Abstract
Using a 3D co-culture model, we identified significant sub-type-specific changes in gene expression, metabolic, and therapeutic sensitivity profiles of breast cancer cells in contact with cancer-associated fibroblasts (CAFs). CAF-induced gene expression signatures predicted clinical outcome and immune-related differences in the microenvironment. We found that fibroblasts strongly protect carcinoma cells from lapatinib, attributable to its reduced accumulation in carcinoma cells and an elevated apoptotic threshold. Fibroblasts from normal breast tissues and stromal cultures of brain metastases of breast cancer had similar effects as CAFs. Using synthetic lethality approaches, we identified molecular pathways whose inhibition sensitizes HER2+ breast cancer cells to lapatinib both in vitro and in vivo including JAK2/STAT3 and hyaluronic acid. Neoadjuvant lapatinib therapy in HER2+ breast tumors lead to a significant increase of phospho-STAT3+ cancer cells and a decrease in the spatial proximity of proliferating (Ki67+) cells to CAFs impacting therapeutic responses. Our studies identify CAF-induced physiologically and clinically relevant changes in cancer cells and offer novel approaches for overcoming microenvironment-mediated therapeutic resistance.
Keywords: breast cancer, microenvironment, lapatinib resistance
Introduction
Cancer associated fibroblasts (CAFs) represent the most abundant non-neoplastic cell type within tumors and are the major source of extracellular matrix (ECM). CAFs have been implicated in tumor initiation, progression, and response to therapies (1,2). Our understanding of mechanisms of interactions between breast carcinoma cells and CAFs remains fragmented in part due to substantial heterogeneity of breast tumors (3,4) and CAFs (1).
Resistance to cytotoxic and targeted therapies is the major obstacle towards improved long-term patient survival. A growing body of evidence suggests that, tumor-associated stroma, and particularly CAFs, play a major role in resistance to both chemo- and targeted therapies, including immunotherapy (5-9). ECM produced by fibroblasts can contribute to interstitial and mechanical pressures within tumors that limit proper blood flow and reduce the delivery of therapeutic agents (10). Stroma-derived gene signatures have also been shown to be strong predictors of clinical outcome of breast cancer patients (11). However, the mechanistic details of this protection, as well as therapeutic options to limit it, remain poorly explored.
Here we found that interactions with CAFs in 3D co-cultures induces substantial sub-type-specific alterations in gene expression and metabolic profiles of breast carcinoma cells. Furthermore, CAFs confer strong resistance to lapatinib, a dual EGFR/HER2 inhibitor (12), attributable to reduced accumulation of the drug within the carcinoma cells and an elevated apoptotic threshold. Interestingly, fibroblasts from normal breast tissues and stromal cultures of brain metastases of breast cancer confer therapeutic resistance similar to CAFs, highlighting a commonality shared by multiple stromal cell types regardless of their tissue of origin. Using synthetic lethality screens and interrogation of the functional relevance of ECM, we identified several signaling pathways and targets whose inhibition overcomes lapatinib resistance, including JAK2/STAT3 and hyaluronan. We validated the physiologic and clinical relevance of our findings obtained from this 3D culture model in vivo using xenografts and primary patient samples. Thus, our study sheds light on the changes induced by the interaction of breast cancer cells with fibroblasts and offers novel approaches to overcoming microenvironmental protection by therapeutically targeting the stroma.
Materials and Methods
Cell lines and tissue culture conditions
Breast cancer cell lines were obtained 1999-2016 from the following sources: MDA-MB-453, HCC1954, MCF7, and T47D cell lines were obtained from ATCC; MCF10DCIS. com from Dr. F. Miller (Karmanos Cancer Institute, Detroit, MI), SUM149PT from Dr. S. Ethier, University of Michigan, Ann Arbor, MI). Identity of the cell lines was confirmed by short tandem repeats (STR) analysis. Normal human astrocytes were purchased from Lonza and Cell Applications. Cells were cultured in media recommended by the provider; regular tests for mycoplasma contamination were performed. Fibroblasts and stroma from brain metastatic lesions of breast cancers were isolated from patient samples as previously described (13,14) and expanded for 3-10 passages prior to the experiments. All human tissue was collected using protocols approved by the Dana-Farber Harvard Cancer Center (DF/HCC) Institutional Review Board. Fibroblasts expansions and all the in vitro experiments were performed in 50/50 mixture of DMEM-F12, 10% FBS/MEGM with supplements. Fluorescently labeled derivatives of carcinoma cell lines and fibroblasts were obtained by lentiviral expression of mCherry/Luciferase (obtained from Dr. C. Mitsiades, DFCI), pLVX-AcGFP or pLVX-dsRED (Clontech Laboratories Inc.). Matrigel 3D culture experiments were performed using on-top method as described previously (15).
Xenograft experiments
All animal procedures were approved by the DFCI IACUC (DFCI protocol#11-023) and followed NIH guidelines. Tumors were induced by bilateral orthotopic injection into 6-weeks old female NOG mice of 1 × 106 carcinoma cells with/without 0.5 × 106 CAFs re-suspended in 50% Matrigel (BD Biosciences) per transplant. Tumor volumes and diameters were monitored bi-weekly with electronic calipers. Lapatinib dissolved in 0.5% HPMC/0.1% Tween 80, and NVP-BK805 (provided by Novartis Oncology) in 50 mM sodium citrate buffer pH 3.0 were administered via daily oral gavages. Hyaluronidase (Vitrase®, Bausch and Lomb) was administered via subcutaneous injection in the vicinity of tumors at 50 μl (10 U) per injection twice per week. PEGPH20 (provided by Halozyme Therapeutics) was intravenously administered at 1 mg/ml/week. Prior to treatment, distribution of tumor sizes was compared between control and experimental groups to ensure uniformity. No blinding was performed during the tumor measurements in live animals. One-two hours prior to euthanasia, animals were injected intraperitoneally with 100 μl of 10 mg/ml BrdU (bromodeoxyuridine) solution to label proliferating cells.
Gene expression profiling
DsRED labeled carcinoma cell lines and GFP expressing CAFs were cultured either separately or together for 17 hours. Colonies were released from Matrigel by 30 min incubation in PBS/EDTA while separately cultured cells were combined to control for cross-contamination. Cell mixtures were washed with PBS, trypsinized for 7 minutes, quenched with serum containing media, washed, and collected by centrifugation. Epithelial cells were selected using Epithelial Enrich Dynabeads (Life Technologies) following manufacturer's protocol. Microscopic examination of captured cells revealed no detectable CAF contamination. 5×105 cells of each sample were used to prepare SAGE-Seq libraries as described before (16). Sequencing tags were mapped and normalized as described previously (17). All genomic profiling data generated in this publication can be found online with GEO Publication Reference ID GSE80333 (http://www.ncbi.nlm.nih.gov/geo/)
Statistical analyses
Statistical analyses of in vitro and in vivo experiments were performed using Graphpad Prism software, using statistical tests indicated in figure legends. Descriptions of analyses of expression and patient data are provided in the supplementary information.
Luciferase reporter assays for cell viability
For cellular viability assays, carcinoma cells expressing lentiviral mCherry/Luciferase were plated with/without unlabeled or GFP expressing CAFs into non-adherent white 96-well plates (Nunc, cat#236105). 2-4 × 104 carcinoma cells with/without 0.5-1 × 104 fibroblast cells were plated per well. After incubating overnight, inhibitors were added to the media. Following 3-day culture in the presence of inhibitor or vehicle control, viability of luciferase-expressing cells was assessed by adding luciferin-D (125 ug/ml) and measuring luminescence using Biotek Synergy plate reader. For synthetic lethality screen, inhibitors of signaling pathways that were either previously implicated in resistance to targeted therapies, or that were identified as altered by interaction with fibroblasts in our analyses, were purchased from Selleck Chemicals, Fisher Scientific, or obtained from manufacturers. Pilot experiments were performed to identify the highest inhibitor doses that display <20% inhibition of luminescence signal compared to vehicle-treated control. Then, sensitivity of separately or co-cultured cells to varying concentrations of lapatinib as a single agent or added with non-toxic or low toxicity doses of inhibitors was assessed as described above. In all cases, lapatinib was added 2-6 hours after the addition of the inhibitors. Fold difference between the expected surviving fraction, based on the additive effect of individual toxicities, and the observed toxicity in the experiment was determined. All of the experiments were performed at least with 3 biological replicates, each containing 3 technical replicates.
Histological, immunohistochemical, and multicolor immunofluorescence analyses
Histologic, immunohistochemical, and multicolor immunofluorescence analyses and image acquisition were performed as previously described (18). Scoring for the expression of each marker was done as follows: the percentage apoptotic cells were estimated by counting an average of 1,000-1,500 cells/sample using ImageJ 1.45s software from 3-4 randomly selected regions of tissue slides stained for cleaved caspase-3. Live colonies were imaged directly by vital microscopy using intrinsic fluorescence with a Nikon Eclipse TE3000 inverted microscope. Single-cell based distance from fibroblast calculation was performed using a custom built macro for ImageJ 1.42r program (code available upon request). The macro was constructed to allow calculation of the closest fibroblast from each cell with particular nuclear staining (BrdU or Ki67). First, the nuclear stain image was used to define nuclei as regions of interest and the coordinates of their centroids were saved, but only if they overlapped with regions containing BrdU/Ki67 staining (positive cells). Then, the fibroblast staining channel was reduced to its maxima points. Finally, for each saved centroid, a distance to every maxima point was measured and the shortest distance for each individual nuclei was saved. This resulted in a table containing the shortest distance between a given positive cell and a fibroblast. Distribution of the distances was plotted using R software.
Intracellular BH3 profiling
Following 24 hours suspension culture with/without CAFs, carcinoma cells were recovered by incubation with 2 mg/ml collagenase for 30 minutes, followed by 10 minute trypsinization and washing with PBS. BH3 profiling was performed essentially as described (19). Results from 4 (HCC1954) or 5 (MDA-MB-453) independent experiments are shown as % cytochrome c release of control DMSO peptide.
Results
3D model of breast carcinoma cells and fibroblasts interaction
Our previous studies have demonstrated that co-culture with breast stromal fibroblasts dramatically alters the morphology of MCF10DCIS cells in organotypic 3D Matrigel cultures (20). We expanded on this observation to analyze the phenotypic consequences of interaction between breast cancer-associated fibroblasts (CAFs) and cell lines representing different breast cancer sub-types, including triple-negative (MCF10DCIS, SUM149PT), luminal estrogen receptor positive (MCF7 and T47D) and HER2 positive (MDA-MB-453). In all cases, fibroblasts engaged in physical interaction with epithelial cells, leading to the formation of larger and more complex colonies (Figure 1A and Figure S1A). We next tested whether this interaction was generalizable to different types of stromal fibroblasts. All of the examined normal breast and breast carcinoma-derived fibroblasts as well as normal human astrocytes and stroma from brain metastases of breast cancer demonstrated strong morphogenic interaction with MCF10DCIS cells (Figure S1B). Heterotypic interactions between carcinoma cells and fibroblasts were disrupted by collagenase (Figure S1C), suggesting collagen dependence. We reasoned that, owing to the relative ease of experimental manipulation, this heterotypic 3D model could be used to gain insights into the molecular changes induced in carcinoma cells by their interaction with stromal fibroblasts. To this end, we examined the functional consequences of the interaction, focusing on changes in gene expression and cellular metabolism (Figure 1B).
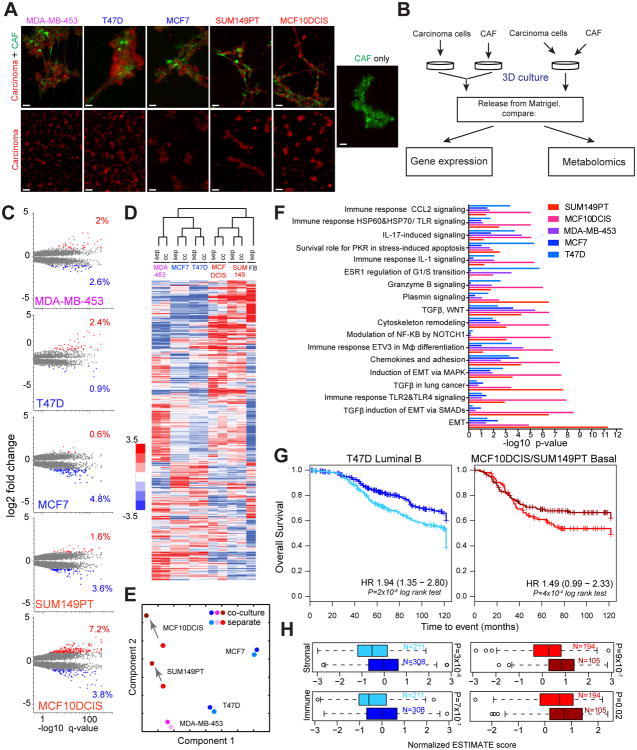
Figure 1.
Phenotypic changes induced by CAFs. A, Vital immunofluorescence images of 3D Matrigel cultures. Breast carcinoma cells and CAFs are labeled with mCherry (red) and GFP (green), respectively. Scale bars represent 100μm. B, Experimental outline Co-cultured cells are compared to separately cultured carcinoma cells and CAFs that are mixed prior to analysis. C, Volcano plots of CAF-induced changes in gene expression. Statistically significantly different genes up- or down-regulated ≥2 fold are indicated by red and blue, respectively, with corresponding percentages. D, Heat map and hierarchical clustering of expression values of genes that are differentially expressed in at least one cell line. E, Principle component analysis of gene expression profiles Lighter and darker shades denote separate and co-cultures, respectively. Arrows indicate change in profiles. F, Molecular pathways significantly affected by interaction with CAFs, cutoff p<10-6. G, Overall survival in the METABRIC patient cohort stratified by the T47D signature in Luminal B patients and a combined MCF10DCIS/SUM149 signature in Basal patients. H, Immune and stromal ESTIMATE scores for the corresponding good and poor prognosis patient groups.
Fibroblasts induce tumor sub-type specific molecular changes in breast cancer cells
To examine changes in gene expression induced in breast carcinoma cells by interaction with stromal fibroblasts, we performed SAGE-seq (16) on tumor epithelial cells purified from separate or CAFs co-cultures. Interaction with fibroblasts induced substantial changes in gene expression leading to a more than 2-fold change in the 3.3-11% of genes analyzed, with more pronounced responses seen in basal-like cell lines (Figure 1C-E and Table S1). Despite the substantial changes in gene expression, cell lines maintained their molecular identity, as judged by hierarchical clustering analysis, and the majority of the fibroblast-induced changes were cell line and sub-type specific (Figure 1D). To gain insights into the biological impact of interaction between carcinoma cells and CAFs, we performed MetaCore analysis (21). Consistent with clustering analysis, most of the changes were sub-type-specific, as exemplified by the induction of epithelial-mesenchymal transition in basal-like cell lines (Figure 1F). Still, some of the changes, such as several immune-response related and cell adhesion and TGF-β signaling pathways, were shared by cell lines representing different molecular sub-types.
To investigate the clinical relevance of these changes, we asked whether the differentially expressed gene sets would subdivide breast tumors into clinically relevant groups using the Metabric cohort (22). We found that luminal T47D and combined MCF10DCIS+SUM149 cell line-derived “basal” gene expression signatures clustered luminal B and basal-like breast tumors into two groups with a statistically significant difference in overall survival (Figure 1G and Table S2). Interestingly, these expression signatures also correlated with estimated stromal and immune cell contents of the tumors and were different between the good and poor outcome groups, with poor outcome associated with lower stromal and immune scores (Figure 1H).
Next, we examined the interaction-induced changes in cellular metabolism. Due to sample size requirements, profiling was performed on mixtures of epithelial cells and fibroblasts. Epithelial cells were co-cultured with CAFs, or cultured separately and then combined with fibroblasts immediately prior to harvesting (Figure 1B). Whereas the majority of co-culture-induced changes were cell line specific, we observed a notable enrichment in metabolites in the glutathione pathway (Figure S2A, B and Table S3). Consistent with these results, we detected a significant reduction in total glutathione levels in co-cultures with all the cell lines (Figure S2C). Since reduced glutathione is a major buffer of reactive oxygen species (ROS), this decrease suggests that epithelial-stromal cell interactions might have a substantial impact on ROS signaling and metabolism, which can affect sensitivity to apoptosis. In summary, our metabolic and expression profiling revealed that despite considerable cell line and sub-type differences, interaction with stromal fibroblasts induces common responses in breast cancer cells that are clinically relevant.
Stromal fibroblasts enhance therapeutic resistance
To interrogate the impact of CAFs on therapy response we stably expressed luciferase in carcinoma cells, enabling us to exclusively monitor the viability of breast cancer cells, as luciferase-negative fibroblasts cannot produce the signal (Figure 2A) (23). Since CAFs degrade Matrigel within 24 hours of co-culture, we performed the assays in suspension cultures where fibroblasts were still capable of engaging in heterotypic interactions with carcinoma cells (Figure S3).
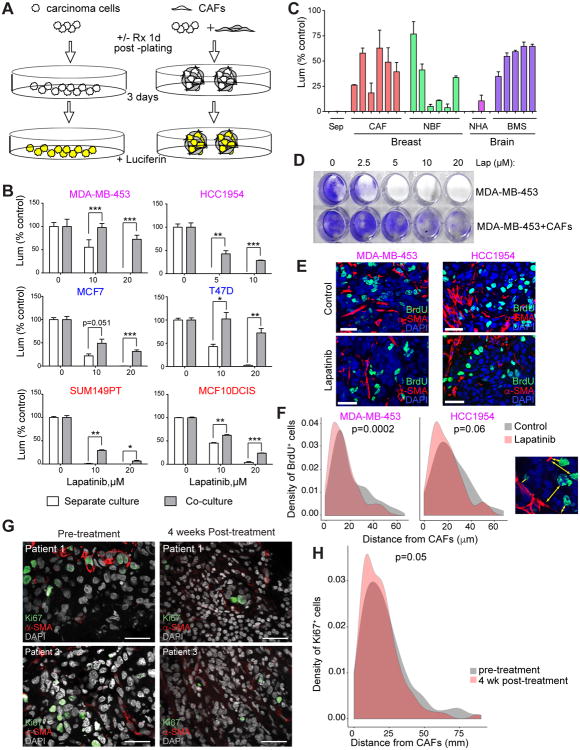
Figure 2.
Fibroblasts protect breast carcinoma cells from lapatinib. A, Experimental outline. Carcinoma cells expressing luciferase are plated with and without fibroblasts. Following overnight incubation, the cultures are treated with a therapeutic agent Luciferase activity is used as a proxy for viable cell numbers. B, Survival of the indicated carcinoma cells in monocultures and CAFs co-cultures with various concentrations of lapatinib. Asterisks indicate statistical significance (* p<005, ** p<001, *** p<0001). C, Protective effect of the indicated stroma on survival of lapatinib-treated MDA-MB-453 cells CAFs – primary breast cancer-associated fibroblasts, NBF – normal breast fibroblasts, NHA- normal human astrocytes, BMS – breast cancer brain metastasis stroma. D, Long-term clonogenic survival of MDA-MB-453 cells following 3 days of lapatinib treatment CAFs were eliminated by puromycin selection and purity of the epithelial cells was confirmed by microscopy. E, Representative images of BrdU+ (green) and α-SMA (red) staining in xenografts treated with vehicle or lapatinib Scale bars correspond to 25μm. F, Distribution of distances between BrdU+ carcinoma cells and nearest CAFs in xenografts treated with vehicle or lapatinib P-values indicate statistical significance (t-test). Right panel depicts a representative picture with distances indicated by arrows. G, Representative images of Ki67+ (green) and α-SMA (red) staining of primary breast tumor samples before and after four weeks of neoadjuvant lapatinib treatment. Scale bars correspond to 25μm. H, Distribution of distances between Ki67+ carcinoma cells and nearest CAFs in breast tumors before and after neoadjuvant lapatinib treatment. P-value indicates statistical significance (two-sided t-test).
We found that co-culture with CAFs resulted in partial protection against commonly used chemotherapeutic agents doxorubicin and taxol, although the effect was relatively modest and variable among cell lines (Figure S4A). Strikingly, CAFs provided an exceptional protection against the dual EGFR/HER2 inhibitor lapatinib in most of the cell lines tested (Figure 2B and Figure S4B). This universal protection was unexpected, as EGFR and HER2 signaling is considered to be characteristic of basal-like and HER2+ breast cancer cells. Nevertheless, we observed lapatinib-sensitive EGFR phosphorylation in all of the cell lines tested, consistent with the panel-wide protection (Figure S4C). For subsequent studies we primarily focused on HER2+ cell lines (HCC1954 and MDA-MB-453), as they better represent clinical targets of lapatinib.
Protection from lapatinib was not limited to a particular type of fibroblast, as stromal fibroblasts from primary breast tumors, normal breast tissues, and brain metastases of breast cancer had similar effects, despite clear differences in gene expression profiles (Figure 2C and Figure S4D). Importantly, carcinoma cells recovered at the end of the assay were capable of clonogenic survival (Figure 2D), demonstrating long-term survival advantage. This protection could not be observed in the 2D co-cultures (Figure S5A). Further CAF conditioned media, spatially separate pre-clustered CAFs or colonies composed of CAFs and unlabeled carcinoma cells failed to confer resistance (Figure S5B, C). Finally, analysis of mCherry fluorescence as a proxy of survival of labeled carcinoma cells revealed that lapatinib-resistant epithelial cells are located in close proximity to the fibroblasts (Figure S5D). These results suggest that protective impact of CAFs require close proximity in 3D context and cannot be attributed to a reduced availability of lapatinib to carcinoma cells due to its uptake by CAFs.
We next evaluated the relevance of our findings in vivo. To this end, we compared the lapatinib sensitivity of carcinoma cells injected with/without CAFs into the contralateral mammary fat pads of the same mouse. We observed rapid colonization of tumors initiated both with/without human CAFs by mouse stromal fibroblasts, thereby complicating a direct assessment of the protective effect of co-injected human fibroblasts, since mouse stromal fibroblasts were also capable of exerting strong protection against lapatinib in 3D cultures (Figure S5E). Therefore, we sought to validate the protective impact of CAFs by examining the distance between CAFs marked by α-SMA staining and proliferating carcinoma cells (marked by BrdU staining), comparing lapatinib-treated and control groups (Figure 2E). Using an automated quantitative image analysis approach, we found that the distance between the infiltrating mouse CAFs and proliferating cancer cells was statistically significantly shorter in the lapatinib treatment group compared to vehicle treated controls (Figure 2F), suggesting that fibroblasts were reducing the cytostatic effects of lapatinib in vivo.
To further validate the physiologic and clinical relevance of our findings, we also performed immunofluorescence analysis for Ki67 (marker of proliferating cells) and α-SMA of pre- and post-treatment biopsies from a cohort of HER2+ breast tumors subjected to neoadjuvant lapatinib therapy (24,25) (Figure 2G). In line with our xenograft data, the distance between SMA+ CAFs and Ki67+ proliferating cancer cells was statistically significantly shorter following 4 weeks of lapatinib treatment compared to pre-treatment biopsies (Figure 2H). Therefore, our results demonstrate that spatial proximity to CAFs enhances the resistance of breast cancer cells to lapatinib in vitro whereas the decrease of distance between proliferating cancer cells and CAFs in xenografts and primary breast tumor samples after lapatinib treatment support the relevance of these findings in vivo. However, alternative explanation of the in vivo data such as altered mobility of cancer cells and loss of CAFs due to lapatinib treatment cannot be excluded.
Mechanisms underlying fibroblast-induced therapy resistance
Despite the sub-type and cell line specificity of gene expression changes, protection against lapatinib in fibroblast co-cultures was observed with most of the cell lines tested. We therefore hypothesized that this protection is likely to involve shared underlying mechanism(s). Hence, we first tested whether the accumulation of lapatinib is reduced in the complex multicellular organoids formed in the presence of fibroblasts. Due to the strong auto-fluorescence of lapatinib, we were able to assess the intracellular accumulation of the drug directly using flow cytometry (FACS). We found that fibroblast co-cultures reduced the accumulation of lapatinib in the epithelial cells (Figure 3A), an effect that was also observed in vivo (Figure S6A). Consistent with the reduced intracellular accumulation of the drug, the effect of lapatinib on the phosphorylation of HER2, EGFR, and AKT was less pronounced in fibroblast co-cultures (Figure 3B). Furthermore, we observed that fibroblast co-culture reduced the levels of cleaved caspase-3, indicative of attenuated apoptosis (Figure 3B).
Figure 3. Mechanisms of stroma-induced lapatinib resistance.
A, Accumulation of lapatinib in MDA-MB-453 cells grown in monocultures and CAFs co-cultures assessed by FACS based on lapatinib autofluorescence. B, Immunoblot analysis of the indicated proteins in HCC1954 and CAFs cultured separately or in post-harvest mixtures, compared to co-cultures. C, Impact of CAFs co-cultures on the ability of indicated pro-apoptotic peptides to induce apoptosis in the indicated cell lines Asterisks indicate results of pairwise t test. D, Sensitization to cytotoxic effect of lapatinib by BCL2/BCL-xl inhibitor ABT737. Asterisks indicate p-values of interaction term by 2-way ANOVA* p<005; ** p<001, *** p<0001.
We next asked if the attenuated apoptotic response is entirely due to the reduced intracellular lapatinib accumulation or it is dependent on some other mechanism. To address this question, we used BH3 profiling (26), a FACS-based assay that measures overall apoptotic sensitivity (19). In this assay, the cells were first dissociated from colonies, then permeabilized and challenged with pro-apoptotic peptides. Therefore, all of the cells were expected to display equal uptake of pro-apoptotic peptides irrespective of previous culture conditions. Nevertheless, we found that cancer cells recovered from fibroblast co-cultures were less responsive to apoptotic stimuli, indicating an elevated apoptotic threshold (Figure 3C). To address the functional relevance of reduced sensitivity to pro-apoptotic stimuli, we asked whether fibroblast-induced resistance could be overcome by targeting the anti-apoptotic machinery. Indeed, a combination of lapatinib with BCL2/BCL-xL inhibitors (e.g., ABT737) reversed the fibroblast-induced protection (Figure 3D), although cancer cells in co-cultures were still less sensitive to this combined treatment than when cultured alone. Therefore, the mechanisms of fibroblast-induced resistance to lapatinib in breast cancer cells involves both reduced accumulation of the drug as well as elevated apoptotic threshold.
Stromal hyaluronan is essential for lapatinib resistance
As demonstrated above, the protection from lapatinib requires close physical proximity between fibroblasts and carcinoma cells. Given that cell adhesion to extracellular matrix (ECM) is essential for survival signaling, and that stromal fibroblasts are the major producer of the ECM, we explored the importance of ECM in the observed protective effect of fibroblasts. First, we asked whether collagen-mediated adhesion, which plays essential pro-survival role in some contexts (27), is involved in stromal protection against lapatinib as seen in our system. We found that collagenase treatment disrupted heterotypic interactions, preventing the formation of complex organoid-like colonies (Figure 4A). Surprisingly, this disruption led to modest, though statistically significant, inhibition of the protection against the drug (Figure 4B). Similarly, inhibition of focal adhesion kinase (FAK), a key mediator of collagen-integrin adhesion signaling led to partial increase in lapatinib sensitivity in fibroblast co-cultures (Figure 4C). We therefore concluded that collagen-mediated adhesion is not an essential component of the pro-survival effects of fibroblasts.
Figure 4.
Hyaluronidase sensitizes CAFs to lapatinib. A, The effect of collagenase and hyaluronidase treatment on the morphology of organoids in co-cultures of MDA-MB-453 cells (mCherry) and CAFs (GFP). Scale bars correspond to 100μm. B-D, Impact of collagenase (B), FAK inhibitor (C) and hyaluronidase (D) treatment on lapatinib sensitivity of MD-MB-453 cells in CAF co-cultures. Asterisks indicate p-values of interaction term in 2-way ANOVA. E, Correlation between production of hyaluronan and protection of MDA-MB-453 cells against 20 μM lapatinib among different fibroblasts. F, Sensitization of CAFs toward lapatinib by hyaluronidase treatment p<0001, interaction term in 2-way ANOVA. G, Tumor weights and H, percentage of cells positive for cleaved caspase-3 staining of xenografts treated with hyaluronidase, lapatinib or both. Treatment was started 10 days post injection and continued for 21 days, at which point tumors were harvested. Asterisks indicate statistical significance (* p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001; unpaired Mann-Whitney test). I, Representative images of cleaved caspase-3 staining. Arrows indicate apoptotic cells. Scale bars correspond to 100μm.
We next asked whether hyaluronic acid (HA), another major component of ECM produced by fibroblasts, is involved in the protection by testing the effect of hyaluronidase. We found that hyaluronidase completely abolished the protective effect of stromal fibroblasts without disaggregating the heterotypic colonies (Figure 4A, D). Remarkably, the extent of HA production by stromal fibroblasts strongly correlated with their ability to protect epithelial cells from lapatinib (Figure 4E), consistent with the hypothesis that hyaluronan is a major fibroblast-produced factor underlying lapatinib resistance. Notably, HA-rich fibroblast-conditioned media did not have any noticeable effect (Figure S5B), suggesting that HA per se is not protective. We therefore asked whether HA might be responsible for the ability of fibroblasts to survive the toxicity arising from the treatment. Indeed, whereas fibroblasts were normally highly resistant to lapatinib, hyaluronidase treatment resulted in their dramatic sensitization to the drug (Figure 4F).
We thus explored the therapeutic utility of sensitization to lapatinib in vivo. To this end, we initiated xenografts, consisting of mixtures of MDA-MB-453 cells and stromal fibroblasts in the mammary fat pads of immune-compromised mice. Following tumor formation, the animals were treated with lapatinib or vehicle control with bi-weekly subcutaneous peritumoral injection of either hyaluronidase or PBS. We found that hyaluronidase monotherapy did not affect tumor size. In contrast, hyaluronidase treatment significantly increased the anti-tumor effects of lapatinib (Figure 4G). Notably, we found that hyaluronidase co-treatment significantly increased apoptosis rates in lapatinib treated animals (Figure 4H, I). Furthermore, we have confirmed that the anti-tumor effects of lapatinib can be enhanced with systemic treatment of stable, PEGylated form of recombinant human hyaluronidase, PEGPH20 (28), which is currently being evaluated in multiple clinical trials (Figure S6B, C). These effects of hyaluronidase were not mediated through CD44, an HA receptor, as its downregulation in breast cancer cells or in CAFs did not impact their survival (Figure S6 D, E and data not shown). Overall these data establish a rationale for combining hyaluronidase with lapatinib to improve therapeutic responses.
Synthetic lethality screen for sensitization to lapatinib
We next sought to define synthetic lethal interactions that may overcome stroma-induced resistance to lapatinib. To this end, we combined lapatinib with small molecule inhibitors of signaling and metabolic pathways that we identified as potential candidates based on our gene expression and metabolic profiling, or those that have been previously implicated in cancer therapy. For this screen, we primarily used MDA-MB-453 cells, with validation of selected inhibitors in additional cell lines (Table S4). In addition to BCL2/BCL-xL inhibitors, resistance to lapatinib was substantially diminished by co-inhibition of PI3K-AKT and JAK-STAT pathways (Figure 5A). This sensitization by PI3K-AKT inhibitors likely reflects increased inhibition of HER2/EGFR signaling, the target of lapatinib. In contrast, the effect of JAK inhibitors might be more specific, since we had observed that lapatinib increased STAT3 phosphorylation and fibroblast co-cultures led to stronger baseline and lapatinib-induced STAT3 phosphorylation (Figure 3B). Given the known anti-apoptotic effects of JAK-STAT3 signaling (29), we hypothesized that the activation of this signaling pathway might be involved in the elevation of apoptotic threshold in the fibroblast co-cultures. Notably, fibroblast-induced protection against lapatinib was reversed by co-treatment with the JAK inhibitor BSK805 (Figure 5B). To confirm the pharmacological data, we performed shRNA-mediated downregulation of GP130, a protein that is part of a number of cytokine receptors and is essential for cytokine-mediated signaling that activates JAK-STAT signaling, and STAT3. Both STAT3 and GP130 downregulation reduced the levels of phosphorylated STAT3 and increased the sensitivity of fibroblast co-cultures to lapatinib (Figure S7A, B). Furthermore, the sensitivity of CAFs to lapatinib was also significantly enhanced by BSK805 (Figure S7C), suggesting that JAK-STAT signaling in both carcinoma cells and CAFs is involved in stromal protection from lapatinib. We further tested the validity of these findings in vivo by co-administering lapatinib with BSK805 in xenograft models. Consistent with the in vitro data, BSK805 significantly increased the anti-tumor activity of lapatinib in vivo (Figure 5C).
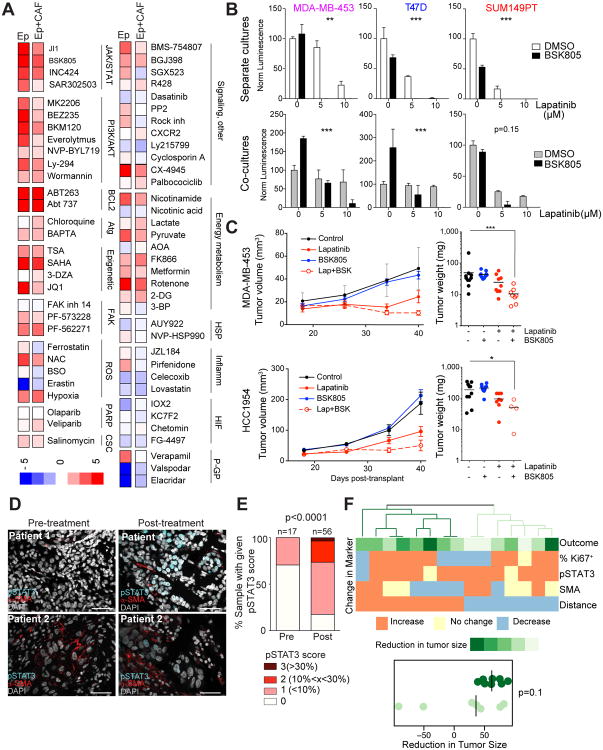
Figure 5. Overcoming CAF-mediated resistance.
A, Heat map of fold differences between observed and expected cytotoxicity of combinations of lapatinib with the indicated inhibitors. B, Sensitization to lapatinib by JAK inhibitor BSK805 in the indicated cell lines. Asterisks indicate p-values of interaction term in 2-way ANOVA. C, Growth of the indicated xenografts treated with lapatinib (100 mg/kg), BSK805 (50 mg/kg), or both compounds starting on day 18 after injection. Right panels display volumes of individual tumors at day 40. Asterisks indicate the p-values of statistical significance (* p<005; ** p<001, *** p<0001, Mann Whitney test). D, Representative images of pSTAT3+ (cyan) and α-SMA (red) staining of primary breast tumor samples before and after neoadjuvant lapatinib treatment. Scale bars correspond to 25μm. E, Bar graph depicting frequency of pSTAT3+ breast tumor cells before and after neoadjuvant lapatinib therapy in all samples combined. P value indicates statistical significance by multiple comparisons (1-way ANOVA). F, Heatmap of differences in pSTAT3, Ki67 and SMA in patients before and after neoadjuvant lapatinib treatment. Hierarchical clustering of patient samples into two groups shows a trend between changes in molecular markers with reduction in tumor size (p=01, Wilcoxon test).
To validate the clinical relevance of these findings, we performed combined pSTAT3 and α-SMA immunofluorescence analysis of HER2+ breast tumors before and after neoadjuvant lapatinib therapy (24,25). Concordant with our xenograft data, we found a significant increase in the fraction of pSTAT3+ breast cancer cells in post-treatment compared to pre-treatment biopsies, and this pattern was consistently observed when analyzing all tumors combined or matched paired samples (Figure 5D,E, Figure S7D,E, and Table S5). These results suggest that combining JAK-STAT inhibitors with HER2-targeting agents could be a more efficacious therapy for HER2+ breast tumors than targeting HER2 alone. We also explored whether changes in the fraction of pSTAT3+ or Ki67+ cancer cells, SMA+ CAFs, or the distance between Ki67+ cancer cells and SMA+ CAFs is associated with response to neoadjuvant lapatinib therapy. Tumors that showed an increase in the distance between Ki67+ cancer cells and SMA+ CAFs as well as a decreased fraction of SMA+ CAFs tended to have better response to lapatinib as assessed by a reduction in tumor size in comparison with tumors where the distance between the two cell types decreased during treatment (Figure 5F and Table S5). These results highlight the importance of CAFs in therapeutic responses and suggest clinical potential of combined targeting of cancer cells and CAFs.
Discussion
Our results demonstrate that the phenotypic response of cancer cells to physical interactions with fibroblasts is, to a large degree, dependent on the initial phenotypic configuration of carcinoma cells, with variability not only between distinct molecular subtypes of breast cancer cells but also with substantial differences among cell lines within one subtype. These findings are in line with prior related studies identifying distinct gene expression changes in luminal and basal-like breast cancer cells in co-cultures with fibroblasts (30,31). Nevertheless, given the substantial inter- and intra-tumor heterogeneity (32), conclusions based on studies involving only a small number of cell lines might offer little room for generalizations, especially regarding detailed characterization of molecular signaling mechanisms.
Yet, the development of viable therapeutic approaches requires the identification of actionable strategies that can be applicable to at least a definable subset of tumors. Our data suggest that protection from the inhibitory effects of the dual HER2/EGFR inhibitor lapatinib might be a common feature resulting from the interaction between fibroblasts and carcinoma cells, and particularly with therapeutic implications for HER2+ breast cancer for which lapatinib is clinically used. This protective effect of CAFs can be overcome by sensitizing them to lapatinib by targeting HA, a major ECM component produced by fibroblasts, and also by combined inhibition of the JAK/STAT pathway. Importantly, testing of HER2+ breast tumor samples subjected to neoadjuvant lapatinib therapy validated the physiologic and clinical relevance of our findings, since it strongly suggests that spatial proximity of breast cancer cells to SMA+ CAFs influences their response to treatment, and this in part might be mediated by the increased pSTAT3 activity.
Stromal protection against anti-cancer therapies has been historically documented for cytotoxic therapies (33). More recently, the involvement of CAFs in the development of resistance has also been documented for therapies targeting certain signaling pathways (2). Generally, CAF-mediated resistance can be attributed to signaling mechanisms arbitrated by adhesion to ECM, as well as paracrine interactions. In our experimental system, protection against the cytotoxic effects of lapatinib was mainly attributable to interactions at close spatial proximity, as fibroblast conditioned media did not confer protection. However, given the induction of STAT3 signaling and activity of JAK inhibitors in vivo and in vitro, paracrine signals are likely to contribute to the protection as well. In contrast to recent findings in stromal resistance against BRAF targeting therapies in melanoma (27), signaling through the collagen-integrin-FAK axis did not appear to be a major mediator of lapatinib resistance in our system, as collagenase treatment and inhibitors of FAK had only a modest impact on the protective effects of CAFs. Instead, targeting HA was able to completely overcome stromal protection. However, the effect is likely to be indirect, as HA-containing CAFs conditioned media or addition of purified external HA failed to protect from lapatinib. Instead, HA removal dramatically increased the sensitivity of fibroblast to lapatinib, thereby affecting all of the protective mechanisms. Enzymatic targeting of HA has recently been shown to enhance the effect of gemcitabine in pancreatic carcinomas (34), and although the authors attributed the effect to the reduction of hydrostatic pressures, a substantial decrease in the numbers of CAFs was also observed, thereby suggesting sensitization of CAFs as a contributing mechanism.
Our studies have validated multiple pathways whose inhibition elevates the sensitivity of cancer cells to lapatinib. However, this sensitization is not specific to cancer cells associated with CAFs. In most cases, CAFs were able to confer relative protection, probably attributable to reduced lapatinib uptake and an elevated apoptotic threshold. Thus, in the absence of the ability to specifically eliminate carcinoma cells within microenvironmental protective niches, targeting CAFs directly might be a more promising approach. Targeting CAFs may also be advantageous since, in contrast to neoplastic cells, CAFs maintain normal genotypes (35), which should limit inter-tumor variability and restrict adaptive potential. In addition, targeting CAFs is less likely to select for resistance compared to targeting cell autonomous pathways within cancer cells (36).
Nevertheless, caveats such as potential toxicity to the normal stroma and different stromal composition within different metastatic sites can present a considerable challenge and has to be considered. Moreover, at least in certain context, tumors developing without CAFs could lead to unintended consequences of accelerated tumor growth and metastasis (37). Similarly, gene expression signatures reflective of stromal composition within or adjacent to breast tumors have been shown to have different impact on clinical outcomes (38-40). Thus, the interaction between CAFs and cancer cells is complex and can have different consequences depending on the stage of tumor development and cancer therapies. However, our data supports that targeting tumor stroma is an attractive therapeutic option, which can be developed in parallel to the mainstream effort of directly targeting tumor cells.
Supplementary Material
Acknowledgments
We thank Drs. David Kang in Halozyme Therapeutics (San Diego, CA) and Thomas Radimerski at Novartis Oncology (Basel, Switzerland) for providing PEGPH20 and BSK805, respectively, and critical reading of our manuscript and useful discussions. We thank the BWH brain tumor bank for the acquisition of brain metastases of breast tumors.
Notes: Financial support: This work was supported by the DBT-Ramalingaswami Fellowship (S. Pyne), CDRMP W81XWH-09-1-0561 (to A. Marusyk), NIH CA116235, Breast Cancer Alliance, and Breast Cancer Research Foundation (to K. Polyak).
List of abbreviations
- CAF
cancer-associated fibroblast
Footnotes
Conflict of Interest: K.P. receives research support from and is a consultant to Novartis Oncology.
References
- 1.Ohlund D, Elyada E, Tuveson D. Fibroblast heterogeneity in the cancer wound. J Exp Med. 2014;211:1503–23. doi: 10.1084/jem.20140692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paraiso KH, Smalley KS. Fibroblast-mediated drug resistance in cancer. Biochem Pharmacol. 2013;85:1033–41. doi: 10.1016/j.bcp.2013.01.018. [DOI] [PubMed] [Google Scholar]
- 3.Polyak K. Heterogeneity in breast cancer. The Journal of clinical investigation. 2011;121:3786–8. doi: 10.1172/JCI60534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–13. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mao Y, Keller ET, Garfield DH, Shen K, Wang J. Stromal cells in tumor microenvironment and breast cancer. Cancer Metastasis Rev. 2013;32:303–15. doi: 10.1007/s10555-012-9415-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hazlehurst LA, Dalton WS. Mechanisms associated with cell adhesion mediated drug resistance (CAM-DR) in hematopoietic malignancies. Cancer Metastasis Rev. 2001;20:43–50. doi: 10.1023/a:1013156407224. [DOI] [PubMed] [Google Scholar]
- 7.Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501:346–54. doi: 10.1038/nature12626. [DOI] [PubMed] [Google Scholar]
- 8.Boelens MC, Wu TJ, Nabet BY, Xu B, Qiu Y, Yoon T, et al. Exosome transfer from stromal to breast cancer cells regulates therapy resistance pathways. Cell. 2014;159:499–513. doi: 10.1016/j.cell.2014.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinez-Outschoorn U, Sotgia F, Lisanti MP. Tumor microenvironment and metabolic synergy in breast cancers: critical importance of mitochondrial fuels and function. Semin Oncol. 2014;41:195–216. doi: 10.1053/j.seminoncol.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 10.Provenzano PP, Hingorani SR. Hyaluronan, fluid pressure, and stromal resistance in pancreas cancer. Br J Cancer. 2013;108:1–8. doi: 10.1038/bjc.2012.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farmer P, Bonnefoi H, Anderle P, Cameron D, Wirapati P, Becette V, et al. A stroma-related gene signature predicts resistance to neoadjuvant chemotherapy in breast cancer. Nat Med. 2009;15:68–74. doi: 10.1038/nm.1908. [DOI] [PubMed] [Google Scholar]
- 12.Johnston SR, Leary A. Lapatinib: a novel EGFR/HER2 tyrosine kinase inhibitor for cancer. Drugs Today (Barc) 2006;42:441–53. doi: 10.1358/dot.2006.42.7.985637. [DOI] [PubMed] [Google Scholar]
- 13.Allinen M, Beroukhim R, Cai L, Brennan C, Lahti-Domenici J, Huang H, et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6:17–32. doi: 10.1016/j.ccr.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 14.Hu M, Yao J, Carroll DK, Weremowicz S, Chen H, Carrasco D, et al. Regulation of in situ to invasive breast carcinoma transition. Cancer Cell. 2008;13:394–406. doi: 10.1016/j.ccr.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee GY, Kenny PA, Lee EH, Bissell MJ. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat Methods. 2007;4:359–65. doi: 10.1038/nmeth1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choudhury S, Almendro V, Merino VF, Wu Z, Maruyama R, Su Y, et al. Molecular Profiling of Human Mammary Gland Links Breast Cancer Risk to a p27(+) Cell Population with Progenitor Characteristics. Cell stem cell. 2013;13:117–30. doi: 10.1016/j.stem.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu ZJ, Meyer CA, Choudhury S, Shipitsin M, Maruyama R, Bessarabova M, et al. Gene expression profiling of human breast tissue samples using SAGE-Seq. Genome Res. 2010;20:1730–9. doi: 10.1101/gr.108217.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Almendro V, Cheng YK, Randles A, Itzkovitz S, Marusyk A, Ametller E, et al. Inference of tumor evolution during chemotherapy by computational modeling and in situ analysis of genetic and phenotypic cellular diversity. Cell reports. 2014;6:514–27. doi: 10.1016/j.celrep.2013.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryan J, Montero J, Rocco J, Letai A. iBH3: simple, fixable BH3 profiling to determine apoptotic priming in primary tissue by flow cytometry. Biological chemistry. 2016 doi: 10.1515/hsz-2016-0107. [DOI] [PubMed] [Google Scholar]
- 20.Kleer CG, Bloushtain-Qimron N, Chen YH, Carrasco D, Hu M, Yao J, et al. Epithelial and stromal cathepsin K and CXCL14 expression in breast tumor progression. Clin Cancer Res. 2008;14:5357–67. doi: 10.1158/1078-0432.CCR-08-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nikolsky Y, Kirillov E, Zuev R, Rakhmatulin E, Nikolskaya T. Functional analysis of OMICs data and small molecule compounds in an integrated “knowledge-based” platform. Methods Mol Biol. 2009;563:177–96. doi: 10.1007/978-1-60761-175-2_10. [DOI] [PubMed] [Google Scholar]
- 22.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012 doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McMillin DW, Delmore J, Weisberg E, Negri JM, Geer DC, Klippel S, et al. Tumor cell-specific bioluminescence platform to identify stroma-induced changes to anticancer drug activity. Nat Med. 2010;16:483–9. doi: 10.1038/nm.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dave B, Migliaccio I, Gutierrez MC, Wu MF, Chamness GC, Wong H, et al. Loss of phosphatase and tensin homolog or phosphoinositol-3 kinase activation and response to trastuzumab or lapatinib in human epidermal growth factor receptor 2-overexpressing locally advanced breast cancers. J Clin Oncol. 2011;29:166–73. doi: 10.1200/JCO.2009.27.7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parma J, Pavlick A, Schiff R, Osborne CK, Chang JC, Rimawi M, et al. Development of acneiform rash does not predict response to lapatinib treatment in patients with breast cancer. Pharmacotherapy. 2013;33:1126–9. doi: 10.1002/phar.1308. [DOI] [PubMed] [Google Scholar]
- 26.Ryan JA, Brunelle JK, Letai A. Heightened mitochondrial priming is the basis for apoptotic hypersensitivity of CD4+ CD8+ thymocytes. Proc Natl Acad Sci U S A. 2010;107:12895–900. doi: 10.1073/pnas.0914878107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirata E, Girotti MR, Viros A, Hooper S, Spencer-Dene B, Matsuda M, et al. Intravital imaging reveals how BRAF inhibition generates drug-tolerant microenvironments with high integrin beta1/FAK signaling. Cancer Cell. 2015;27:574–88. doi: 10.1016/j.ccell.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson CB, Shepard HM, O'Connor PM, Kadhim S, Jiang P, Osgood RJ, et al. Enzymatic depletion of tumor hyaluronan induces antitumor responses in preclinical animal models. Mol Cancer Ther. 2010;9:3052–64. doi: 10.1158/1535-7163.MCT-10-0470. [DOI] [PubMed] [Google Scholar]
- 29.Al Zaid Siddiquee K, Turkson J. STAT3 as a target for inducing apoptosis in solid and hematological tumors. Cell Res. 2008;18:254–67. doi: 10.1038/cr.2008.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Casbas-Hernandez P, Fleming JM, Troester MA. Gene expression analysis of in vitro cocultures to study interactions between breast epithelium and stroma. J Biomed Biotechnol. 2011;2011:520987. doi: 10.1155/2011/520987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Camp JT, Elloumi F, Roman-Perez E, Rein J, Stewart DA, Harrell JC, et al. Interactions with fibroblasts are distinct in Basal-like and luminal breast cancers. Mol Cancer Res. 2011;9:3–13. doi: 10.1158/1541-7786.MCR-10-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: a looking glass for cancer? Nat Rev Cancer. 2012;12:323–34. doi: 10.1038/nrc3261. [DOI] [PubMed] [Google Scholar]
- 33.Meads MB, Gatenby RA, Dalton WS. Environment-mediated drug resistance: a major contributor to minimal residual disease. Nature reviews Cancer. 2009;9:665–74. doi: 10.1038/nrc2714. [DOI] [PubMed] [Google Scholar]
- 34.Provenzano PP, Cuevas C, Chang AE, Goel VK, Von Hoff DD, Hingorani SR. Enzymatic targeting of the stroma ablates physical barriers to treatment of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;21:418–29. doi: 10.1016/j.ccr.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Campbell I, Polyak K, Haviv I. Clonal mutations in the cancer-associated fibroblasts: the case against genetic coevolution. Cancer Res. 2009;69:6765–8. doi: 10.1158/0008-5472.CAN-08-4253. discussion 69. [DOI] [PubMed] [Google Scholar]
- 36.Pepper JW. Drugs that target pathogen public goods are robust against evolved drug resistance. Evolutionary applications. 2012;5:757–61. doi: 10.1111/j.1752-4571.2012.00254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ozdemir BC, Pentcheva-Hoang T, Carstens JL, Zheng X, Wu CC, Simpson TR, et al. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell. 2014;25:719–34. doi: 10.1016/j.ccr.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buess M, Nuyten DS, Hastie T, Nielsen T, Pesich R, Brown PO. Characterization of heterotypic interaction effects in vitro to deconvolute global gene expression profiles in cancer. Genome Biol. 2007;8:R191. doi: 10.1186/gb-2007-8-9-r191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roman-Perez E, Casbas-Hernandez P, Pirone JR, Rein J, Carey LA, Lubet RA, et al. Gene expression in extratumoral microenvironment predicts clinical outcome in breast cancer patients. Breast Cancer Res. 2012;14:R51. doi: 10.1186/bcr3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Troester M, Hoadley K, D'Arcy M, Cherniack A, Stewart C, Koboldt D, et al. DNA defect, epigenetics, and gene expression in cancer-adjacent breast: a study from The Cancer Genome Atlas. Breast Cancer. 2016;2:16007. doi: 10.1038/npjbcancer.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.