Abstract
Background
The prominent role of mitochondria in the generation of reactive oxygen species, cell death, and energy production contributes to the importance of this organelle in the intracellular mechanism underlying the progression of the common sensory disorder of the elderly, presbycusis. Reduced mitochondrial DNA (mtDNA) gene expression and coding region variation have frequently been reported as being associated with the development of presbycusis. The mtDNA control region regulates gene expression and replication of the genome of this organelle. To comprehensively understand of the role of mitochondria in the progression of presbycusis, we compared variations in the mtDNA control region between subjects with presbycusis and controls.
Methods
A total of 58 presbycusis patients and 220 control subjects were enrolled in the study after examination by the otolaryngologist and audiology tests. Variations in the mtDNA control region were investigated by polymerase chain reaction and Sanger sequencing.
Results
A total of 113 sequence variants were observed in mtDNA, and variants were detected in 100% of patients, with 84% located in hypervariable regions. The frequencies of the variants, 16,223 C>T, 16,311 T>C, 16,249 T>C, and 15,954 A>C, were significantly different between presbycusis and control subjects.
Conclusion
The statistically significant difference in the frequencies of four nucleotide variants in the mtDNA control region of presbycusis patients and controls is in agreement with previous experimental evidence and supports the role of mitochondria in the intracellular mechanism underlying presbycusis development. Moreover, these variants have potential as diagnostic markers for individuals at a high risk of developing presbycusis. The data also suggest the possible presence of changes in the mtDNA control region in presbycusis, which could alter regulatory factor binding sites and influence mtDNA gene expression and copy number.
Keywords: age-related hearing impairment, presbycusis, mtDNA control region, audiology
Introduction
Presbycusis, or age-related hearing impairment, is the most prevalent sensory disorder in aging people.1,2 Presbycusis is defined as the progressive decline of hearing ability during aging and causes symmetrical, bilateral, high-frequency sensorineural hearing impairment.3 As a multifactorial disorder, presbycusis is associated with a combination of environmental factors, medical history, and nuclear and mitochondrial genome variation.4–6 Presbycusis has a great impact on the quality of life of patients as it impairs communication skills and independent daily activity and, therefore, gradually diminishes psychosocial functioning.7 Patients with the disorder frequency experience decreased self-esteem and increased dependency, isolation, and frustration. A study by Lin et al8 using the definition of the World Health Organization and information from the National Health and Nutritional Examination Survey demonstrated that 63.1% of the population more than 70 years old suffer from a hearing impairment of ≥25 dB. Currently, socioeconomic development and health care improvement have led to increased longevity. In contrast, while the elderly population exhibits exponential growth, fertility has reduced. Due to the growing elderly population, the prevalence of age-related disorders, such as presbycusis, is also increasing.9
Mitochondria are considered one of the main factors in the progression of presbycusis;4 these organelles are responsible for vital cellular functions, including energy production, apoptosis, cell signaling, and calcium storage. Variants in the mtDNA control region are associated with different disorders, including cancer, Huntington disease, and β-thalassemia.10–12 The control region is composed of three hypervariable regions (HVR), HVR-I at 16,024−16,383, HVR-II at 57−372, and HVR-III at 438−574 (numbering according to NCBI Accession No NC_0122920.1).13 A strong relationship between mtDNA coding region variation and presbycusis has also been reported in the literature;14–16 however, little is known about the relationship between variation in the mtDNA control region and the progression of presbycusis. Advances in the understanding of the intracellular mechanisms underlying presbycusis could lead to the development of diagnostic markers and therapies to slow or reverse the adverse changes in the auditory system characteristic of this disorder. In the present study, to determine the role of mtDNA in the intracellular mechanism underling the development of presbycusis, we compared the frequency of variants in the mtDNA control region sequences of presbycusis patients with those of control subjects.
Materials and methods
Patients and samples
Presbycusis and control subjects were enrolled in this study by ear-nose-and-throat (ENT) specialists at the ENT and Head & Neck Research Center of Iran University of Medical Sciences. For all participants, ENT examination, conventional pure-tone audiometry, tympanometry, Speech Discrimination Scores, and Speech Reception Thresholds were carried out, as described previously.17 Briefly, an Amplaid 319 audiometer (Amplaid Inc., Milan, Italy) was used for pure-tone audiometry. Air and bone conduction thresholds were measured at 250–8,000 and 250–4,000 Hz, respectively. Calibration was performed according to the American National Standards Institute. A multifrequency admittance meter (Amplaid 728) was used for tympanometry.
Patients with symmetric, bilateral, sensorineural hearing impairment with pure-tone average (PTA) frequencies of ≥30 dB HL at 1, 2, 4, and 8 kHz were defined as having presbycusis and were included in the study. Control subjects had normal ear examinations and PTA frequencies of <25 dB HL.
Exclusion criteria were history of early sensorineural hearing impairment, any ear disease or otological surgery, conductive hearing impairment, exposure to noise or ototoxic drugs, and a medical history that could have affected hearing sensitivity, such as brain tumor, kidney or liver failure, cardiovascular disease, or stroke.18 Finally, 58 presbycusis and 220 control subjects were enrolled in this study.
This study was approved by the Ethics Committee of Iran University of Medical Sciences. Participants signed informed consent letters, according to the Declaration of Helsinki. Peripheral blood samples (5 mL) were collected from all participants in EDTA collection tubes.
DNA extraction and genotyping
Total DNA samples were extracted from peripheral blood using a QIAamp DNA mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The quality and purity of extracted DNA were measured using a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
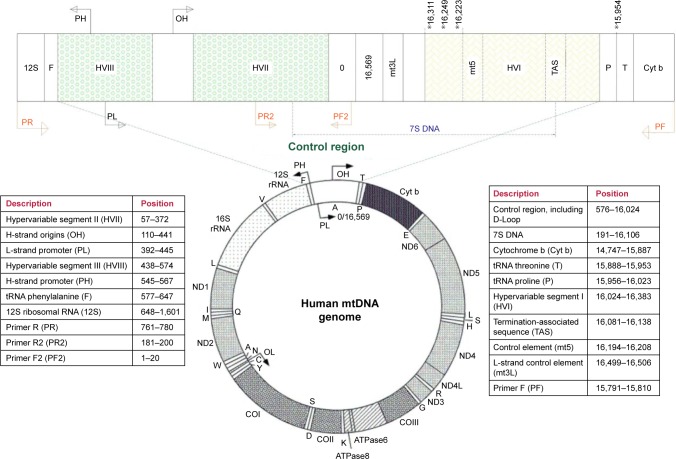
The mtDNA control region was amplified using a set of specific primers with sequences as follows (numbering according to NCBI Accession No NC_0122920.1): PF, 5′-ATCATTGGACAAGTAGCATC-3′ (15,791−15,810 bp) and PR, 5′-GAGCTGCATTGCTGCGTGCT-3′ (780−761 bp) (Figure 1).12 Polymerase chain reaction (PCR) amplification was carried out using TEMPase Hot Start 2x Master Mix A BLUE (Ampliqon, Odense, Denmark) in a final reaction volume of 50 µL, containing 100 ng of DNA, 0.32 µL of each primer (10 pmol), 25 µL of TEMPase 2x Master Mix, and 23.2 µL RNase-free water.
Figure 1.
The genomic structure of human mitochondrial DNA.
Notes: The control region is expanded and shown. The MT-RNR (12S) genes, tRNA phenylalanine, hypervariable I, II, III (HVI, HVII, HVIII), control elements mt5 and mt3L, 7S DNA, termination-associated sequence, tRNA threonine, tRNA proline, and cytochrome b sequences are indicated in the expanded control region diagram. The L- and H-strand promoters and the origin of H-strand replication are represented by PL, PH, and OH, respectively. The positions of the two primer pairs (PF, PR and PF2, PR2) used to amplify and sequence the control region are indicated. Four variant positions that were statistically significant in presbycusis patients than controls are indicated in the expanded control region diagram by asterisks.
PCR amplification was performed with the following program: pre-PCR incubation at 95°C for 15 min, 35 cycles of 95°C for 20 s, annealing at 60°C for 45 s, and extension at 72°C for 30 s, with a final extension at 72°C for 5 min. The specific amplification of a 1,550 bp fragment was confirmed by 1.5% agarose gel electrophoresis (Figure 2).
Figure 2.
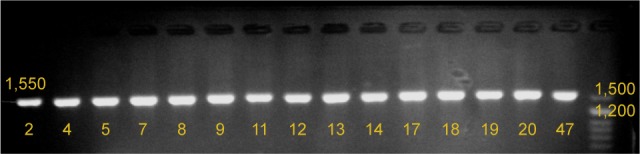
Results of agarose gel electrophoresis of mtDNA control region PCR products.
Notes: The lanes 2 to 47 show PCR products of study participants. A DNA molecular weight marker (50 bp DNA marker [Sinaclon, Iran]) was run in the last lane.
Abbreviations: mtDNA, mitochondrial DNA; PCR, polymerase chain reaction.
Next, PCR products were sequenced by direct DNA sequencing (Bioneer, South Korea). Due to a cytosine nucleotide repeat in the target region, to obtain good quality sequence two further primers were used in addition to those used for PCR amplification, with sequences as follows: PF2, 5′-GAT CAC AGG TCT ATC ACC CT-3′ (1−20 bp) and PR2, 5′-TAG TAA GTA TGT TCG CCT GT-3′ (200−181 bp) (Figure 1). Sequencing results were analyzed using Codon Code Aligner 6.0.2 software (Codon Code, Centerville, MA, USA). Sequences were compared to the revised Cambridge Reference Sequence (rCRS) (Accession No NC_012920.1) using the BLAST sequence analysis tool (NCBI, Bethesda, MD, USA). The Mitomap (http://www.mitomap.org) and mtDB (http://www.mtdb.igp.uu.se/) databases were also checked for mitochondrial genome sequence variants.
Statistical analyses
χ2 and Fisher’s exact tests were used to determine the significance of differences in mtDNA variant frequencies between presbycusis patient and control subjects. Statistical analyses were performed using SPSS 22 for Windows (IBM, Armonk, NY, USA). Statistical significance was defined as P<0.05.
Results
In this study, presbycusis subjects had a mean age of 66±9 years, while that of controls had 65±8 years; there was no statistically significant difference in age between presbycusis and control subjects (P>0.05).
A 1,550 bp fragment of the mtDNA control region was amplified by PCR from all presbycusis and control subjects (Figure 2).
Comparison of the results of sequencing of the mtDNA control region from presbycusis patients with the rCRS revealed 113 variants in patient DNA. mtDNA variants were identified in all presbycusis patients, indicating a 100% mutation rate.
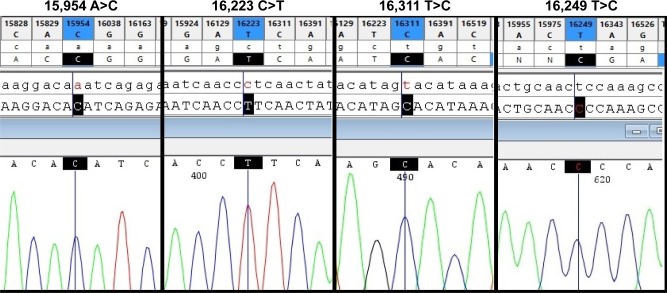
Among the identified variants, 84% were located in the HVRs, with the highest number in HVR-I (42%), followed by HVR-II (33%). The most common nucleotide substitution was T>C (31%), followed by C>T (28.5%), A>G (22.6%), and G>A (15%). Four variants (16,223 C>T, 16,311 T>C, 16,249 T>C, and 15,954 A>C) were present at statistically significant different frequencies in presbycusis patients and healthy subjects, all of which were more frequent in presbycusis subjects (Table 1). Figure 3 shows the 16,223T, 16,311C, 16,249C, and 15,954 sequence variants in comparison to the wild type sequences in the equivalent positions in the rCRS. Four novel variants, not previously reported in mitomap or the mtDB database, were identified in the control subjects and were submitted to mitomap. These variants were 96 C>A (previously reported as 96 C>T),19 15,864 A>G,20 15,906 A>T (previously reported as 15,906 A>C),21 and 16,046 T>G (previously reported as 16,046 T>C).22
Table 1.
The position of nucleotides in the mtDNA control region is significantly more frequent in patients with presbycusis than control subjects
| Nucleotide position | rCRS | Variant | Presbycusis (%) | Control (%) | P-value |
|---|---|---|---|---|---|
| 15,954 | A | C | 5.2 | 0 | 0.009 |
| 16,223 | C | T | 31 | 17.7 | 0.026 |
| 16,249 | T | C | 8.6 | 1.8 | 0.02 |
| 16,311 | T | C | 27.6 | 14.1 | 0.01 |
Abbreviations: rCRS, revised Cambridge Reference Sequence; mtDNA, mitochondrial DNA.
Figure 3.
Four nucleotide variants that show statistically significant differences in frequency between presbycusis and control subjects.
Notes: The panels show comparisons of each nucleotide variant with the rCRS using Codon Code Aligner. The upper sequences represent rCRS and the lower sequences are those amplified from study participants. The black lines and squares indicate the nucleotide positions and alternations.
Abbreviations: rCRS, revised Cambridge Reference Sequence; mtDNA, mitochondrial DNA.
Discussion
Mitochondria are well-characterized as cellular powerhouses that participate in the most fundamental functions of cells. Those organelles contribute to the regulation of apoptosis and calcium storage and participate in energy production and oxidative stress regulation through their role in the electron transport chain. Previous studies have demonstrated the role of oxidative stress and excessive reactive oxygen species (ROS) in presbycusis development.23 Changes in the coding region of the mitochondrial genome have been frequently reported as key factors in the development of presbycusis. Initially, a 4,977 bp deletion in mtDNA, known as the common deletion, was reported by two different groups in the temporal bone of presbycusis patients.14,24 Subsequently, Markaryan et al25 described three extra big deletions of 5,354, 5,142, and 9,682 bp in the mitochondrial cytochrome c oxidase subunit III gene (MT-CO3), in the structural element of the cochlea in patients with presbycusis. Further investigations revealed a significant relationship between the level of deletion and the severity of hearing loss in presbycusis.15,16 In addition to large deletions, point mutations in the mitochondrial genome have been determined as important factors in presbycusis.24,26 In the present study, the control region of mtDNA was compared between Iranian presbycusis patients and control subjects for the first time, with the aim of completing a compressive analysis of the different aspects of mtDNA in presbycusis progression. Previous studies have confirmed the relationship between variants in the mtDNA control region and the pathophysiology of various disorders.10,27
This results of this study revealed that the mtDNA control region contained four variants (16,223 C>T, 16,249 T>C, 16,311 T>C, and 15,954 A>C) that were significantly more frequent in presbycusis patients than control subjects (Table 1; Figure 3). The minimum and maximum numbers of nucleotide changes in presbycusis patients were 2 and 18, respectively, while the equivalent values were 2 and 14 for control subjects.
Previous studies have reported that the 16,223 C>T variant is a risk factor for repeated miscarriage, along with sudden infantile death syndrome and Huntington disease.10,28,29 Investigation of the mtDNA control region also indicated a relationship between 16,311 T>C and both prostate cancer and acute myeloid leukemia.30–32 Pliss et al33 identified 16,311 T>C as the most frequent variant in HRV-I among genetically unrelated Latvians during the aging process. Manwaring et al34 reported an association between haplogroup K and presbycusis.34 The 16,311 T>C variant is one of the polymorphisms that contribute to haplogroup K. The variant 16,249 T>C was also reported in prostate cancer.30 Variants 16,223 C>T, 16,311 T>C, and 16,249 T>C are mapped to mtDNA loci HRV-I, ATT, D-LOOP, and 7S DNA; however, the 15,954 A>C is a noncoding nucleotide located between mtDNA sequences encoding the threonine and proline tRNA molecules (Figure 1). In this study, this variant was observed only in the presbycusis subjects. Previously, the 15,954 A>C variant has been reported as being associated with both mitochondrial encephaloneuromyopathies and dilated cardiomyopathy.35–37
The mtDNA control region contains the origin of replication for the heavy strand and the promoter for transcription of both the heavy and light strands (Figure 1). Therefore, the control region participates in the replication, organization, and regulation of gene expression of the mitochondrial genome.
The significant variations identified in this study were located close to the control elements, mt5 and mt3L, 7S DNA, the termination-associated sequence (TAS), and the binding site for the mtSSB (mitochondrial single strand DNA-binding) protein, which contributes to stabilization of the control region and maintenance of the mtDNA (Figure 1).38,39 Since these variants may change functional protein binding sites in mtDNA, they have the potential to influence gene expression levels from mtDNA. The reduced expression of the mitochondrial gene MT-CO3 in presbycusis patients supports this hypothesis.40
Moreover, variation in the control region can also alter the function of the mitochondrial electron transport chain, enhance ROS generation and oxidative stress, and thereby induce the mitochondrial intrinsic apoptotic pathway. Our previous study demonstrated elevated expression of BAK1, a proapoptotic member of the intrinsic pathway, in patients with presbycusis.17
Jemt et al41 demonstrated that the TAS of the control region is crucial for mtDNA replication. Moreover, the significantly associated variants identified in this study were located in a cluster identified by Yasukawa et al42 as being a potential site for bidirectional mtDNA replication initiation; therefore, these variants could influence the development of presbycusis by altering mtDNA copy number. This hypothesis is supported by our previous observation of lower mtDNA copy numbers in presbycusis.43
Although studies have shown that the levels of large deletions in mtDNA increase during aging,44 these deletions are never observed in the mtDNA control region, and their breakpoints are generally located outside of this region.13 Moreover, the mtDNA control region exhibited a very low mutation rate in the PolgAmut mouse model,13,45 in which the numbers of point mutations are increased as a result of impairment in the proofreading subunit of DNA polymerase gamma.46 All of these facts highlight the role of the mtDNA control region in the regulation of transcription, replication, and stability of the genome of this organelle.
Conclusion
In conclusion, the statistically significant differences in the frequencies of variants in the mitochondrial control region sequence between presbycusis patients and controls at four positions provide new evidence and contribute to the construction of a comprehensive picture of the role of the mitochondria in the progression of presbycusis. The results of this study, together with those of previous reports, demonstrate the pressing need for further functional investigation of the mitochondrial genome to identify novel therapies for prevention, delay, or treatment of this common impairment. Our data also demonstrate that mitochondrial research could enable development of biomarkers to assist in identification of individual at high risk of developing presbycusis.
Acknowledgments
The authors appreciate all the volunteers for participating in this research.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Yang CH, Schrepfer T, Schacht J. Age-related hearing impairment and the triad of acquired hearing loss. Front Cell Neurosci. 2015;9:276. doi: 10.3389/fncel.2015.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohlemiller KK, Frisina RD. Age-related hearing loss and its cellular and molecular bases. In: Schacht J, Popper AN, Fay RR, editors. Auditory Trauma, Protection, and Repair. Boston, MA: Springer US; 2008. pp. 145–194. [Google Scholar]
- 3.Huang Q, Tang J. Age-related hearing loss or presbycusis. Eur Arch Otorhinolaryngol. 2010;267(8):1179–1191. doi: 10.1007/s00405-010-1270-7. [DOI] [PubMed] [Google Scholar]
- 4.Chen H, Tang J. The role of mitochondria in age-related hearing loss. Biogerontology. 2014;15(1):13–19. doi: 10.1007/s10522-013-9475-y. [DOI] [PubMed] [Google Scholar]
- 5.Falah M, Houshmand M, Farhadi M. Presbycusis: from current knowledge to future treatment prospects. JIsfahan Med School. 2016;34(382):526–535. [Google Scholar]
- 6.Newman DL, Fisher LM, Ohmen J, et al. GRM7 variants associated with age-related hearing loss based on auditory perception. Hear Res. 2012;294(1–2):125–132. doi: 10.1016/j.heares.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ciorba A, Bianchini C, Pelucchi S, Pastore A. The impact of hearing loss on the quality of life of elderly adults. ClinInterv Aging. 2012;7:159–163. doi: 10.2147/CIA.S26059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin FR, Thorpe R, Gordon-Salant S, Ferrucci L. Hearing loss prevalence and risk factors among older adults in the United States. J Gerontol A BiolSci Med Sci. 2011;66(5):582–590. doi: 10.1093/gerona/glr002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Administration on Aging (AoA) Available from: http://www.aoa.acl.gov/Aging_Statistics/index.aspx.
- 10.Mousavizadeh K, Rajabi P, Alaee M, Dadgar S, Houshmand M. Usage of mitochondrial D-loop variation to predict risk for Huntington disease. Mitochondrial DNA. 2015;26(4):579–582. doi: 10.3109/19401736.2013.878902. [DOI] [PubMed] [Google Scholar]
- 11.Shakhssalim N, Houshmand M, Kamalidehghan B, et al. The mitochondrial C16069T polymorphism, not mitochondrial D310 (D-loop) mononucleotide sequence variations, is associated with bladder cancer. Cancer Cell Int. 2013;13(1):120. doi: 10.1186/1475-2867-13-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jamali L, Banoei MM, Khalili E, Dadgar S, Houshmand M. Association of genetic variations in the mitochondrial D-loop with beta-thalassemia. Mitochondrial DNA. 2016;27(3):1693–1696. doi: 10.3109/19401736.2014.958730. [DOI] [PubMed] [Google Scholar]
- 13.Nicholls TJ, Minczuk M. In D-loop: 40 years of mitochondrial 7S DNA. ExpGerontol. 2014;56:175–181. doi: 10.1016/j.exger.2014.03.027. [DOI] [PubMed] [Google Scholar]
- 14.Bai U, Seidman MD, Hinojosa R, Quirk WS. Mitochondrial DNA deletions associated with aging and possibly presbycusis: a human archival temporal bone study. Am J Otol. 1997;18(4):449–453. [PubMed] [Google Scholar]
- 15.Markaryan A, Nelson EG, Hinojosa R. Quantification of the mitochondrial DNA common deletion in presbycusis. Laryngoscope. 2009;119(6):1184–1189. doi: 10.1002/lary.20218. [DOI] [PubMed] [Google Scholar]
- 16.Liu H, Han Y, Wang S, Wang H. Association between the mitochondrial DNA 4977 common deletion in the hair shaft and hearing loss in presbycusis. Mol Med Rep. 2015;11(2):1127–1131. doi: 10.3892/mmr.2014.2877. [DOI] [PubMed] [Google Scholar]
- 17.Falah M, Najafi M, Houshmand M, Farhadi M. Expression levels of the BAK1 and BCL2 genes highlight the role of apoptosis in age-related hearing impairment. ClinInterv Aging. 2016;11:1003–1008. doi: 10.2147/CIA.S109110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Eyken E, Van Laer L, Fransen E, et al. KCNQ4: a gene for age-related hearing impairment? Hum Mutat. 2006;27(10):1007–1016. doi: 10.1002/humu.20375. [DOI] [PubMed] [Google Scholar]
- 19.Falah M, Farhadi M, Houshmand M. Submitted variant 20160806004. Found in an Iranian individual (J1d): blood sample, 2016. Available from: http://www.mitomap.org/foswiki/bin/view/MITOMAP/Submissions/20160806004.
- 20.Falah M, Farhadi M, Houshmand M. Submitted variant 20160806005. Found in an Iranian individual (H2a): blood sample, 2016. Available from: http://www.mitomap.org/foswiki/bin/view/MITOMAP/Submissions/20160806005.
- 21.Falah M, Farhadi M, Houshmand M. Submitted variant 20160806006. Found in an Iranian individual (H20a): blood sample, 2016. Available from: http://www.mitomap.org/foswiki/bin/view/MITOMAP/Submissions/20160806006.
- 22.Farhadi M, Houshmand M, Falah M. Submitted variant 20160806007. Found in an Iranian individual (U1): blood sample, 2016. Available from: http://www.mitomap.org/foswiki/bin/view/MITOMAP/Submissions/20160806007.
- 23.Fujimoto C, Yamasoba T. Oxidative stresses and mitochondrial dysfunction in age-related hearing loss. Oxid Med Cell Longev. 2014;2014:582849. doi: 10.1155/2014/582849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fischel-Ghodsian N, Bykhovskaya Y, Taylor K, et al. Temporal bone analysis of patients with presbycusis reveals high frequency of mitochondrial mutations. Hear Res. 1997;110(1–2):147–154. doi: 10.1016/s0378-5955(97)00077-4. [DOI] [PubMed] [Google Scholar]
- 25.Markaryan A, Nelson EG, Hinojosa R. Detection of mitochondrial DNA deletions in the cochlea and its structural elements from archival human temporal bone tissue. Mutat Res. 2008;640(1–2):38–45. doi: 10.1016/j.mrfmmm.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 26.Zhu Y, Zhao J, Feng B, et al. Mutations in the mitochondrial 12S rRNA gene in elderly Chinese people. ActaOtolaryngol. 2015;135(1):26–34. doi: 10.3109/00016489.2014.949849. [DOI] [PubMed] [Google Scholar]
- 27.Maggrah A, Robinson K, Creed J, et al. Paired ductal carcinoma in situ and invasive breast cancer lesions in the D-loop of the mitochondrial genome indicate a cancerization field effect. Biomed Res Int. 2013;2013:379438. doi: 10.1155/2013/379438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seyedhassani SM, Houshmand M, Kalantar SM, Modabber G, Aflatoonian A. No mitochondrial DNA deletions but more D-loop point mutations in repeated pregnancy loss. J Assist Reprod Genet. 2010;27(11):641–648. doi: 10.1007/s10815-010-9435-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arnestad M, Opdal SH, Musse MA, Vege A, Rognum TO. Are substitutions in the first hypervariable region of the mitochondrial DNA displacement-loop in sudden infant death syndrome due to maternal inheritance? ActaPaediatr. 2002;91(10):1060–1064. doi: 10.1080/080352502760311557. [DOI] [PubMed] [Google Scholar]
- 30.Chen JZ, Gokden N, Greene GF, Mukunyadzi P, Kadlubar FF. Extensive somatic mitochondrial mutations in primary prostate cancer using laser capture microdissection. Cancer Res. 2002;62(22):6470–6474. [PubMed] [Google Scholar]
- 31.Sharawat SK, Bakhshi R, Vishnubhatla S, Bakhshi S. Mitochondrial D-loop variations in paediatric acute myeloid leukaemia: a potential prognostic marker. BrJHaematol. 2010;149(3):391–398. doi: 10.1111/j.1365-2141.2010.08084.x. [DOI] [PubMed] [Google Scholar]
- 32.Silkjaer T, Norgaard JM, Aggerholm A, et al. Characterization and prognostic significance of mitochondrial DNA variations in acute myeloid leukemia. Eur J Haematol. 2013;90(5):385–396. doi: 10.1111/ejh.12090. [DOI] [PubMed] [Google Scholar]
- 33.Pliss L, Brakmanis A, Ranka R, et al. The link between mitochondrial DNA hypervariable segment I heteroplasmy and ageing among genetically unrelated Latvians. ExpGerontol. 2011;46(7):560–568. doi: 10.1016/j.exger.2011.02.016. [DOI] [PubMed] [Google Scholar]
- 34.Manwaring N, Jones MM, Wang JJ, et al. Mitochondrial DNA haplogroups and age-related hearing loss. Arch Otolaryngol Head Neck Surg. 2007;133(9):929–933. doi: 10.1001/archotol.133.9.929. [DOI] [PubMed] [Google Scholar]
- 35.Da Pozzo P, Cardaioli E, Radi E, Federico A. Sequence analysis of the complete mitochondrial genome in patients with mitochondrial encephaloneuromyopathies lacking the common pathogenic DNA mutations. BiochemBiophys Res Commun. 2004;324(1):360–364. doi: 10.1016/j.bbrc.2004.09.058. [DOI] [PubMed] [Google Scholar]
- 36.Ruppert V, Nolte D, Aschenbrenner T, et al. Novel point mutations in the mitochondrial DNA detected in patients with dilated cardiomyopathy by screening the whole mitochondrial genome. BiochemBiophys Res Commun. 2004;318(2):535–543. doi: 10.1016/j.bbrc.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 37.Sternberg D, Danan C, Lombes A, et al. Exhaustive scanning approach to screen all the mitochondrial tRNA genes for mutations and its application to the investigation of 35 independent patients with mitochondrial disorders. Hum Mol Genet. 1998;7(1):33–42. doi: 10.1093/hmg/7.1.33. [DOI] [PubMed] [Google Scholar]
- 38.Roberti M, Musicco C, Polosa PL, et al. Multiple protein-binding sites in the TAS-region of human and rat mitochondrial DNA. BiochemBiophys Res Commun. 1998;243(1):36–40. doi: 10.1006/bbrc.1997.8052. [DOI] [PubMed] [Google Scholar]
- 39.Park KS, Chan JC, Chuang LM, et al. A mitochondrial DNA variant at position 16189 is associated with type 2 diabetes mellitus in Asians. Diabetologia. 2008;51(4):602–608. doi: 10.1007/s00125-008-0933-z. [DOI] [PubMed] [Google Scholar]
- 40.Markaryan A, Nelson EG, Hinojosa R. Major arc mitochondrial DNA deletions in cytochrome c oxidase-deficient human cochlear spiral ganglion cells. ActaOtolaryngol. 2010;130(7):780–787. doi: 10.3109/00016480903397702. [DOI] [PubMed] [Google Scholar]
- 41.Jemt E, Persson O, Shi Y, et al. Regulation of DNA replication at the end of the mitochondrial D-loop involves the helicase TWINKLE and a conserved sequence element. Nucleic Acids Res. 2015;43(19):9262–9275. doi: 10.1093/nar/gkv804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yasukawa T, Yang MY, Jacobs HT, Holt IJ. A bidirectional origin of replication maps to the major noncoding region of human mitochondrial DNA. Mol Cell. 2005;18(6):651–662. doi: 10.1016/j.molcel.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 43.Falah M, Houshmand M, Najafi M, et al. The potential role for use of mitochondrial DNA copy number as predictive biomarker in presbycusis. TherClin Risk Manag. 2016;12:1573–1578. doi: 10.2147/TCRM.S117491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.ZabihiDiba L, Mohaddes Ardebili SM, Gharesouran J, Houshmand M. Age-related decrease in mtDNA content as a consequence of mtDNA 4977 bp deletion. Mitochondrial DNA. 2016;27(4):3008–3012. doi: 10.3109/19401736.2015.1063046. [DOI] [PubMed] [Google Scholar]
- 45.Bailey LJ, Cluett TJ, Reyes A, et al. Mice expressing an error-prone DNA polymerase in mitochondria display elevated replication pausing and chromosomal breakage at fragile sites of mitochondrial DNA. Nucleic Acids Res. 2009;37(7):2327–2335. doi: 10.1093/nar/gkp091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trifunovic A, Wredenberg A, Falkenberg M, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429(6990):417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]