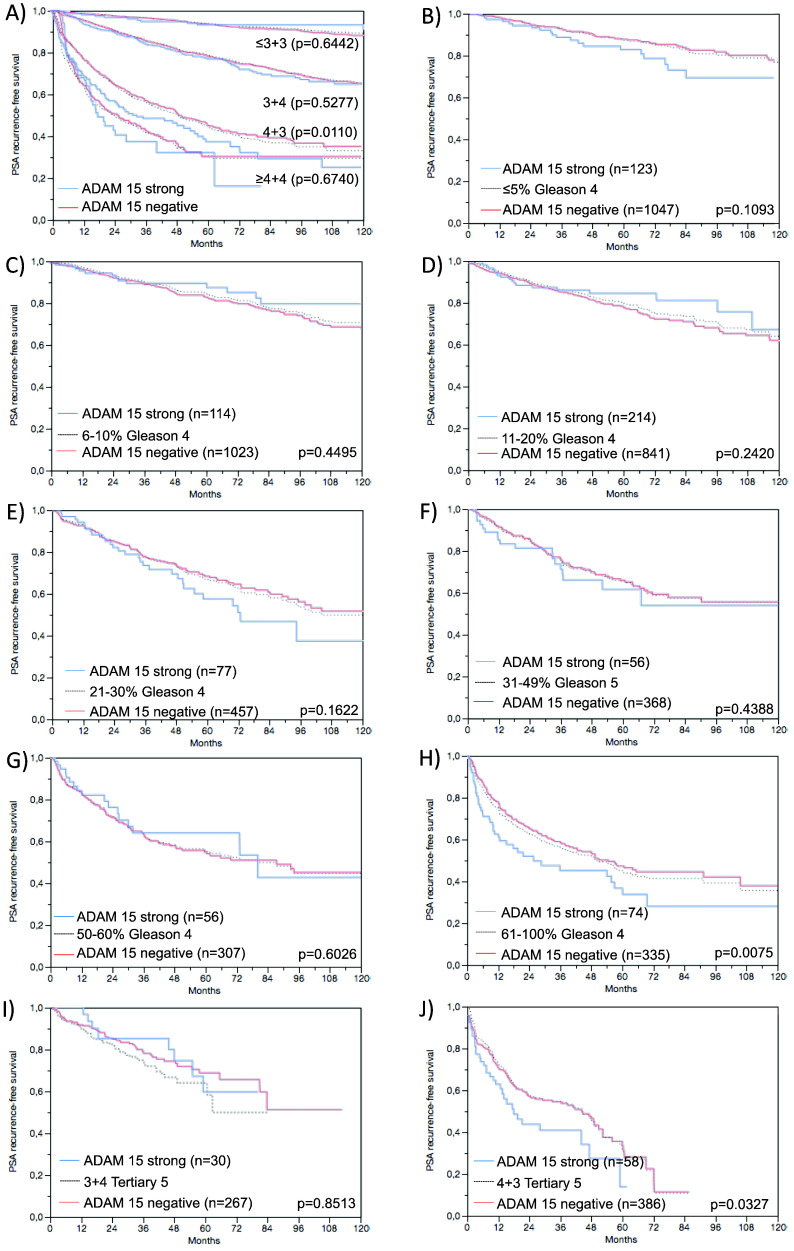
Follow-up data were available for 8890 patients with interpretable ADAM15 immunostaining on the TMA. A highly significant association was seen between early PSA recurrence and elevated ADAM15 expression (Figure 4A, P < .0001) in all tumors and also in subgroups of ERG fusion negative and positive cancers (P < .0001, P = .0002); (Figure 4B and C). Because of the strong association between ADAM15 expression and PTEN deletions, we sought to clarify the prognostic value of coalterations. To facilitate the analysis, tumors with negative, weak or moderate ADAM15 expression were grouped together based on their comparable prognosis (see Figure 4, A–C). It showed that the prognostic value of ADAM15 was limited to cancers lacking PTEN deletion. However, it was remarkable that tumors with strong ADAM15 expression had a comparably poor prognosis than cancers harboring PTEN deletions (Figure 4D). Because of the strong association between ADAM15 expression and Gleason score we performed additional subset analyses in cancers with identical classical and quantitative Gleason grade [16]. These analyses showed that ADAM15 provided independent prognostic impact in subsets of cancers with extended Gleason 4 patterns, including classical 4 + 3 cancers (Figure 5A) and also 4 + 3/4 + 4 cancers with 61–100% Gleason 4 patterns and Gleason 4 + 3 tertiary grade 5 cancers according to the quantitative Gleason grade (5 h, P = .0195; 5 j, P = .0327).
Figure 4.
(A) Relationship of Gleason categories with Gleason 7 separated into 3 + 4 and 4 + 4 with biochemical recurrence, (B) “quantitative” Gleason with patient groups defined by the fraction of Gleason 4. Association between ADAM15 expression and biochemical recurrence in (C) all cancers, (D) ERG fusion negative cancers and (C) ERG fusion positive cancers. (D) Prognostic impact depending on patterns of coalterations of ADAM15 and PTEN. ADAM15 “low” indicates tumors with negative, weak or moderate expression; ADAM15 “high” indicates tumors with strong expression.
Figure 5.
Prognostic impact of ADAM 15 expression in subsets of cancers defined by the Gleason score. (A) Impact of negative (red line) and strongly positive (blue line) ADAM 15 expression as compared to the classical Gleason score categories (B–H) (indicated by black dotted lines). Although survival curves of cancers with weak and moderate ADAM15 are not displayed to facilitate inspection of the figure, these cancers were included to calculate the indicated p-values. Impact of negative (red line) and strongly positive (blue line) ADAM 15 expression as compared to the quantitative Gleason score categories (black dotted lines) defined by subsets of cancers with (B) ≤5% Gleason 4 patterns, (C) 6–10% Gleason4 patterns, (D) 11–20% Gleason 4 patterns, (E) 21–30% Gleason 4 patterns, (F) 31–49% Gleason 4 patterns, (G) 50–60% Gleason 4 patterns, (H) 61–100% Gleason 4 patterns. (I and J) Impact of negative (red line) and strongly positive (blue line) ADAM 15 expression in cancers with a tertiary Gleason 5 pattern, including (I) 3 + 4 tertiary grade 5 and (J) 4 + 3 tertiary grade 5.
Multivariate Analysis
Four different types of multivariate analyses were performed evaluating the clinical relevance of ADAM15 expression in different scenarios (Table 3). Scenario 1 evaluated all postoperatively available parameters including pathological tumor stage, pathological lymph node stage (pN), surgical margin status, preoperative PSA value and pathological Gleason grade obtained after the morphological evaluation of the entire resected prostate. In scenario 2, all postoperatively available parameters with exception of nodal status were included. The rational for this approach was that the indication and extent of lymph node dissection is not standardized in the surgical therapy of prostate cancer and that excluding pN in multivariate analysis can markedly increase case numbers. Two additional scenarios had the purpose to model the preoperative situation as much as possible. Scenario 3 included ADAM15 expression, preoperative PSA, clinical tumor stage (cT stage) and Gleason grade obtained on the prostatectomy specimen. Since postoperative determination of a tumors Gleason grade is “better” than the preoperatively determined Gleason grade (subjected to sampling errors and consequently under-grading in more than one third of cases [26]), another multivariate analysis was added. In scenario 4, the preoperative Gleason grade obtained on the original biopsy was combined with preoperative PSA, cT stage and ADAM15 expression. These analyses suggested that ADAM15 expression might have prognostic value in preclinical scenarios, especially in ERG-negative cancers (Table 3). The overwhelming prognostic impact of the Gleason grade in comparison with ADAM15 expression is demonstrated in Figs. 5 a-l. These panels indicate that ADAM15 expression has no relevant prognostic effect in cancers with identical classical or quantitative Gleason grades.
Table 3.
Multivariate Cox Regression Analysis Including Established Prognostic Parameters and the ADAM15 Localization in All Prostate Cancers, the ERG-Negative- and ERG-Positive Subset. Scenario 1 Includes all Postoperatively Available Parameters (Pathological Tumor (pT) Stage, Lymph Node (pN), Surgical Margin (R) Status, Preoperative PSA Value and Gleason Grade Obtained After the Morphological Evaluation of the Entire Resected Prostate. Scenario 2 Excluded the Nodal Status from Analysis. Scenario 3 Included Preoperative PSA, Clinical Tumor (cT) Stage and Gleason Grade Obtained on the Prostatectomy Specimen. In Scenario 4, the Preoperative Gleason Grade Obtained on the Original Biopsy was Combined with Preoperative PSA, and cT Stage. P Values in Brackets Indicate That the Quantitative Gleason was Used Instead of the Classical Gleason for Multivariate Modeling
| Tumor subset | Scenario | N analyzable (analyzable qGleason) |
P value |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| preoperative PSA-Level | pT Stage | cT Stage | Gleason grade RPE | Gleason grade biopsy | pN Stage | R Stage | ADAM 15 Expression | |||
| all cancers | 1 | 5273 (4796) | <.0001 (<.0001) | <.0001 (<.0001) | - | <.0001 (<.0001) | - | <.0001 (.0019) | .0019 (.0013) | .4609 (.5599) |
| 2 | 8682 (7920) | <.0001 (<.0001) | <.0001 (<.0001) | - | <.0001 (<.0001) | - | - | <.0001 (<.0001) | .1800 (.3601) | |
| 3 | 8542 (7846) | <.0001 (<.0001) | - | <.0001 (<.0001) | <.0001 (<.0001) | - | - | - | .0015 (.0178) | |
| 4 | 8239 (1146) | <.0001 (.1273) | - | <.0001 (.7135) | - | <.0001 (<.0001) | - | - | <.0001 (.8263) | |
| ERG-negative cancers | 1 | 2642 (2422) | .0061/(.0091) | <.0001 (<.0001) | - | <.0001 (<.0001) | - | .0003 (.0157) | .0751 (.3424) | .1976 (.5569) |
| 2 | 4267 (3901) | <.0001 (<.0001) | <.0001 (<.0001) | - | <.0001 (<.0001) | - | - | .0008 (.0082) | .0999 (.4653) | |
| 3 | 4223 (3881) | <.0001 (<.0001) | - | <.0001 (<.0001) | <.0001 (<.0001) | - | - | - | .0331 (.1945) | |
| 4 | 4158 (591) | <.0001 (.5560) | - | <.0001 (.6150) | - | <.0001 (<.0001) | - | - | .0035 (.4171) | |
| ERG-positive cancers | 1 | 2108 (1909) | .0043/(.0085) | <.0001 (<.0001) | - | <.0001 (<.0001) | - | .0499 (.1997) | .0033 (.0024) | .6137 (.7265) |
| 2 | 3421 (3091) | .0001/(.0009) | <.0001 (<.0001) | - | <.0001 (<.0001) | - | - | <.0001 (<.0001) | .4555 (.5605) | |
| 3 | 3338 (3040) | <.0001 (<.0001) | - | <.0001 (.0010) | <.0001 (<.0001) | - | - | - | .0608 (.1960) | |
| 4 | 3282 (565) | <.0001 (.4325) | - | <.0001 (.6217) | - | <.0001 (<.0001) | - | - | .1105 (.3435) | |
Radical prostatectomy RPE.
Discussion
The results of this study demonstrate that high level overexpression of ADAM15 identifies a small subset of aggressive prostate cancers with increased risk for PSA recurrence.
Our immunohistochemical analysis revealed membranous ADAM15 staining of variable intensity - typically located at the apical cell pole - in a small fraction (12%) of our 9826 analyzable prostate cancers, while normal prostate epithelium and other non-malignant cells were negative under the selected experimental conditions. Absence of ADAM15 expression in normal prostatic epithelium was also described in one earlier study [15]. High-level ADAM15 staining was strongly linked to adverse phenotypical features of prostate cancer and early PSA recurrence in our study. Similar findings were made by Kuefer et al. [15] using a different scoring scheme that does not disclose the fraction of positive cancer samples. That a statistical association was not found in this study might be due to a relatively low number of cases (n = 167).
Overall, these findings from our study provide strong in vivo evidence for a role of ADAM15 expression in development and progression of prostate cancer. This assumption is strongly supported by earlier in vivo evidence from functional studies using prostate cancer cell lines. These data had suggested that ADAM15 overexpression contributes to the metastatic cascade by prostate tumor cell interaction with vascular endothelium and thus facilitating blood vessel invasion [14].
The large number of cancers included in our study enabled us to investigate possible associations of ADAM15 expression with previously established molecular key features of prostate cancer. Our data demonstrate a strong link of ADAM15 expression to ERG activation. In about 50% of tumors, fusion of the androgen receptor-regulated TMPRSS2 gene with ERG leads to AR-stimulated overexpression of ERG, an ETS-transcription factor [27]. ERG is a pioneering factor that modulates transcription of more than 1600 genes [28], including many AR regulated genes as it opens cryptic AR binding sites in the vicinity of its own recognition site when bound to gene promoters [29]. Cell line models suggesting AR-dependent ADAM15 expression [30] provide a possible mechanistic explanation for the strong association between ADAM15 expression and ERG fusion, which was also observed by others [31].
Deletions of certain small and large chromosomal regions represent another hallmark of prostate cancer. Data from next generation sequencing studies demonstrate that such deletions are more prevalent than any mutations of specific coding genes and many of these deletions have been linked to either ERG-positive (i.e. PTEN and 3p13) or ERG-negative cancers (i.e. 6q15 and 5q23). That high ADAM15 expression is tightly linked to PTEN deletions, but not to any other of the studied deletions suggests a specific functional relationship of ADAM15 and PTEN. It is tempting to speculate that a PTEN/ADAM15 interaction could affect cell growth control as both proteins are involved in this process. ADAM15 is believed to liberate membrane bound growth factors such as Heparin-binding EGF-like growth factor and amphiregulin [13], [32], as well as extracellular fragments of E-cadherin that function as activators of the EGF growth factor signaling pathways including Akt/PTEN [14]. Interestingly, some data from mouse models suggest that loss of PTEN alone is not sufficient for invasive prostate cancer growth, but requires up-regulation of additional mechanisms driving cell motility [33], [34]. It appears thus possible, that the combination of PTEN loss and ADAM15 overexpression contributes to both endogenous growth stimulation and increased invasiveness.
ADAMs gained substantial interest as targets for anti-cancer therapies. Several first- and second-generation inhibitors, such as marimastat, prinomastat, tanomastat, or batimastat, are active against a broad spectrum of matrix metalloproteases (MMPs) and ADAMs. However, disappointing results were made in clinical trials, where severe side effects were observed [35], [36], [37], [38]. These might be connected to the fact that MMPs and ADAMs are involved in many physiological processes involving remodeling of the extracellular matrix, e. g. inflammation, wound healing, tissue repair or the menstrual cycle [39], [40], [41], [42]. It is hoped that better results could be obtained with more specific inhibitors. For example, an inhibitory humanized monoclonal antibody targeting ADAM17, a potent and independent predictor of disease outcome in patients with breast cancer [43], showed promising anticancer effects in vitro [44]. A dual ADAM10/ADAM17 inhibitor was well tolerated in a phase I trial in patients with HER2-positive advanced breast cancer. In particular, there was no evidence of musculoskeletal side effects (which were previously found with matrix metalloproteinase inhibitors), increased release of liver enzymes, bone marrow toxicity or increase in cardiomyopathy [45], [46]. However, ADAM15-specific inhibitors have not been reported as to yet.
Although ADAM15 expression was a highly significant prognostic factor – comparable to PTEN deletion - in univariate calculations, its prognostic impact was lost in most multivariate analyses. The power of morphological methods competing with biomarkers for predicting prostate cancer aggressiveness is best demonstrated by the separate analysis of different prognostic Gleason groups. Already within traditional grade groups, a prognostic impact of ADAM15 expression was only found in Gleason 4 + 3 tumors. Based on the large cohort of prostate cancers available at our institution, we had recently shown, that prognostic Gleason Grade information can be refined by using the percentage of Gleason 4 grades as a continuous variable. Both in biopsies and in prostatectomy samples, prostate cancer prognosis continuously deteriorates with increasing percentage of Gleason 4 pattern (quantitative Gleason Grade) [16]. That ADAM15 expression lacks significant prognostic impact in almost all subgroups defined by a comparable quantitative Gleason grade demonstrates how difficult it is for biomarkers to beat morphological malignancy parameters in prostate cancer.
Conclusion
ADAM15 is overexpressed in a small fraction of prostate cancers, and linked to unfavorable histological features and poor outcome of the disease. Our multivariate modeling approach suggests, however, that the prognostic value of ADAM15 may be limited to clinical situations were definite histological parameters (such as the tumor stage and Gleason score after radical prostatectomy) are not available.
Acknowledgement
We thank Janett Lüttgens, Sünje Seekamp, and Inge Brandt for excellent technical assistance.
Footnotes
Disclosure/Conflict of interest: There are no proprietary interests and no financial support was received. No conflicts of interest regarding the article exist.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.neo.2017.01.005.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
Apical cell membrane staining of ADAM15 at 400× magnification in (A) moderate and (B) strongly stained spot.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Wilt TJ, Brawer MK, Jones KM, Barry MJ, Aronson WJ, Fox S, Gingrich JR, Wei JT, Gilhooly P, Grob BM. Radical prostatectomy versus observation for localized prostate cancer. N Engl J Med. 2012;367(3):203–213. doi: 10.1056/NEJMoa1113162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thompson IM, Jr., Tangen CM. Prostate cancer--uncertainty and a way forward. N Engl J Med. 2012;367(3):270–271. doi: 10.1056/NEJMe1205012. [DOI] [PubMed] [Google Scholar]
- 4.Lucas N, Day ML. The role of the disintegrin metalloproteinase ADAM15 in prostate cancer progression. J Cell Biochem. 2009;106(6):967–974. doi: 10.1002/jcb.22087. [DOI] [PubMed] [Google Scholar]
- 5.Edwards DR, Handsley MM, Pennington CJ. The ADAM metalloproteinases. Mol Aspects Med. 2008;29(5):258–289. doi: 10.1016/j.mam.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baren JP, Stewart GD, Stokes A, Gray K, Pennington CJ, O'Neill R, Deans DA, Paterson-Brown S, Riddick AC, Edwards DR. mRNA profiling of the cancer degradome in oesophago-gastric adenocarcinoma. Br J Cancer. 2012;107(1):143–149. doi: 10.1038/bjc.2012.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carl-McGrath S, Lendeckel U, Ebert M, Roessner A, Rocken C. The disintegrin-metalloproteinases ADAM9, ADAM12, and ADAM15 are upregulated in gastric cancer. Int J Oncol. 2005;26(1):17–24. [PubMed] [Google Scholar]
- 8.Schutz A, Hartig W, Wobus M, Grosche J, Wittekind C, Aust G. Expression of ADAM15 in lung carcinomas. Virchows Arch. 2005;446(4):421–429. doi: 10.1007/s00428-004-1193-z. [DOI] [PubMed] [Google Scholar]
- 9.Dong DD, Zhou H, Li G. ADAM15 targets MMP9 activity to promote lung cancer cell invasion. Oncol Rep. 2015;34(5):2451–2460. doi: 10.3892/or.2015.4203. [DOI] [PubMed] [Google Scholar]
- 10.Toquet C, Colson A, Jarry A, Bezieau S, Volteau C, Boisseau P, Merlin D, Laboisse CL, Mosnier JF. ADAM15 to alpha5beta1 integrin switch in colon carcinoma cells: a late event in cancer progression associated with tumor dedifferentiation and poor prognosis. Int J Cancer. 2012;130(2):278–287. doi: 10.1002/ijc.25891. [DOI] [PubMed] [Google Scholar]
- 11.Ungerer C, Doberstein K, Burger C, Hardt K, Boehncke WH, Bohm B, Pfeilschifter J, Dummer R, Mihic-Probst D, Gutwein P. ADAM15 expression is downregulated in melanoma metastasis compared to primary melanoma. Biochem Biophys Res Commun. 2010;401(3):363–369. doi: 10.1016/j.bbrc.2010.09.055. [DOI] [PubMed] [Google Scholar]
- 12.White JM. ADAMs: modulators of cell–cell and cell-matrix interactions. Curr Opin Cell Biol. 2003;15(5):598–606. doi: 10.1016/j.ceb.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 13.Schafer B, Marg B, Gschwind A, Ullrich A. Distinct ADAM metalloproteinases regulate G protein-coupled receptor-induced cell proliferation and survival. J Biol Chem. 2004;279(46):47929–47938. doi: 10.1074/jbc.M400129200. [DOI] [PubMed] [Google Scholar]
- 14.Najy AJ, Day KC, Day ML. ADAM15 supports prostate cancer metastasis by modulating tumor cell-endothelial cell interaction. Cancer Res. 2008;68(4):1092–1099. doi: 10.1158/0008-5472.CAN-07-2432. [DOI] [PubMed] [Google Scholar]
- 15.Kuefer R, Day KC, Kleer CG, Sabel MS, Hofer MD, Varambally S, Zorn CS, Chinnaiyan AM, Rubin MA, Day ML. ADAM15 disintegrin is associated with aggressive prostate and breast cancer disease. Neoplasia. 2006;8(4):319–329. doi: 10.1593/neo.05682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sauter G, Steurer S, Clauditz TS, Krech T, Wittmer C, Lutz F, Lennartz M, Janssen T, Hakimi N, Simon R. Clinical Utility of Quantitative Gleason Grading in Prostate Biopsies and Prostatectomy Specimens. Eur Urol. 2016;69(4):592–598. doi: 10.1016/j.eururo.2015.10.029. [DOI] [PubMed] [Google Scholar]
- 17.Schlomm T, Iwers L, Kirstein P, Jessen B, Kollermann J, Minner S, Passow-Drolet A, Mirlacher M, Milde-Langosch K, Graefen M. Clinical significance of p53 alterations in surgically treated prostate cancers. Mod Pathol. 2008;21(11):1371–1379. doi: 10.1038/modpathol.2008.104. [DOI] [PubMed] [Google Scholar]
- 18.Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4(7):844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 19.Weischenfeldt J, Simon R, Feuerbach L, Schlangen K, Weichenhan D, Minner S, Wuttig D, Warnatz HJ, Stehr H, Rausch T. Integrative genomic analyses reveal an androgen-driven somatic alteration landscape in early-onset prostate cancer. Cancer Cell. 2013;23(2):159–170. doi: 10.1016/j.ccr.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 20.Minner S, Wittmer C, Graefen M, Salomon G, Steuber T, Haese A, Huland H, Bokemeyer C, Yekebas E, Dierlamm J. High level PSMA expression is associated with early PSA recurrence in surgically treated prostate cancer. Prostate. 2011;71(3):281–288. doi: 10.1002/pros.21241. [DOI] [PubMed] [Google Scholar]
- 21.Burkhardt L, Fuchs S, Krohn A, Masser S, Mader M, Kluth M, Bachmann F, Huland H, Steuber T, Graefen M. CHD1 is a 5q21 tumor suppressor required for ERG rearrangement in prostate cancer. Cancer Res. 2013;73(9):2795–2805. doi: 10.1158/0008-5472.CAN-12-1342. [DOI] [PubMed] [Google Scholar]
- 22.Kluth M, Hesse J, Heinl A, Krohn A, Steurer S, Sirma H, Simon R, Mayer PS, Schumacher U, Grupp K. Genomic deletion of MAP3K7 at 6q12-22 is associated with early PSA recurrence in prostate cancer and absence of TMPRSS2:ERG fusions. Mod Pathol. 2013;26(7):975–983. doi: 10.1038/modpathol.2012.236. [DOI] [PubMed] [Google Scholar]
- 23.Krohn A, Diedler T, Burkhardt L, Mayer PS, De Silva C, Meyer-Kornblum M, Kotschau D, Tennstedt P, Huang J, Gerhauser C. Genomic deletion of PTEN is associated with tumor progression and early PSA recurrence in ERG fusion-positive and fusion-negative prostate cancer. Am J Pathol. 2012;181(2):401–412. doi: 10.1016/j.ajpath.2012.04.026. [DOI] [PubMed] [Google Scholar]
- 24.Krohn A, Seidel A, Burkhardt L, Bachmann F, Mader M, Grupp K, Eichenauer T, Becker A, Adam M, Graefen M. Recurrent deletion of 3p13 targets multiple tumour suppressor genes and defines a distinct subgroup of aggressive ERG fusion-positive prostate cancers. J Pathol. 2013;231(1):130–141. doi: 10.1002/path.4223. [DOI] [PubMed] [Google Scholar]
- 25.Minner S, Enodien M, Sirma H, Luebke AM, Krohn A, Mayer PS, Simon R, Tennstedt P, Muller J, Scholz L. ERG status is unrelated to PSA recurrence in radically operated prostate cancer in the absence of antihormonal therapy. Clin Cancer Res. 2011;17(18):5878–5888. doi: 10.1158/1078-0432.CCR-11-1251. [DOI] [PubMed] [Google Scholar]
- 26.Epstein JI, Feng Z, Trock BJ, Pierorazio PM. Upgrading and downgrading of prostate cancer from biopsy to radical prostatectomy: incidence and predictive factors using the modified Gleason grading system and factoring in tertiary grades. Eur Urol. 2012;61(5):1019–1024. doi: 10.1016/j.eururo.2012.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310(5748):644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 28.Brase JC, Johannes M, Mannsperger H, Falth M, Metzger J, Kacprzyk LA, Andrasiuk T, Gade S, Meister M, Sirma H. TMPRSS2-ERG -specific transcriptional modulation is associated with prostate cancer biomarkers and TGF-beta signaling. BMC Cancer. 2011;11:507. doi: 10.1186/1471-2407-11-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cai C, Wang H, He HH, Chen S, He L, Ma F, Mucci L, Wang Q, Fiore C, Sowalsky AG. ERG induces androgen receptor-mediated regulation of SOX9 in prostate cancer. J Clin Invest. 2013;123(3):1109–1122. doi: 10.1172/JCI66666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garritano S, Romanel A, Ciribilli Y, Bisio A, Gavoci A, Inga A, Demichelis F. In-silico identification and functional validation of allele-dependent AR enhancers. Oncotarget. 2015;6(7):4816–4828. doi: 10.18632/oncotarget.3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18(1):11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hart S, Fischer OM, Prenzel N, Zwick-Wallasch E, Schneider M, Hennighausen L, Ullrich A. GPCR-induced migration of breast carcinoma cells depends on both EGFR signal transactivation and EGFR-independent pathways. Biol Chem. 2005;386(9):845–855. doi: 10.1515/BC.2005.099. [DOI] [PubMed] [Google Scholar]
- 33.King JC, Xu J, Wongvipat J, Hieronymus H, Carver BS, Leung DH, Taylor BS, Sander C, Cardiff RD, Couto SS. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet. 2009;41(5):524–526. doi: 10.1038/ng.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carver BS, Tran J, Gopalan A, Chen Z, Shaikh S, Carracedo A, Alimonti A, Nardella C, Varmeh S, Scardino PT. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41(5):619–624. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore MJ, Hamm J, Dancey J, Eisenberg PD, Dagenais M, Fields A, Hagan K, Greenberg B, Colwell B, Zee B. Comparison of gemcitabine versus the matrix metalloproteinase inhibitor BAY 12-9566 in patients with advanced or metastatic adenocarcinoma of the pancreas: a phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J Clin Oncol. 2003;21(17):3296–3302. doi: 10.1200/JCO.2003.02.098. [DOI] [PubMed] [Google Scholar]
- 36.Nuti E, Tuccinardi T, Rossello A. Matrix metalloproteinase inhibitors: new challenges in the era of post broad-spectrum inhibitors. Curr Pharm Des. 2007;13(20):2087–2100. doi: 10.2174/138161207781039706. [DOI] [PubMed] [Google Scholar]
- 37.Mant TG, Bradford D, Amin DM, Pisupati J, Kambayashi Y, Yano Y, Tanaka K, Yamada-Sawada T. Pharmacokinetics and safety assessments of high-dose and 4-week treatment with S-3304, a novel matrix metalloproteinase inhibitor, in healthy volunteers. Br J Clin Pharmacol. 2007;63(5):512–526. doi: 10.1111/j.1365-2125.2006.02794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nuti E, Cantelmo AR, Gallo C, Bruno A, Bassani B, Camodeca C, Tuccinardi T, Vera L, Orlandini E, Nencetti S. N-O-Isopropyl Sulfonamido-Based Hydroxamates as Matrix Metalloproteinase Inhibitors: Hit Selection and in Vivo Antiangiogenic Activity. J Med Chem. 2015;58(18):7224–7240. doi: 10.1021/acs.jmedchem.5b00367. [DOI] [PubMed] [Google Scholar]
- 39.van der Vorst EP, Keijbeck AA, de Winther MP, Donners MM. A disintegrin and metalloproteases: molecular scissors in angiogenesis, inflammation and atherosclerosis. Atherosclerosis. 2012;224(2):302–308. doi: 10.1016/j.atherosclerosis.2012.04.023. [DOI] [PubMed] [Google Scholar]
- 40.Schonefuss A, Abety AN, Zamek J, Mauch C, Zigrino P. Role of ADAM-15 in wound healing and melanoma development. Exp Dermatol. 2012;21(6):437–442. doi: 10.1111/j.1600-0625.2012.01490.x. [DOI] [PubMed] [Google Scholar]
- 41.Moali C, Hulmes DJ. Extracellular and cell surface proteases in wound healing: new players are still emerging. Eur J Dermatol. 2009;19(6):552–564. doi: 10.1684/ejd.2009.0770. [DOI] [PubMed] [Google Scholar]
- 42.Thathiah A, Carson DD. MT1-MMP mediates MUC1 shedding independent of TACE/ADAM17. Biochem J. 2004;382(Pt 1):363–373. doi: 10.1042/BJ20040513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGowan PM, Duffy MJ. Matrix metalloproteinase expression and outcome in patients with breast cancer: analysis of a published database. Ann Oncol. 2008;19(9):1566–1572. doi: 10.1093/annonc/mdn180. [DOI] [PubMed] [Google Scholar]
- 44.Caiazza F, McGowan PM, Mullooly M, Murray A, Synnott N, O'Donovan N, Flanagan L, Tape CJ, Murphy G, Crown J. Targeting ADAM-17 with an inhibitory monoclonal antibody has antitumour effects in triple-negative breast cancer cells. Br J Cancer. 2015;112(12):1895–1903. doi: 10.1038/bjc.2015.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Infante JR, Yardley DA, Burris HA, 3rd, Greco FA, Farley CP, Webb C, Spigel DR, Hainsworth JD. Phase II trial of weekly docetaxel, vinorelbine, and trastuzumab in the first-line treatment of patients with HER2-positive metastatic breast cancer. Clin Breast Cancer. 2009;9(1):23–28. doi: 10.3816/CBC.2009.n.004. [DOI] [PubMed] [Google Scholar]
- 46.Fridman JS, Caulder E, Hansbury M, Liu X, Yang G, Wang Q, Lo Y, Zhou BB, Pan M, Thomas SM. Selective inhibition of ADAM metalloproteases as a novel approach for modulating ErbB pathways in cancer. Clin Cancer Res. 2007;13(6):1892–1902. doi: 10.1158/1078-0432.CCR-06-2116. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Apical cell membrane staining of ADAM15 at 400× magnification in (A) moderate and (B) strongly stained spot.