Abstract
Klebsiella pneumoniae is a common opportunistic pathogen causing nosocomial infections. One of the main virulence determinants of K. pneumoniae is the type 3 pilus (T3P). T3P helps the bacterial interaction to both abiotic and biotic surfaces and it is crucial for the biofilm formation. T3P is genetically organized in three transcriptional units: the mrkABCDF polycistronic operon, the mrkHI bicistronic operon and the mrkJ gene. MrkH is a regulatory protein encoded in the mrkHI operon, which positively regulates the mrkA pilin gene and its own expression. In contrast, the H-NS nucleoid protein represses the transcriptional expression of T3P. Here we reported that MrkH and H-NS positively and negatively regulate mrkJ expression, respectively, by binding to the promoter of mrkJ. MrkH protein recognized a sequence located at position -63.5 relative to the transcriptional start site of mrkJ gene. Interestingly, our results show that, in addition to its known function as classic transcriptional activator, MrkH also positively controls the expression of mrk genes by acting as an anti-repressor of H-NS; moreover, our results support the notion that high levels of MrkH repress T3P expression. Our data provide new insights about the complex regulatory role of the MrkH protein on the transcriptional control of T3P in K. pneumoniae.
Introduction
Klebsiella pneumoniae is an opportunistic Gram-negative bacterium causing nosocomial infections ranging from pneumonia and urinary tract infections to septicemia and pyogenic liver abscesses [1–6]. Several virulence determinants of K. pneumoniae have been described: capsular polysaccharide, lipopolysaccharide, siderophores and pili [1, 7, 8]. Different types of pili are encoded in the genome of K. pneumoniae such as Type 1 pilus (T1P), Type 3 pilus (T3P) and E. coli common pilus (ECP) [9–12]. In particular, K. pneumoniae T3P mediates adherence to renal tubular cells and cells of the respiratory tract such as tracheal epithelial cells, and basolateral surfaces of lung tissue, which is crucial for biofilm formation [13–17].
T3P is genetically organized in three transcriptional units: the mrkABCDF polycistronic operon, the mrkHI bicistronic operon and the mrkJ gene. The biogenesis of T3P is dependent on the mrkABCDF operon expression [18, 19]. The filament is composed of the major pilus subunit MrkA and the tip adhesion protein MrkD [8]. MrkH is a regulatory protein encoded in the mrkHI operon, which positively regulates the mrkA pilin gene and its own expression [20–22]. MrkH protein contains a PilZ domain, whose interaction with c-di-GMP is crucial for its role as a transcriptional activator [23]. The mrkHI operon also codes for MrkI, a LuxR-type transcriptional regulator reported to act as a co-activator for the expression of mrkA [20, 24]. The mrkJ gene encodes a phosphodiesterase that degrades c-di-GMP, which in turn, controls the MrkH activity [25].
In addition to MrkH, global regulators such as the H-NS nucleoid protein also control the T3P expression [26]. H-NS is a DNA-binding protein, which plays a dual role as an architectural protein component of the nucleoid and as a global regulator of bacterial gene expression [27, 28]. H-NS affects bacterial evolution by directly repressing the expression of AT-rich DNA (i.e. pathogenicity islands) acquired by horizontal transfer events, thus facilitating tolerance of these foreign sequences, which allows their integration into pre-existing regulatory networks [29–31]. H-NS differentially regulates the transcriptional expression of T3P: represses mrkHI/mrkJ and activates mrkA [26].
In this work we reported that the mrkJ gene is directly activated and repressed by MrkH and H-NS, respectively. A sequence located at position -63.5 relative to the transcriptional start site of mrkJ gene was recognized by the MrkH protein. Furthermore, we found that MrkH induces the expression of mrkJ, as well as that of mrkI, by dual regulation: it antagonizes H-NS-mediated repression on these genes and also acts as a transcriptional activator. Moreover, our results support the notion that MrkH can also act as a transcriptional repressor of mrk genes. Overall, our data provides new insights on the complex regulatory function of MrkH protein on the transcriptional control of T3P in K. pneumoniae.
Materials and methods
Bacterial strains and culture conditions
Bacterial strains and plasmids used in this study are listed in Table 1. Bacterial cultures were grown in Luria-Bertani (LB) broth until exponential phase (OD600nm = 0.8) was reached. Cultures were grown overnight at 37°C shaken at 160 rpm, with or without antibiotics [200 μg/ml (ampicillin), 50 μg/ml (kanamycin), 30 μg/ml (chloramphenicol) or 10 μg/ml (tetracycline)]. MrkH production from pT6-MrkH plasmid was induced with different L(+)-arabinose concentrations under several genetic backgrounds.
Table 1. Bacterial strains and plasmids used in this study.
| Strain or plasmid | Genotype or description | Reference or source |
|---|---|---|
| K. pneumoniae strains | ||
| Kpn 123/01 | WT, serotype K39 | [26] |
| Kpn hns | Δhns::KmR | [26] |
| Kpn mrkH | ΔmrkH::KmR | This study |
| Kpn mrkH | ΔmrkH::FRT | This study |
| Kpn hns mrkH | Δhns::KmR ΔmrkH::CmR | This study |
| Kpn mrkJ | ΔmrkJ::KmR | This study |
| Kpn hns mrkJ | Δhns::KmR ΔmrkJ::CmR | This study |
| Kpn mrkI | ΔmrkI::KmR | This study |
| Kpn mrkJ* | mrkJ- ΔMrkHbox::FRT | This study |
| Plasmids | ||
| pMPM-T6 | p15A derivative cloning vector, pBAD (ara) promoter, TcR | [45] |
| pT6-MrkH | pMPM-T6 derivative expressing MrkH-His6 from the pBAD (ara) promoter | [26] |
| pKK-mrkJ-wt | pKK232-9 derivative containing a mrkJ-cat transcriptional fusion from nucleotides -352 to +33 | This study |
| pKK-mrkJ-mut | pKK-mrkJ-wt derivative containing three point mutations in the TAT motif of the MrkH-binding box | This study |
| pKD119 | pINT-ts derivative containing the λ Red recombinase system under an arabinose-inducible promoter, TcR | [32] |
| pKD4 | pANTsγ derivative template plasmid containing the kanamycin cassette for λ Red recombination, ApR | [32] |
| pKD3 | pANTsγ derivative template plasmid containing the chloramphenicol cassette for λ Red recombination, ApR | [32] |
ApR, ampicillin resistance; KmR, kanamycin resistance; CmR, chloramphenicol resistance; TcR, tetracycline resistance.
Construction of K. pneumoniae mutants and transcriptional fusions
Construction of single and double mutants was performed as previously described [26]. We generated a ΔmrkH mutant, by amplifying a PCR product containing mrkH sequence flanking a kanamycin cassette using the pKD4 plasmid, and using gene-specific primer pairs (Table 2). Kpn mrkJ* mutant was obtained by deletion of the MrkH-box on the mrkJ promoter (mrkJ-ΔMrkHbox::KmR) using Kpn-mrkJΔMrkH-H1P1 and Kpn-mrkJΔMrkH-H2P2 primers (Table 2). The FRT-flanked Km cassette was excised from strains ΔmrkH and ΔmrkJ* after transformation with pCP20, as described previously [32]. For Δhns ΔmrkH and Δhns ΔmrkJ double mutants, K. pneumoniae Δhns was targeted to carry out the mutagenesis of mrkH and mrkJ, amplifying a PCR fragment containing mrkH and mrkJ sequences flanking a chloramphenicol cassette using the pKD3 plasmid. The corresponding mutations were confirmed by PCR and sequencing.
Table 2. Primers used in this study.
| Primer | Sequence (5’-3’) | Target gene |
|---|---|---|
| For qPCR | ||
| cat-F | TGGCAATGAAAGACGGTGAG | cat |
| cat-R | AGAAACTGCCGGAAATCGTC | |
| For mutagenesis | ||
| Kpn-mrkH-H1P1 | CACGACAACTATTTACAAGGGATGCA TATGACAGAGGGAACGATATGTAGGC TGGAGCTGCTTCG | mrkH |
| Kpn-mrkH-H2P2 | GCAATATACTGTCCAAGGTTGTCAGA TTCTCTTTTTGCGCTTGGCCATATGA ATATCCTCCTTAG | |
| Kpn-mrkI-H1P1 | CAAAAAGAGAATCTGACAACCTTGGA CAGTATATTGCTGTACACCTGTAGGC TGGAGCTGCTTCG | mrkI |
| Kpn-mrkI-H2P2 | ACTGATTTACCGGGAGAACATTTAGC ATTGATGGAGAGCGGCAATCATATGA ATATCCTCCTTAG | |
| Kpn-mrkJ-H1P1 | CTAACCTCGTGAAGAGGGATAATGAA CACTAAAATATTCGAAGACTGTAGGC TGGAGCTGCTTCG | mrkJ |
| Kpn-mrkJ-H2P2 | GCCGGGAATTCCCGGCTTTGTTTACA TGGCAATATCATCGGCGACCATATGA ATATCCTCCTTAG | |
| Kpn-mrkJΔMrkH-H1P1 | ATGCTAAATGTTCTCCCGGTAAATCA GTAGCGGATAAAGCGTACTTGTAGGC TGGAGCTGCTTCG | mrkJ |
| Kpn-mrkJΔMrkH-H2P2 | ACCTGATGATTAATGGGAATGGCGGG AAATGTAAATCAACAGCGACATATGA ATATCCTCCTTAG | |
| For mutants characterization | ||
| Kpn-mrkH-F | CTATTGCTATAAGAAAAATCAAAC | mrkH |
| Kpn-mrkH-R | TGATAGATTGAGTGACCAATGAGA | |
| Kpn-mrkI-F | TAGAGAAGATACTGCTGGACCTGA | mrkI |
| Kpn-mrkI-R | GGAATGGCGGGAAATGTAAATCA | |
| Kpn-mrkJ-F | CGCCATTCCCATTAATCATCAGG | mrkJ |
| Kpn-mrkJ-R | TACCAGCTGGGCAACGTG | |
| For constructions | ||
| mrkJ-BamHI-F | ACTGGATCCTCATCTATCGTCCAGCGCGCC | mrkJ |
| mrkJ-BamHI-R | CATAAGCTTTCTTCACGAGGTTAGTCAGAC | |
| mrkJ-mut-F | TACTCGCTCGCTGTTGATTTACATTTCCCGC | |
| mrkJ-mut-R | CGGGAAATGTAAATCAACAGCGAGCGAGTA | |
| For EMSA | ||
| mrkJ-EM-F | ACTGGCCCAGACGATTATTTTC | mrkJ |
| mrkJ-EM-R | TAAAATGTTGTCTTCGAATATTTTAG | |
| mrkH-EM-F | AGGCGCAGGAGTTGAACGAGGTC | mrkH |
| mrkH-EM-R | GGTCTTTATCGTTCCCTCTGTCATATG | |
| mrkA-EM-F | ATGGCGGTTTGATGGCGTAAAC | mrkA |
| mrkA-EM-R | TGCTGCAGAGAGAAGAACCTTTTTC | |
| fbpA-EM-F | TTCCTGACCAGCGAGCTGCCG | fbpA |
| fbpA-EM-R | CCCCAGTACTCCCAGCTGTGC |
Italic letters indicate the respective restriction enzyme site in the primer. The sequence corresponding to the template plasmids pKD4 or pKD3 is underlined.
Regulatory region of mrkJ was amplified using primers mrkJ-BamHI-F and mrkJ-HindIII-R (Table 2). This product was digested with BamHI and HindIII and then ligated into pKK-232-8 (ApR), previously digested with the same restriction enzymes. This plasmid was digested with BamHI and NcoI and the insert was subcloned into pKK-232-9 plasmid (KmR) [33] generating pKK-mrkJ-wt construct. Site-directed mutagenesis was carried out on the pKK-mrkJ-wt plasmid by overlapping PCR with specific primers (mrkJ-mut-F and mrkJ-mut-R) to obtain the pKK-mrkJ-mut using the primers mrkJ-mut-F and mrkJ-mut-R (Table 2). Plasmids were sequenced to verify the integrity of the inserts and the introduction of the point mutations.
Quantitative RT-PCR
Total RNA extraction was performed using the hot phenol method [34]. Purification of RNA and qRT-PCR were performed as previously reported [26]. 16S rRNA was used as a reference gene for normalization and the relative gene expression was calculated using the 2-ΔCt method [35]. Primers for qPCR experiments were previously reported [26], except for cat quantification (Table 2).
MrkH-His6 purification
Purification of MrkH-His6 protein was performed with Ni-nitrilotriacetic acid. Briefly, K. pneumoniae carrying the pT6-MrkH (Table 1) was grown to mid-logarithmic phase. L(+)-arabinose (Sigma-Aldrich) was added to a final concentration of 0.1%, and bacteria were grown for 6 h at 30°C. Cells were then pelleted by centrifugation, resuspended in urea buffer [8M urea, 100mM NaH2PO4, 10mM Tris-HCl (pH 8.0)] and disrupted by sonication. The suspension was centrifuged, and the supernatant was filtered through a Ni-nitrilotriacetic acid agarose column (QIAExpress, Qiagen) preequilibrated with urea buffer. After an extensive washing with binding buffer containing 50mM imidazole (100 ml), protein was eluted with 500mM imidazole. Fractions were analyzed by SDS-PAGE, and protein concentration was determined by the Bradford procedure. Aliquots of the purified protein were stored at -70°C until used.
Electrophoretic Mobility Shift Assay (EMSA)
EMSA experiments were performed as previously described [36, 37]. PCR products corresponding to the mrk promoter regions were amplified using specific primers (Table 2). PCR products (100 ng) were mixed with increasing concentrations of H-NS-Myc-His6 or MrkH-His6 in the presence of the binding buffer 10X (400mM HEPES, 80mM MgCl2, 500mM KCl, 10mM DTT, 0.5% NP40 and 1 mg/ml BSA). fbpA coding region of Mycobacterium tuberculosis was used as negative control. The reactions were incubated during 30 min at room temperature (for H-NS) and 4°C (for MrkH), and then separated in 6% SDS-PAGE gels in Tris-Borate-EDTA buffer. The DNA bands were visualized by the ethidium bromide staining.
Assay for biofilm formation on abiotic surface
Adhesion to abiotic surface (polystyrene) was analyzed using 96-well plates as described previously [26]. Overnight cultures of bacteria grown in LB broth (10 μl) were added to 1 ml of LB. This volume was distributed in quintuples (200 μl per well) into a 96-well plate and incubated at room temperature for 24 h. Unbound bacteria were removed by washing the wells three times with PBS, and bound bacteria were stained with 1% violet crystal (CV) for 20 min. Wells were thoroughly rinsed thrice with PBS, and the dye in the adhered bacteria was solubilized with 100 μl of ethanol 70%. Finally, the amount of extracted violet crystal was determined using an enzyme-linked immunosorbent assay (ELISA) and measuring the OD600 in a multiskan plate reader (Thermo Scientific).
Statistical analysis
For statistical differences, one-way ANOVA followed by the Tukey’s comparison test was performed using Prism 5.0 (GraphPad Software Inc., San Diego, CA, USA). p≤0.05 was considered statistically significant.
Results
mrkJ promoter is directly regulated by MrkH
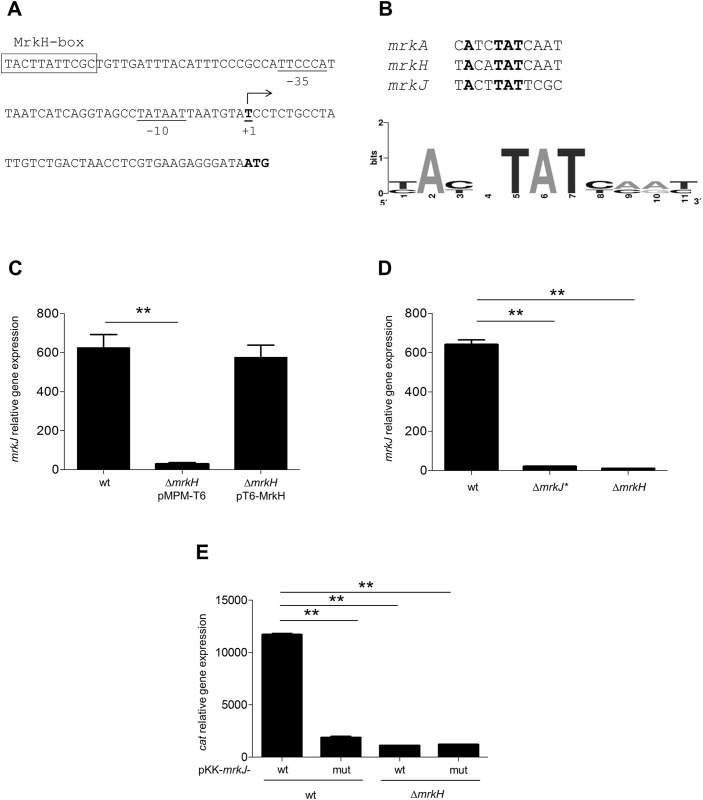
Although MrkH protein has been reported as a master regulator of the T3P [20–22], there are no reports of its effect on mrkJ expression; thus we studied whether the MrkH DNA-binding sequence, previously reported for mrkA and mrkH [21, 22] was present in the regulatory region of mrkJ. We found a putative MrkH-box on the mrkJ promoter, located at position -63.5 relative to its transcriptional start site (Fig 1A). This putative MrkH binding sequence presented the TAT motif conserved in the MrkH binding sites on mrkA and mrkH (Fig 1B). Then, to know whether MrkH regulates the expression of mrkJ, we determined the expression of this gene in the wild-type (WT) K. pneumoniae strain and its isogenic ΔmrkH mutant. As shown in Fig 1C, the transcription level of mrkJ was drastically decreased in the ΔmrkH mutant, with respect to the WT strain. The complemented ΔmrkH mutant had expression levels similar to the WT strain. To demonstrate that the putative MrkH-box was essential for MrkH-mediated mrkJ activation, the TACTTATTCGC sequence (Fig 1A) was deleted from the K. pneumoniae chromosome to generate a mutant strain, Kpn mrkJ* (mrkJ-ΔMrkHbox::FRT). We found that this deletion caused a severe reduction in the transcription of mrkJ gene (Fig 1D). Moreover, this reduction was similar to that observed in the absence of MrkH, supporting the notion that the deleted sequence is essential for the MrkH-mediated activation of the mrkJ promoter. In addition, using transcriptional reporters, we cloned the regulatory region of mrkJ and introduced a three nucleotides change in the TAT conserved motif into MrkH box (TAT to CGC). This mutant construction presented a reduction in MrkH-mediated mrkJ activation (Fig 1E), corroborating the relevance of the TAT motif in the MrkH-mediated mrkJ positive regulation. These results indicate that, similarly to other mrk genes, MrkH positively regulates the expression of mrkJ.
Fig 1. MrkH regulates the mrkJ promoter.
A. Schematic representation of the mrkJ promoter. The panel shows the nucleotide sequence of the regulatory region, showing the previously reported transcription start site (+1) [24]. The -35 and -10 promoter sequences and the transcription start site are underlined. Putative MrkH-binding site is boxed. B. Logo motif analysis using the MrkH-binding sites for mrkA, mrkH and mrkJ promoter regions. C. Transcriptional expression (qRT-PCR) of mrkJ gene in WT, ΔmrkH and complemented ΔmrkH backgrounds. D. mrkJ expression (qRT-PCR) in the wild-type (WT) and mrkJΔMrkHbox::FRT (mrkJ*). E. qRT-PCR assays determining the cat expression of mrkJ (pKK-mrkJ-wt) and a mutant variant within the MrkH-binding box (pKK-mrkJ-mut). Results represent mean and standard deviations of three independent experiments. ns, not significant; **, statistically significant with respect to the WT strain (p<0.01).
MrkH induces the expression of mrkJ by antagonizing H-NS-mediated repression and by acting as a transcriptional activator
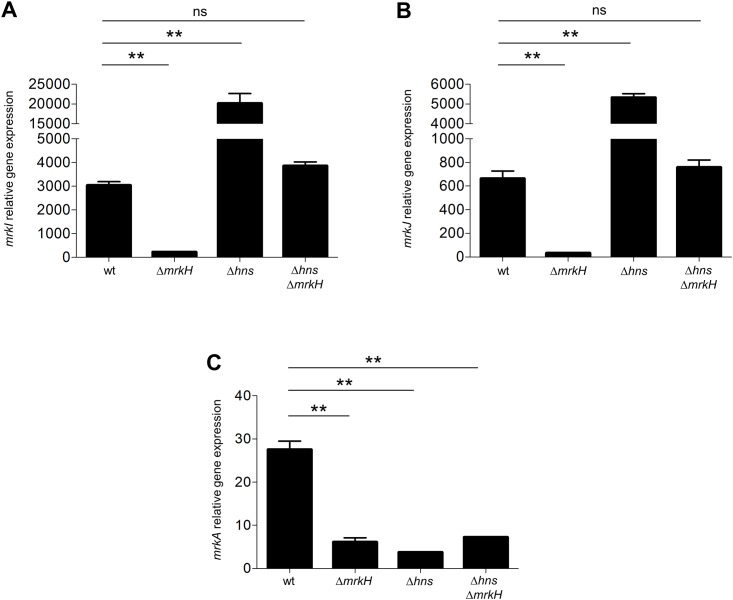
We have previously reported that H-NS represses the transcription of mrkH, mrkI and mrkJ [26]. Since our results indicated that the expression of the mrkJ gene is positively and negatively controlled by MrkH and H-NS, respectively, we hypothesized that MrkH induces the expression of these genes by counteracting the H-NS mediated repression. To investigate this, we determined the expression of the mrkI and mrkJ genes in a Δhns ΔmrkH double mutant, by qRT-PCR (Fig 2A and 2B). The expression of the mrkA gene was also tested as a control. As expected, the expression of the mrkA gene was reduced in the Δhns ΔmrkH double mutant, at levels similar to those observed in the Δhns and ΔmrkH single mutants (Fig 2C). Interestingly, the expression of mrkI and mrkJ was restored in the Δhns ΔmrkH double mutant, to a level similar to that in the WT strain, which supports that MrkH induces the expression of both genes by antagonizing their H-NS-mediated repression. However, the expression levels of the mrkI and mrkJ genes in the Δhns ΔmrkH double mutant were lower than those observed in the Δhns mutant (Fig 2A and 2B), suggesting that MrkH further activates the expression of these genes in the absence of H-NS, showing a dual activator/anti-repressor activity.
Fig 2. MrkH acts as anti-repressor and activator of mrk genes.
Transcriptional expression (qRT-PCR) of mrkI (A), mrkJ (B) and mrkA (C) genes in the WT, ΔmrkH mutant, Δhns mutant and Δhns ΔmrkH double mutant. Results represent the mean and standard deviations of three independent experiments. ns, not significant; **, statistically significant with respect to the WT strain (p<0.01).
MrkH and H-NS proteins directly bind to the mrk promoters
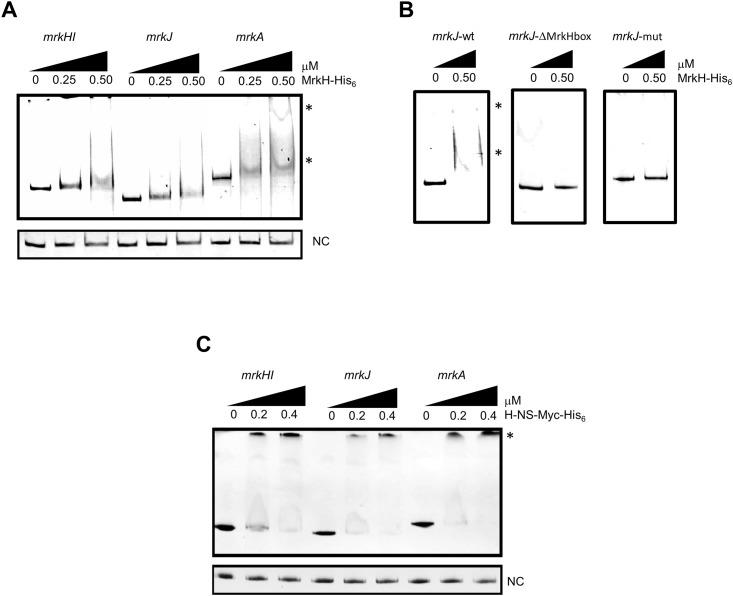
To determine whether MrkH and H-NS directly regulates mrkI and mrkJ, the interaction of purified MrkH-His6 and H-NS-Myc-His6 proteins and DNA fragments carrying the regulatory regions of the mrk genes was analyzed by EMSA. The interaction of MrkH-6XHis with the regulatory regions of mrkA/mrkH and with a DNA fragment of M. tuberculosis fbpA, were also analyzed as positive and negative controls, respectively. MrkH-His6 recombinant protein specifically bound to the regulatory regions of mrkA and mrkH as previously described [21, 23] (Fig 3A). Interestingly, MrkH-His6 protein also interacted with the mrkJ regulatory region, supporting that MrkH directly activates mrkJ expression (Fig 3A). Both, deletion of MrkH-box or the nucleotide change of the TAT conserved motif affected the binding of MrkH-His6 protein on the mrkJ promoter (Fig 3B). These results indicate that MrkH recognizes a site located at position -63.5 relative to the transcriptional start site of mrkJ gene.
Fig 3. MrkH and H-NS directly bind to mrk genes.
EMSA experiments were performed to test the binding of purified recombinant MrkH-His6 (A and B) or H-NS-Myc-His6 (C) proteins to the corresponding amplified DNA fragment from mrkHI, mrkJ (wt, ΔMrkHbox and mut) and mrkA regulatory regions. One hundred nanograms of the PCR product of each regulatory region was mixed, incubated with increasing concentrations (μM) of purified H-NS-Myc-His6 and MrkH-His6, and subsequently separated in 6% polyacrylamide gels. DNA-protein complexes stained with ethidium bromide are indicated (*). fbpA coding region of M. tuberculosis was used as negative control (NC).
In addition to MrkH-His6 protein, we analyze whether H-NS-Myc-His6 recombinant protein could bind to the upstream region of these three promoters. We found that H-NS-Myc-His6 protein specifically bound to the mrk promoters (Fig 3B). These observations strongly suggest that MrkH and H-NS are regulators that directly bind to mrk promoters and that both proteins have antagonistic functions.
MrkH differentially regulates mrk promoters
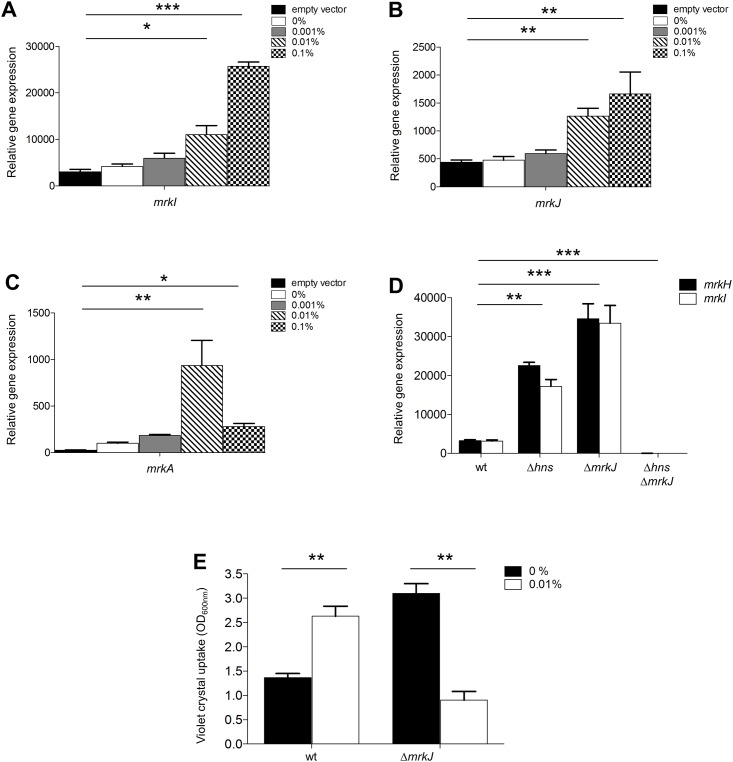
It has been reported that at high concentrations, antagonists of H-NS can act as transcriptional repressors [36, 38]. In order to investigate whether an over-production of MrkH protein could repress mrk genes, we introduced the pT6-MrkH plasmid, which expresses MrkH under an arabinose-inducible promoter, into the WT K. pneumoniae strain, to induce the expression of different amounts of MrkH with distinct concentrations of arabinose. MrkH increased the expression of the mrkI and mrkJ genes at all arabinose concentrations tested, although this was not observed for mrkA (Fig 4A–4C). Interestingly, mrkA expression reached a peak of induction at an arabinose concentration of 0.01%, while diminished at 0.1% (Fig 4C). Therefore, our results indicate that high expression levels of MrkH can repress T3P. Since Johnson and Clegg (2010) and Ares et al (2016) have shown that MrkJ and H-NS are the main negative regulators of mrkH, we evaluated the expression of mrkH and mrkI in several K. pneumoniae backgrounds: WT, Δhns mutant, ΔmrkJ mutant and Δhns ΔmrkJ double mutant. Transcriptional expressions of mrkH and mrkI were derepressed in the absence of H-NS or MrkJ, supporting the negative role of these two proteins (Fig 4D). In contrast, both mrkH and mrkI genes were repressed in the Δhns ΔmrkJ double mutant (Fig 4D). Since MrkH autoregulates its own expression [21], our observations demonstrate that in the absence of both H-NS and MrkJ, mrkH expression is repressed, suggesting that high levels of MrkH could repress mrkH gene and subsequently the MrkH-dependent mrk genes. To confirm the negative role of MrkH at functional level, we evaluated its overexpression on the biofilm formation. Since a Δhns mutant does not form biofilm [26], MrkH protein was overexpressed in the WT and ΔmrkJ mutant. The induction of MrkH stimulated the biofilm formation in the WT strain, while in the absence of MrkJ, this phenomenon was diminished (Fig 4E). These observations support the repressor activity of MrkH protein on T3P expression.
Fig 4. MrkH acts as repressor of mrk genes.
Transcriptional expression (qRT-PCR) of mrkI (A), mrkJ (B) and mrkA (C) genes overexpressing the MrkH protein at different L(+)-arabinose concentrations in the WT strain. D. Transcriptional expression (qRT-PCR) of mrkH and mrkI in the WT, Δhns, ΔmrkJ and Δhns ΔmrkJ backgrounds. E. Quantification of biofilm formation by measuring violet crystal uptake under overexpression of MrkH in the WT and ΔmrkJ mutant. Results represent the mean and standard deviations of three independent experiments performed. Statistically significant with respect to the WT strain: *p<0.05; **p<0.01; ***p<0.001.
MrkI does not affect the expression of T3P
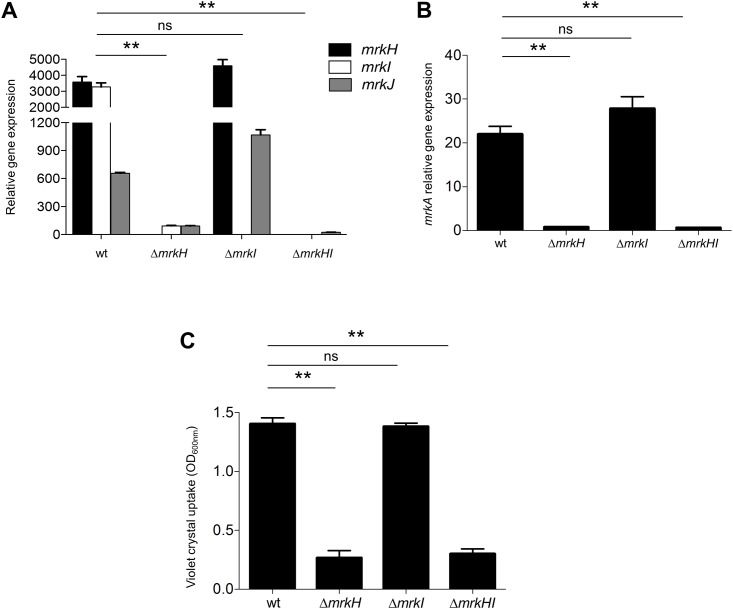
mrkI is found adjacent to mrkH, and codes for a LuxR-type regulator [20]. In order to investigate whether MrkI is somehow involved in the MrkH-mediated regulation of the mrk genes, we determined the transcriptional expression of mrk genes in several backgrounds: WT, ΔmrkH mutant, ΔmrkI mutant and ΔmrkHI double mutant. Transcription of mrkJ gene was down-regulated in both ΔmrkH and ΔmrkHI mutants but not in ΔmrkI (Fig 5A). In addition, MrkI did not affect the expression of either mrkH regulatory or mrkA pilin genes (Fig 5A and 5B), supporting the crucial role of MrkH regulatory protein on mrk genes [20, 23]. In order to show that MrkI does not affect the T3P at functional level, we performed assays of biofilm formation using the same mutant strains. We found that MrkH but not MrkI affected the biofilm formation of K. pneumoniae (Fig 5C), showing the same phenotype observed in the transcriptional expression of mrkA. Overall, this set of data may suggest that MrkI protein is not involved in the regulation of T3P.
Fig 5. The absence of MrkI does not affect the T3P.
Transcriptional expression (qRT-PCR) of mrkH-I-J (A) and mrkA (B) genes in WT, ΔmrkH mutant, ΔmrkI mutant and ΔmrkHI double mutant. C. Quantification of biofilm formation by measuring violet crystal uptake in WT, ΔmrkH mutant, ΔmrkI mutant and ΔmrkHI double mutant. Results shown represent the mean and standard deviations of three independent experiments. ns, not significant; **, statistically significant with respect to the WT strain (p<0.01).
Discussion
K. pneumoniae is a well-established opportunistic pathogen causing nosocomial infections. One of its most studied virulence factors is the T3P, that helps this bacterium to be adhered to both biotic and abiotic surfaces, and therefore, to establish a successful colonization in host tissues. Because of this relevant function for the pathogenesis of K. pneumoniae, this pilus must be subject to a fine regulation, not only at transcriptional level, but also at post-transcriptional and post-translational levels. In this sense, MrkH has been reported to be the master regulator of T3P. Since MrkH has a c-di-GMP-binding domain [23], its activity is controlled by fluctuations in concentrations of this second messenger. The transcriptional control of T3P is driven by three promoters located upstream from the coding regions of mrkA, mrkH and mrkJ genes. MrkH protein activates both mrkABCDF and mrkHI promoters, functioning as a classic transcriptional activator interacting with the α-CTD of RNA polymerase [21, 22]. Our data show that MrkH also positively regulates the mrkJ gene (Fig 1C), by binding to its regulatory region (Fig 3A), probably by a mechanism similar to that in the other mrk promoters. In terms of consensus sequence, we found that the putative MrkH-binding box on mrkJ promoter presented homology to that reported for mrkH in its auto-regulation [21]. Interestingly, a TAT motif located in the center of the MrkH-binding boxes (Fig 1B) has been reported to participate in the recognition of MrkH protein to both mrkH and mrkA promoters [21, 22]. Both MrkH-binding box deletion and site-directed mutagenesis experiments corroborated the relevance of this putative box on the MrkH-mediated mrkJ regulation and the in vitro binding of MrkH-His6 on DNA. Our assays of DNA-protein interaction did not include c-di-GMP; however, it has been reported that MrkH can bind in vitro to the promoter region of mrk genes in the absence of c-di-GMP [21, 23].
Whereas MrkJ indirectly represses the transcription of mrk genes by degrading c-di-GMP, H-NS directly silences them [25, 26], and these two proteins are considered to be the main negative regulators of T3P. Indeed, we have shown that H-NS protein binds to the three mrk promoters silencing their expression (Fig 3B). We showed that MrkH has a dual function: it acts as an anti-repressor of H-NS protein, antagonizing its negative effect on mrk genes and as a classic transcriptional activator because it is necessary to activate the expression of mrk genes (Fig 2). Similar to MrkH, Vibrio cholerae VpsT protein has also been reported to be an anti-repressor of H-NS that binds c-di-GMP, overcoming H-NS-mediated repression of biofilm genes [39, 40]. Accordingly, MrkH and VpsT are required for biofilm formation in both pathogens and they have anti-repressor/activator dual activity. Our data suggest a mechanism of competition between MrkH and H-NS on the mrkJ promoter, similar to what has been reported for other antagonists such as SlyA, RovA, Ler, and LeuO [36, 41–44]. We are currently trying to define the mechanistic details of how MrkH protein counteracts the H-NS-mediated repression of mrk promoters.
Some anti-repressors of H-NS have a negative role on several genes, and it is thought that this function may be relevant to maintain optimal levels of the proteins coded by such genes, probably to control their toxic effects. In our work, we detected high levels of mrkH transcription in Δhns and ΔmrkJ single mutants; in contrast, mrkH expression was abolished in the Δhns ΔmrkJ double mutant (Fig 4D). These results may suggest that the high transcriptional expression together to high concentrations of c-di-GMP may provoke that MrkH negatively auto-regulates its own expression. A similar repression pattern was detected for mrkI in the Δhns ΔmrkJ double mutant (Fig 4D). Moreover, the overexpression of MrkH diminished the biofilm formation in an mrkJ mutant (Fig 4E), since the biofilm formation is T3P-dependent. Our experiments showed that in addition to its role as an anti-repressor and transcriptional activator, MrkH acts also as a repressor. Antagonists of H-NS such as Ler in enteropathogenic E. coli and LeuO in Salmonella enterica, function as concentration-dependent transcriptional repressors. In the case of Ler, a negative effect of this protein on its own auto-regulation in LEE1 promoter has been reported [38]. LeuO, a LysR-type regulator, activates ompS2 gene at low concentrations and represses it at high concentrations [36], where the negative effect is suggested to occur by the competition of LeuO with the OmpR transcriptional activator for the binding site. In fact, previous reports have demonstrated by EMSA experiments that different DNA-MrkH complexes are formed, suggesting that MrkH can oligomerize on the promoter region of mrkA [23]. According to our results, we hypothesized that high level of MrkH may cause the binding of this protein close to the -35 and -10 boxes, blocking the interaction of RNA polymerase with the promoter. Footprinting experiments will elucidate the nucleotides recognized by MrkH on mrk promoters.
In previous experiments our group has shown that the absence of H-NS up-regulated the expression of mrkH, mrkI and mrkJ, while the expression of mrkA is down-regulated [26]. Thus, MrkH induced in the WT strain had positive and negative effects on mrkA expression as compared to mrkI and mrkJ genes (Fig 4A and 4C). This repression may be due to a greater affinity of MrkH on mrkA regulatory region as compared to mrkHI and mrkJ promoters observed by EMSA (Fig 3A) [21–23]. However, Surface Plasmon Resonance (SPR) analysis or fluorescence anisotropy would be necessary to determine the dissociation constant (Kd) between MrkH protein and mrk promoters.
In addition to MrkH, MrkI protein is also coded in the mrk cluster adjacent to mrkH, forming a bicistronic operon [20]. Controversial results regarding the involvement of MrkI on both mrk expression and biofilm formation in K. pneumoniae have been reported [20, 23, 24]. While Johnson et al (2011) have reported that the absence of MrkI has significantly reduced levels of mrkA transcription, Wilksch et al (2011) have reported that the ΔmrkI mutant appears to express more mrkA than the WT strain. The conditions evaluated in these studies as well as the differences between K. pneumoniae strains, could possibly explain the discrepancies in the phenotypes. However, our results show that MrkI does not participate neither in the T3P expression nor in biofilm formation of K. pneumoniae (Fig 5).
In conclusion our work provides new insights into the complex regulatory functions of MrkH protein on the transcriptional control of T3P in K. pneumoniae. This wide range of MrkH function would explain the importance of intracellular concentrations of this protein to regulate K. pneumoniae virulence functions such as biofilm formation, adherence to eukaryotic cells and colonization of its host.
Acknowledgments
We thank Dr Victor H. Bustamante for the critical reading of the manuscript. We thank Jessica Martínez-Cruz, and Mariana Ramírez-Etienne for technical assistance.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998;11(4):589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Han SH. Review of hepatic abscess from Klebsiella pneumoniae. An association with diabetes mellitus and septic endophthalmitis. West J Med. 1995;162(3):220–4. [PMC free article] [PubMed] [Google Scholar]
- 3.Schelenz S, Bramham K, Goldsmith D. Septic arthritis due to extended spectrum beta lactamase producing Klebsiella pneumoniae. Joint Bone Spine. 2007;74(3):275–8. 10.1016/j.jbspin.2006.08.007 [DOI] [PubMed] [Google Scholar]
- 4.Chang CM, Lee HC, Lee NY, Lee IW, Wu CJ, Chen PL, et al. Community-acquired Klebsiella pneumoniae complicated skin and soft-tissue infections of extremities: emphasis on cirrhotic patients and gas formation. Infection. 2008;36(4):328–34. 10.1007/s15010-008-7272-3 [DOI] [PubMed] [Google Scholar]
- 5.Tsai FC, Huang YT, Chang LY, Wang JT. Pyogenic liver abscess as endemic disease, Taiwan. Emerg Infect Dis. 2008;14(10):1592–600. 10.3201/eid1410.071254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fung CP, Chang FY, Lee SC, Hu BS, Kuo BI, Liu CY, et al. A global emerging disease of Klebsiella pneumoniae liver abscess: is serotype K1 an important factor for complicated endophthalmitis? Gut. 2002;50(3):420–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brisse S, Fevre C, Passet V, Issenhuth-Jeanjean S, Tournebize R, Diancourt L, et al. Virulent clones of Klebsiella pneumoniae: identification and evolutionary scenario based on genomic and phenotypic characterization. PLoS One. 2009;4(3):e4982 10.1371/journal.pone.0004982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerlach GF, Clegg S, Allen BL. Identification and characterization of the genes encoding the type 3 and type 1 fimbrial adhesins of Klebsiella pneumoniae. J Bacteriol. 1989;171(3):1262–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alcantar-Curiel MD, Blackburn D, Saldana Z, Gayosso-Vazquez C, Iovine NM, De la Cruz MA, et al. Multi-functional analysis of Klebsiella pneumoniae fimbrial types in adherence and biofilm formation. Virulence. 2013;4(2):129–38. 10.4161/viru.22974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allen BL, Gerlach GF, Clegg S. Nucleotide sequence and functions of mrk determinants necessary for expression of type 3 fimbriae in Klebsiella pneumoniae. J Bacteriol. 1991;173(2):916–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schurtz TA, Hornick DB, Korhonen TK, Clegg S. The type 3 fimbrial adhesin gene (mrkD) of Klebsiella species is not conserved among all fimbriate strains. Infect Immun. 1994;62(10):4186–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Struve C, Bojer M, Krogfelt KA. Identification of a conserved chromosomal region encoding Klebsiella pneumoniae type 1 and type 3 fimbriae and assessment of the role of fimbriae in pathogenicity. Infect Immun. 2009;77(11):5016–24. 10.1128/IAI.00585-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langstraat J, Bohse M, Clegg S. Type 3 fimbrial shaft (MrkA) of Klebsiella pneumoniae, but not the fimbrial adhesin (MrkD), facilitates biofilm formation. Infect Immun. 2001;69(9):5805–12. 10.1128/IAI.69.9.5805-5812.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sebghati TA, Korhonen TK, Hornick DB, Clegg S. Characterization of the type 3 fimbrial adhesins of Klebsiella strains. Infect Immun. 1998;66(6):2887–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jagnow J, Clegg S. Klebsiella pneumoniae MrkD-mediated biofilm formation on extracellular matrix- and collagen-coated surfaces. Microbiology. 2003;149(Pt 9):2397–405. 10.1099/mic.0.26434-0 [DOI] [PubMed] [Google Scholar]
- 16.Tarkkanen AM, Virkola R, Clegg S, Korhonen TK. Binding of the type 3 fimbriae of Klebsiella pneumoniae to human endothelial and urinary bladder cells. Infect Immun. 1997;65(4):1546–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schroll C, Barken KB, Krogfelt KA, Struve C. Role of type 1 and type 3 fimbriae in Klebsiella pneumoniae biofilm formation. BMC Microbiol. 2010;10:179 10.1186/1471-2180-10-179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang YJ, Liao HW, Wu CC, Peng HL. MrkF is a component of type 3 fimbriae in Klebsiella pneumoniae. Res Microbiol. 2009;160(1):71–9. 10.1016/j.resmic.2008.10.009 [DOI] [PubMed] [Google Scholar]
- 19.Hornick DB, Thommandru J, Smits W, Clegg S. Adherence properties of an mrkD-negative mutant of Klebsiella pneumoniae. Infect Immun. 1995;63(5):2026–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson JG, Murphy CN, Sippy J, Johnson TJ, Clegg S. Type 3 fimbriae and biofilm formation are regulated by the transcriptional regulators MrkHI in Klebsiella pneumoniae. J Bacteriol. 2011;193(14):3453–60. 10.1128/JB.00286-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan JW, Wilksch JJ, Hocking DM, Wang N, Srikhanta YN, Tauschek M, et al. Positive autoregulation of mrkHI by the cyclic di-GMP-dependent MrkH protein in the biofilm regulatory circuit of Klebsiella pneumoniae. J Bacteriol. 2015;197(9):1659–67. 10.1128/JB.02615-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang J, Wilksch JJ, Tan JW, Hocking DM, Webb CT, Lithgow T, et al. Transcriptional activation of the mrkA promoter of the Klebsiella pneumoniae type 3 fimbrial operon by the c-di-GMP-dependent MrkH protein. PLoS One. 2013;8(11):e79038 10.1371/journal.pone.0079038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilksch JJ, Yang J, Clements A, Gabbe JL, Short KR, Cao H, et al. MrkH, a novel c-di-GMP-dependent transcriptional activator, controls Klebsiella pneumoniae biofilm formation by regulating type 3 fimbriae expression. PLoS Pathogens. 2011;7(8):e1002204 10.1371/journal.ppat.1002204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu CC, Lin CT, Cheng WY, Huang CJ, Wang ZC, Peng HL. Fur-dependent MrkHI regulation of type 3 fimbriae in Klebsiella pneumoniae CG43. Microbiology. 2012;158(Pt 4):1045–56. 10.1099/mic.0.053801-0 [DOI] [PubMed] [Google Scholar]
- 25.Johnson JG, Clegg S. Role of MrkJ, a phosphodiesterase, in type 3 fimbrial expression and biofilm formation in Klebsiella pneumoniae. J Bacteriol. 2010;192(15):3944–50. 10.1128/JB.00304-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ares MA, Fernandez-Vazquez JL, Rosales-Reyes R, Jarillo-Quijada MD, von Bargen K, Torres J, et al. H-NS Nucleoid Protein Controls Virulence Features of Klebsiella pneumoniae by Regulating the Expression of Type 3 Pili and the Capsule Polysaccharide. Front Cell Infect Microbiol. 2016;6:13 10.3389/fcimb.2016.00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dorman CJ. H-NS: a universal regulator for a dynamic genome. Nature reviews Microbiology. 2004;2(5):391–400. 10.1038/nrmicro883 [DOI] [PubMed] [Google Scholar]
- 28.Tendeng C, Bertin PN. H-NS in Gram-negative bacteria: a family of multifaceted proteins. Trends Microbiol. 2003;11(11):511–8. [DOI] [PubMed] [Google Scholar]
- 29.Navarre WW, McClelland M, Libby SJ, Fang FC. Silencing of xenogeneic DNA by H-NS-facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. Genes Dev. 2007;21(12):1456–71. 10.1101/gad.1543107 [DOI] [PubMed] [Google Scholar]
- 30.Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, et al. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science. 2006;313(5784):236–8. 10.1126/science.1128794 [DOI] [PubMed] [Google Scholar]
- 31.Dorman CJ. H-NS, the genome sentinel. Nat Rev Microbiol. 2007;5(2):157–61. 10.1038/nrmicro1598 [DOI] [PubMed] [Google Scholar]
- 32.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97(12):6640–5. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gallego-Hernandez AL, Hernandez-Lucas I, De la Cruz MA, Olvera L, Morett E, Medina-Aparicio L, et al. Transcriptional regulation of the assT-dsbL-dsbI gene cluster in Salmonella enterica serovar Typhi IMSS-1 depends on LeuO, H-NS, and specific growth conditions. J Bacteriol. 2012;194(9):2254–64. 10.1128/JB.06164-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jahn CE, Charkowski AO, Willis DK. Evaluation of isolation methods and RNA integrity for bacterial RNA quantitation. J Microbiol Methods. 2008;75(2):318–24. 10.1016/j.mimet.2008.07.004 [DOI] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 36.De la Cruz MA, Fernandez-Mora M, Guadarrama C, Flores-Valdez MA, Bustamante VH, Vazquez A, et al. LeuO antagonizes H-NS and StpA-dependent repression in Salmonella enterica ompS1. Mol Microbiol. 2007;66(3):727–43. 10.1111/j.1365-2958.2007.05958.x [DOI] [PubMed] [Google Scholar]
- 37.De la Cruz MA, Zhao W, Farenc C, Gimenez G, Raoult D, Cambillau C, et al. A toxin-antitoxin module of Salmonella promotes virulence in mice. PLoS Pathogens. 2013;9(12):e1003827 10.1371/journal.ppat.1003827 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Berdichevsky T, Friedberg D, Nadler C, Rokney A, Oppenheim A, Rosenshine I. Ler is a negative autoregulator of the LEE1 operon in enteropathogenic Escherichia coli. J Bacteriol. 2005;187(1):349–57. 10.1128/JB.187.1.349-357.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ayala JC, Wang H, Silva AJ, Benitez JA. Repression by H-NS of genes required for the biosynthesis of the Vibrio cholerae biofilm matrix is modulated by the second messenger cyclic diguanylic acid. Mol Microbiol. 2015;97(4):630–45. 10.1111/mmi.13058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zamorano-Sanchez D, Fong JC, Kilic S, Erill I, Yildiz FH. Identification and characterization of VpsR and VpsT binding sites in Vibrio cholerae. J Bacteriol. 2015;197(7):1221–35. 10.1128/JB.02439-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stoebel DM, Free A, Dorman CJ. Anti-silencing: overcoming H-NS-mediated repression of transcription in Gram-negative enteric bacteria. Microbiology. 2008;154(Pt 9):2533–45. 10.1099/mic.0.2008/020693-0 [DOI] [PubMed] [Google Scholar]
- 42.Lithgow JK, Haider F, Roberts IS, Green J. Alternate SlyA and H-NS nucleoprotein complexes control hlyE expression in Escherichia coli K-12. Mol Microbiol. 2007;66(3):685–98. 10.1111/j.1365-2958.2007.05950.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cathelyn JS, Ellison DW, Hinchliffe SJ, Wren BW, Miller VL. The RovA regulons of Yersinia enterocolitica and Yersinia pestis are distinct: evidence that many RovA-regulated genes were acquired more recently than the core genome. Mol Microbiol. 2007;66(1):189–205. 10.1111/j.1365-2958.2007.05907.x [DOI] [PubMed] [Google Scholar]
- 44.Bustamante VH, Santana FJ, Calva E, Puente JL. Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: Ler antagonizes H-NS-dependent repression. Mol Microbiol. 2001;39(3):664–78. [DOI] [PubMed] [Google Scholar]
- 45.Mayer MP. A new set of useful cloning and expression vectors derived from pBlueScript. Gene. 1995;163(1):41–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.