Abstract
Background
Leptospirosis is an important re-emerging infectious disease that affects humans worldwide. Infection occurs from indirect environment-mediated exposure to pathogenic leptospires through contaminated watered environments. The ability of pathogenic leptospires to persist in the aqueous environment is a key factor in transmission to new hosts. Hence, an effort was made to detect pathogenic leptospires in complex environmental samples, to genotype positive samples and to assess leptospiral viability over time.
Methodology/Principal findings
We focused our study on human leptospirosis cases infected with the New Caledonian Leptospira interrogans serovar Pyrogenes. Epidemiologically related to freshwater contaminations, this strain is responsible for ca. 25% of human cases in New Caledonia. We screened soil and water samples retrieved from suspected environmental infection sites for the pathogen-specific leptospiral gene lipL-32. Soil samples from all suspected infection sites tested showed detectable levels of pathogenic leptospiral DNA. More importantly, we demonstrated by viability qPCR that those pathogenic leptospires were viable and persisted in infection sites for several weeks after the index contamination event. Further, molecular phylogenetic analyses of the leptospiral lfb-1 gene successfully linked the identity of environmental Leptospira to the corresponding human-infecting strain.
Conclusions/Significance
Altogether, this study illustrates the potential of quantitative viability-PCR assay for the rapid detection of viable leptospires in environmental samples, which might open avenues to strategies aimed at assessing environmental risk.
Author summary
Leptospirosis is an emerging zoonotic disease caused by infection with pathogenic strains of Leptospira. Most human infections arise from environmental exposure to contaminated freshwater environments or watered soils where pathogenic Leptospira are considered as able to survive for prolonged periods. Therefore, a good understanding of Leptospira survival strategy in the environment is a key step to identifying crucial factors amenable to interventions and public health actions to lower leptospirosis burden. In this study, we investigated the environmental presence and survival of pathogenic leptospires in areas where recent human leptospirosis cases had been reported. Although detection of Leptospira from complex environmental samples is difficult, we successfully detected the presence of pathogenic Leptospira in soils of suspected infection sites. In addition, we showed that these pathogenic leptospires were alive and present in soils several weeks after the infecting event. Typing of leptospiral DNA retrieved from the environment revealed identities between environmental pathogenic Leptospira and the causative strains involved in human leptospirosis index cases. Interestingly, we also identified yet unreported genotypes. Altogether, our work illustrates the potential of quantitative molecular assays for the rapid detection and typing of viable leptospires in environmental samples, which could prove useful to assess the risk of environmental exposure.
Introduction
Leptospirosis is an acute febrile disease caused by pathogenic spirochetes of the genus Leptospira. It is considered an important re-emerging infectious disease that affects more than 1 million humans worldwide [1]. The spectrum of human disease caused by leptospires is extremely wide, ranging from subclinical infection to a severe syndrome of multiorgan infection with high mortality. Leptospira transmission from the urine of reservoir hosts to incidental hosts, including humans, usually occurs through the contamination of skin lesions or mucosae with contaminated surface water or soil [2]. The incidence of such infections depends on several factors including the density of the reservoir species and its Leptospira carriage prevalence, the dilution into watered environment and the survival time of the leptospires into possibly nutrient-poor and adverse environmental conditions. Estimation of survival time and virulence preservation of pathogenic Leptospira spp. after excretion into the environment is becoming a crucial challenge to determine the environmental risk and to adopt preventive measures. The duration of Leptospira survival in natural habitat is affected by many factors including abiotic and biotic factors. The persistence of pathogenic Leptospira in moist soil and freshwater for long periods of time is thought to depend on a slightly alkaline pH, high oxygen, and low salt concentrations [3–5]. The classical assumption is that slightly higher alkalinity (up to pH 8.0) allows for longer survival. Under laboratory conditions, a strain of serovar Javanica was reported to survive in distilled water (pH 7.8) for 152 days [6]. More recently, Andre-Fontaine et al. [7] showed that pathogenic Leptospira can survive for months in mineral water. Interestingly, Leptospira were reported to survive as long as 10 months in adverse conditions (4°C) and up to 20 months when stored at 30°C.
Interactions of Leptospira spp. with the environmental microbiota also begin to be examined. Environmental microbial blooms alter the concentration of oxygen, minerals, and other nutrients in the water and favor either multiplication or destruction of some species of pathogenic Leptospira [8]. Several common bacterial genera including Azospirillum and Sphingomonas were found along with pathogenic and saprophytic Leptospira spp. in biofilms formed in freshwater or in dental water unit systems [9,10]. Co-incubation with a Sphingomonas spp. increased Leptospira growth rate [8], suggesting possible syntrophic interactions. When incubated with Azospirillum brasilense, viability of pathogenic Leptospira was enhanced at high temperature and extended under UV radiation or exposure to penicillin G, tetracycline or ampicillin. In addition, soil adsorption, thought to be an important step that favors leptospire persistence in the environment, was greatly increased in the presence of A. brasilense [8].
A major impediment to assess environmental risk for leptospirosis has been the difficulty to isolate pathogenic Leptospira from environmental samples, attributable in part to the fact that non-pathogenic leptospires outgrow pathogenic strains in culture. Other methods including direct animal inoculation are time-consuming, ethically questionable and have a low analytical sensitivity. However, the increasing use of molecular methods overcomes some limitations inherent to culture- and animal-based methods and provides quantitative information about the concentration of leptospires in contaminated waters [11–13].
New Caledonia provides an ideal location for studying environmental risk factors of leptospirosis because of its high leptospirosis incidence, on average 45 cases per 100,000 inhabitants, and the presence of known hot spots where annual incidence reaches up to 500 cases/100,000 population. Based on data of leptospirosis surveillance in New Caledonia, serogroup Icterohaemorrhagiae is the dominant serogroup involved in ca. 60% of human cases. Other serogroups involved in human leptospirosis include Pyrogenes (18–25%), Ballum, Australis and Pomona. Interestingly, the New Caledonian L. interrogans serovar Pyrogenes was formerly shown to be epidemiologically related to freshwater contaminations. Therefore, human leptospirosis cases infected with this strain provide opportunities to investigate the persistence and survival of pathogenic Leptospira in natural habitats.
The purpose of the present study was to assess the presence of pathogenic leptospires in environmental samples and to estimate their viability over time. Using a TaqMan-based real time quantitative polymerase chain reaction, we screened 73 environmental samples retrieved from 4 suspected environmental infection sites for the pathogen-specific leptospiral gene lipL-32. This study found that a large proportion of soil samples were positive for pathogenic leptospiral DNA, suggesting that repeated exposure to Leptospira may be occurring in these high-risk areas. Herein, we report findings from retrospective investigations of environmental contaminated areas to assess the presence of pathogenic Leptospira in order to better delineate and monitor high risk areas.
Materials and methods
Ethics statement
Patient samples, contact and authorization for interview
Institut Pasteur in New Caledonia is the country reference and only laboratory for the biological diagnosis of human leptospirosis. Biological diagnosis relies on qPCR using serum or urine as well as the reference Microagglutination Technique (MAT) using a panel of serovars of epidemiological relevance. Over the last decade, more patients have been diagnosed by qPCR, probably reflecting higher awareness and earlier medical consultation [14]. The patients were identified by a positive diagnostic qPCR targeting lipL-32 [15]. Notification of leptospirosis to the New Caledonian Health Authority is compulsory and the infecting strain is routinely identified using a lfb-1-derived phylogeny of New Caledonian isolates [16] as part of the surveillance system, which also investigates cases through interviews. Only patients infected by the L. interrogans Pyrogenes were included in the study. Oral consent was obtained by the Health Authority to meet with the patient (or his parents for minors) and collect environmental samples in the suspected infection sites. Because no human sample was collected as part of the study, no written consent was required. The oral consent led to organize a site investigation. The study was approved by Institut Pasteur in New Caledonia and the ethics clearance was granted by the New Caledonian Health Authority (Direction des Affaires Sanitaires et Sociales de la Nouvelle-Calédonie).
Study sites
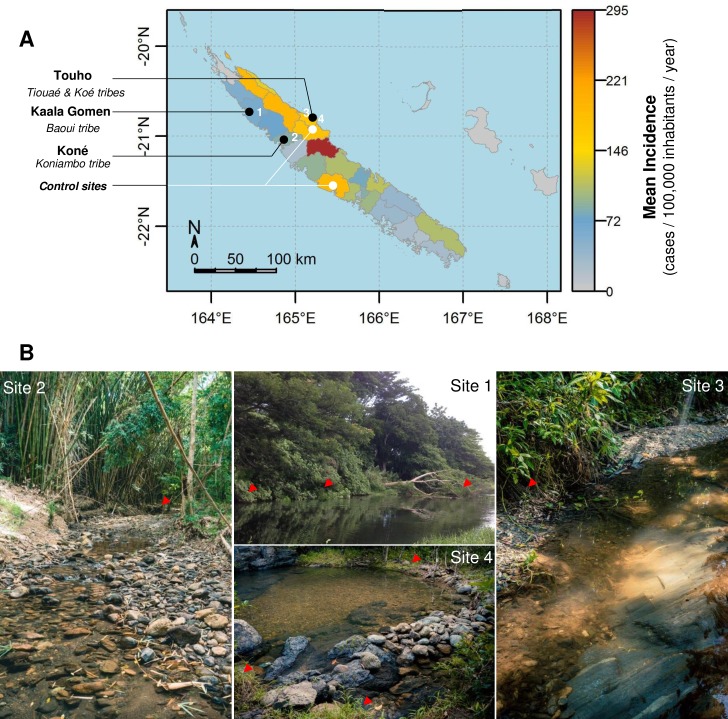
Four sites were identified according to the infectious strain and the good acceptance of the project by the patients and custom chiefdom (Kaala-Gomen, Koné, Touho (2 sites), Fig 1). All four study sites were within Melanesian tribes, where many outdoor activities are part of the everyday life, including fishing and bathing in freshwater streams, maintenance of backyard pig pens, hunting (deer and wild hogs). In addition, two extra sites where L. interrogans Pyrogenes was known to have been involved in former cases but where no recent contamination were reported were chosen as control sites and investigated according to the same sampling procedure. Most of the investigated sites were located in the North province of the main island where climate is sub-tropical and oceanic with a hot and rainy season from December to March (average temperature 28°C) and a cooler season from June to September (average temperature 20°C). Annual cumulative rainfall is 2400 mm on average but can range from 1460 mm to 3550 mm. Daily rainfall data for each site were obtained from the Météo France free online public database, using the nearest meteorological station for each study site.
Fig 1. Localization of the 6 investigated environmental sites.
A. Mapping of the 6 environmental sites investigated in this study. The map legend shows the incidence rate of leptospirosis per municipality, calculated over the 2007–2013 period. B. Photography showing rivers associated with activities of daily living where patients reported to be contaminated. Red arrowhead shows collected samples positive for pathogenic Leptospira DNA.
Collection and processing of environmental samples
Environmental investigations were started 6 to 10 weeks after the supposed infection date. Between March and June 2016, a total of 73 environmental samples were collected: 10 water samples, 52 soil samples and 11 other samples (vegetal floating debris, algae) were analyzed. Water and soil sample collections were carried out as follow: For water samples, 10 mL of subsurface water (stream or river) were collected at a 10–30 cm depth every 10 meters, alongside the water body directly into 15-mL sterile Falcon tubes, stored on ice and transported to the laboratory. For soil samples, approximately 50 g topsoil was collected from river banks (from 10 cm below to 1 meter above water level) in shaded areas using a core drilling (3 cm large by 5–7 cm height). Each soil sample was immediately placed into a 50-mL sterile Falcon tube. Water quality and environmental parameters were collected at the time of sampling (apparent meteorological and hydrological conditions, presence of iridescences, debris, foam or stagnant fludge, water color, clarity, turbidity, salinity, temperature, dissolved oxygen, pH, UV radiation, altitude). The location of sampling sites was taken with a Garmin GPS. All samples were transported to the laboratory and processed within 48 hours of collection.
DNA isolation from environmental samples
Each water sample (10 mL) was centrifuged at 8000 × g for 10 min. The pellets were resuspended to a total volume of 200 μL in the original water and immediately lysed to begin the extraction process using a commercial kit (QIAamp DNA Mini Kit, Qiagen, Australia) according to the manufacturer’s instructions. DNA elution was performed with 50 μL of buffer AE. The quantity of DNA was measured by NanoDrop (Thermo Fisher Scientific). Soil samples were submitted to DNA extraction using the PowerSoil DNA Isolation kit (MO BIO), shown in preliminary experiments to be the most efficient to extract leptospiral DNA from New Caledonian soils.
Briefly, 250 mg of soil is poured in a PowerSoil bead tube before addition of 60μL suspension Buffer C1. This suspension is shaken for 5 minutes at 2,000 rpm using a MagNA Lyser (Roche). The supernatant is lysed at 4°C for 5 min with 250 μL lysis buffer C2. Up to 600 μL of supernatant is transferred in a new tube before addition of 200 μL of Inhibitory Removal Technology solution C3 before incubation at 4°C for 5 min. This step is essential for the final DNA quality as it allows the cationic flocculation of humic substances which usually account for low DNA recovery and qPCR inhibition. Up to 750 μL of the supernatant is transferred in a new tube and gently mixed with 1,200 μL of DNA binding solution C4 prior to be loaded into a Spin Filter and centrifuged at 10,000x g for 1 minute at room temperature. After washing the precipitated DNA with 500 μL of wash buffer C5 through the spin filter membrane, the DNA is eluted with 100 μL elution buffer C6.
PCR detection of pathogenic Leptospira spp
Soil and water samples were tested for the presence of pathogenic Leptospira DNA using the real-time PCR targeting lipL-32 [15]. The reactions were performed in a final volume of 20 μL containing 1X LightCycler 480 probes Master (Roche Applied Science, New Zealand), 0.4 μM each primer and 0.13 μM probe, and 2 μL template DNA. The cycling conditions were as described in the original publication in a LightCycler 480 (Roche Applied Science) [15].
Leptospira viability qPCR assay
Samples with a positive lipL-32 qPCR were investigated with BLU-V Viability PMA Kit (Qiagen) to evaluate the presence of viable pathogenic leptospires, except for site 1. Briefly, 5 g of soil were gently resuspended in 5 mL of 1X Phosphate Buffered Saline and let to settle down for 1 hour. Then 100 μL of this soil suspension supernatant was mixed with 2 μL of propidium monoazide (PMA; 50 μM final concentration) in light-transparent 1.5 mL microcentrifuge tubes. Following a 10 min incubation in the dark, samples were exposed for 10 min to a 3-watt LED light (460-470 nm) with gentle homogenization every 2 minutes. The sample tubes were laid horizontally under the light source to ensure optimal PMA/DNA cross-linking, thus avoiding false positive results. In order to test the efficiency of PMA treatment of membrane-compromised bacterial cells, duplicate tubes of the same soil solution supernatant were heated at 80°C for 10 min. The heat-treated samples were then cooled to room temperature before PMA addition, incubation and photoactivation. In addition, a control tube without PMA was included to determine the presence of total pathogenic Leptospira (both dead and live) in the soil sample. After photoinduced cross-linking, samples were treated for DNA isolation using QIAamp DNA Mini kit (Qiagen). The corresponding DNA extracts were used as templates for qPCR targeting the lfb-1 gene [17] in order to subsequently phylogenetically identify the viable pathogenic Leptospira present in the sample. This qPCR was run on a LightCycler LC 2.0 using the LightCycler FastStart DNA Master SYBR Green I kit (Roche Applied Science, New Zealand) as described before [17].
Gene sequence determination of qPCR amplicons, phylogenetic analysis and Accession Numbers
The lfb-1 sequence polymorphism was used as a molecular phylogenetic target to link the identity of environmental leptospiral sequences to the corresponding human infecting strain. Amplified lfb-1 DNA products obtained from environmental samples were identified by DNA sequencing. The amplicons were purified using a DNA purification kit (Qiagen, Australia) and sequenced directly as described before [16]. The resulting DNA sequence data were compared with sequences retrieved from the patient’s sample and with the GenBank database using the BLAST algorithm. The sequences obtained in this research were deposited in GenBank under the Accession Numbers: KY052025; KY052026; KY052027; KY052028; KY052029; KY052030; KY052031; KY052032; KY052033; KY052034; KY052035; KY052036; KY052037; KY052038; KY052039; KY052040.
Results
Pathogenic Leptospira are widespread in soil samples months after the index contamination event
Located in the north of New Caledonia, in places were leptospirosis is endemic, 4 different sites distributed over 3 municipalities were investigated (Fig 1). For sites 1, 2 and 4, the infections supposedly occurred on February 2nd, during the same heavy rain event which hit New Caledonia the same day (S1 Fig). All patients were swimming in a freshwater stream when the rain started to fall and all 3 reported an increase of the water flow and a change in water color and turbidity in their respective bathing sites. For site 3, the patient probably got infected on February 9th when fishing freshwater shrimp using a mask and snorkel. For site 1, only one investigation was performed 6 weeks after the contamination event. Sites 2, 3 and 4 were investigated twice with 7 weeks (site 3 and 4) or 10 weeks (site 2) between investigations (S1 Fig).
The overall results for detection of pathogenic leptospires from these 4 sites are summarized in Table 1.
Table 1. Summary of sampling data and results for the six areas investigated.
| Weeks Post Infection | City | Tribe | Coordinates | Infection date | Investigation date | lipL-32 positive samples | |||
|---|---|---|---|---|---|---|---|---|---|
| Water | Soil | Other | |||||||
| Site 1 | 6 | Kaala Gomen | Baoui | 20°40.823S 64°26.895E | 02/02/2016 | 17/03/2016 | 0 (4) | 6 (6) | 0 (5) |
| Site 2 | 9 | Koné | Koniambo | 20°59.988S | 02/02/2016 | 06/04/2016 | 0 (1) | 4 (6) | 0 (2) |
| 19 | 164°52.415E | 13/06/2016 | - | 5* (7) | - | ||||
| Control site 1 | - | Bourail | Pouéo | 21°30.835S 165°30.436E | - | 06/04/2016 | 0 (1) | 0 (1) | - |
| Control site 2 | - | Touho | Pombei | 20°54.220S 165°08.927E | - | 11/04/2016 | 0 (2) | 0 (2) | - |
| Site 3 | 9 | North Touho | Tiouaé | 20°47.572S | 09/02/2016 | 11/04/2016 | - | 4 (9) | 1 (2) |
| 16 | 165°08.896E | 31/05/2016 | 0 (2) | 2* (6) | 0 (1) | ||||
| Site 4 | 10 | South Touho | Koé | 20°49.090S | 02/02/2016 | 12/04/2016 | - | 8* (9) | 1 (1) |
| 17 | 165°14.702E | 31/05/2016 | - | 1 (6) | - | ||||
| total | 0 (10) | 30 (52) | 2 (11) | ||||||
| % positive | 0% | 57.69% | 12.20% | ||||||
Bold numbers represent positive samples for lipL-32 qPCR. Numbers in brackets represent the total number of samples analyzed.
* indicates the presence of a soil sampled above water level and positive for lipL-32 qPCR.
Interestingly, of the 10 water samples collected, none were positive for the presence of pathogenic Leptospira DNA by qPCR, either at the early or late investigation time point. Contrarily, soil samples were mostly positive: 58% of soil samples (30/52) were positive using lipL-32 qPCR. It is worth to note that among soil samples investigated, we were able to amplify pathogenic Leptospira DNA from the river bank up to 1 meter above the water level. In such a core soil sample, DNA from pathogenic Leptospira was amplified from all 1-cm thick slices down to a 5-cm depth. In addition, 2 samples mostly made of benthic algae collected on the bottom of the streams were also positive using lipL-32-qPCR.
Despite a decreasing number of leptospiral DNA-positive soil samples in sites 3 and 4, we still successfully detected pathogenic Leptospira DNA in the late samples collected 4 months after the index infection event, although the qPCR Cycle Thresholds (Ct) slightly increased (S1 Fig). In contrast, in the two control sites where L. interrogans Pyrogenes was known to have been involved in human cases in previous years but without recent contamination reported (> 1 year), none of the samples collected was positive.
Viability-PCR combined with lfb-1 phylogenetic analysis successfully linked the presence of viable environmental leptospires to the corresponding human cases
For each soil sample positive for lipL-32 by qPCR, we further investigated the genotype of these pathogenic leptospires using the lfb-1 phylogenetic scheme used for patients. Viability-PCR (v-PCR) and qPCR targeting lfb-1 using the v-PCR treated DNA as a matrix were performed subsequently when possible (for sites 2–4). The overall results for v-PCR and lfb-1-derived phylogenetic analysis from these 6 sites are summarized in Table 2.
Table 2. qPCR and v-PCR results and lfb-1 sequence analysis for the six investigated areas.
| Weeks Post Infection | PCR lipL-32 | PCR lfb-1 | v-PCR | lfb-1 Sequence | |
|---|---|---|---|---|---|
| Site 1 | 6 | 6 (15) | 5 (6) | n.d. | L. interrogans Pyrogenes |
| Site 2 | 9 | 4 (9) | 2 (2) | + | L. interrogans Pyrogenes; L. interrogans Australis |
| 19 | 5 (7) | 0 (5) | - | n.d. | |
| Control site 1 | - | 0 (2) | n.d. | n.d. | n.d. |
| Control site 2 | - | 0 (4) | n.d. | n.d. | n.d. |
| Site 3 | 9 | 5 (11) | 2 (2) | + | L. interrogans Pyrogenes |
| 16 | 2 (9) | 2 (2) | -; - | Leptospira spp. | |
| Site 4 | 10 | 8 (10) | 3 (3) | -; + | L. interrogans Pyrogenes; Leptospira spp. |
| 17 | 1 (6) | 1 (1) | - | Leptospira spp. |
n.d.: not determined. Bold numbers represents positive samples for the corresponding qPCR. Numbers in brackets represent the total number of analyzed samples. Underlined results highlight the lfb-1 sequences obtained using v-PCR-treated DNA as template.
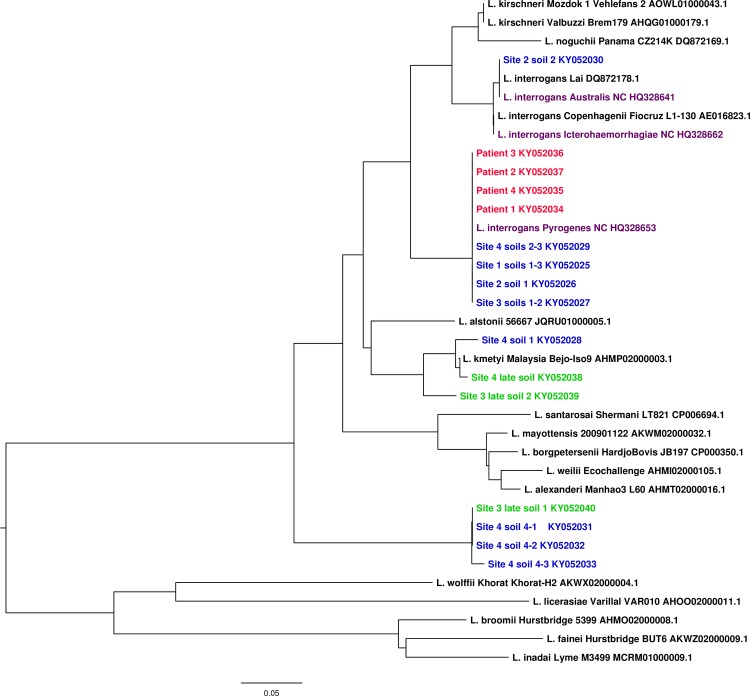
In all 4 sites investigated, we were able to amplify DNA from L. interrogans Pyrogenes, respectively at 6; 9; 9 and 10 weeks post-infection (WPI) for site 1, 2, 3 and 4. To further assess if this L. interrogans Pyrogenes DNA derived from live cells, we performed a viability-PCR (for sites 2–4). Two sites (2 and 3) out of the 3 investigated were positive for v-PCR and phylogenetic analyses of the amplified DNA matched to L. interrogans Pyrogenes. Interestingly, a lfb-1 sequence identical to a L. interrogans from serogroup Australis, involved in other human cases in New Caledonia, was also detected in site 2, concomitantly with Pyrogenes. v-PCR was also positive in site 4, but the phylogenetic analysis of the amplified lfb-1 sequences did not match the infecting strain nor any other reported isolate (except one sequence displaying 96% identity with L. kmetyi; Fig 2).
Fig 2. Phylogenetic analysis of leptospiral lfb-1 gene sequences.
Sequences from the 4 patients are shown in red while clones from environmental soils samples obtained during the retrospective study are shown in blue (first environmental sampling) or green (late environmental sampling). Note the presence of the L. interrogans Pyrogenes NC lfb-1 sequence from the patient in all the corresponding investigated soils. lfb-1 sequence from Site 2 soil 2 was identify as L. interrogans Australis and indeed cluster with L. interrogans Australis NC. Several sequences for Site 4 either cluster with L. kmetyi (Soil1) or form a new branch within pathogenic Leptospira species (Soil 4 sequence 1, 2 and 3). Phylogenetic tree was built using Phylo-win program with 500 bootstrap replicates applying Neighbour Joining method and Kimura’s 2-parameter distances.
To clarify whether the pathogenic leptospires could be detected over a longer period, soil samples were collected again 19 (site 2), 16 (site 3) or 17 WPI (site 4). All the samples investigated from these 3 sites were negative using v-PCR. However, we were still able to amplify a few lfb-1 sequences using direct qPCR for site 3 and 4. Phylogenetic analysis of these lfb-1 sequences appeared to differ from any known strain or species, though some were similar to those amplified during our first investigation (Fig 2).
Finally, it is interesting to note that temporal analysis of our results seems to highlight dynamic changes of the pathogenic leptospires in environmental sites. Indeed, when sequences identical to L. interrogans Pyrogenes or Australis were found during our first investigation, they were either not re-detected (site 2 and 4) or substituted by other unknown pathogenic leptospires upon our 2nd investigation (site 3).
Discussion
Infected mammals by shedding large amounts of virulent leptospires in their urine, massively contaminate their surrounding environment [5,18–20]. These pathogens eventually get drained in freshwater systems upon heavy rain episodes. This dispersion through freshwater not only participates to substantial contamination of large areas but also brings the threat right in human influence area. As environmental contamination is the major source of human leptospirosis, we attempted in this study, to evidence the presence of virulent leptospires in natural habitats in New Caledonia. Using molecular-based methods, we investigated the presence and viability of pathogenic leptospires in area believed to be contamination sites. Although the use of qPCR is becoming frequent for diagnostic purpose [14], the use of this technique on environmental samples is not commonplace, mainly because of the presence of inhibitors impairing qPCR efficiency [13].
We applied this methodology to complex environmental samples from places selected as putatively involved in human cases and we successfully amplified pathogenic Leptospira DNA in all the sites investigated. Phylogenetic analysis based on the lfb-1 gene sequence successfully linked the identity of environmental leptospiral sequences to the corresponding human cases. More importantly, we demonstrated by viability qPCR that these pathogenic leptospires were viable and probably persisted in infection sites weeks after the contamination event. Notwithstanding that little is known regarding the mechanisms regulating the persistence of pathogenic leptospires in natural habitats, outside a mammalian host, general agreement in the scientific community agrees on the fact that pathogenic Leptospira can survive for long periods in freshwater [7] and a few studies exploring the survival of Leptospira in soils successfully reported re-isolation of the same Leptospira isolate 5 months later [21]. Our results show, that when performing retrospective investigations, pathogenic leptospires could be evidenced only in soils samples, up to 4 months after the index contamination. Though repeated contaminations from an animal source might occur, our results strongly suggest a prolonged survival in river banks and soils. Moreover, to our knowledge, this study reports the first successful viability-PCR performed on Leptospira from complex soil samples in combination with molecular-based typing to identify environmental leptospires involved in human cases. We formally established that up to 9 weeks after infection, the pathogenic Leptospira strain involved in human cases was viable in environmental soil samples, and thus potentially infectious, suggesting an ongoing risk for humans. Using samples collected 4 months after the contamination event, we were not able to evidence the presence of this particular virulent strain, therefore suggesting a decrease in Leptospira viability over time, a hypothesis supported by higher Ct values in qPCR from late samples (S1 Fig). These 2nd investigations were performed during the cool season, also assumed to be detrimental for Leptospira survival. Whether this change in temperature contributed to the decrease in environmental contamination remains unknown but would be in good agreement with empirical knowledge as well as the experimental results reported by Andre-Fontaine and collaborators [7].
Interestingly, we have not evidenced pathogenic leptospires in any of our water samples, contrasting with previous observations [22,23]. Following patients’ interviews, we have investigated the suspected flowing water bodies (streams and rivers) at their normal flow rate and weeks after the index contamination event. Oppositely, stagnant water sources (gutters, wells, puddles, reservoirs) considered in other studies [22,23] were not mentioned in the interviews and therefore were not investigated. In addition, we have processed a relatively small volume of water for DNA extraction (10 mL), contrasting with larger volumes (50–1,000 mL) in other studies. Lastly, we also have investigated a smaller number of water samples compared with soils. Taken together, these facts may explain the differences observed.
The detection (or absence of detection) of live L. interrogans Pyrogenes after long periods should be interpreted with caution because of possible limitations of the v-PCR methodology, especially in environmental samples [24]. Although selective nucleic acid intercalating dyes, like propidium monoazide used in this study, represent one of the most successful recent approaches to detect viable cells (as defined by an intact cell membrane) by PCR and have been effectively evaluated in different microorganisms, major drawbacks have also been reported [24]. When applied to complex environmental samples the dye may be able to penetrate into viable or reversibly damaged cells, leading to false negative results. Further, bacteria might not all be transferred from their substratum to the supernatant or be damaged during the initial steps of the v-PCR protocol. Considering that Leptospira concentrations in our samples were low, v-PCR most probably underestimated bacterial viability in our experimental procedure.
Interestingly, the control sites, defined by the former presence of L. interrogans Pyrogenes but without human contamination reported over more than one year, showed no positive sample for lipL-32 qPCR detection. This not only suggests that the index patients actually got infected in the sites investigated, but also that targeting human contamination areas is a valuable strategy to properly identify Leptospira-contaminated areas, notably for research purpose.
Dynamic changes in Leptospira population in environmental samples seem to have occurred over the time course of this study. While the infecting L. interrogans Pyrogenes could only be detected by qPCR during the first investigation, our study revealed the presence of other pathogenic Leptospira DNA not associated to any known species (site 4). In sites 2 and 3, L. interrogans Pyrogenes was detected alongside with other pathogenic Leptospira spp. It is well known that microorganisms can cooperate in complex assemblages to better exploit nutritional resources and resist to stressful environmental conditions. Because leptospires are thought to be highly susceptible to adverse environmental stresses, they could have promoted a unique microbial interaction, by which leptospires would successfully survive and persist in the environment. This emerging idea has been highlighted by the recent discovery of biofilm produced by pathogenic leptospires [25,26]. Whether multispecific biofilms either produced by Leptospira spp. or formed by other environmental bacteria and providing shelter to leptospires, might be present in natural habitats, contribute to the persistence and allow long-term survival of pathogenic leptospires in nutrient-poor or adverse aqueous environments deserves consideration. Recent work in other settings where leptospirosis is highly endemic supports this hypothesis [9].
Interestingly, during the flooding event which occurred on February 2nd, people who got infected at sites 1, 2 and 3 were bathing with at least 2 other persons who were exposed similarly to the Leptospira environmental risk, but did not develop a clinical form of leptospirosis. This observed low attack rate raises many questions including asymptomatic leptospirosis. A recent sero-incidence study in Brazil has shown that only a very small proportion of infections actually leads to clinical disease [27].
Overall, this study revealed that pathogenic Leptospira are widespread in river soils in places associated with recent human cases. The infecting strain was evidenced in all the investigated sites and viable leptospires were still detected 9 weeks after the contamination event. These observations are particularly interesting especially if they are analyzed in regards of daily rainfall data (S1 Fig). Analysis of the daily rainfall shows that in all 4 study sites, several episodes of heavy rain occurred over the 6-month period when this study was performed. But consistently with our qPCR results these rain events have probably not been a major source of re-contamination with human threatening strains, as supported by (i) the fact that a similar sampling strategy failed to evidence an environmental contamination with L. interrogans Pyrogenes in late samples and (ii) an increase in qPCR Ct values (S1 Fig), suggesting a decrease of the environmental Leptospira load over time. Therefore, it is likely that the Leptospira DNA that was detected over this 4-month study corresponded to leptospires deposited by the flooding event of February 2nd.
Still, temporal investigations evidenced changes in leptospiral diversity and revealed the presence of yet unreported strains in soil samples and never evidenced in any mammal in New Caledonia despite long research, suggesting that soil might act as an environmental reservoir of pathogenic leptospires offering them a protective atmosphere. v-PCR coupled to molecular-based typing on soil samples proved effective at confirming infection sites and investigating the leptospiral risk over time. Because soil DNA extraction only uses small amounts of soil, the use of this approach for risk evaluation should consider the possibility of false negative results. Still, the assessment and quantification of the leptospiral burden in environmental samples might prove valuable to guide public health interventions. To help expand the current knowledge about the leptospirosis environmental cycle and the spatial and temporal distribution of leptospires in the environment, further studies will also characterize the physicochemical characteristics of soils shown to support or oppositely compromise the survival of pathogenic leptospires. Furthermore, determination of the environmental burden may help inform health authorities before adopting preventive measures such as access restrictions to contaminated areas during heavy rainfall events. Finally, evaluation of the environmental leptospiral load through quantitative methods can be a useful method to monitor high risk areas and help protect local populations, but also to discover an unexplored biodiversity of pathogenic leptospires.
Supporting information
(TIF)
Acknowledgments
Many thanks go to Dr Magali Teurlai who gratefully generated for us, using “R” software, the Leptospirosis incidence map of New Caledonia over the 2007–2013 period. We gratefully acknowledge Dr Anne Pfannstiel for her generous help regarding the coordination of patients interviews. CG is a member of GLEAN (Global Leptospirosis Environmental Action Network), which main purpose is to help countries in developing and implementing policies and tools for leptospirosis outbreak prediction, prevention, detection and intervention. Sequence reads were obtained on the ABI 3730 xl at the regional genomic core research facilities for life science in New-Caledonia “Plate-Forme du Vivant de Nouvelle-Calédonie PFV-NC”.
Data Availability
All relevant data including NCBI Accession Numbers for DNA sequences are within the paper and its Supporting Information files.
Funding Statement
This study was funded by Axa Research Fund and Institut Pasteur in New Caledonia. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Costa F, Hagan JE, Calcagno J, Kane M, Torgerson P, et al. (2015) Global Morbidity and Mortality of Leptospirosis: A Systematic Review. PLoS Negl Trop Dis 9: e0003898 10.1371/journal.pntd.0003898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McBride AJ, Athanazio DA, Reis MG, Ko AI (2005) Leptospirosis. Curr Opin Infect Dis 18: 376–386. [DOI] [PubMed] [Google Scholar]
- 3.Chang SL, Buckingham M, Taylor MP (1948) Studies on Leptospira icterohaemorrhagiae; survival in water and sewage; destruction in water by halogen compounds, synthetic detergents, and heat. J Infect Dis 82: 256–266. [DOI] [PubMed] [Google Scholar]
- 4.Karaseva EV, Chernukha YG, Piskunova LA (1973) Results of studying the time of survival of pathogenic leptospira under natural conditions. J Hyg Epidemiol Microbiol Immunol 17: 339–345. [PubMed] [Google Scholar]
- 5.Levett PN (2001) Leptospirosis. Clin Microbiol Rev 14: 296–326. 10.1128/CMR.14.2.296-326.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith CEG, Turner LH (1961) The effect of pH on the survival of leptospires in water. Bulletin WHO 24: 35–43. [PMC free article] [PubMed] [Google Scholar]
- 7.Andre-Fontaine G, Aviat F, Thorin C (2015) Waterborne Leptospirosis: Survival and Preservation of the Virulence of Pathogenic Leptospira spp. in Fresh Water. Current Microbiology 71: 136–142. 10.1007/s00284-015-0836-4 [DOI] [PubMed] [Google Scholar]
- 8.Barragan VA, Mejia ME, Travez A, Zapata S, Hartskeerl RA, et al. (2011) Interactions of Leptospira with Environmental Bacteria from Surface Water. Curr Microbiol 62: 1802–1806. 10.1007/s00284-011-9931-3 [DOI] [PubMed] [Google Scholar]
- 9.Vinod Kumar K, Lall C, Vimal Raj R, Vedhagiri K, Vijayachari P (2015) Co-existence and survival of pathogenic leptospires by formation of biofilm with Azospirillum. FEMS Microbiol Ecol 91: pii:fiv051. [DOI] [PubMed] [Google Scholar]
- 10.Singh R, Stine OC, Smith DL, Spitznagel JK Jr., Labib ME, et al. (2003) Microbial diversity of biofilms in dental unit water systems. Appl Environ Microbiol 69: 3412–3420. 10.1128/AEM.69.6.3412-3420.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rawlins J, Portanova A, Zuckerman I, Loftis A, Ceccato P, et al. (2014) Molecular detection of leptospiral DNA in environmental water on st. Kitts. Int J Environ Res Public Health 11: 7953–7960. 10.3390/ijerph110807953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Viau EJ, Boehm AB (2011) Quantitative PCR-based detection of pathogenic Leptospira in Hawai'ian coastal streams. J Water Health 9: 637–646. 10.2166/wh.2011.064 [DOI] [PubMed] [Google Scholar]
- 13.Riediger IN, Hoffmaster AR, Casanovas-Massana A, Biondo AW, Ko AI, et al. (2016) An Optimized Method for Quantification of Pathogenic Leptospira in Environmental Water Samples. PLoS One 11: e0160523 10.1371/journal.pone.0160523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goarant C, Laumond-Barny S, Perez J, Vernel-Pauillac F, Chanteau S, et al. (2009) Outbreak of leptospirosis in New Caledonia: diagnosis issues and burden of disease. Trop Med Int Health 14: 926–929. 10.1111/j.1365-3156.2009.02310.x [DOI] [PubMed] [Google Scholar]
- 15.Stoddard RA, Gee JE, Wilkins PP, McCaustland K, Hoffmaster AR (2009) Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the LipL32 gene. Diagn Microbiol Infect Dis 64: 247–255. 10.1016/j.diagmicrobio.2009.03.014 [DOI] [PubMed] [Google Scholar]
- 16.Perez J, Goarant C (2010) Rapid Leptospira identification by direct sequencing of the diagnostic PCR products in New Caledonia. BMC Microbiol 10: 325 10.1186/1471-2180-10-325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merien F, Portnoi D, Bourhy P, Charavay F, Berlioz-Arthaud A, et al. (2005) A rapid and quantitative method for the detection of Leptospira species in human leptospirosis. FEMS Microbiol Lett 249: 139–147. 10.1016/j.femsle.2005.06.011 [DOI] [PubMed] [Google Scholar]
- 18.Muñoz-Zanzi C, Mason MR, Encina C, Astroza A, Romero A (2014) Leptospira Contamination in Household and Environmental Water in Rural Communities in Southern Chile. Int J Environ Res Public Health 11: 6666–6680. 10.3390/ijerph110706666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calderon A, Rodriguez V, Mattar S, Arrieta G (2014) Leptospirosis in pigs, dogs, rodents, humans, and water in an area of the Colombian tropics. Trop Anim Health Prod 46: 427–432. 10.1007/s11250-013-0508-y [DOI] [PubMed] [Google Scholar]
- 20.Adler B, de la Pena Moctezuma A (2009) Leptospira and leptospirosis. Vet Microbiol 140: 287–296. 10.1016/j.vetmic.2009.03.012 [DOI] [PubMed] [Google Scholar]
- 21.Saito M, Villanueva SY, Chakraborty A, Miyahara S, Segawa T, et al. (2013) Comparative analysis of Leptospira strains isolated from environmental soil and water in the Philippines and Japan. Appl Environ Microbiol 79: 601–609. 10.1128/AEM.02728-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mason MR, Encina C, Sreevatsan S, Muñoz-Zanzi C (2016) Distribution and Diversity of Pathogenic Leptospira Species in Peri-domestic Surface Waters from South Central Chile. PLoS Negl Trop Dis 10: e0004895 10.1371/journal.pntd.0004895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ganoza CA, Matthias MA, Collins-Richards D, Brouwer KC, Cunningham CB, et al. (2006) Determining risk for severe leptospirosis by molecular analysis of environmental surface waters for pathogenic Leptospira. PLoS Med 3: e308 10.1371/journal.pmed.0030308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fittipaldi M, Codony F, Adrados B, Camper AK, Morato J (2011) Viable real-time PCR in environmental samples: can all data be interpreted directly? Microb Ecol 61: 7–12. 10.1007/s00248-010-9719-1 [DOI] [PubMed] [Google Scholar]
- 25.Ristow P, Bourhy P, Kerneis S, Schmitt C, Prevost MC, et al. (2008) Biofilm formation by saprophytic and pathogenic leptospires. Microbiology 154: 1309–1317. 10.1099/mic.0.2007/014746-0 [DOI] [PubMed] [Google Scholar]
- 26.Trueba G, Zapata S, Madrid K, Cullen P, Haake D (2004) Cell aggregation: a mechanism of pathogenic Leptospira to survive in fresh water. Int Microbiol 7: 35–40. [PubMed] [Google Scholar]
- 27.Felzemburgh RD, Ribeiro GS, Costa F, Reis RB, Hagan JE, et al. (2014) Prospective study of leptospirosis transmission in an urban slum community: role of poor environment in repeated exposures to the leptospira agent. PLoS Negl Trop Dis 8: e2927 10.1371/journal.pntd.0002927 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
Data Availability Statement
All relevant data including NCBI Accession Numbers for DNA sequences are within the paper and its Supporting Information files.