Abstract
The Y145Stop prion protein (PrP23–144), which has been linked to the development of a heritable prionopathy in humans, is a valuable in vitro model for elucidating the structural and molecular basis of amyloid seeding specificities. Here we report the sequential backbone and side-chain 13C and 15N assignments of mouse and Syrian hamster PrP23-144 amyloid fibrils determined by using 2D and 3D magic-angle spinning solid-state NMR. The assigned chemical shifts were used to predict the secondary structures for the core regions of the mouse and Syrian hamster PrP23-144 amyloids, and the results compared to those for human PrP23-144 amyloid, which has previously been analyzed by solid-state NMR techniques.
Keywords: prion protein, amyloid, magic-angle spinning, solid-state NMR
Biological Context
Transmissible spongiform encephalopathies (TSEs) are a class of fatal mammalian neurodegenerative diseases, which include Creutzfeldt-Jakob disease in humans, mad cow disease in cattle, and scrapie in sheep (Sejvar et al., 2008). TSEs result from conformational conversion of cellular prion protein (PrPC), a glycoprotein expressed within the central nervous system, into its infectious, amyloid-like isoform (PrPSc) (Prusiner, 1998; Caughey and Chesebro, 2001; Collinge, 2001; Aguzzi and Polymenidou, 2004). While the molecular mechanisms of mammalian prion propagation are, at present, not understood in atomistic detail, the transmissibility of infectious prions appears to be intimately linked to the three-dimensional structures adopted by the PrPSc aggregates (Collinge and Clarke, 2007; Aguzzi et al., 2008; Cobb and Surewicz, 2009).
We have previously shown that a C-truncated Y145Stop prion protein mutant (PrP23-144), which is associated with a hereditary PrP cerebral amyloidosis in humans (Ghetti et al., 1996), is a useful in vitro model for investigating in-depth the structural basis of amyloid transmissibility barriers (Surewicz et al., 2006). More recently, we also demonstrated that amyloid fibrils generated from mouse PrP23-144 are bona fide prions, causing transmissible prion diseases in mice (Choi et al., 2016). The key insight furnished by initial studies which relied on low-resolution biochemical and biophysical approaches (Kundu et al., 2003; Vanik et al., 2004; Jones and Surewicz, 2005) was that the cross-seeding specificities of human, mouse and Syrian hamster PrP23-144 amyloids (also referred to as [hu], [mo] and [Sha], respectively, for brevity) were encoded in their molecular conformations, with the latter notably found to be distinct in spite of high levels of sequence identity between the different PrP23-144 variants. Building upon these findings, we have subsequently commenced atomic level structural and dynamic studies of recombinant 13C,15N-enriched huPrP23-144 amyloid fibrils by using magic-angle spinning (MAS) solid-state nuclear magnetic resonance (NMR) techniques (Helmus et al., 2008; Helmus et al., 2010; Helmus et al., 2011; Jones et al., 2011). Our initial solid-state NMR analysis revealed that [hu] amyloid consists of an immobilized parallel in-register β-core of ~30 C-terminal amino acid (aa) residues and a dynamically disordered ~90-residue N-terminal tail. These results have also set the stage for the determination of a high-resolution structural model for the huPrP23-144 fibrils, which is currently near completion and will be reported elsewhere. Concurrently, in order to ultimately be able to elucidate the structural basis for the observed PrP23-144 seeding specificities, we have undertaken analogous solid-state NMR structural studies of moPrP23-144 and ShaPrP23-144 amyloid fibrils. The sequential backbone and side-chain 13C and 15N chemical shift assignments and secondary structure analysis reported here for the [mo] and [Sha] amyloids serve as an important first step in this direction.
Methods and Experiments
Protein Expression and Purification
Human, mouse and Syrian hamster PrP23-144 were expressed and purified according to previously published protocols (Morillas et al., 1999; Kundu et al., 2003; Vanik et al., 2004; Helmus et al., 2008). Briefly, E. coli BL21 (DE3) cells were transformed with plasmid encoding for huPrP23-144, moPrP23-144 or ShaPrP23-144 and initially grown at 37 °C in Luria-Bertani medium to OD600 of ~1.0. The cells were then centrifuged and resuspended in the same volume of M9 minimal medium containing 1 g/L 15N ammonium chloride and 3 g/L of 13C glucose as the sole nitrogen and carbon sources. Protein expression was induced by adding 1 mM isopropyl β-D-thiogalactoside and cells were grown at 37 °C and 250 rpm for 16 hours. The cell pellet from a 3 L culture was resupended in 120 mL of lysis buffer (6 M GdmCl, 10 mM Tris-HCl, 100 mM potassium phosphate, pH 8.0) and sonicated for 30 minutes, followed by centrifugation at 20,000 rpm for 30 minutes. PrP23-144 in the supernatant was purified by using nickel-nitrilotriacetic acid (Ni-NTA) superflow resin (Qiagen). The N-terminal His6-tag was cleaved with biotinylated thrombin (Novagen), followed by removal of the thrombin using streptavidin-agarose beads (Novagen) and the free His6-tag by dialysis against ultrapure Milli-Q water. Purified proteins, which were >95% pure as assessed by SDS/PAGE, were lyophilized and stored at −20 °C.
Preparation of PrP23-144 Amyloid Fibrils for Solid-State NMR
Lyophilized huPrP23-144, moPrP23-144 or ShaPrP23-144 was dissolved in ultrapure Milli-Q water at a concentration of 400 μM. Amyloid fibril formation was initiated by adding 1 M potassium phosphate pH 6.4 buffer to a final concentration of 50 mM, followed by incubation at 25 °C. During incubation the fibril suspensions were left largely undisturbed, except for gentle inversion of the sample tubes every 12 hours to ensure thorough mixing. The quantitative conversion of monomers into mature fibrils, as assessed by monitoring of the supernatant UV spectra following sample centrifugation, required ~2 days for huPrP23-144 and ~7 days for moPrP23-144 and ShaPrP23-144. Prior to packing into solid-state NMR rotors the fibrils were routinely analyzed by atomic force microscopy as described below. Fibrils were then washed with several aliquots of 50 mM potassium phosphate pH 6.4 buffer containing 20% (v/v) sodium azide, and transferred to 3.2 mm limited-speed Agilent/Varian zirconia rotors via centrifugation. The rotors were sealed using custom-made spacers (Revolution NMR) to prevent sample dehydration during the solid-state NMR measurements.
Atomic Force Microscopy
Prior to the solid-state NMR experiments, atomic force microscopy (AFM) was used to confirm the presence of huPrP23-144, moPrP23-144 and ShaPrP23-144 amyloid fibrils. For each PrP23-144 variant, a small aliquot of the fibril suspension was diluted 10-fold with ultrapure Milli-Q water, deposited onto a freshly cleaved mica substrate (Ted Pella, Inc.), incubated for 2 minutes, rinsed with two 50 μl aliquots of ultrapure water to remove salts and unbound protein, and air dried. Imaging was performed in tapping mode in air using a Bruker Dimension Icon AFM and high-sensitivity silicone Bruker RTESPA MPP-11120-10 probes with a nominal spring constant of 40 N/m and nominal tip radius of 8 nm. The images were processed using interactive plane fitting and low-pass Gaussian filtering within the Bruker NanoScope Analysis software.
Solid-State NMR Spectroscopy
NMR spectra were recorded on a 500 MHz Varian spectrometer equipped with a 3.2 mm BioMAS 1H-13C-15N probe. For all experiments the magic angle spinning (MAS) frequency was set to 11.111 kHz ± 3 Hz, and the sample temperature was maintained at ~5 °C by using a stream of compressed air at 0 °C delivered to the sample through a variable-temperature (VT) stack. The sequential 13C and 15N resonance assignments were established using the following chemical shift correlation experiments: 2D 15N-13Cα (NCA), 2D 15N-13Cα-13CX (NCACX), 3D 15N-13Cα-13CX (NCACX), 3D 15N-13C′-13CX (NCOCX), and 3D 13C′-15N-13Cα (CONCA), with parameters similar to those used in our previous solid-state NMR analysis of huPrP23-144 amyloid fibrils (Helmus et al., 2008). Data were processed using NMRPipe (Delaglio et al., 1995) and analyzed in Sparky (Goddard and Kneller, 2006). For each PrP23-144 variant, NMR spectra were recorded for several independent fibril preparations to confirm sample reproducibility.
Assignments and data deposition
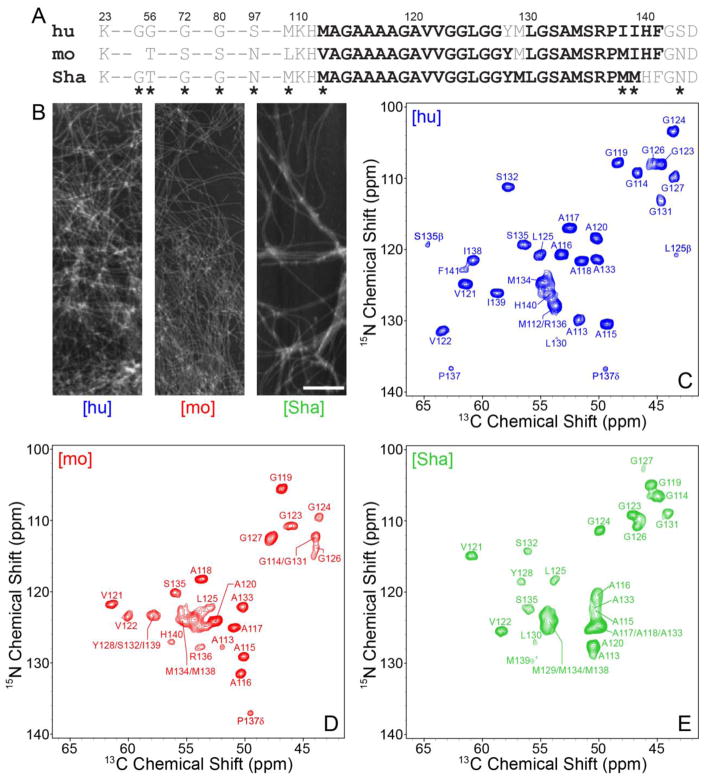
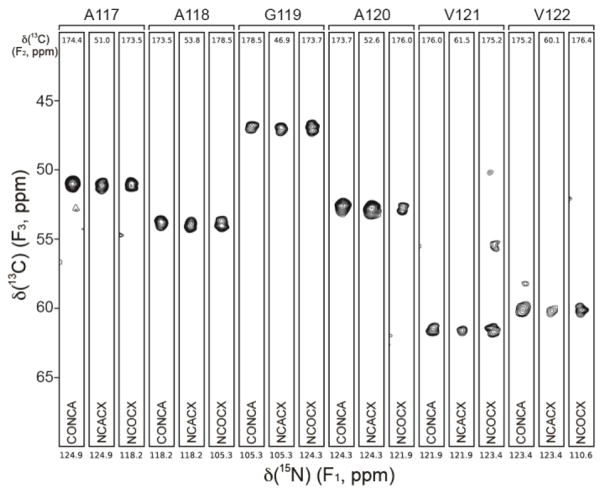
As previously reported (Jones and Surewicz, 2005) and illustrated in the AFM images in Fig. 1, hu, mo and ShaPrP23-144 were all found to readily assemble into micron-length amyloid fibrils. Also shown in Fig. 1 are the 2D fingerprint 15N-13Cα solid-state NMR spectra of uniformly 13C,15N-labeled [hu], [mo] and [Sha] amyloids. In analogy to published data for huPrP23-144 fibrils (Helmus et al., 2008), the 15N-13Cα spectra for [mo] and [Sha] fibrils each display a limited set of relatively narrow resonances, with typical 13C and 15N linewidths on the order of ca. 0.75 and 1 ppm, respectively, pointing to the presence of compact and well-ordered amyloid core regions for both proteins. A cursory comparison of the 15N-13Cα fingerprint spectra also reveals significant overall chemical shift differences among all the PrP23-144 variants, in spite of nearly identical amino acid sequences, indicative of huPrP23-144, moPrP23-144 and ShaPrP23-144 adopting distinct three-dimensional structures in the amyloid state. Sequential resonance assignments of the core residues for the [mo] and [Sha] amyloids were unambiguously established by using a set of 3D 15N-13C-13C correlation spectra listed in the Methods and Experiments section, with the distinctive 13C chemical shift patterns for Ala, Ile, Ser and Val serving as convenient handles in the assignment process. Representative strips from 3D NCACX, NCOCX, and CONCA datasets, illustrating the sequential backbone connectivity of residues A117-V122 for the [mo] fibrils, are shown in Fig. 2.
Fig. 1.
(A) Amino acid sequences of hu, mo and ShaPrP23-144. Immobile amyloid core residues detectable in the solid-state NMR spectra are shown in bold black font and conformationally flexible residues are shown in grey font. Residues that are not conserved between the hu, mo and ShaPrP23-144 sequences are indicated by asterisks. (B) Representative atomic force microscopy images of [hu], [mo], and [Sha] amyloid fibrils used for the solid-state NMR measurements. The scale bar corresponds to 1 μm. (C–E) Assigned two-dimensional 500 MHz 15N-13C solid-state NMR spectra of (C) [hu], (D) [mo] and (E) [Sha] amyloid fibrils recorded at 11.111 kHz MAS. Each spectrum was recorded with acquisition times of 15 ms (t1, 15N) and 28 ms (t2, 13C), and a total measurement time of ~24 hours. Cross-peaks are drawn with the lowest contour at ~15 times the root mean square (rms) noise level.
Fig. 2.
Representative strips from 500 MHz 3D CONCA, NCACX, and NCOCX spectra for moPrP23-144 amyloid fibrils, showing sequential connectivity for residues A117-V122. Strips are labeled by residue number according to the 15N frequency (F1), with the 13C frequencies (F2) indicated to the top of each strip. The figure was generated using the nmrglue software (Helmus and Jaroniec, 2013).
The nearly complete backbone and side-chain 13C and 15N chemical shift assignments reveal that in conventional solid-state NMR spectra based on dipolar coupling-driven magnetization transfers signals for 29 and 28 residues (out of ~122 total) are observed for [mo] and [Sha] fibrils, respectively. Specifically, the rigid core region for moPrP23-144 amyloid spans residues V112 to F141, while the large N-terminal domain (aa 23-111), three C-terminal amino acids, G142-D144, and residue M129 located within the core are not detected due to increased conformational flexibility; for ShaPrP23-144 amyloid, the core region consists of residues M112 through M139, with amino acids 23-111 and 140-144 not detected. Remarkably, the approximate sizes and locations within the protein sequence of the immobilized core regions for [mo] and [Sha] fibrils closely mirror those reported for [hu] amyloid in our earlier study (Helmus et al., 2008), where the rigid core was mapped to aa 112-141 and found to contain two conformationally dynamic residues, Y128 and M129 (see Fig. 1A for a summary of these results).
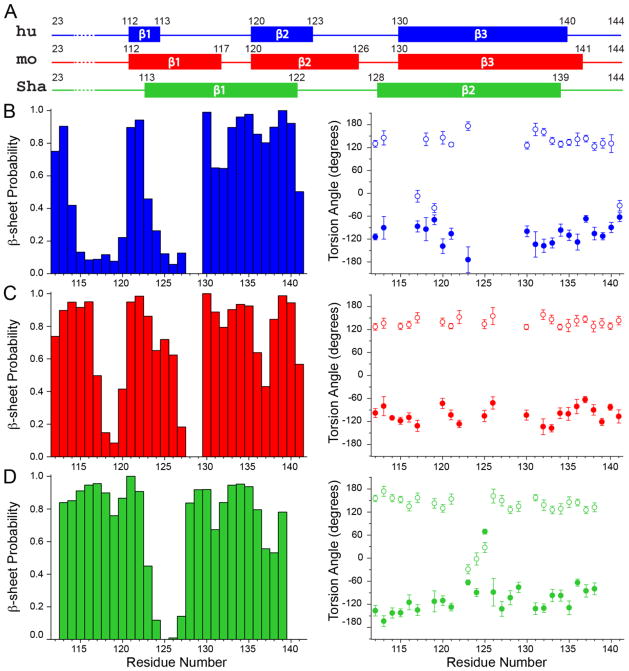
The secondary structures for the [hu], [mo] and [Sha] amyloid core residues were assessed based on the 13C and 15N chemical shifts as follows. The TALOS-N program (Shen and Bax, 2013) was used to predict the residue-specific ϕ and ψ backbone dihedral angles and β-sheet probabilities (Fig. 3), and all residues having β-sheet probabilities of 50% or higher were classified as making up the β-strands. Note that a few additional residues with β-sheet probabilities below 50% were also included as part of the assigned β-strand regions provided that: (1) all of their ϕ and ψ predictions fell into the β-strand region of the Ramachandran plot and (2) they were directly adjacent to a set of residues having 50% or higher β-sheet probabilities. This analysis, summarized in Fig. 3A, indicates that the secondary structures for [hu] and [mo] amyloids show a degree of similarity in spite of the considerable differences in the residue-specific 13C and 15N chemical shifts between the two proteins (e.g., the 13Cα chemical shift rms deviations for the common residues among the PrP23-144 variants corresponding to different species were all on the order of ~2 ppm). Namely, the huPrP23-144 fibrils are predicted to contain three segments with particularly high β-strand propensity (aa ~112-113, ~120-123 and ~130-140) flanked by two largely non-β stretches (aa ~114-119 and ~124-129), the latter of which includes the flexible Y128-M129 fragment, while the moPrP23-144 fibrils also consist of three β-strands (aa ~112-117, ~120-126 and ~130-141) separated by two non-β segments (aa ~118-119 and ~127-129), and include the flexible M129 residue. In contrast, the backbone conformation for ShaPrP23-144 fibrils (two extended β-strands, aa ~113-122 and ~128-139, separated by a short non-β segment) appears to differ rather substantially from those for [hu] and [mo] amyloids. Altogether, the findings that [hu] and [mo] amyloids adopt similar secondary structures at the level of individual residues while the [Sha] fibril conformation is clearly distinct are consistent with the results of the earlier biochemical and biophysical studies (Vanik et al., 2004; Jones and Surewicz, 2005), which showed the absence and presence of cross-seeding barriers between [hu] and [mo] amyloids and between [hu] and [Sha] amyloids, respectively, while correlating the observed seeding profiles to the global protein conformations in the fibrillar state.
Fig. 3.
(A) Summary of the secondary structure analysis for hu, mo and ShaPrP23-144 amyloid fibrils based on the assigned 13C and 15N chemical shifts (see text for details). (B) (Left) Normalized β-sheet probability and (right) backbone ϕ (filled circles) and ψ (open circles) torsion angle values for [hu] amyloid fibrils as a function of residue number predicted using the TALOS-N software (Shen and Bax, 2013). The ϕ/ψ predictions include the reported estimated errors and only predictions classified within TALOS-N as “strong” or “generous” are included, with the overwhelming majority of the predictions falling into the “strong” category. (C, D) Same as panel (B), but for (C) [mo] and (D) [Sha] amyloid fibrils.
Acknowledgments
This research was supported by NIH (grants R01GM094357 and S10OD012303 to C.P.J. and P01AI106705 and R01NS083687 to W.K.S.) and the Camille & Henry Dreyfus Foundation (Camille Dreyfus Teacher-Scholar Award to C.P.J.).
Footnotes
Accession Numbers
Chemical shift assignments for huPrP23-144, moPrP23-144 and ShaPrP23-144 amyloid fibrils have been deposited in the BioMagResBank (http://www.bmrb.wisc.edu) under accession numbers 26925, 26924, and 26926, respectively.
References
- Aguzzi A, Polymenidou M. Mammalian prion biology: one century of evolving concepts. Cell. 2004;116:313–327. doi: 10.1016/s0092-8674(03)01031-6. [DOI] [PubMed] [Google Scholar]
- Aguzzi A, Sigurdson C, Heikenwaelder M. Molecular mechanisms of prion pathogenesis. Annu Rev Pathol. 2008;3:11–40. doi: 10.1146/annurev.pathmechdis.3.121806.154326. [DOI] [PubMed] [Google Scholar]
- Caughey B, Chesebro B. Transmissible spongiform encephalopathies and prion protein interconversions. Adv Virus Res. 2001;56:277–311. doi: 10.1016/s0065-3527(01)56031-5. [DOI] [PubMed] [Google Scholar]
- Choi JK, Cali I, Surewicz K, Kong Q, Gambetti P, Surewicz WK. Amyloid fibrils from the N-terminal prion protein fragment are infectious, Proc. Natl Acad Sci USA. 2016;113:13851–13856. doi: 10.1073/pnas.1610716113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb NJ, Surewicz WK. Prion diseases and their biochemical mechanisms. Biochemistry. 2009;48:2574–2585. doi: 10.1021/bi900108v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinge J. Prion diseases of humans and animals: Their causes and molecular basis. Annu Rev Neurosci. 2001;24:519–550. doi: 10.1146/annurev.neuro.24.1.519. [DOI] [PubMed] [Google Scholar]
- Collinge J, Clarke AR. A general model of prion strains and their pathogenicity. Science. 2007;318:930–936. doi: 10.1126/science.1138718. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Ghetti B, Piccardo P, Spillantini MG, Ichimiya Y, Porro M, Perini F, Kitamoto T, Tateishi J, Seiler C, Frangione B, Bugiani O, Giaccone G, Prelli F, Goedert M, Dlouhy SR, Tagliavini F. Vascular variant of prion protein cerebral amyloidosis with τ-positive neurofibrillary tangles: The phenotype of the stop codon 145 mutation in PRNP. Proc Natl Acad Sci USA. 1996;93:744–748. doi: 10.1073/pnas.93.2.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Kneller DG. SPARKY 3. University of California; San Francisco: 2006. [Google Scholar]
- Helmus JJ, Surewicz K, Nadaud PS, Surewicz WK, Jaroniec CP. Molecular conformation and dynamics of the Y145Stop variant of human prion protein in amyloid fibrils. Proc Natl Acad Sci USA. 2008;105:6284–6289. doi: 10.1073/pnas.0711716105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmus JJ, Surewicz K, Surewicz WK, Jaroniec CP. Conformational flexibility of Y145Stop human prion protein amyloid fibrils probed by solid-state nuclear magnetic resonance spectroscopy. J Am Chem Soc. 2010;132:2393–2403. doi: 10.1021/ja909827v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmus JJ, Surewicz K, Apostol MI, Surewicz WK, Jaroniec CP. Intermolecular alignment in Y145Stop human prion protein amyloid fibrils probed by solid-state NMR spectroscopy. J Am Chem Soc. 2011;133:13934–13937. doi: 10.1021/ja206469q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmus JJ, Jaroniec CP. Nmrglue: an open source Python package for the analysis of multidimensional NMR data. J Biomol NMR. 2013;55:355–367. doi: 10.1007/s10858-013-9718-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones EM, Surewicz WK. Fibril conformation as the basis of species- and strain-dependent seeding specificity of mammalian prion amyloids. Cell. 2005;121:63–72. doi: 10.1016/j.cell.2005.01.034. [DOI] [PubMed] [Google Scholar]
- Jones EM, Wu B, Surewicz K, Nadaud PS, Helmus JJ, Chen S, Jaroniec CP, Surewicz WK. Structural polymorphism in amyloids: New insights from studies with Y145Stop prion protein fibrils. J Biol Chem. 2011;286:42777–42784. doi: 10.1074/jbc.M111.302539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu B, Maiti NR, Jones EM, Surewicz KA, Vanik DL, Surewicz WK. Nucleation-dependent conformational conversion of the Y145Stop variant of human prion protein: Structural clues for prion propagation. Proc Natl Acad Sci USA. 2003;100:12069–12074. doi: 10.1073/pnas.2033281100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillas M, Swietnicki W, Gambetti P, Surewicz WK. Membrane environment alters the conformational structure of the recombinant prion protein. J Biol Chem. 1999;274:36859–36865. doi: 10.1074/jbc.274.52.36859. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. Prions. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sejvar JJ, Schonberger LB, Belay ED. Transmissible spongiform encephalopathies. J Am Vet Med Assoc. 2008;233:1705–12. doi: 10.2460/javma.233.11.1705. [DOI] [PubMed] [Google Scholar]
- Shen Y, Bax A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J Biomol NMR. 2013;56:227–241. doi: 10.1007/s10858-013-9741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surewicz WK, Jones EM, Apetri AC. The emerging principles of mammalian prion propagation and transmissibility barriers: Insight from studies in vitro. Acc Chem Res. 2006;39:654–662. doi: 10.1021/ar050226c. [DOI] [PubMed] [Google Scholar]
- Vanik DL, Surewicz KA, Surewicz WK. Molecular basis of barriers for interspecies transmissibility of mammalian prions. Mol Cell. 2004;14:139–145. doi: 10.1016/s1097-2765(04)00155-8. [DOI] [PubMed] [Google Scholar]