Abstract
Iron (Fe) deficiency is a critical agricultural problem, especially in calcareous soil, which is distributed worldwide. Rice plants take up Fe(II) from soil through a OsIRT1 transporter (Strategy I-related system) and also take up Fe(III) via a phytosiderophore-based system (Strategy II system). However, rice plants are susceptible to low-Fe conditions because they have low Fe(III) reduction activity and low-level phytosiderophore secretion. Previously, we produced transgenic rice plants expressing a mutationally reconstructed yeast ferric chelate reductase, refre1/372, under the control of the OsIRT1 promoter. This transgenic rice line exhibited higher Fe(III) chelate reductase activity and tolerance to Fe deficiency. In addition, we produced transgenic rice overexpressing the Fe deficiency-inducible transcription factor, OsIRO2, which regulates the expression of various genes involved in the strategy II Fe(III) uptake system, including OsNAS1, OsNAAT1, OsDMAS1, OsYSL15, and TOM1. This transgenic rice exhibited improved phytosiderophore secretion ability and tolerance to Fe deficiency. In the present research, transgenic rice plants that possess both the OsIRT1 promoter-refre1/372 and the 35S promoter-OsIRO2 (RI lines) were produced to enhance both Strategy I Fe(II) reductase ability and Strategy II phytosiderophore productivity. RI lines exhibited enhanced tolerance to Fe-deficient conditions at the early and middle-late stages of growth in calcareous soil, compared to both the non-transgenic line and lines harboring either OsIRT1 promoter-refre1/372 or 35S promoter-OsIRO2 alone. RI lines also exhibited a 9-fold higher yield than the non-transgenic line. Moreover, we successfully produced Fe-deficiency-tolerant Tachisugata rice, which is a high-biomass variety used as fodder. Collectively, our results demonstrate that combined enhancement of two Fe uptake systems in rice is highly effective in conferring tolerance to low Fe availability in calcareous soil.
Introduction
Iron (Fe) deficiency is a widespread agricultural problem that is common in calcareous soils, which cover more than 30% of the earth’s surface. Although there are abundant minerals in soil, Fe is sparingly soluble under aerobic conditions, in particular at high pH in calcareous soil [1]. Consequently, plants often exhibit Fe-deficiency symptoms, such as chlorosis, leading to reduced crop yield and quality. Thus, Fe is a key determinant of biomass production and of plant product quality [2]. Development of crops tolerant to low Fe availability is important to meet the increased demand for food caused by rapidly increasing populations. Moreover, cultivation of high-biomass crops tolerant to low Fe availability in calcareous soil would reduce the carbon dioxide concentration in the atmosphere and, consequently, ameliorate global warming.
Higher plants use two major Fe uptake strategies—Strategy I and Strategy II [3]. Under conditions of low Fe availability, non-graminaceous plants use the Strategy I system, which involves induction of two major processes. First, Fe(III)-chelate reductase (FRO) is induced in roots and reduces Fe(III)-chelate to Fe2+ [4], which is assumed to be the rate-limiting step for Fe acquisition from soil [5]. Next, the Fe2+ is absorbed via the Fe-regulated transporter (IRT) [6], which is the major Fe2+ transporter in plant roots.
Although rice is a graminaceous plant, rice also possesses a direct Fe2+ uptake system mediated by the ferrous transporter OsIRT1. This is a part of the Strategy I system [7–8]. In paddy fields (where rice plants are normally grown), Fe2+ is abundant because of the low redox potential, which is assumed to be the reason why rice plants possess Fe2+ uptake systems [7–8]. However, rice plants have a low Fe(III)-chelate-reductase activity [8], and thus lack a complete Strategy I system and show limited Fe uptake efficiency.
The Strategy II mechanism is specific to graminaceous plants, which synthesize and secrete mugineic acid family phytosiderophores (MAs) into soil; these compounds chelate sparingly soluble Fe(III) [9]. Secretion of MAs from roots to the rhizosphere is mediated by the “transporter of mugineic acid 1” (TOM1:Nozoye et al. [10]). The resulting Fe(III)–MAs complexes are solubilized and absorbed into the root by the Fe(III)-MAs transporters termed Yellow Stripe 1 (YS1) and Yellow Stripe-Like (YSL) of the plasma membrane, such as maize ZmYS1 [11], rice OsYSL15 [12–13], and barley HvYS1 [14]. The biosynthetic pathway of MAs from methionine, via nicotianamine (NA) as an intermediate, has been elucidated [15–23]. Nicotianamine synthase (NAS) catalyzes S-adenosyl-L-methionine to form NA [17], which is then converted into the 3″-oxo intermediate via transfer of an amino group by nicotianamine aminotransferase (NAAT) [19]. Subsequently, deoxymugineic acid synthase (DMAS) produces 2′-deoxymugineic acid (DMA) from the 3″-oxo intermediate [23]. All the MAs share the biosynthetic pathway from S-adenosyl-L-methionine to DMA, which is then converted to other MAs in several graminaceous plants via deoxygenases, such as IDS3 and IDS2 [20–21]. Rice plants also synthesize and secrete MAs under conditions of Fe deficiency. Nozoye et al. [24] demonstrated that OsNAS2 was localized to vesicles in Fe-deficient roots; transport of these vesicles is a crucial step in NA synthesis by rice, in turn leading to DMA synthesis and secretion. However, rice secretes lower levels of MAs than do other graminaceous crops, such as barley [25]. This is one of the reasons why rice, among the graminaceous plants tested to date, is the most susceptible to Fe deficiency.
To produce Fe-deficiency-tolerant rice, the first approach used was to enhance biosynthesis of MAs in the plant. Takahashi et al. [26] produced transgenic rice carrying a barley genome fragment containing the HvNAAT-A and HvNAAT-B genes, and demonstrated that the transgenic rice was tolerant to Fe deficiency in a calcareous soil. Suzuki et al. [27] carried out field experiments with transgenic rice lines harboring the barley genomic fragment of either HvNAS1 or HvNAS1 plus HvNAAT-A and HvNAAT-B, or IDS3; all rice lines were tolerant to Fe deficiency. These results support the notion that rice with enhanced phytosiderophore productivity exhibits tolerance to Fe deficiency.
The second approach used was to enhance the Fe(III)-chelate reductase activity in rice. To this end, Oki et al. [28] artificially reconstructed and mutagenized the yeast Fe(III)-chelate reductase gene FRE1, to generate refre1/372, whose encoding protein exhibits enhanced enzymatic activity at high pH, to facilitate growth in calcareous soils. Ishimaru et al. [29] introduced refre1/372 into rice plants under the control of the OsIRT1 promoter, which drives gene expression predominantly in the root epidermis in response to Fe deficiency [8]. Such transgenic rice plants exhibited higher Fe(III)-chelate reductase activity and enhanced tolerance to low Fe availability in calcareous soils [29]. The grain yield was 7.9-fold higher than that of non-transformants (NT).
The third approach used was to enhance expression of transcription factors that control Fe homeostasis-related genes in rice. Ogo et al. [30] identified an Fe deficiency-inducible basic helix–loop–helix (bHLH) transcription factor, OsIRO2, in rice. OsIRO2 is responsible for regulation of the key genes involved in Strategy II-related Fe uptake; e.g., OsNAS1, OsNAS2, OsNAAT1, OsDMAS1, TOM1, and OsYSL15 [31–32]. Ogo et al. [31] introduced OsIRO2 under the control of the constitutive cauliflower mosaic virus 35S promoter (35S promoter) into rice. OsIRO2-overexpressing rice secreted a greater quantity of DMA than did NT plants and exhibited enhanced tolerance to Fe deficiency in calcareous soils [31–32]. Based on these results, we hypothesize that transgenic rice plants with combined enhancement of refre1/372 (second approach) and OsIRO2 (third approach) would be more tolerant to low Fe availability, because refre1/372 and OsIRO2 enhance the activities of two different Fe acquisition systems, Strategy I and Strategy II, respectively.
In the present study, firstly, we produced transgenic rice lines using the Tsukinohikari cultivar with Fe deficiency-inducible expression of refre1/372 under the control of the OsIRT1 promoter, with constitutive overexpression of OsIRO2, and evaluated their tolerance to low Fe availability in calcareous soil. Introduction of both refre1/372 and OsIRO2 was more effective than single introduction of either gene under water-submerged conditions. Additionally, Ohta et al. [33] developed a high-biomass rice variety, Tachisugata, the entire silage of which is used as fodder. Thus secondly, we also produced Fe-deficiency-tolerant Tachisugata rice by introducing the refre1/372 and OsIRO2 genes.
Materials and methods
Production of refre1/372-OxOsIRO2 lines (RI lines)
The plasmid pIG121Hm containing the 35S promoter-OsIRO2 ORF [31] was used as a PCR template. The 35S promoter-OsIRO2 ORF fragment lacking a HindIII site at the 3′ end was amplified using the primers 5′-TAT AAG CTT GCA TGC CTG CAG GTC-3′ and 5′-TTA GAG TTT TGC TTT GTT CCT GAC G-3′. The amplified fragment was ligated to the nopaline synthase terminator (AF485783) fragment, excised from pE7133-GUS [34] by Ecl136II and EcoRI, using T4 DNA ligase (TaKaRa, Japan) and subsequently amplified again, using the primers 5′-TAT AAG CTT GCA TGC CTG CAG GTC-3′ and 5′-ATA AAG CTT CCG ATC TAG TAA CAT AGA TG -3′, to append an HindIII site to the 3′ end of the nopaline synthase terminator. This amplified fragment was subcloned into the pTA2 vector (TOYOBO, Japan), verified by DNA sequencing, and finally inserted into pIG121Hm containing the OsIRT1 promoter-refre1/372 [29] at the HindIII site to construct pIG121Hm-Refre1/372-OxOsIRO2. Agrobacterium tumefaciens strain EHA105 transformed with pIG121Hm-Refre1/372-OxOsIRO2 was used to transform two rice cultivars (Oryza sativa L. cv. Tsukinohikari and Tachisugata).
Transformation of the rice cultivar Tsukinohikari was performed using the method outlined by Sallaud et al. [35] and Terada et al. [36]. Most transgenic procedures for transformation of the Tachisugata rice cultivar are based on the same method, with the following exceptions. Calli were cultivated at 30°C throughout all rice transformation steps except co-cultivation with Agrobacterium at 23°C. For regeneration, Murashige and Skoog (MS) salts and vitamins (PhytoTechnology Laboratories, Kansas, USA), 30 g/L sorbitol (Wako, Japan), 30 g/L maltose (Wako, Japan), 2 g/L casamino acids (Nihon Pharmaceutical, Japan), 2 mg/L kinetin (Wako, Japan), 2 μg/L NAA (Wako, Japan), 10 mg/L hygromycin B (Wako, Japan), and 5 g/L gelrite (San-Ei Gen F.F.I., Japan) formed the regeneration medium. Hygromycin B at 15 and 10 mg/L was used for selection, and regeneration and rooting, respectively.
After confirmation of gene insertion into regenerated plants by genomic PCR as described below, the transformants (RI lines) were cultivated in a greenhouse at 28°C under natural light until mature T1 seeds were obtained.
Detection of inserted genes of transgenic lines by genomic PCR
Genomic DNA was extracted from the leaves of Tsukinohikari-RI T0 transgenic rice plants using the method of Thomson and Henry [37], and introduction of OsIRO2 and refre1/372 was confirmed using KOD FX (TOYOBO, Japan). The primers used for genomic PCR of OsIRO2 were 5′-ATG GAG CAG CTG TTC GTC GAC G-3′ and 5′-TTA GAG TTT TGC TTT GTT CCT GAC G-3′. The primers used for genomic PCR of refre1/372 were 5′-TAA CAA GAC TCT GGA CTC CGC TTT G-3′ and 5′-TAG AAC CAG GCT GAT TTT GGT GAA A-3′. The pIG121Hm-Refre1/372-OxOsIRO2 plasmid was used as a PCR template for confirmation of band size.
RNA preparation and quantitative RT-PCR
NT and T1 seeds of Tsukinohikari-RI transgenic rice were germinated for 13 days on MS medium (sucrose 30 g/L, NH4NO3 1.65 g/L, KNO3 1.9 g/L, CaCl2•2H2O 440 mg/L, MgSO4•7H2O 370 mg/L, KH2PO4 170 mg/L, Fe(III)-EDTA 42.1 mg/L, H3BO3 6.2 mg/L, MnSO4•4H2O 22.3 mg/L, ZnSO4•7H2O, 8.6 mg/L, KI 0.83 mg/L, Na2MoO4•2H2O 250 ng/L, CuSO4•5H2O 25 ng/L, CoCl2•6H2O 25 ng/L, thiamine-HCl 100 ng/L, nicotinic acid 500 ng/L, pyridoxine-HCl 500 ng/L, glycine 2 mg/L, myo-inositol 100 mg/L, and agar 8 g/L, pH 5.8) with hygromycin (50 mg/L, for transgenic plants) or without hygromycin (for NT plants) at 28°C under 24 h-light conditions. Then, plantlets were acclimated for 3 days and transferred to a 15 L plastic container containing a hydroponic culture solution of the following composition: K2SO4 122 mg/L, KCl 7.5 mg/L, KH2PO4 14 mg/L, Ca(NO3)2•4H2O 472 mg/L, MgSO4•7H2O 123 mg/L, H3BO3 0.62 mg/L, MnSO4•5H2O 0.12 mg/L, ZnSO4•7H2O 0.14 mg/L, CuSO4•5H2O 0.05 mg/L, (NH4)6Mo7O24 0.012 mg/L and Fe(III)-EDTA 42 mg/L. The nutrient solution was adjusted to pH 5.5 with 1 M HCl every 2 days and renewed weekly. After 2 weeks, plants were transferred to a hydroponic culture solution without Fe(III)-EDTA and grown for a further 7 days for exposure to Fe-deficiency. Total RNA was extracted from roots using an RNeasy Plant Mini Kit (QIAGEN, Germany). First-strand cDNA was synthesized from extracted total RNA using a Rever Tra Ace qPCR RT Kit (TOYOBO, Japan). Quantitative RT-PCR was performed using a 7300 Real-Time PCR system (Applied Biosystems, California, USA) and GoTaq® qPCR Master Mix (Promega, Wisconsin, USA). The primers used were as follows: 5′-GGC ATG GCT CCC ATC GT-3′ and 5′-AAC AAG CTG ACC TGA ACC ATG A-3′ (OsIRO2), and 5′-TCA CGC CGT GCT GAC TTG-3′ and 5′-TCC GGA TAC CGA AAA GGT ACA-3′ (refre1/372). Transcript levels were normalized to the expression levels of alpha-Tubulin, as determined using the primers 5′-GCA ACT CTC TGT TGC CGA GAT-3′ and 5′-GTC GCA CTT GGC CAT CAT G-3′. The sizes of the amplified fragments were confirmed by agarose gel electrophoresis.
T3 seeds of Tsukinohikari-RI line Nos. 21 and 22, seeds of Tsukinohikari-Refre1 line No. 7 [29], seeds of Tsukinohikari-IRO2 line No. 2 (Ogo et al. [31], OX2), and non-transgenic (NT) Tsukinohikari seeds were also germinated and cultivated in hydroponic culture solution as described above for 10 days under Fe-sufficient conditions, and then grown in Fe-deficient hydroponic culture without Fe(III)-EDTA for 1 day. After RNA extraction and cDNA synthesis as described above, quantitative RT-PCR was performed using StepOnePlus™ Real-Time PCR System (Life technologies, Tokyo, Japan) using primers used in [10] for OsTOM1, those in [12] for OsYSL15 (5′-ACT GGT ACC CTG CAA ACA TAC-3′ and 5′-GCA ATG ATG CTT AGC AAG AAG-3′), and those in [38] for OsNAS1, OsNAS2, OsNAAT1 and OsDMAS1.
Root Fe(III)-chelate reductase activity assay
Tsukinohikari-RI plants were germinated on MS medium. Two weeks later, plantlets were cultivated in hydroponic culture solution as described above for 10 days under Fe-sufficient conditions, and then cultured in Fe-deficient culture solution without any form of Fe. Root Fe(III)-chelate reductase activities in whole intact root systems were determined as described previously [29] at 3, 5, 7, and 10 days after onset of Fe-deficient cultivation. Roots were rinsed with water and submerged in 40 mL of assay solution (0.2 mM CaSO4, 5 mM 2-[4-(2-hydroxyethyl)-1-piperazinyl] ethanesulfonic acid at pH 5.5, 0.1 mM Fe(III)-EDTA, and 0.2 mM bathophenanthroline disulfonic acid disodium salt) (Wako, Japan). After incubation for 1 h at 25°C, an aliquot of the assay solution was collected, and the absorbance of the solution at 535 nm was determined using a UV-2450 spectrophotometer (SHIMADZU, Japan). The amount of Fe2+ produced was calculated from a standard curve prepared using standard solutions of Fe2+.
Growth test of RI lines on calcareous soil and yield analysis
T2 seeds of Tsukinohikari-RI line Nos. 21 and 22, T2 seeds of Tsukinohikari-Refre1 line No. 7 [29], T2 seeds of Tsukinohikari-IRO2 line No. 2 (Ogo et al. [31], OX2), and non-transgenic (NT) Tsukinohikari seeds were used for growth testing in calcareous soil. Seeds were germinated on MS medium with hygromycin 50 mg/L (for transgenic seeds) or without hygromycin (for NT seeds) for 20 days. Then, each plantlet was acclimated for 3 days and then transferred to a pot containing 1 kg calcareous soil obtained from Takaoka City, Toyama, Japan (Nihonkai Kougyou, Japan; pH 8.9, soluble CaO: 39.6%, Fe2O3: 1.7%); the soil was autoclaved just before transplanting to prevent pest contamination, then supplied with the slow-release fertilizers Eco long total 70 and Eco long total 140 containing N:P:K = 13:11:13, Fe 0.20% as EDTA-Na-Fe(III), Cu 0.050%, Zn 0.015%, and Mo 0.020% (JCAM AGRI, Japan) 3.5 g/pot. A netting sheet was placed on the base of each pot to prevent the roots from growing outside the pots. The plants were grown constantly submerged in water in a greenhouse at 28°C under natural light conditions. Shoot height and the SPAD value of the newest leaf were measured once every three days using a SPAD-502 chlorophyll meter (Konika Minolta, Japan) until 58 days after transplanting (DAT), thus to the panicle initiation stage. After seed maturation, water submergence was continued until 151 DAT. Tiller numbers were counted at 151 DAT. The water supply then ceased for 28 days, then plants were harvested on 179 DAT. Plant heights, and the dry weights of straws and panicles, were measured on the day of harvesting. The number of grains per panicle, the number of panicles per plant, and the total number of grains were measured. Fully matured grains (specific gravity >1.06) were selected by salty water. The filled grain rate, the 1,000-grain weight, and total grain weight of fully matured grains were calculated.
T1 seeds of Tachisugata RI lines were germinated on MS medium with hygromycin 30 mg/L without Fe, and incubated at 13°C for 1 week. Next, the seeds were incubated at 32°C for 1 day. After germination, each plantlet was grown in 400 mL of calcareous soil in a pot. Ion-exchanged water (50 mL) and half-concentration Fe-deficient hydroponic culture solution (50 mL; pH 8.0) described above were supplied to each pot on alternating days.
Measurement of metal content
Plants grown in calcareous soil were dried for 2 days at 60°C. Metal concentration analysis was carried out according to the method of Masuda et al. [39]. Ten brown-rice grains were digested with 1 mL HNO3 and 1 mL H2O2 for 20 min at 200°C in a Mars Xpress oven (CEM, NC, USA). After digestion, the samples were collected in a tube and the volume made up to 5 mL with 3 mL 0.1 M HCl. Next, the metal content was measured using an ICPS-8100 (SHIMADZU, Kyoto, Japan) at wavelengths of 238.204 nm (Fe), 257.610 nm (Mn), 202.551 nm (Zn), and 324.754 nm (Cu). Straw was digested with 2 mL of HNO3 and 2 mL of H2O2, and the metal contents measured using the method described above.
Statistical analysis
For the Fe(III)-chelate reductase activity assay, we grew three plants of each variety (NT, IRO2, Refre1, and RI lines 21 and 22) as biological replicates (n = 3) and for calcareous cultivation, we grew four plants of each variety (n = 4). Analysis of variance (ANOVA) via Student’s t-test was used to examine experimental data from both the Fe(III)-chelate reductase activity assay and calcareous soil cultivation, such as plant height, tiller number, weight of straw, weight of panicle yield index, and Fe content per plant; we employed JMP9 software (SAS Institute, Cary, NC, U.S.A.) to this end and p < 0.05 was considered statistically significant.
Results
Selection of transgenic lines with higher expression levels of OsIRO2 and refre1/372 in roots under Fe-deficiency conditions
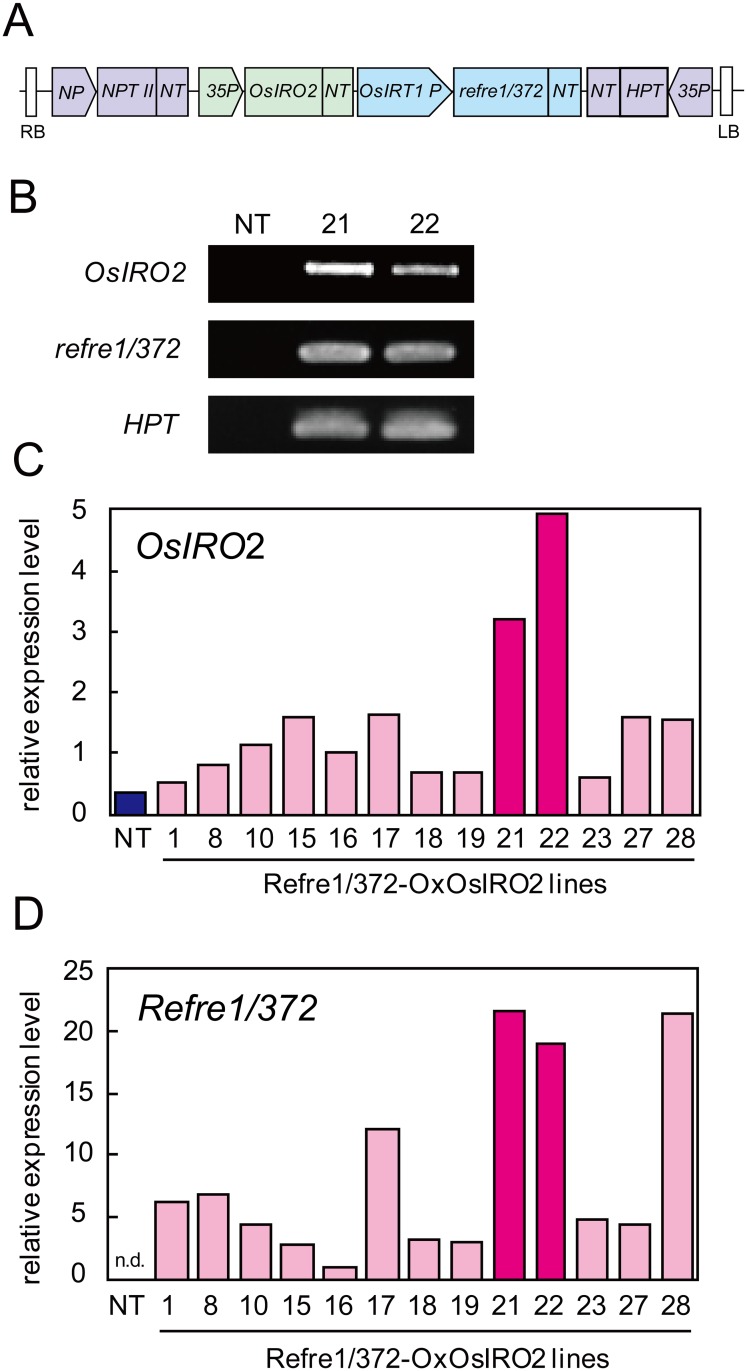
A construct harboring the 35S promoter-OsIRO2 and the OsIRT1 promoter-refre1/372 was introduced into rice plants by Agrobacterium-mediated transformation (Fig 1A). We used genomic PCR to confirm the introduction of OsIRO2 and refre1/372 into 28 regenerated plant lines. Among these, 24 lines were found to include both inserted genes and we termed these lines ‘RI lines’. As representatives, the genomic PCR results of line numbers 21 and 22 compared with NT are shown in Fig 1B. T1 plants were cultivated under Fe-deficient conditions, and OsIRO2 and refre1/372 expression levels in roots were analyzed by quantitative RT-PCR (Fig 1C and 1D). The expression levels of OsIRO2 in the RI lines were higher than those in the NT line (Fig 1C). refre1/372 expression was detected in RI lines but not in the NT line (Fig 1D). The expression levels of OsIRO2 and refre1/372 were higher in RI lines 21 and 22 than in all or most of the other transgenic lines and the NT line. Therefore, we selected RI lines 21 and 22 to obtain T2 and T3 seeds, which were then used for further analysis.
Fig 1. Production of RI rice.
(A) The gene cassette introduced into rice to produce RI lines. LB, left border; RB, right border; NP, nopaline synthase promoter; NPT II, neomycin phosphotransferase II; NT, nopaline synthase terminator; 35P, cauliflower mosaic virus 35S promoter; OsIRO2, ORF region of the Fe deficiency-inducible bHLH transcription factor gene OsIRO2 [31]; OsIRT1 P, 0.8-kb 5' upstream region of the OsIRT1 gene [8]; refre1/372, the mutationally reconstructed ferric-chelate reductase gene refre1/372 from yeast [28]. HPT, hygromycin phosphotransferase. The plasmid backbone was that of the pIG121Hm binary vector [40]. (B) Confirmation of gene insertion in RI lines by genomic PCR. NT, non-transgenic line; 21 and 22, RI lines 21 and 22. Gene insertions in the other RI lines and the pIG121Hm-refre1/372-OxOsIRO2 plasmid template were confirmed (the band sizes were identical). (C, D) OsIRO2 and refre1/372 expression levels as determined by quantitative RT-PCR. (C) OsIRO2 expression. (D) refre1/372 expression. T1 transgenic rice and NT lines were grown in hydroponic culture solution for 2 weeks and transferred to Fe-deficient culture solution for 7 days. Total RNA was extracted from roots of rice plants and gene expression levels were analyzed. n.d., not detected.
We confirmed the expression levels of OsIRO2, refre1/372 and representative genes regulated by OsIRO2 such as OsNAS1, OsNAS2, OsNAAT1, OsDMAS1, OsYSL15 and TOM1 in hydroponically grown roots after 1-day Fe deficiency cultivation (S1 Fig). Among these genes, OsNAS1, OsNAS2, OsYSL15 and TOM1 showed higher expression level in IRO2 line compared to NT, and in RI lines 21 and 22 compared to Refre1 line. OsNAAT1 and OsDMAS1 were expressed at higher levels in RI lines 21 and 22 compared to Refre1 line.
Fe-deficient hydroponic culture and root Fe(III)-chelate reductase activity
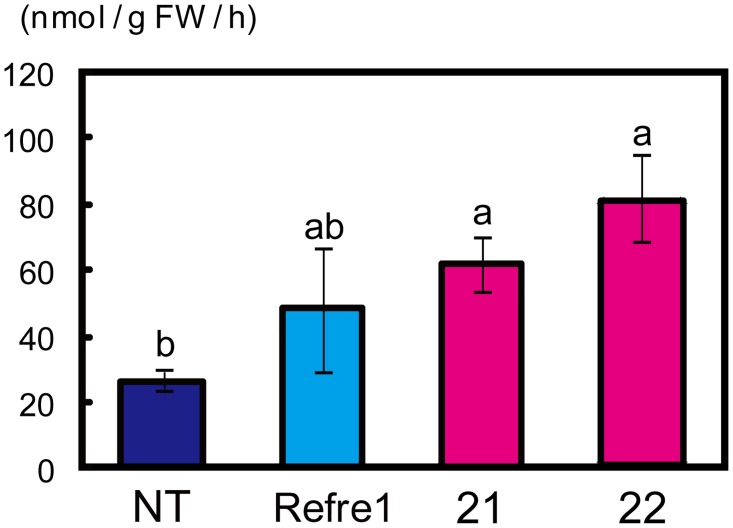
T3 plants of RI lines 21 and 22 and the NT line were grown for 10 days in Fe-sufficient hydroponic culture solution and then transferred to Fe-deficient hydroponic culture solution. Relative chlorophyll contents (measured as SPAD values) of leaves and Fe(III)-chelate reductase activities were determined. T3 plants of the OsIRO2-overexpressing transgenic rice (IRO2 line, [31]) and T3 plants of transgenic rice expressing the OsIRT1 promoter-refre1/372 (Refre1 line, [29]) were also grown and analyzed. The SPAD values of IRO2 and RI lines 21 and 22 were higher than those of the Refre1 and NT lines at 3–10 DAT to Fe-deficient hydroponic culture solution (S2A Fig). At 3 DAT, Fe(III)-chelate reductase activities in RI lines 21 and 22 were 2.8- and 3.1-fold that of the NT line, respectively, and were comparable to the Refre1 line (Fig 2). However, the activities of RI lines 21 and 22 decreased to become lower than that of the Refre1 line at 5 and 7 DAT (S2B Fig).
Fig 2. Assay of Fe(III)-chelate reductase activity.
The non-transgenic line (NT), transgenic rice lines harboring OsIRT1 promoter-refre1/372’ (Refre1), and RI lines 21 and 22 were grown in hydroponic culture solution with Fe for 10 days and then transferred to hydroponic culture solution without Fe. Fe(III)-chelate-reductase activity of roots growing under Fe-deficiency conditions for 3 days were measured (means ± standard error, n = 3). Values with different letters were significantly different by Student’s t-test (p < 0.05).
Tolerance of RI lines to Fe deficiency in calcareous soil
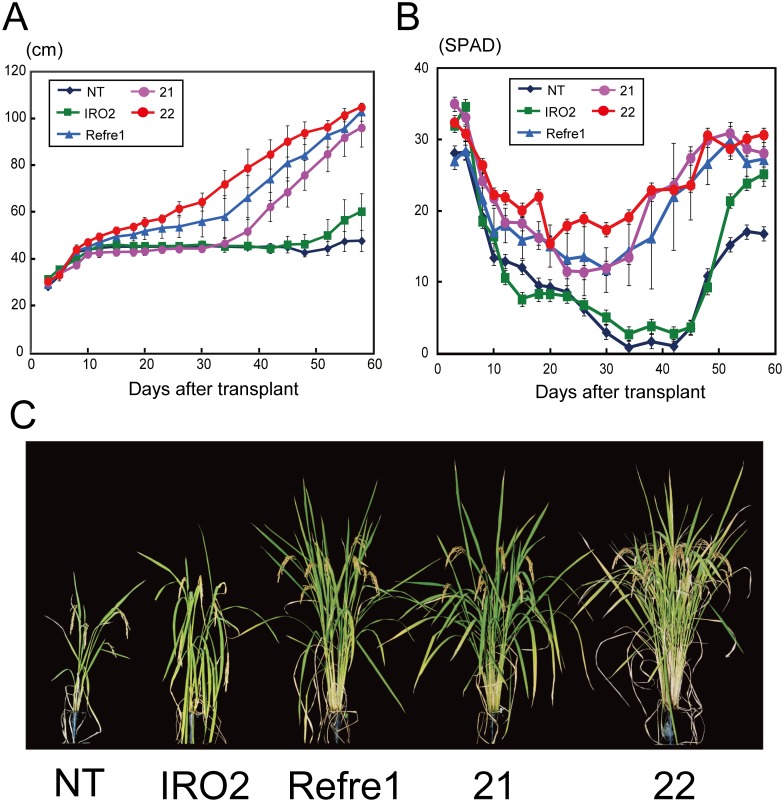
T3 plants of the IRO2 and Refre1 lines, T2 plants of RI lines 21 and 22, and the NT line were grown in calcareous soil (pH = 8.9) under water submerged conditions (S3 Fig). Four plants were cultivated per line. Plant heights and SPAD values of the new leaves were measured (Fig 3A and 3B). The plant growth of RI line 22 and the Refre1 line increased continuously (Fig 3A), while those of the NT and IRO2 lines and RI line 21 did not increase after 10 DAT. The plant height of RI line 21 increased again from 30 DAT, as it gradually recovered from chlorosis. The SPAD values of all plants decreased after transplantation to calcareous soil (Fig 3B). During the first week after transplantation, RI line 21, RI line 22, and the IRO2 line showed markedly higher SPAD values and higher tolerance to low Fe availability compared to the NT and Refre1 lines. After 1 week, the SPAD value of the IRO2 line decreased markedly, whereas those of RI lines 21 and 22, and Refre1 lines decreased to a lesser extent, compared to the NT and IRO2 lines, and then recovered gradually beginning at 30 DAT (Fig 3B). The SPAD value of RI line 22 was higher than those of the other lines at 20–40 DAT. At 41 DAT, newest leaves of the NT and IRO2 lines displayed severe Fe-deficiency symptoms caused by low Fe availability, while RI line 22 grew better than the other lines and exhibited a greater number of tillers (S4 Fig). The SPAD values of NT and IRO2 also increased gradually from 40 DAT, and leaf color recovered, although the recovery was less marked than that of RI lines 21 and 22, and Refre1 lines. At 144 DAT, RI line 22 set more matured grains than did the other transgenic lines and the NT line (Fig 3C).
Fig 3. Growth test of Tsukinohikari-RI rice lines in calcareous soil.
(A) Plant heights after transplantation to calcareous soil. (B) SPAD values (chlorophyll content) of the newest leaves after transplantation to calcareous soil. Means ± standard error, n = 4. (C) Photograph of plants at 144 DAT. NT, non-transgenic line; IRO2, OsIRO2-overexpressing transgenic rice; Refre1, OsIRT1 promoter-refre1/372’ transgenic rice; 21 and 22, RI transgenic rice lines 21 and 22.
Yields of RI lines
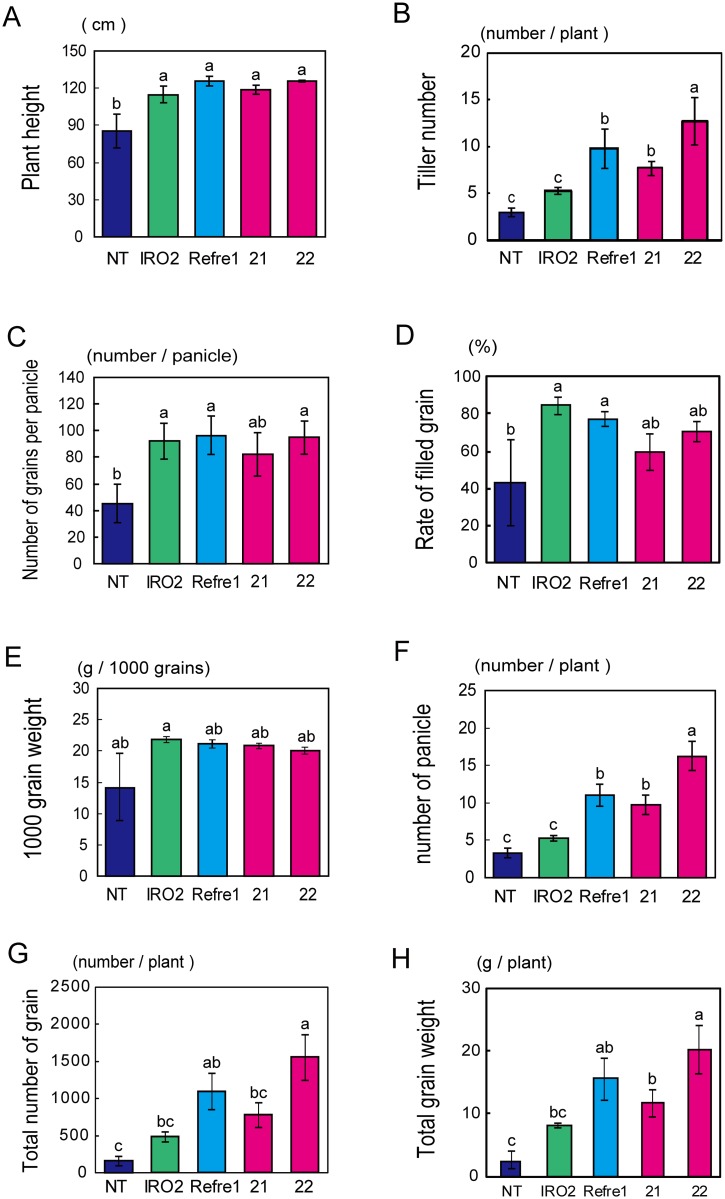
We stopped the water supply at 151 DAT, and harvested the plants at 179 DAT. The average plant heights of the IRO2, Refre1, and RI lines were ~120 cm, but that of the NT line was ~90 cm (Fig 4A). The tiller number of RI line 22 was ~4-fold higher than that of the NT line and 2.4-fold higher than that of the IRO2 line (Fig 4B). Both the straw and panicle dry weights (DW) of RI line 22 were ~10-fold higher than that of NT (S5A and S5B Fig). In terms of yield components, the number of grains per panicle, the rate of filled grains, and the 1,000-grain weight of the IRO2 Refre1 line, and RI lines 21 and 22, were similar, and higher than those of the NT line (Fig 4C–4E). The number of grains per panicle of transgenic rice plants was 2-fold higher than that of the NT line (Fig 4C). The rate of filled grains of transgenic plants was 60–80%, while that of the NT line was around 40% (Fig 4D). The 1,000-grain weight of transgenic rice plants tended to be higher than that of the NT line (Fig 4E). Moreover, the number of panicles per plant, the total number of grains, and the total grain weight were higher in RI line 22 than in the NT and OsIRO2 lines (Fig 4F–4H). The total grain weight of RI line 22 was about 9-fold higher than that of the NT line, and 2.5-fold higher than that of the IRO2 line (Fig 4H). NT plants exhibited severely impaired growth, and produced fewer panicles (S6A Fig). The total number of grains and total grain weight were also very low in NT plants (S6B and S6C Fig). In contrast, all transgenic lines, especially RI line 22, produced higher numbers of panicles and total numbers of grains.
Fig 4. Grain yield and yield components of Tsukinohikari-RI lines cultivated in calcareous soil.
(A) Plant height. (B) Tiller number per plant. (C) Number of grains per panicle. (D) Rate of filled grain. (E) 1000-grain weight. (F) Number of panicles per plant. (G) Total number of grains per plant. (H) Total grain weight per plant. Means ± standard error, n = 4. Non-transgenic line (NT), OsIRO2-overexpressing transgenic rice (IRO2), transgenic rice possessing OsIRT1 promoter-refre1/372’ (Refre1), and RI lines 21 and 22 (21 and 22) were transplanted to calcareous soil and harvested at 179 DAT. Values with different letters were significantly different by Student’s t-test (p < 0.05).
Fe content per plant in RI lines
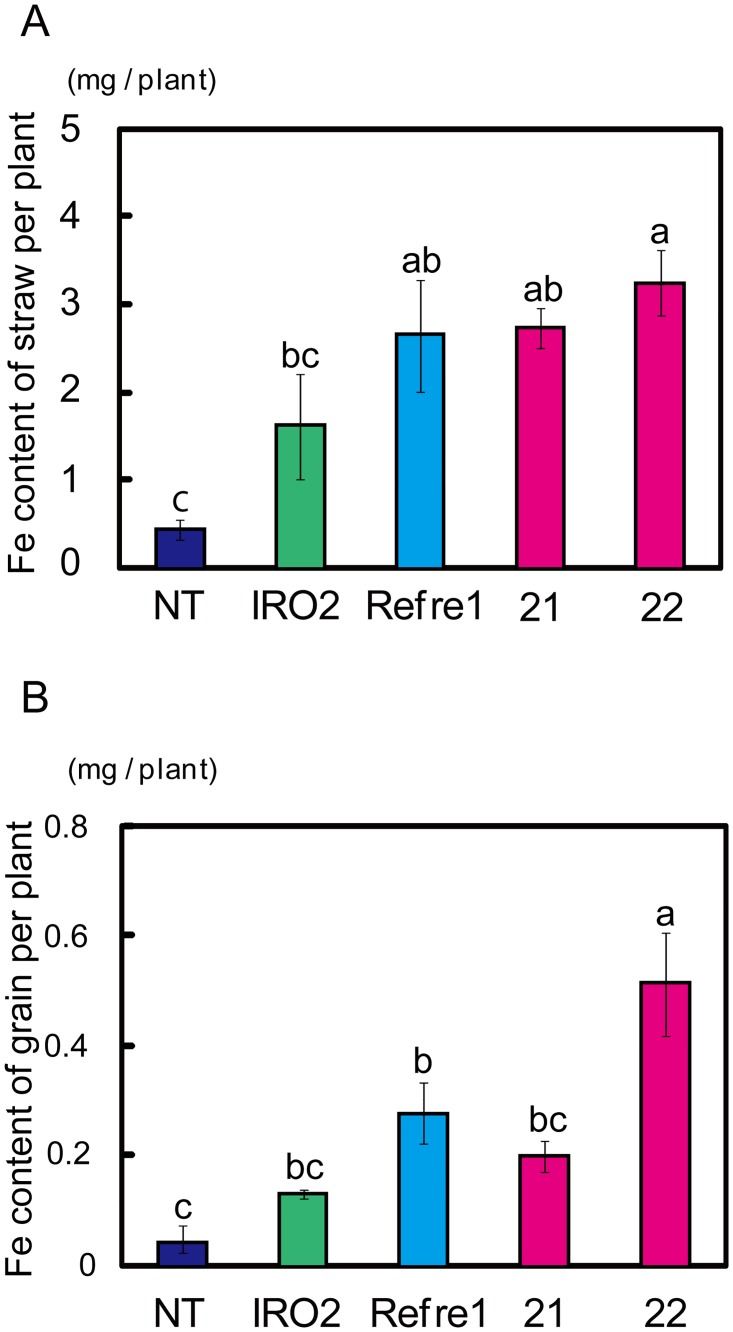
To examine Fe accumulation in RI lines, Fe contents in straw and grain were measured after harvesting. The Fe content in straw of RI line 22 was 3.3 mg/plant, which was 7.3-fold higher than that of the NT line, 2-fold higher than that of the IRO2 line, and 1.2-fold higher than that of the Refre1 line (Fig 5A). Moreover, the Fe content in grain of RI line 22 was around 510 μ g/plant, which was 12-fold higher than that of the NT line, 4-fold higher than that of the IRO2 line, and 2-fold higher than that of the Refre1 line (Fig 5B). Thus, RI line 22 accumulated more Fe in grain and straw than did NT or other transgenic lines.
Fig 5. Fe content of Tsukinohikari-RI seeds and straw per plant.
(A) Fe content of straw per plant. (B) Fe content of grain (brown rice) per plant. Means ± standard error, n = 4. Values with different letters were significantly different by Student’s t-test (p < 0.05). 21 and 22, RI transgenic rice lines 21 and 22.
Production of Fe-deficiency-tolerant high-biomass rice
The construct harboring the 35S promoter-OsIRO2 and OsIRT1 promoter-refre1/372 was also introduced into a high-biomass rice cultivar, Tachisugata [33]. Tachisugata-RI lines were produced and T1 plants were cultivated in calcareous soil. The shoot lengths and SPAD values of the newest leaves of all cultivated lines were measured at 21 DAT (S7A and S7B Fig). Most of the transgenic lines showed higher heights or SPAD values than regular Tachisugata-NT plants. We selected line 39 as it exhibited the greatest value of plant height multiplied by SPAD value among all transgenic lines (S7C Fig). We confirmed that OsIRO2 expression in Fe-deficient roots of Tachisugata-RI line 39 was 4-fold higher than in the NT line (data not shown). Refre1/372 expression was also observed in Fe-deficient roots of line 39 (data not shown).
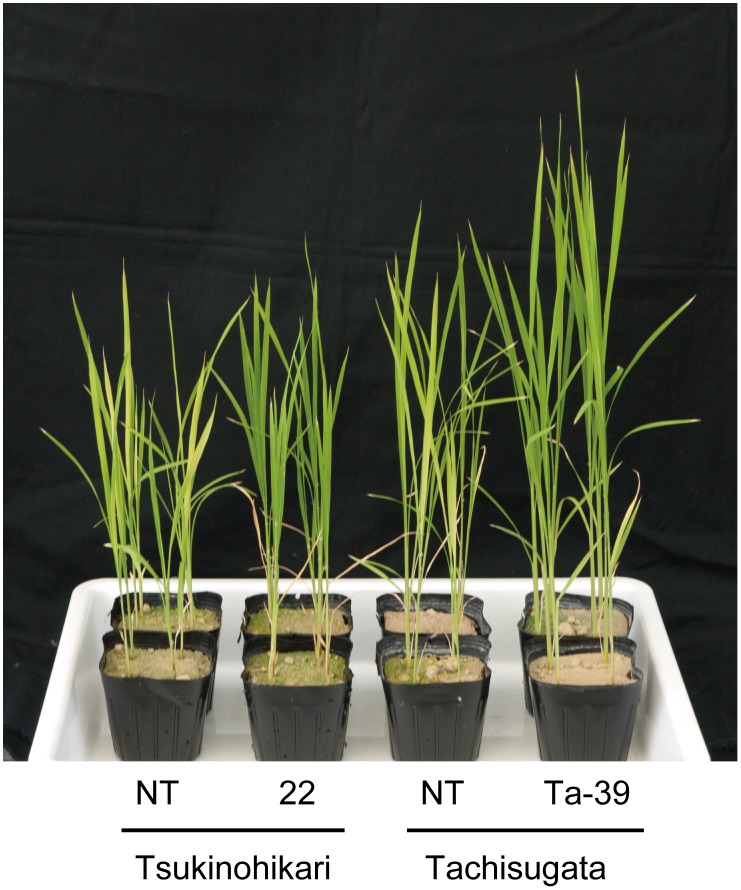
The Tsukinohikari NT line, Tsukinohikari-RI line 22, Tachisugata NT line, and Tachisugata-RI line 39 were cultivated in calcareous soil. Plant heights and SPAD values were measured at 41 DAT. The Tsukinohikari-RI line 22 and the Tachisugata-RI line 39 exhibited better growth, and higher plant heights and SPAD values, compared to the non-transgenic plants (Fig 6 and S8 Fig). Thus, we have successfully produced Fe deficiency-tolerant and high-biomass Tachisugata-RI rice.
Fig 6. Fe-deficiency-tolerant Tachisugata-RI rice.
Photograph taken at 41 DAT to calcareous soil. 22, Tsukinohikari-RI line 22; Ta-39, Tachisugata-RI line 39.
Discussion
RI rice exhibits enhanced growth in calcareous soil
This study is the first to report a combination of both the reduction and transcription factor strategies to achieve greater tolerance to Fe deficiency than either strategy alone. In calcareous soil, transgenic rice line 22, which harbored both OsIRO2 and refre1/372, grew better than the NT, IRO2, and Refre1 lines, with higher shoot heights and SPAD values (Fig 3 and S4 Fig).
Role of OsIRO2 overexpression in Fe-deficiency tolerance
RI lines 21 and 22 and the IRO2 line exhibited higher SPAD values than the NT and Refre1 lines after 10 DAT of Fe deficiency in hydroponic culture (S2A Fig) and within 1 week after transplanting to calcareous soil (Fig 3B). Thus, overexpression of OsIRO2 was confirmed to improve Fe deficiency tolerance at the early stages of growth. OsIRO2 plays a central role as a regulator of genes involved in strategy II-related Fe uptake and translocation [31–32]. OsIRO2 positively regulates the expression of genes involved in the biosynthesis of NA and DMA, as well as those for DMA and Fe(III)-DMA transporters, under both Fe-deficient and -sufficient conditions, and OsIRO2 overexpression results in increased DMA secretion under conditions of Fe deficiency [31–32]. We also confirmed higher expression of OsIRO2-downstream genes involved in DMA biosynthesis, such as OsNAS1, OsNAS2, OsNAAT1 and OsDMAS1, in OsIRO2 lines at an early stage of Fe deficiency (S1 Fig). Thus, the IRO2 line is assumed to be ever ready for strategy II-based tolerance to Fe deficiency before Fe deficiency occurs. Expressions of some of OsIRO2-downstream genes were not higher in RI lines compared to NT, but were higher than Refre1 line (S1 Fig). This might be the effect of enhanced Fe deficiency tolerance conferred by refre1/372.
Nishiyama et al. [41] reported the presence of Fe(III)-DMA in rice phloem sap. The abundant DMA in OsIRO2-overexpressing lines may contribute not only to Fe uptake but also to Fe translocation within rice plants. In contrast, refre1/372 was expressed under the control of the OsIRT1 promoter, which induces gene expression mainly under Fe-deficient conditions in roots of the Refre1 line. Therefore, the Refre1 line likely must encounter Fe-deficient conditions before tolerance can be induced. This might account for the relatively weak tolerance of the Refre1 line to Fe-deficient conditions during the first week of growth in calcareous soil (Fig 3B) and after 10 days of hydroponic culture under Fe-deficient conditions (S2A Fig).
OsIRO2 expression in the NT line is very low under Fe-sufficient conditions [30]. During 10 days of growth on calcareous soil, OsIRO2 expression increases gradually in the NT line, and expression of downstream genes, such as OsNAS1, OsNAS2, OsNAAT1, OsDMAS1, TOM1, and OsYSL15, also increases [32]. Due to this induction in NT, the expression levels of these downstream genes become similar in the NT and IRO2 lines after 10 days of growth in calcareous soil [32]. This Fe deficiency-induced function of endogenous OsIRO2 might account for the relatively small contribution of OsIRO2 overexpression in the IRO2 compared to the NT line during the mid-stage of growth in calcareous soil (Fig 3A and 3B). Nevertheless, the plant height and SPAD value of RI line 22 were higher than that of the Refre1 line in the mid-stage of growth in calcareous soil (Fig 3A and 3B, S4 Fig), which could have resulted from improved efficiency of Fe availability by enhanced DMA biosynthesis.
The Fe(III)-chelate reductase activity in Tsukinohikari-RI lines 21 and 22 was higher at 3 DAT and decreased at 5 and 7 DAT of Fe-deficient hydroponic culture (S2B Fig). At 5, 7, and 10 DAT, the SPAD values were higher in RI lines 21 and 22 than in the NT and Refre1 lines (S2A Fig). It is assumed that improved Fe nutrition in RI lines 21 and 22 resulted in lower induction of Fe-deficiency signal than in the Refre1 line. Several Fe uptake-related genes in rice, including OsIRO2 and OsIRT1, are positively regulated by transcription factor IDEF1, which is also proposed as a possible Fe sensor in rice cells [38, 42]. Thus, induction of Fe(III)-chelate reductase activity by OsIRT1 promoter-driven refre1/372 in RI lines 21 and 22 might not be induced at a high level compared to the Refre1 line at the early Fe-deficient stage because of improved Fe nutrition.
Role of refre1/372 expression in Fe-deficiency tolerance
During the middle and late stages of cultivation on calcareous soil, RI lines and the Refre1 line exhibited superior tolerance compared to the NT and IRO2 lines (Fig 3A and 3B). The Refre1 line had a 2-fold greater Fe(III)-chelate reductase activity than did the NT line under Fe-deficient conditions (Fig 2 and S2B Fig). Because of this advantage, Refre1 line may be more resilient than the NT line in the mid- and late stages of growth in calcareous soil. In contrast, the IRO2 line exhibited only slight tolerance during these growth stages. Ogo et al. [32] and Ishimaru et al. [29] grew the IRO2 and Refre1 lines, respectively, in calcareous soils with low water levels (always less than half of the pot height), which led to significant tolerance compared to the NT line [29, 32, personal communications]. The calcareous soil test of the present study featured a higher water level (continuous submergence). Diffusion of MAs might be increased under such conditions, possibly accounting for the relatively weak contribution of OsIRO2 overexpression. Araki et al. [43] reported dose-dependent effects of DMA application to hydroponic culture solution on improvement of Fe nutrition. Thus, enhancement of the reduction strategy mediated by refre1/372 would be particularly effective in supporting Fe uptake when strategy II-based uptake is insufficient during the mid- and late stages of submerged growth.
In summary, RI lines 21 and 22 were tolerant to Fe deficiency during the early and mid-late stages of cultivation in calcareous soil because of a combination of OsIRT1 promoter-induced refre1/372 expression and overexpression of OsIRO2.
Higher plant survival rate and increased tiller number were the main factors improving grain yield in RI lines
The total grain weight of RI line 22 was 9-fold higher than that of the NT line (Fig 4H). All four plants of the IRO2, Refre1, and RI transgenic lines developed seeds and attained a respectable total grain weight during calcareous soil cultivation (Fig 3C, S6B and S6C Fig). In contrast, in the NT line, the total number of grains and total grain weight were low in two of the four plants and one plant had no filled grains (S6B and S6C Fig). NT plants did not survive well in calcareous soil until the end of their life cycle. The number of surviving plants per unit area in calcareous soil is key to achieve high yields.
The number of grains per panicle, the rate of filled grains, and the 1,000-grain weight were similar among the transgenic lines (Fig 4C–4E). In contrast, the tiller number and number of panicles were higher in RI line 22 than in the other transgenic lines (Fig 4B and 4F). The total number of grains and the total grain weight were also higher in RI line 22 (Fig 4G and 4H). This suggests that tiller number and the number of panicles might affect grain yields in calcareous soil. The number of panicles is determined by the tiller number, and reflected Fe-deficiency-tolerance during early growth. In fact, RI line 22 exhibited a higher plant height and SPAD value during the early and mid-stages of growth in calcareous soil (Fig 3A, 3B and S4 Fig). Therefore, it is crucial for rice cultivated on calcareous soil to survive the early stage of growth. The panicle initiation date was 3 or 4 days earlier in line 22 than in the other transgenic lines, and 1 week earlier than in the NT line (data not shown). Such early panicle initiation may also have contributed to the higher yield of line 22.
Enhanced Fe uptake and translocation to grains in RI lines
The per-plant Fe contents of straw and grain were markedly higher in RI lines compared to the NT line (Fig 5A and 5B). This was likely due to a combination of enhanced Fe uptake ability from soil by refre1/372 induction and OsIRO2 overexpression, and enhanced Fe translocation mediated by increased NA and DMA levels caused by OsIRO2 overexpression. Masuda et al. [39] and Lee et al. [44] showed that enhancement of NA and DMA productivity enhances Fe translocation in rice plants, yielding grain of high Fe content. Masuda et al. [45] also showed that MA production increased the Fe content of rice grains. In the present calcareous test, the mean Fe concentrations in brown seeds were 20, 18, 19, 19, and 30 μg/g in the NT, IRO2, Refre1, RI 21, and RI 22 lines, respectively; these levels did not differ significantly (data not shown). Takahashi et al. [46] showed that NA-chelated metals, such as Fe and Zn, are essential, especially during grain maturation. Rice plants may have a system that mediates accumulation of a certain amount of Fe in each grain. Under Fe-limited conditions, NT plants may set fewer seeds to ensure distribution of the limited Fe available to all seeds. Therefore, NT plants have many unfilled grains (Fig 4D). In contrast, transgenic lines with improved Fe uptake and translocation likely produce and distribute Fe to, a greater number of grains, but accumulate only the required amount of Fe in each seed. Ogo et al. [32] reported higher Fe concentration in brown seeds of the IRO2 line (up to 20 μg/g) than that of the NT line (6 μg/g) which is lower than that in ordinary grains of this cultivar (Tsukinohikari). This might account for the 3-fold higher Fe concentration in grains of the IRO2 line by Ogo et al. [32].
Irrespective of the similar Fe concentrations in each grain of the NT, IRO2, Refre1 and RI lines in the present report, the markedly increased grain weight in the IRO2, Refre1, and RI lines resulted in higher Fe accumulation in grains per plant (Figs 4H and 5B). Enhanced Fe(III)-reduction and transcription factor strategies may thus be used for Fe biofortification of rice seeds cultivated under low Fe conditions. On the other hand, other strategies are also required in combination to consistently accumulate more Fe in seeds [47].
Production of an Fe-deficiency-tolerant, high-biomass rice line
We successfully produced an Fe-deficiency-tolerant high-biomass Tachisugata rice cultivar (Fig 6, S7 and S8 Figs). This cultivar has long and thick culms, is highly resistant to lodging, and is adapted to direct-sowing cultivation. Thus, this cultivar is suitable for production of biomass or whole-crop silage for use as fodder [33]. Moreover, an efficient on-site ethanol production system using Tachisugata rice is under development [48]. Therefore, the Fe deficiency-tolerant Tachisugata rice produced in this study may be useful. Moreover, Fe deficiency-tolerant lines of other high-biomass crops—such as maize, sugarcane, or sorghum—could be produced using Fe(III)-chelate reductase genes and enhancement of the expression of Fe homeostasis-related transcription factors, such as OsIRO2 homologs.
Other approaches to production of Fe-deficiency-tolerant rice lines
Transgenic rice plants harboring IDEF1 under the control of the barley Fe deficiency-inducible IDS2 promoter exhibited improved Fe-deficiency tolerance during early growth, but not during the mid-late stages in calcareous soils [42, 49]. Ishimaru et al. [50] identified the rice phenolic efflux transporter, PEZ1, and reported that PEZ1-overexpressing rice showed enhanced growth in calcareous soil. Kobayashi et al. [49] reported that the knock-down of Fe-binding hemerythrin RING ubiquitin ligases, OsHRZ1 and OsHRZ2, in rice resulted in enhanced expression of Fe deficiency-inducible genes involved in Fe utilization, and enhanced tolerance to Fe deficiency during growth in calcareous soil. This HRZ-knockdown rice also showed markedly higher accumulation of Fe in grains and shoots under both Fe-sufficient and -deficient conditions [49], and thus is a promising candidate for both improved production in problem soils and biofortification. Introduction of barley MA biosynthesis genes (HvNAS1, HvNAAT-A,B, or IDS3) also rendered rice plants tolerant to Fe deficiency [26,27,51]. These reports suggest that rice lines with enhanced tolerance to Fe deficiency could be produced using a combination of these various transgenic approaches.
Conclusion
RI rice lines had higher yields than the NT line because of the enhanced tolerance to low Fe availability at both the early and mid-late stages of growth in calcareous soil. All plants survived and tiller number increased at all growth stages. Also, Fe was effectively transported to seeds. Thus, the rate of filled grains improved at the grain-maturation stage. Therefore, we have successfully produced transgenic rice lines affording increased grain yields in calcareous soils by means of both enhanced reduction (Strategy I) and chelation (Strategy II). We also successfully produced a high-biomass rice variety with increased tolerance to Fe deficiency for increased grain yield and higher biomass productivity.
Supporting information
Non-transgenic line (NT), and transgenic rice expressing the 35S promoter-OsIRO2 (IRO2), the OsIRT1 promoter-refre1/372’ (Refre1), and the RI lines (21 and 22) were grown in hydroponic culture solution for 10 days and transferred to Fe-deficient culture solution for 1 day. Total RNA was extracted from roots and gene expression levels were analyzed by quantitative RT-PCR. Data are shown as copies of each gene / OsTublin1 copies [means ± standard error of technical replication, n = 3 except for OsNAAT1 (n = 1)]. n.d., not detected.
(PDF)
(A) SPAD value of the newest leaves. (B) Fe(III)-chelate reductase activity. Non-transgenic line (NT), and transgenic rice expressing the 35S promoter-OsIRO2 (IRO2), the OsIRT1 promoter-refre1/372’ (Refre1), and the RI lines (21 and 22) were grown in hydroponic culture solution for 10 days and then transferred to hydroponic culture solution without Fe. SPAD values and Fe(III)-chelate reductase activities of roots were measured at the indicated days after onset of Fe-deficiency treatment (means ± standard error, n = 4).
(JPG)
(JPG)
NT, non-transformant; IRO2, OsIRO2-overexpressing transgenic rice; Refre1, OsIRT1 promoter-refre1/372’ transgenic rice; 21 and 22, RI transgenic rice lines 21 and 22.
(JPG)
(A) Dry weight of straw per plant. (B) Dry weight of panicle per plant. Plants were harvested after 179 days of growth in calcareous soil, and the straw and panicle weight per plant was analyzed (means ± standard error, n = 4). Values followed by different letters were significantly different according by Tukey–Kramer’s HSD test (p < 0.05). NT, non-transformant; IRO2, OsIRO2-overexpressing transgenic rice; Refre1, OsIRT1 promoter-refre1/372’ transgenic rice; 21 and 22, RI lines 21 and 22.
(JPG)
(A) Number of panicles per plant. (B) Total number of grains per plant. (C) Total grain weight per plant. Each bar shows the value of a single plant (T2 sub-lines of transgenic plants or NT plants) cultivated in calcareous soil. NT, non-transformant; IRO2, OsIRO2-overexpressing transgenic rice; Refre1, OsIRT1 promoter-refre1/372’ transgenic rice; 21 and 22, RI lines 21 and 22.
(JPG)
(A) Plant height of individual lines. (B) SPAD value of the newest leaves of individual lines. (C) Value of plant height multiplied by the SPAD value of each plant used for line selection. Plant heights and SPAD values were measured at 21 DAT to calcareous soil. We selected Tachisugata-RI line 39 as it yielded the highest value of (C). A–E on the X-axis indicate non-transgenic Tachisugata plants. Numbers on the X-axis represent individual Tachisugata-RI lines. White circles on each line are data from individual plants. Red circles on the NT lines are the mean values of eight NT plants. Black circles are the mean values of eight plants of each Tachisugata-RI line. The red dotted line shows the mean values of all 40 NT plants.
(JPG)
(A) Height of individual plants. (B) SPAD value of individual plants. TK-NT, Tsukinohikari non-transgenic plants; 22, Tsukinohikari-RI line 22; Ta-NT, Tachisugata non-transgenic plants; Ta-39, Tachisugata-RI line 39. Values for eight individual plants of each non-transgenic and transgenic line are shown in individual bars.
(JPG)
Acknowledgments
We thank Dr. Kazuhiko Sugimoto and Dr. Kazuko Ono of the Rice Applied Genomics Center, National Institute of Agrobiological Sciences, Tsukuba, Ibaraki, Japan for advice on the Tachisugata rice transformation method. We are grateful to Nihonkai Kougyou, Toyama, Japan for kindly providing calcareous soil. We also thank Mr. Keisuke Minamide of Ishikawa Prefectural University, Nonoichi, Ishikawa, Japan for advice on rice cultivation in calcareous soil.
Data Availability
All relevant data are within the paper and its Supporting Infromation files.
Funding Statement
This work was supported by the Advanced Low Carbon Technology Research and Development Program (ALCA) of the Japan Science and Technology Agency (JST) to NK Nishizawa and a Grant-in-Aid for Scientific Research (A) from the Japan Society for the Promotion of Sciences (JSPS) to NK Nishizawa.
References
- 1.Marschner H. 1995. Mineral Nutrition of Higher Plants. 2nd Edition Academic Press, London. [Google Scholar]
- 2.Briat JF, Dubos C, Gaymard F. Iron nutrition, biomass production, and plant product quality. Trends Plant Sci. 2015;20: 33–40 10.1016/j.tplants.2014.07.005 [DOI] [PubMed] [Google Scholar]
- 3.Römheld V, Marschner M. Evidence for a specific uptake system for iron phytosiderophores in roots of grasses. Plant Physiol. 1986;80: 175–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robinson NJ, Procter CM, Connolly EL, Guerinot ML. A ferric-chelate reductase for iron uptake from soils. Nature. 1999;397: 694–697. 10.1038/17800 [DOI] [PubMed] [Google Scholar]
- 5.Connolly EL, Campbell NH, Grotz N, Prichard CL, Guerinot ML. Overexpression of the FRO2 ferric chelate reductase confers tolerance to growth on low iron and uncovers posttranscriptional control. Plant Physiol. 2003;113: 1102–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eide D, Brodrius M, Fett J, Guerinot ML. A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Pro Nat Aca Sci USA. 1996;93: 5624–5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bughio N, Yamaguchi H, Nishizawa NK, Nakanishi H, Mori S. Cloning an iron-regulated metal transporter from rice. J Exp Bot. 2002;53: 1677–1682. [DOI] [PubMed] [Google Scholar]
- 8.Ishimaru Y, Suzuki M, Tsukamoto T, Suzuki K, Nakazono M, Kobayashi T, et al. Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+. Plant J. 2006;45: 335–346. 10.1111/j.1365-313X.2005.02624.x [DOI] [PubMed] [Google Scholar]
- 9.Takagi S. Naturally occurring iron-chelating compounds in oat- and rice-root washings. I. Activity measurement and preliminary characterization. Soil Sci Plant Nutr. 1976;22: 423–433. [Google Scholar]
- 10.Nozoye T, Nagasaka S, Kobayashi T, Takahashi M, Sato Y, Sato Y, et al. Phytosiderophore Efflux Transporters Are Crucial for Iron Acquisition in Graminaceous Plants. J Biol Chem. 2011;286: 5446–5454. 10.1074/jbc.M110.180026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curie C, Panaviene Z, Loulergue C, Dellaporta SL, Briat JF, Walker EL. Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature. 2001;409: 346–349. 10.1038/35053080 [DOI] [PubMed] [Google Scholar]
- 12.Inoue H, Kobayashi T, Nozoye T, Takahashi M, Kakei Y, Suzuki K, et al. Rice OsYSL15 is an iron-regulated iron(III)-deoxymugineic acid transporter expressed in the roots and is essential for iron uptake in early growth of the seedlings. J Biol Chem. 2009;284: 3470–3479. 10.1074/jbc.M806042200 [DOI] [PubMed] [Google Scholar]
- 13.Lee S, Chiecko JC, Kim SA, Walker EL, Lee Y, Guerinot ML, et al. Disruption of OsYSL15 leads to iron inefficiency in rice plants. Plant Physiol. 2009;150: 786–800. 10.1104/pp.109.135418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murata Y, Ma JF, Yamaji N, Ueno D, Nomoto K, Iwashita T. A specific transporter for iron(III)-phytosiderophore in barley roots. Plant J. 2006;46: 563–572. 10.1111/j.1365-313X.2006.02714.x [DOI] [PubMed] [Google Scholar]
- 15.Mori S, Nishizawa N. Methionine as a dominant precursor of phytosiderophore in graminaceae plant. Plant Cell Physiol. 1987;28: 1081–1092. [Google Scholar]
- 16.Shojima S, Nishizawa NK, Fushiya S, Nozoe S, Irifune T, Mori S. Biosynthesis of phytosiderophores—invitro biosynthesis of 2'-deoxymugineic acid from l-methionine and nicotianamine. Plant Physiol. 1990;93: 1497–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Higuchi K, Suzuki K, Nakanishi H, Yamaguchi H, Nishizawa NK, Mori S. Cloning of nicotianamine synthase genes, novel genes involved in the biosynthesis of phytosiderophores. Plant Physiol. 1999;119: 471–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma JF, Taketa S, Chang YC, Iwashita T, Matsumoto H, Takeda K, et al. Genes controlling hydroxylations of phytosiderophores are located on different chromosomes in barley (Hordeum vulgare L.). Planta. 1999;207: 590–596. [Google Scholar]
- 19.Takahashi M, Yamaguchi H, Nakanishi H, Shioiri T, Nishizawa NK, Mori S. Cloning two genes for nicotianamine aminotransferase, a critical enzyme in iron acquisition (Strategy II) in graminaceous plants. Plant Physiol. 1999;121: 947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakanishi H, Yamaguchi H, Sasakuma T, Nishizawa NK, Mori S. Two dioxygenase genes, Ids3 and Ids2, from Hordeum vulgare are involved in the biosynthesis of mugineic acid family phytosiderophores. Plant Mol. Biol. 2000;44: 199–207. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi T, Nakanishi H, Takahashi M, Kawasaki S, Nishizawa NK, Mori S. In vivo evidence that Ids3 from Hordeum vulgare encodes a dioxygenase that converts 2’-deoxymugineic acid to mugineic acid in transgenic rice. Planta. 2001;212: 864–871. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi T, Suzuki M, Inoue H, Itai RN, Takahashi M, Nakanishi H, et al. Expression of iron-acquisition-related genes in iron-deficient rice is co-ordinately induced by partially conserved iron-deficiency-responsive elements. J Exp Bot. 2005;56: 1305–1316. 10.1093/jxb/eri131 [DOI] [PubMed] [Google Scholar]
- 23.Bashir K, Inoue H, Nagasaka S, Takahashi M, Nakanishi H, Mori S, et al. Cloning and characterization of deoxymugineic acid synthase genes from graminaceous plants. J Biol Chem. 2006;281: 32395–32402. 10.1074/jbc.M604133200 [DOI] [PubMed] [Google Scholar]
- 24.Nozoye T, Nagasaka S, Takahashi M, Bashir K, Takahashi M, Kobayashi T et al. Nicotianamine synthase 2 localizes to the vesicles of iron-deficient rice roots, and its mutation in the YXXΦ or LL motif causes the disruption of vesicle formation or movement in rice. Plant J. 2014; 77:246–260. 10.1111/tpj.12383 [DOI] [PubMed] [Google Scholar]
- 25.Marschner H, Römheld V, Kissel M. Different strategies in higher plants in mobilization and uptake of iron. J Plant Nut. 1986;9: 695–713. [Google Scholar]
- 26.Takahashi M, Nakanishi H, Kawasaki S, Nishizawa NK, Mori S. Enhanced tolerance of rice to low iron availability in alkaline soils using barley nicotianamine aminotransferase genes. Nat Biotech. 2001;19: 466–469. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki M, Morikawa C, Nakanishi H, Takahashi M, Saigusa M, Mori S, et al. Transgenic rice lines that include barley genes have increased tolerance to low iron availability in a calcareous paddy soil. Soil Sci Plant Nutr. 2008;54: 77–85. [Google Scholar]
- 28.Oki H, Kim S, Nakanishi H, Takahashi M, Yamaguchi H, Mori S, et al. Directed evolution of yeast ferric reductase to produce plants with tolerance to iron deficiency in alkaline soils. Soil Sci Plant Nut. 2004;50: 1159–1165. [Google Scholar]
- 29.Ishimaru Y, Kim S, Tsukamoto T, Oki H, Kobayashi T, Watanabe S, et al. Mutational reconstructed ferric chelate reductase confers enhanced tolerance in rice to iron deficiency in calcareous soil. Pro Nat Aca Sci USA. 2007;104: 7373–7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogo Y, Itai RN, Nakanishi H, Inoue H, Kobayashi T, Suzuki M, et al. Isolation and characterization of IRO2, a novel iron-regulated bHLH transcription factor in graminaceous plants. J Exp Bot. 2006;57: 2867–2878. 10.1093/jxb/erl054 [DOI] [PubMed] [Google Scholar]
- 31.Ogo Y, Itai RN, Nakanishi H, Kobayashi T, Takahashi M, Mori S, et al. The rice bHLH protein OsIRO2 is an essential regulator of the genes involved in Fe uptake under Fedeficient conditions. Plant J. 2007;51: 366–377. 10.1111/j.1365-313X.2007.03149.x [DOI] [PubMed] [Google Scholar]
- 32.Ogo Y, Itai RN, Kobayashi T, Aung MS, Nakanishi H, Nishizawa NK. OsIRO2 is responsible for iron utilization in rice and improves growth and yield in calcareous soil. Plant Mol Biol. 2011;75: 593–605. 10.1007/s11103-011-9752-6 [DOI] [PubMed] [Google Scholar]
- 33.Ohta H, Nemoto H, Ando I, Kato H, Sato H, Hirabayashi H, et al. "Tachisugata", a new rice cultivar for whole crop silage use. Bull Natl Inst Crop Sci. 2010;11: 67–8 (Japanese). [Google Scholar]
- 34.Mitsuhara I, Ugaki M, Hirochika H, Ohshima M, Murakami T, Gotoh Y, et al. Efficient Promoter Cassettes for Enhanced Expression of Foreign Genes in Dicotyledonous and Monocotyledonous Plants. Plant Cell Physiol. 1996; 37: 49–59. [DOI] [PubMed] [Google Scholar]
- 35.Sallaud C, Meynard D, van Boxtel J, Gay C, Bès M, Brizard JP, et al. Highly efficient production and characterization of T-DNA plants for rice (Oriza sativa L.) functional genomics. Theor Appl Genet. 2003;106: 1396–1408. 10.1007/s00122-002-1184-x [DOI] [PubMed] [Google Scholar]
- 36.Terada R, Asao H, Iida S. A large-scale Agrobacterium-mediated transformation procedure with a strong positive-negative selection for gene targeting in rice (Oryza sativa L.). Plant Cell Rep. 2004;22: 653–659. 10.1007/s00299-003-0752-0 [DOI] [PubMed] [Google Scholar]
- 37.Thomson D, Henry R. Single-step protocol for preparation of plant tissue for analysis by PCR. BioTechniques. 1995;19: 394–397. [PubMed] [Google Scholar]
- 38.Kobayashi T, Itai RN, Ogo Y, Kakei Y, Nakanishi H, Takahashi M, et al. The rice transcription factor IDEF1 is essential for the early response to iron deficiency, and induces vegetative expression of late embryogenesis abundant genes. Plant J. 2009;60: 948–961. 10.1111/j.1365-313X.2009.04015.x [DOI] [PubMed] [Google Scholar]
- 39.Masuda H, Usuda K, Kobayashi T, Ishimaru Y, Kakei Y, Takahashi M, et al. Overexpression of the barley nicotianamine synthase gene HvNAS1 increase iron and zinc concentrations in rice grains. Rice. 2009;2: 155–166. [Google Scholar]
- 40.Hiei Y., Ohta S., Komari T., Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the bounderies of the T-DNA. Plant J. 1994;6:271–282. [DOI] [PubMed] [Google Scholar]
- 41.Nishiyama R, Kato M, Nagata S, Yanagisawa S, Yoneyama T (2012) Identification of Zn-nicotianamine and Fe-2′-Deoxymugineic acid in the phloem sap from rice plants (Oryza sativa L.). Plant Cell Physiol 53:381–390 10.1093/pcp/pcr188 [DOI] [PubMed] [Google Scholar]
- 42.Kobayashi T, Ogo Y, Itai RN, Nakanishi H, Takahashi M, Mori S, et al. The transcription factor IDEF1 regulates the response to and tolerance of iron deficiency in plants. Pro Nat Aca Sci USA. 2007;104: 19150–19155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Araki R, Kousaka K, Namba K, Murata Y, Murata J. 2'-Deoxymugineic acid promoters growth of rice (Oryza sativa L.) by orchestrating iron and nitrate uptake processes under high pH conditions. 2015; 81: 233–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee S, Jeon US, Lee SJ, Kim YK, Persson DP, Husted S, et al. Iron fortification of rice seeds through activation of the nicotianamine synthase gene. Proc Natl Acad Sci USA. 2009;106: 22014–22019. 10.1073/pnas.0910950106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Masuda H, Suzuki M, Morikawa KC, Kobayashi T, Nakanishi H, Takahashi M, et al. Increase in iron and zinc concentrations in rice grains via the introduction of barley genes involved in phytosiderophore synthesis. Rice. 2008;1: 100–108. [Google Scholar]
- 46.Takahashi M, Terada Y, Nakai I, Nakanishi H, Yoshimura E, Mori S, et al. Role of nicotianamine in the intracellular delivery of metals and plant reproductive development. Plant Cell. 2003;15: 1263–1280. 10.1105/tpc.010256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Masuda H, Aung May Sann, Nishizawa NK. Iron biofortification of rice using different transgenic approaches. Rice. 2013;6: 40 10.1186/1939-8433-6-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Horita M, Kitamoto H Kawaide T Tachibana Y Shinozaki Y. On-farm solid state simultaneous saccharification and fermentation of whole crop forage rice in wrapped round bale for ethanol production. Biotech Biofuels. 2015;8: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kobayashi T, Nagasaka S, Senoura T, Itai RN, Nakanishi H, Nishizawa NK. Iron-binding haemerythrin RING ubiquitin ligases regulate plant iron responses and accumulation. Nat Commun 2013;4: 2792 10.1038/ncomms3792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ishimaru Y, Kakei Y, Shimo H, Bashir K, Sato Y, Sato Y et al. A Rice Phenolic Efflux Transporter is essential for solubilizing precipitated apoplasmic iron in the plant stele. J Biol Chem. 2011. 286 24649–24655 10.1074/jbc.M111.221168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Masuda H, Kobayashi T, Ishimaru Y, Takahashi M, Aung MS, Nakanishi H, et al. Iron-biofortification in rice by the introduction of three barley genes participated in mugineic acid biosynthesis with soybean ferritin gene. Front Plant Sci. 2013;4: 132 10.3389/fpls.2013.00132 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Non-transgenic line (NT), and transgenic rice expressing the 35S promoter-OsIRO2 (IRO2), the OsIRT1 promoter-refre1/372’ (Refre1), and the RI lines (21 and 22) were grown in hydroponic culture solution for 10 days and transferred to Fe-deficient culture solution for 1 day. Total RNA was extracted from roots and gene expression levels were analyzed by quantitative RT-PCR. Data are shown as copies of each gene / OsTublin1 copies [means ± standard error of technical replication, n = 3 except for OsNAAT1 (n = 1)]. n.d., not detected.
(PDF)
(A) SPAD value of the newest leaves. (B) Fe(III)-chelate reductase activity. Non-transgenic line (NT), and transgenic rice expressing the 35S promoter-OsIRO2 (IRO2), the OsIRT1 promoter-refre1/372’ (Refre1), and the RI lines (21 and 22) were grown in hydroponic culture solution for 10 days and then transferred to hydroponic culture solution without Fe. SPAD values and Fe(III)-chelate reductase activities of roots were measured at the indicated days after onset of Fe-deficiency treatment (means ± standard error, n = 4).
(JPG)
(JPG)
NT, non-transformant; IRO2, OsIRO2-overexpressing transgenic rice; Refre1, OsIRT1 promoter-refre1/372’ transgenic rice; 21 and 22, RI transgenic rice lines 21 and 22.
(JPG)
(A) Dry weight of straw per plant. (B) Dry weight of panicle per plant. Plants were harvested after 179 days of growth in calcareous soil, and the straw and panicle weight per plant was analyzed (means ± standard error, n = 4). Values followed by different letters were significantly different according by Tukey–Kramer’s HSD test (p < 0.05). NT, non-transformant; IRO2, OsIRO2-overexpressing transgenic rice; Refre1, OsIRT1 promoter-refre1/372’ transgenic rice; 21 and 22, RI lines 21 and 22.
(JPG)
(A) Number of panicles per plant. (B) Total number of grains per plant. (C) Total grain weight per plant. Each bar shows the value of a single plant (T2 sub-lines of transgenic plants or NT plants) cultivated in calcareous soil. NT, non-transformant; IRO2, OsIRO2-overexpressing transgenic rice; Refre1, OsIRT1 promoter-refre1/372’ transgenic rice; 21 and 22, RI lines 21 and 22.
(JPG)
(A) Plant height of individual lines. (B) SPAD value of the newest leaves of individual lines. (C) Value of plant height multiplied by the SPAD value of each plant used for line selection. Plant heights and SPAD values were measured at 21 DAT to calcareous soil. We selected Tachisugata-RI line 39 as it yielded the highest value of (C). A–E on the X-axis indicate non-transgenic Tachisugata plants. Numbers on the X-axis represent individual Tachisugata-RI lines. White circles on each line are data from individual plants. Red circles on the NT lines are the mean values of eight NT plants. Black circles are the mean values of eight plants of each Tachisugata-RI line. The red dotted line shows the mean values of all 40 NT plants.
(JPG)
(A) Height of individual plants. (B) SPAD value of individual plants. TK-NT, Tsukinohikari non-transgenic plants; 22, Tsukinohikari-RI line 22; Ta-NT, Tachisugata non-transgenic plants; Ta-39, Tachisugata-RI line 39. Values for eight individual plants of each non-transgenic and transgenic line are shown in individual bars.
(JPG)
Data Availability Statement
All relevant data are within the paper and its Supporting Infromation files.