Abstract
The presence of intact ligand-binding domain (LBD) ensures the strict androgen-dependent regulation of androgen receptor (AR): binding of androgen induces structural reorganization of LBD resulting in release of AR from HSP90, suppression of nuclear export which otherwise dominates over import and nuclear translocation of AR as a transcription factor. Thus, loss or defects of the LBD abolish constraint from un-liganded LBD as exemplified by constitutively active AR variants (AR-Vs), which are associated with emerging resistance mechanism to anti-AR therapy in castration-resistant prostate cancer (mCRPC). Recent analysis of the AR splicing landscapes revealed mCRPC harboring multiple AR-Vs with diverse patterns of inclusion/exclusion of exons (exons 4–8) corresponding to LBD to produce namely exon-skipping variants. In silico construction for these AR-Vs revealed four novel AR-Vs having unique features: Exclusion of specified exons introduces a frameshift in variants v5es, v6es and v7es. ARv56es maintains the reading frame resulting in the inclusion of the C-terminal half of the LBD. We systematically characterized these AR-Vs regarding their subcellular localization, affinity for HSP90 and transactivation capability. Notably, ARv5es was free from HSP90, exclusively nuclear, and constitutively active similarly as previously reported for v567es. In contrast, v6es and v7es were similar in that they are cytoplasmic, transcriptionally inactive and bind HSP90, ARv56es was present in both nucleus and cytoplasm, does not bind HSP90 and is transcriptionally inactive. Converting these transcriptionally inactive AR-Vs into active forms, we identified the two separate elements that allosterically suppress otherwise constitutively active AR-Vs; one in exon 5 for v6es and v7es and the other in exon 8 for v56es. Our findings identify a novel constitutively active AR-V, ARv5es and establish a method to predict potential activities of AR-Vs carrying impaired LBD.
Introduction
Androgen receptor (AR) signaling is arguably pivotal to not only hormone-sensitive but also advanced castration-resistant prostate cancer (CRPC).1, 2 AR is comprised of four functional domains with the N-terminal transactivation domain encoded by exon 1, the DNA-binding domain by exons 2 and 3, the hinge domain (HD) by exon 4 and the ligand-binding domain (LBD) by the remaining exons 4–8.3, 4, 5 The structural integrity in the LBD permits a strict ligand-dependency of AR's release from cytoplasmic HSP90 and thus nuclear translocation where AR acts as a transcription factor.6 However, ligand-independent AR activation can occur by overactivation of kinase signaling, amplification of the AR gene and promiscuous mutations in LBD accounting for sustained AR signaling even during the mainstay treatment of prostate cancer, resulting in low androgen or the inhibition of ligand binding.5, 6, 7, 8, 9
On the other hand, AR variants (AR-Vs), which commonly harbor exons 1–3, undergo either insertion of a cryptic exon after exon 2 or 3 or alternative inclusion/exclusion of exons 4–8, and thus have a deletion in the LBD.8, 9, 10, 11 Generally, the former and the latter groups are termed truncated and exon-skipping variants, respectively.12, 13, 14 Two clinically relevant AR variants V7 (aka AR3) and v567es (aka AR-V12) represent each group and display constitutive activity and thus drive growth and proliferation of cellular models of CRPC even during the most advanced anti-androgen treatment.15, 16 In fact, their elevated expression is closely associated with worse prognosis and shorter cancer-specific survival17, 18
Numerous other AR-Vs have been identified and predicted, but these are less characterized regarding their biochemical functions and clinical relevance.12, 19 This prompted us to perform systematic and non-biased analysis of clinical samples in relation to the expression patterns of AR-Vs.20, 21 The resultant splicing landscape revealed that mCRPC harbored multiple forms of exon-skipping variants including some previously unreported ones. Notably many of these AR-Vs have diverse patterns of inclusion/exclusion of exons corresponding to the LBD and thus some may show constitutive activity and lose the target for the antiandrogen drugs such as abiraterone and enzalutamide.2, 7, 22 We sought to determine which of these AR-Vs can contribute to CRPC progression and thereby identify the novel AR-V that potentially shows resistance to the current anti-AR therapy. In this study we characterized and categorized these exon-skipping AR variants and found the novel constitutively active variant namely ARv5es that structurally and functionally resembles v567es. We constructed a cellular model to characterize ARv5es gene product. Furthermore, by generating and systematically analyzing a series of deletion and point mutants, we examined what elements intrinsically determine constitutive activity of exon-skipping AR variants.
Results
Characterization of novel AR-Vs constructed in silico and in vitro
Recent analysis of the AR splicing landscape in the TCGA prostate and SU2C data sets revealed that both truncated and exon-skipping variants of AR were present at varying levels.20, 21 Although AR-V7 is the predominant variant in SU2C metastases, many patients have more than one variant (Supplementary Figure S1). With special focus on the LBD, we constructed in silico transcripts for the AR-Vs detected in the data sets and found four novel exon-skipping variants that have unique features based on their predicted amino acid sequences (Figure 1a and Supplementary Figure S2). Exclusion of specified exons introduces a frameshift in variants 5es, 6es and 7es. On the other hand, ARv56es still maintains the reading frame resulting in the inclusion of the C-terminal half of the LBD.
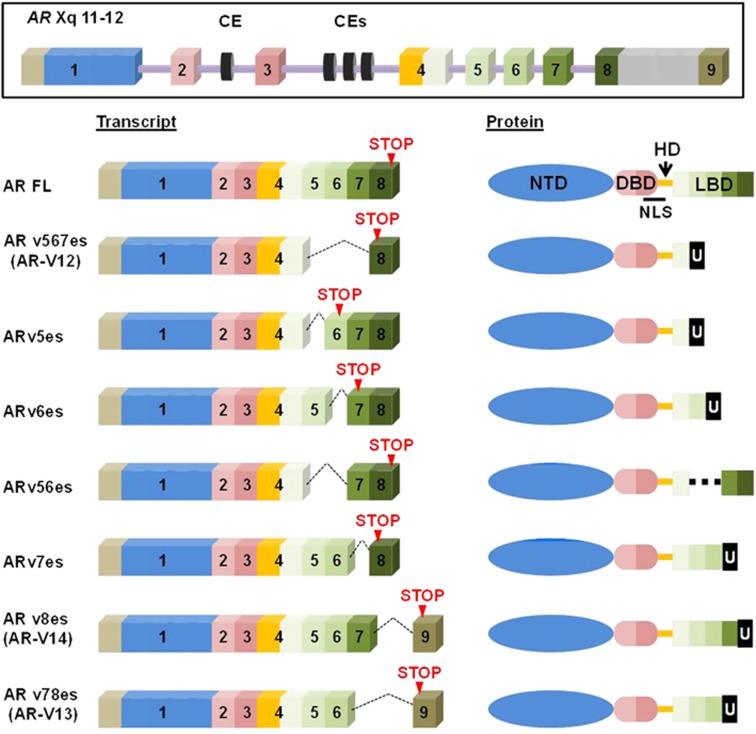
Figure 1.
Schematic representation of AR-FL and AR-Vs. AR gene structure with canonical and cryptic exons. Normal splicing produces the transcript for AR-FL protein (GenBank accession no. NP_000035.2) consisting of four domains with the N-terminal transactivation domain (NTD) encoded by exon 1, the DNA-binding domain (DBD) by exons 2 and 3, the hinge domain (HD) by exon 4 and the ligand-binding domain (LBD) by the remaining exons 4–8. The exons in transcript and the corresponding protein regions are shown in matched colors. Here, exon 9 is defined as the exon located far downstream of exon 8. As exemplified by the transcript (GenBank accession No. NM_000044.3), the eighth exon corresponds to the unspliced long exon covering exon 8, intron 8 and exon 9. Skipping the specified exons results in the frameshift, by which the respective AR-Vs have their unique C-terminal sequences (U). Note that ARv5es, v6es, v56es and v7es are newly discovered AR-Vs through construction of splicing landscapes of AR in mCRPC.20, 21
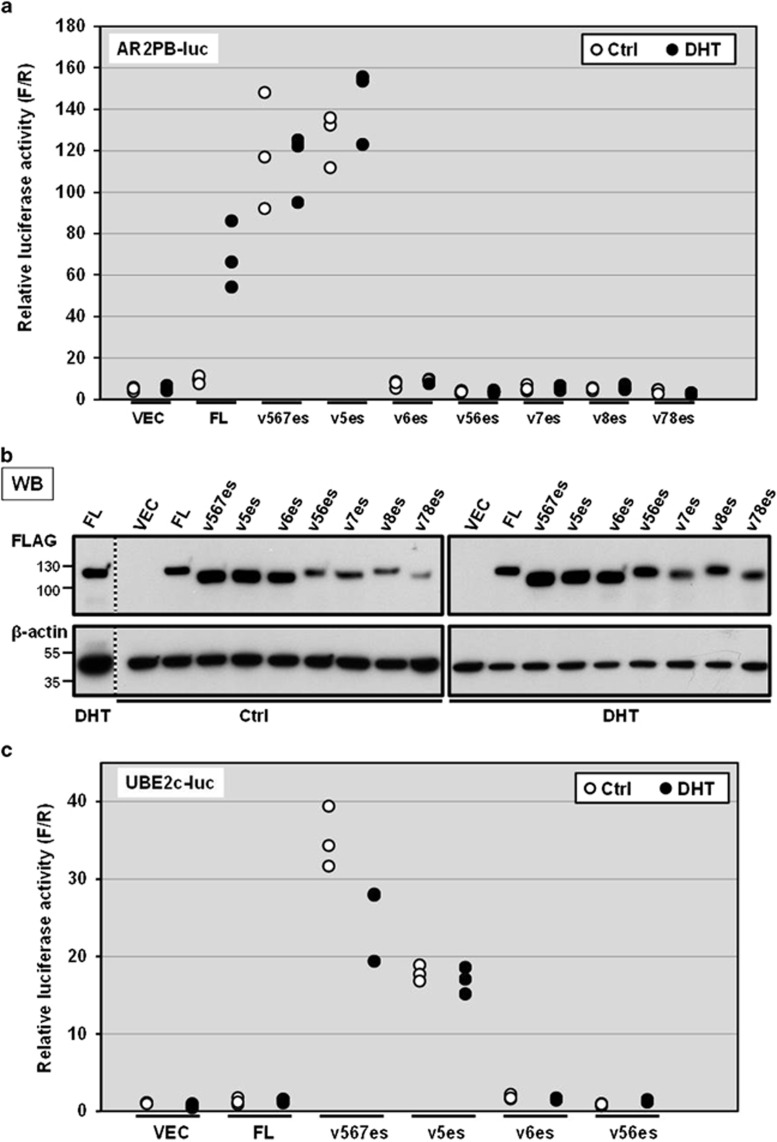
As the first step, we took a well-established approach to evaluating the basic properties of these previously uncharacterized variants. The previously described AR-Vs, ARv567es, v8es and v78es were also used for the purpose of comparative studies.11, 19, 23 To evaluate their transactivation activities, we used the canonical AR activity reporter whose expression is under the control of a small composite rat probasin promoter containing two androgen response elements.23 AR-negative M12 human prostate cancer cells were co-transfected with this reporter and a construct expressing one of the AR-Vs or AR-FL (Figure 2a). Expression levels were comparable in the presence and absence of the ligand dihydrotestosterone (DHT) (Figure 2b). ARv5es markedly increased the reporter activity similar to ARv567es, irrespective of a ligand status. DHT activated AR-FL, but we failed to detect transactivation activities for ARv56es, v6es, v7es, v8es and v78es under either condition or even when v56es or v6es were expressed in far excess over v567es (Supplementary Figure S3). Similar results were obtained with the AR-negative PC-3 and APIPC cell lines (Supplementary Figure S4). In addition to the canonical AR response elements that are shared with AR-FL, v567es and V7 reportedly display unique binding specificities toward regulatory elements of a subset of genes including the G2–M phase gene UBE2C.24, 25 As shown in Figure 2c, similarly to ARv567es, ARv5es transactivated a luciferase construct driven by three repeats of the respective UBE2C promoter element which is known to be specifically activated by constitutively active AR-Vs (AR-V7 and ARv567es) but not AR-FL.25 This suggests that v5es has some commonality in DNA binding preference with v567es and V7. Likewise, ARv5es was resistant to enzalutamide, further suggesting that ARv5es and v567es share similar properties despite their different C-terminal sequences (Figure 1b and Supplementary Figure S5).26 Enzalutamide reduced DHT-induced AR-FL activation, as expected.
Figure 2.
Transactivation activities of AR-Vs on the canonical and AR-V-specific luciferase reporters. (a) Androgen-depleted M12 cells were triply transfected with an AR-expression vector encoding AR-FL or AR-V, the AR activity reporter plasmid in which the firefly luciferase gene is coupled to a small composite rat probasin promoter containing two androgen response elements, and a plasmid for constitutive expression of Renilla luciferase. At 16 h after transfection, cells were treated with 1 nm DHT (closed circle) or vehicle control (ctrl: open circle). Luciferase activities were measured 24 h later. The firefly luciferase activity of each sample was normalized to the Renilla luciferase activity. All individual data points were plotted. Similar results were obtained from three independent experiments. (b) Western blot analysis confirmed comparable expression of FLAG tagged AR-FL and AR-Vs in the presence or absence of DHT. The common cell lysates of 'AR-FL treated with DHT' sample were loaded on two different gels entitled 'Ctrl' (control) and DHT as the control to compare the intensities of the bands on the different gels. The vertical dotted line indicates western blot image cropped from the same gel. (c) Transactivation activities of AR-FL and AR-Vs on the reporter under the control of AR-V-specific binding sites. Transient transfection and luciferase assays were carried out in the same manner as described in (a).
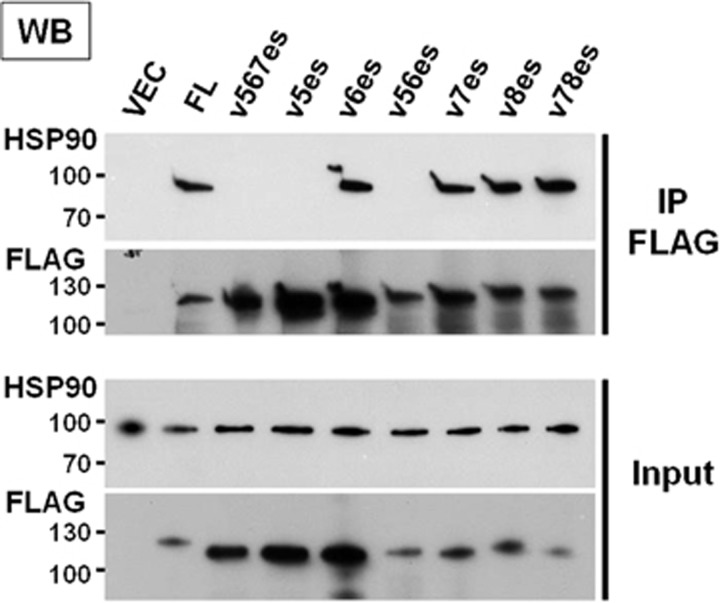
It has been well accepted that HSP90 keeps AR in the cytosol and facilitates maturation and conformational maintenance of the LBD. Upon binding to DHT, AR is released from HSP90 to exert its nuclear actions as a transcription factor.6 Because the AR-Vs cannot bind androgen ligand, HSP90 may tether AR-Vs in the cytoplasm. We tested whether AR-V and HSP90 protein complexes exist in M12 cells cultured in DHT-free media. Cells were transfected with the plasmid for expression of FLAG-tagged AR-FL or AR-V (Figure 3). Heavy metal molybdate was included to stabilize ATP-bound HSP90 during the co-immunoprecipitation (co-IP) procedure.27 We failed to detect HSP90 complexed with either ARv5es or v56es as has been reported for ARv567es.28 On the other hand, AR-FL and other AR-Vs, including v7es, v8es, and v78es, formed a complex with HSP90. This interaction profile is in agreement with the report that an HSP90-binding site defined based on capacities of deletion constructs for forming 8 s complex, which lies in exon 5 and 5' end of exon 6 (amino acids 722–776 equivalent to 704–758 in rat AR) (Figure 4).27 Except for ARv56es, which does not bind HSP90 and remains transcriptionally inactive, reactivity toward HSP90 appears to be a predictive determinant for constitutive activity of AR-V.
Figure 3.
HSP90 binds to the specified AR-Vs. M12 cells grown in androgen-depleted media were transfected with FLAG-tagged AR-FL or AR-V. Following immunoprecipitation with FLAG antibody, immune complexes were analyzed by western blot analysis using anti-HSP90 and FLAG antibodies. An empty vector served as a negative control.
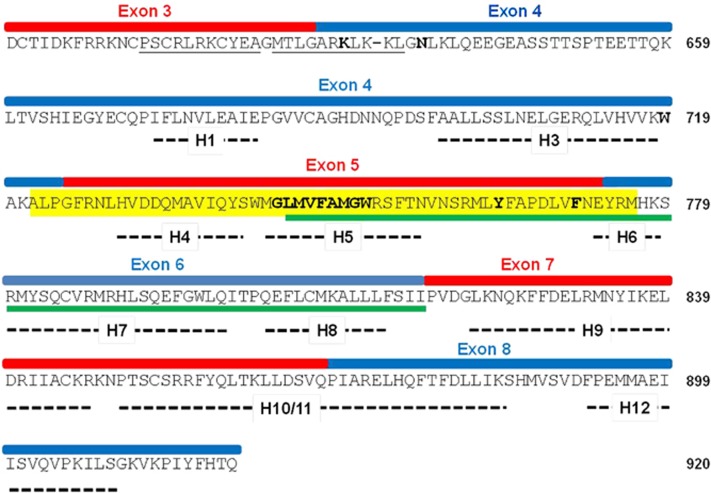
Figure 4.
The primary and secondary structures of LBD of AR. The LBDs of the steroid receptors fold into 12 helices, of which positions in AR protein are indicated by thick dashed lines.34 Note that AR LBD lacks an identifiable helix 2. Protein regions and the corresponding exons are shown; the amino acid residues composing the NES are underlined in green and the HSP90-binding site is highlighted in yellow27, 29 the bipartite NLS is underlined in black. Letters in bold represent the positions where the termination codons were introduced to create a series of deletion mutants. V717 and E898 which were manipulated to disrupt N–C interaction of AR are located in exon 4 and exon 8, respectively.
Subcellular localization of AR-Vs
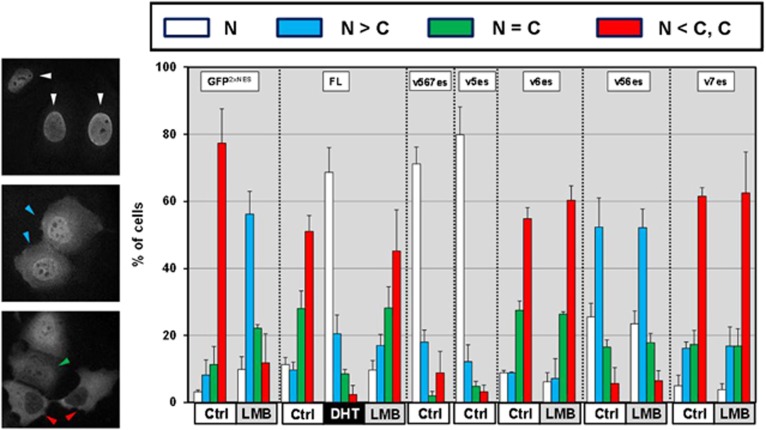
Nucleocytoplasmic trafficking of AR is tightly regulated through nuclear localization and export signals. AR has a proposed nuclear export signal (NES) (marked by a green line in Figure 4) that is independent of the canonical CRM1 nuclear export pathway.29 Inactive AR-Vs have variables in this region by inclusion/exclusion of the specified exons while ARv5es completely lacks this NES region (Figures 1, 4 and Supplementary Figure S1). Therefore, we analyzed the subcellular distribution of AR-Vs and AR-FL in a parallel experiment. Left panels are the fluorescence microscopy images of FLAG-tagged AR-Vs (Figure 5). In the top panel, signals were exclusively detected in the nucleus. Typically we see this type of distribution when ARv5es and v567es are expressed or when AR-FL is expressed in the presence of DHT. In the middle panel, nuclear staining is stronger than cytosolic. ARv56es variant shows this distribution. In the bottom panel, the cells show equal nucleocytosolic staining and stronger cytosolic staining. The typical images of ARv5es and v7es can also be seen in Supplementary Figure S6. In none of the cases did leptomycin B, which inhibits CRM1-dependent nuclear export,30 significantly alter the subcellular distribution of AR-Vs and AR-FL. As a control, in M12 cells, this chemical completely inhibited nuclear export of GFP C-terminally fused with two repeats of NES from inhibitor of cAMP-dependent protein kinase.31 Thus subcellular localization of the AR-Vs is not dependent on the CRM1 export pathway. Importantly, as observed with AR-FL, for the AR-Vs 6es and 7es the NES dominates over import and it is suppressed by ligand DHT. ARv56es displayed partial nuclear localization, which is similar to the localization patterns of NES-deficient AR and markedly different from other inactive variants such as ARv6es and v7es.
Figure 5.
Subcellular localization of AR-Vs. Androgen-depleted M12 were transfected with the expression vectors for FLAG-tagged AR-FL or AR-V. The transfectants with GFP-2xNESPKI were analyzed in parallel as positive controls for leptomycin B (LMB)-sensitive pathway. The cells were treated 24 h later with DHT (1 nm), LMB (10 ng/ml) or vehicle control for 4 h. Representative immunofluorescence images for ARv5es (top panel), v56es (middle panel) and v6es (bottom panel). Subcellular localization of AR-FL and AR-Vs was determined in 200 cells according to the following criteria. Cells display predominant nuclear FLAG immunofluorescence (white triangles); nuclear>cytosolic staining (blue); nuclear=cytosolic (green); cytoplasmic staining (red). Similar results were obtained from three independent experiments.
ARv5es transcript is a potential substrate for nonsense-mediated decay (NMD)
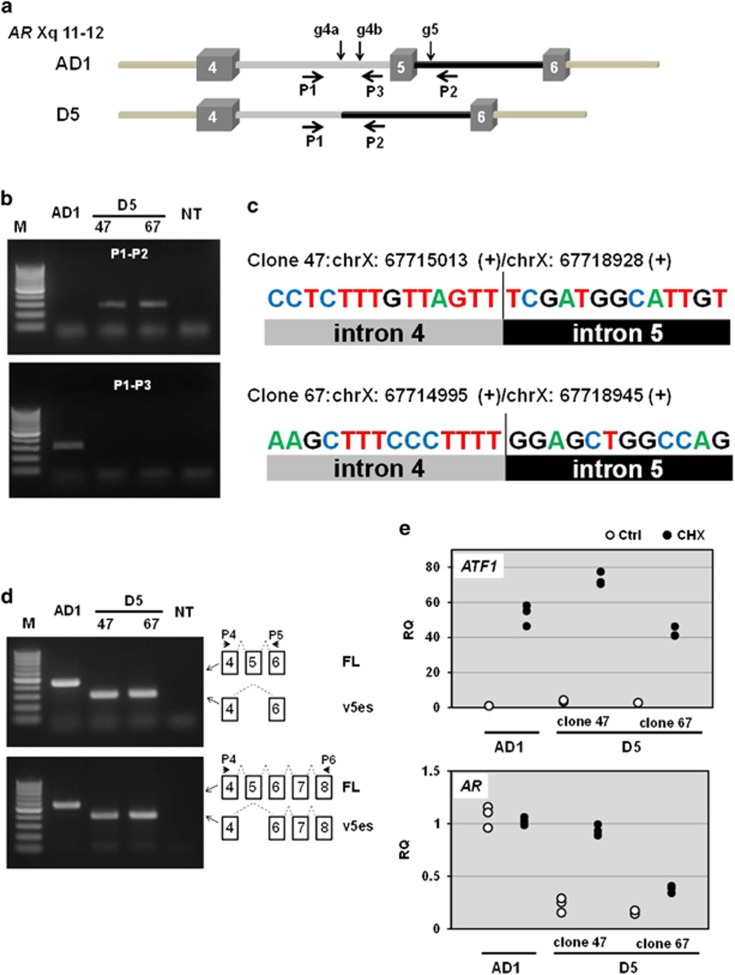
In the AR, skipping exon 5 introduces a stop codon in exon 6, which is located 242 bp upstream of the last exon–exon junction (E7-8) (Figure 1). This distance is long enough to elicit NMD that rapidly eliminates aberrant transcripts containing premature termination codons.32 Thus, we examined whether ARv5es transcript can be a NMD target as follows: CRISPR/Cas9 system was employed in the CWR-R1-AD1 prostate cancer cell line that harbors one intact AR gene copy and expresses AR-FL.33, 41 The guide RNAs were designed to target introns 5 and 6 for a selective elimination of exon 5 from AR genomic DNA (Figure 6a), by which the resulting cells can endogenously produce ARv5es transcript. We screened cells by PCR to identify the clones that miss exon 5 by deletion-specific PCR, selected two cell lines and determined the breakpoints by direct Sanger DNA sequencing of the deletion-specific PCR products (Figures 6b and c). The conventional (non-quantitative) RT–PCR confirmed expression of ARv5es transcript in each clone but not in the parental cell line (Figure 6d). NMD requires translation for nonsense codon recognition. Thus, we treated the cells with the protein synthesis inhibitor cycloheximide, which blocks translation and thereby inactivates the NMD pathway. Indeed, cycloheximide increased a well-documented NMD substrate ATF1 in the parental AD1 cells and D5 clones 47 and 67 (Figure 6e). Whereas AR-FL message in AD1 cells was not largely affected in both conditions, cycloheximide increased D5 message in both D5 clones by about fourfold, suggesting that v5es is a potential NMD target (Figure 6e). Indeed, v5es protein was barely detectable by western blotting with AR441 antibody directed against N-terminal domain (Supplementary Figure S7). AR-V7 protein, however, was present in varying amounts across the samples.
Figure 6.
ARv5es transcript may elicit NMD. (a) AR genomic structures featuring exons 4, 5 and 6 and the corresponding introns are drawn. Activation of non-homologous end joining pathway by CRISPER/Cas9 system results in deletion of exon 5 in AR gene in the parental AD1 cell to create cell line D5. While co-targeting with gRNAs g4a and g5 resulted in production of D5 clone 47, g4b and g5 were used to obtain clone 67. The information about gRNA is provided in Table 1. (b) PCR analysis of exon 5 deletion of AR gene. Genomic DNA isolated from AD1 and D5 clone 47 and 67 were used for PCR with primers P1 and P2 to test the absence of exon 5, and P1 and P3 primers were used as described in Table 1 to examine the absence of the targeted segment of intron 4 in D5 clones and its presence in AD1. M, DNA size marker (O'RangeRuler 100 bp DNA Ladder: Thermo Fisher Scientific). NT, non-template. (c) Direct Sanger DNA sequencing of the deletion-specific PCR products determined the breakpoints and confirmed deletion of exon 5. (d) Conventional RT–PCR analysis confirmed expression of ARv5es transcript in D5 cells (upper panel). ARv5es is the major exon-skipping variant in D5 clones (lower panel). PCR products were individually subjected to DNA sequencing to further confirm that AR transcript in D5 is v5es at the nucleotide levels. (e) Total RNA was isolated from the specified cells treated with 100 μg/ml cycloheximide (CHX) for 7 h. H2O was used as a vehicle control (Ctrl). The primers P4 and P5 were used as described in Table 1 to assess AR-FL and v5es transcripts. Relative levels of AR and ATF1 transcripts were compared with the levels of the corresponding genes in 'Ctrl'-treated AD1 cell using RPL13A as a normalizer. All individual data points were plotted as the relative quantity (RQ). Similar results were obtained from three independent experiments. NMD inhibition uniformly increased ATF1 levels in all of the cell lines. ARv5es transcript responded much better to NMD inhibition than AR-FL counterpart.
Helix 5 in LBD allosterically suppresses otherwise transcriptionally active ARv6es and v7es
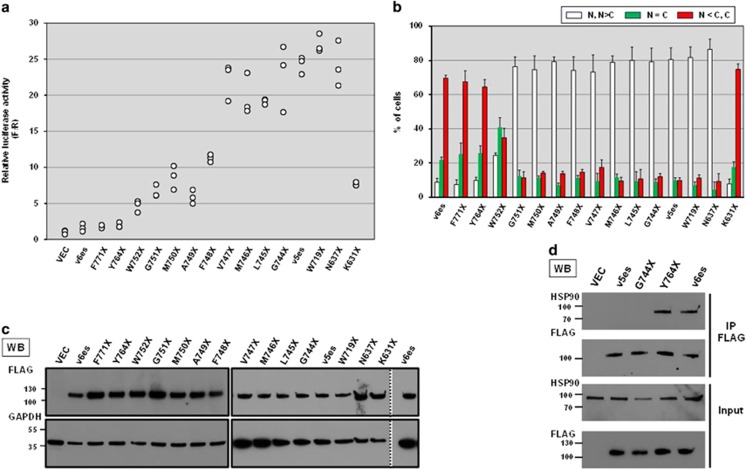
Next we attempted to gain insight into what aspects are different between active and inactive exon-skipping AR-Vs. Both active and inactive AR-Vs commonly possess exon 1 through exon 4. Accordingly, amino acid residues corresponding to exon 5 appear to blunt transactivation capability of ARv6es while those corresponding to both/either of two exons 7 and 8 suppress the activity of ARv56es. We generated and systematically analyzed a series of mutants with deletion of exon 5 in ARv6es toward exon 4–5 junction (Figure 4). LBD of type I steroid receptors fold into 12 helices (H1–H12) (Figure 4).34 Among them, H4 and H5 helices reside in exon 5. The deletion mutants with intact H5 (F771X, in which codon 771 is converted to stop codon, and Y764X) still resembled ARv6es with respect to transactivation capacity and subcellular localization (Figures 7a and b). A subset of mutants with compromised H5 (W752X, G751X, M750X, A749X, and F748X) exhibited exclusive nuclear localization and displayed significant transactivation effects. Notably, V747X was predominantly nuclear and its transactivation activity was comparable to that of v5es, which implies that removing a complete H5 sequence is not necessary for AR to gain full constitutive activity. We confirmed comparable expression levels among the variety of deletion mutants (Figure 7c). Integrity of H5 appears to determine subcellular distribution of AR, which is consistent with the report that the proposed NES includes a major part of H5 (Figure 4).29 Manipulation of the exon 5 sequence presumably affects the binding affinity of AR to HSP90 (Figure 4).27 H5-less mutant G744X largely lost HSP90-binding capability while comparable amount of HSP90 was co-immunoprecipitated with ARv6es and the H5-intact mutant Y764 (Figure 7d). Thus, we conclude that the presence or absence of intact H5 determines AR's subcellular localization, HSP90-binding affinity and transactivation activity (G744X immunofluorescence image is provided in Supplementary Figure S6). It is not well documented what factors require the H5 segment for exporting AR from nucleus. Therefore, it still remains to be examined whether HSP90 determines H5-dependent subcellular localization of AR. On the other hand, as confirmed by the characteristic difference between N637X and K631X and the measurable transactivation activity of K631X, the bipartite nuclear localization signal encoded by exons 3 and 4 is important but not essential for nuclear localization of exon-skipping AR-Vs and thus transactivation activity as well (Figures 7a and b). This is in agreement with the observation done with a synthetic truncated AR variant composed of only the exons 1 through 3 displaying nuclear/cytoplasmic distribution as well as significant ligand-independent transcriptional activity.35
Figure 7.
Characterization of synthetic mutants with partial deletion of exons 4 and 5. (a) Probasin-based reporter was used to evaluate transactivation activities of a series of deletion mutants. Luciferase activities were measured as described in Figure 2a. All individual data points were plotted. Similar results were obtained from three independent experiments. (b) Western blot analysis confirmed comparable expression of FLAG AR constructs. The common cell lysates of v6es sample were loaded on two different gels loaded with the samples including F771X and those including V747X as the control to compare the intensities of the bands on the different gels. The vertical dotted line indicates western blot image cropped from the same gel. (c) Binding affinity of deletion mutants to HSP90. FLAG-tagged AR mutants expressed in M12 cells were subjected to co-IP assay as described in Figure 4. (d) Subcellular localization of AR deletion mutants FLAG-tagged constructs were evaluated in M12 as described in Figure 5. Localization was classified following the similar criteria. N and N>C, white bar; N=C, green; N<C and C, red. Three hundred cells were assessed in each well. Similar results were obtained from three independent experiments.
The determinants for transcriptional inactivity of ARv56es
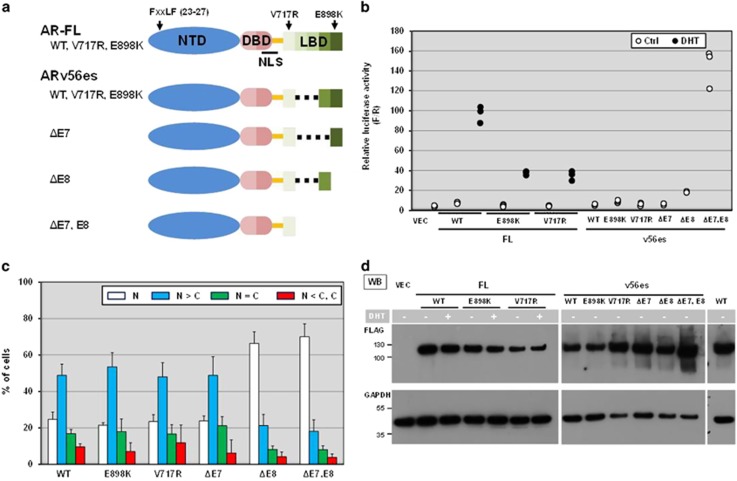
AR N-terminal/C-terminal (N–C) interaction is the key step for dimerization initiation of transcription of AR-FL. N–C interaction is mediated by the FXXLF motif (FQNLF) in the NTD and the activation function 2 (AF2) which is formed by ligand-induced conformational change in the LBD (Figure 8a, AR-FL).3, 5, 36 ARv56es still maintains the in-frame open reading frame corresponding to exon 7 and thus retains many AF2-forming amino acids including V717 in exon 4 and E898 in exon 8 (Figure 8a, ARv56es).34, 37, 38 Accordingly, ARv56es is structurally similar to ARv5es and ARv567es except for C-terminal extension of incomplete LBD. We introduced V717R and E898K substitutions which reportedly disrupt the interaction between AF-2 and AR NTD without compromising ligand binding.37, 38 As expected, DHT-induced transactivation activities were markedly lowered by these substitutions in AR-FL (Figure 7b). Nevertheless, introduction of the same substitutions into ARv56es did not affect either their transactivation activities or subcellular localization (Figures 8b and c). This rules out the possibility that aberrant N–C interaction suppresses otherwise constitutive transcriptional activity in ARv56es. When inhibitory effects of exons 7 and 8 were taken into account (Figure 8a), v56es without exon 7 sequence (v56esΔE7) and v56es were similar in their subcellular locations as well as transactivation capacities (Figures 8b and c). In contrast, v56es missing exon 8 (v56esΔE8) was exclusively localized in the nucleus and displayed significant but much lower transcriptional activity than that of v5es and the v56es derivative with simultaneous deletion of exons 7 and 8 (v56esΔE7,E8) (Figures 8b and c). We confirmed comparable expression levels among the variety of deletion and point mutants (Figure 8d). Amino acid residues corresponding to exon 8 control the subcellular localization of ARv56es while those in exon 7 alone or together with exon 8 negatively modulate transactivation activity of v56es.
Figure 8.
Characterization of AR-FL and ARv56es derivatives. (a) The diagram depicts the domain organization of AR-FL and the corresponding exons. Arrows indicate the relative positions of FXXLF motif (amino acid 23–27) and two residues V717 and E898 involving formation of AF2. ARv56esΔE7 misses the amino acid sequence for exon 7, but maintains that for exon 8. ARv56esΔE8 and ARv56esΔE7, E8 are truncated mutants of ARv56es that lack the amino acid sequence corresponding to missing exons. (b) Probasin-based reporter was used to evaluate transactivation activities of AR-FL, V56es and their point and deletion mutants. Luciferase activities were measured as described in Figure 2a. All individual data points were plotted. Similar results were obtained from three independent experiments. (c) Western blot analysis confirmed comparable expression of FLAG AR constructs. The common cell lysates of 'AR-FL treated w/o DHT' sample were loaded on two different gels loaded with FL derivatives and v56es derivatives as the control to compare the intensities of the bands on the different gels. The vertical dotted line indicates western blot image cropped from the same gel. (d) Subcellular localization of ARv56es and its point and deletion mutants. FLAG-tagged constructs were evaluated in M12 cells as described in Figure 5. Note that deletion of exon 8 markedly promoted nuclear localization as opposed to no obvious effect with loss of exon 7.
Discussion
To precisely translate the increasing deluge of genomic data into precision treatment plans, it is urgent to predict and determine the functional outcomes causally associated with genomic mutations and aberrant gene expression. This notion holds true for AR, particularly in advanced CRPC patients who express multiple forms of its splicing variants and in whom the variants appear in more complex manners than previously thought. For instance, AR-V7 detection represents a predictive biomarker for response to the antiandrogen enzalutamide and abiraterone.17 Taking into account the less abundance of most AR-Vs compared with AR-FL and AR-V7, one may wonder whether these variants are simply noise of alternative splicing events or potential drivers in progression of CRPC. As suggested in our genomic approach for v5es (Figure 6), the presence of premature termination codons in some exon-skipping variants could render their transcripts susceptible to NMD.32 A number of the corresponding exon–exon junction reads, however, suggest the presence of a substantial amount of transcript in the specimen at steady-state levels.20, 21 De-regulated activities of splicing factors in cancer could generate multiple splicing variants. NMD inhibition is known to be caused by hypoxic conditions or other stresses of tumor microenvironment.39 Collectively, these factors may contribute to diversity and stability of alternative splicing variants. Moreover, despite extremely low abundance in TCGA and SU2C tumor samples, substantial evidence has already demonstrated that the presence of any detectable ARv567es in CRPC is associated with worse prognosis and shorter cancer-specific survival.18 In this regard, among the multiple variants characterized in this study, ARv5es stands out as a novel constitutively active variant. Importantly, it is far more abundantly and frequently expressed than ARv567es in TCGA and SU2C tumor samples.20, 21 AR-Vs including ARv567es and V7 are reportedly generated as a result of mis-splicing that is caused or permitted by genomic alterations such as intragenic rearrangement, deletion and mutations in the exon–intron junctions.12, 40, 41 Notably, multiple forms of AR-Vs can be present in the same patient and same cell (Supplementary Figure S1).42 Accordingly, it would be well reasoned that these variants arose by virtue of alternative splicing from the same genomic DNA event where certain alterations are introduced in the course of CRPC progression.
AR-FL, v5es and v567 are sub-grouped together by the presence of the hinge domain (HD), which is missing in AR-V7. The hinge domain not only bridges between the DBD and LBD but also serves as a determinant for physical interactions with many regulatory factors including microtubules and the E3 ubiquitin ligase SPOP.43, 44 The anti-microtubule agent taxane targets microtubule-dependent trafficking for AR nuclear import.45 Therefore, a patient expressing ARv5es may display resistance to antiandrogen therapy but benefit from taxane-based chemotherapy.46 Microtubule-dependency of AR-Vs needs to be examined more rigorously because the cross-resistance between taxanes and antiandrogens is a matter of debate.46, 47 On the other hand, cancer-associated mutations in SPOP would most likely increase the stability and activity of ARv5es as observed for AR-FL and v567es.43
In the cytoplasm, binding of androgen to the unliganded AR in complex with HSP90 exposes its otherwise masked NLSs, thereby allowing AR entry into the nucleus.6 Additionally, non-canonical NES, which is mapped to a stretch in LBD (amino acids 745–819 in human AR), dominates over NLS and it is suppressed by ligand binding (Figure 5).29 Thus, it stands to reason that v5es, which lacks both the NES and HSP90-binding sites, exhibits predominantly nuclear localization. Our deletion study related to ARv6es revealed nucleocytoplasmic localization is in part determined by the presence or absence of a sequence of amino acids situated in helix 5 of the LBD. It is noteworthy that both the G751X and V747X mutants are exclusively localized in the nucleus but G751X mutant shows significantly lower transcriptional activities than V747X and v5es (Figures 7a and b), suggesting possible inhibitory roles of peptide 'MAFV' in transactivation capacity. Further, binding capacity to HSP90 appears to determine constitutive activity of the corresponding deletion mutants. This holds true for other type I steroid receptors. For instance, loss of binding affinity to HSP90 is closely associated with constitutively active forms of glucocorticoid receptor (GR) and requires removal of H3 and H4 together with H5.49, 50 HSP90-binding profiles and subcellular localization patterns were not easily separable in our study (Figures 7a, b and d). Though AR and GR are highly homologous in the LBD (Supplementary Figure S8), they display differential preference for substructures of the LBD in HSP90 binding.49 A GR deletion mutant (1-615 in rat GR), which is structurally and functionally equivalent to truncated AR mutant (1-739 in human AR), displays predominantly nuclear localization but is nevertheless transcriptionally inactive.50, 51 GR1–615 may form a complex with HSP90 in the nucleus, which does not contradict with the recent demonstration that steroid receptors remain associated with HSP90 during nuclear import.52
Splicing factor Prp8 was recently identified as a plausible candidate that regulates AR's nuclear export in an NES-dependent manner.53 It would be intriguing to examine whether two LBD-binding proteins, Prp8 and HSP90, compete for the binding to AR or collaboratively work to keep inactive AR away from the nucleus. Expression of Prp8 is reportedly downregulated in castration-resistant prostate tumor xenograft.53 ARv56es, which also lacks the entire NES, still shows considerable cytoplasmic distribution while the synthetic ARΔNES (G744X) is predominantly localized to the nucleus.29 This discrepancy may arise as results of inclusion/exclusion of the region covering amino acids 725–744 or usage of different types of tags that might affect conformations of their corresponding proteins. In the case of v56es, elimination of the sequence corresponding to exon 8 leads to exclusive nuclear localization of the corresponding deletion mutant. There must be additional factors regulating localization of AR.
In this study, we took a step further to define intrinsic determinants for constitutive activity of AR. This now allows us to predict whether cancer-driving activities are associated not only with exon-skipping AR-Vs but also LBD-defective ARs that are generated through somatic nonsense mutations in the LBD. Such mutations are reported to occur at the positions of Q641 (exon 4), W742 (exon 5), W752 (exon 5), R787 (exon 6) and Q868 (exon 7)54, 55 (Figure 4). As a proof of principle, ARQ641X is purported to be constitutively active.54 ARW742X is presumably as potent as that of v5es and ARQ641X. ARW752X is a probable driver on the basis of our observation. On the other hand, ARR787X and ARQ868X resemble v6es and v7es, respectively, so they are predicted to be inactive. We propose that our rules can be applied to determine the potential activity of AR nosnsense mutants to be identified through analysis of genomic DNA in liquid biopsies.
To reduce the inherent complexity, we aimed to characterize the individual AR-Vs separately in this study and discovered a novel constitutively active variant, namely ARv5es. Future studies will be aimed at deepening our knowledge about the functional aspects of AR-Vs, their interaction with one another or AR-FL, and potential transcriptomes to develop the future AR-targeted therapeutics.
Materials and methods
Chemicals and antibodies
Enzalutamide (Enza) and leptomycin B were obtained from Selleck Chemicals (Houston, TX, USA) and EMD Millipore (Billerica, MA, USA), respectively. Sodium molybdate and cycloheximide were from Sigma (St Louis, MO, USA). All other chemicals were of analytical grade. Antibodies and their dilutions used for western blot analysis were FLAG M2 monoclonal (F1804; Sigma; 1:2000), FLAG polyclonal (F7425; Sigma; 1:1000), HSP90 (C45G5; Cell Signaling Technology, Danvers, MA, USA; 1:1000), β-actin (13E5; Cell Signaling Technology; 1:1000), androgen receptor (441; Santa Cruz Biotechnology, Dallas, TX, USA; 1:2000) and GAPDH (14C10; Cell Signaling Technology; 1:5000). Horseradish peroxidase-conjugated goat anti-mouse or rabbit IgG were from Santa Cruz Biotechnology. For immunofluorescence studies, FLAG M2 antibody (1:1000) was used in combination with Alexa Fluor 488 Goat Anti-Mouse IgG (H+L) antibody (A11001; Thermo Fisher Scientific, Waltham, MA, USA; 1:1000) and DAPI (4',6-diamidino-2-phenylindole) for nuclear staining.
Bioinformatics
Collected SU2C RNA-seq reads20 were further processed for analysis of co-occurrence of AR-Vs across the patient samples.
CRISPR/Cas9-mediated genome editing in CWR-R1-AD1
The 20-nt guide sequences targeting intron 4 and intron 5 were designed using the CRISPR DESIGN software (http://crispr.mit.edu/). The guide DNA (gDNA) sequences encoding gRNA targeting intron 4 (g4a and g4b) and intron 5 (g5) were individually cloned into lentiCRISPR v2 vector (Addgene, Cambridge, MA, USA; 52961) which co-expresses human codon–optimized Cas9 and purormycin N-acetyltransferase through P2A bicistronic linker.57 CWR-R1-AD1 cells were transfected with the plasmid g5 in combination with either of the plasmid g4a or g4b. Transiently transfected cells were selected in medium supplemented with 1 μg/ml puromycin for 36 h, subsequently washed with fresh medium and then re-plated on new dishes. The cultures were maintained in medium without puromycin until visible colonies appeared. The deletion-positive colonies were selected by PCR with two primers that flank the target sites by gRNAs (P1 and P2) using genomic DNA isolated from the individual colonies. The selected positive clones were further subjected to PCR with primers that flank the targeted sites in intron 4 (P1 and P3) to verify the purity of the clones. Phenotypically, the positive clones were characterized by reverse transcription–PCR with two different sets of primers: primers targeting exons 4 (P4) and 8 (P5) or primers targeting exons 4 (P4) and 6 (P6).
Reporter assay
AR negative cell lines (M12, PC3 and LNCaPAPIPC) (see the Supplementary Data for details) were co-transfected with the p3XFLAG-CMV10-based expression vector and the reporter plasmid. Specifically, the reporter assay was done with either the luciferase reporter driven by canonical androgen responsive regions (pARR2PB-luc),23 or a luciferase construct under the control of AR-V-specific binding sites derived from the promoter element of the ubiquitin-conjugating enzyme E2C (UBE2C) gene (UBE2C-luc).24, 25, 58 Renilla luciferase expression plasmid was used as an internal standard in conjunction with pARR2PB-luc. At 16 h after transfection, cells were treated with 1 nm 5α-dihydrotestosterone (DHT) or vehicle control. As necessary, 10 μm enzalutamide was added 2 h prior to DHT treatment. Luciferase assay was done at 24 h after DHT treatment using the Dual-Glo luciferase assay system (Promega, Madison, WI, USA) with GloMax Multi Detection System for detection (Promega). Firefly luciferase luminescence was normalized to that of the co-expressed Renilla luciferase or protein concentration per each sample.
Cell lysis and co-immunoprecipitation
M12 transfectants were lysed in M-Per lysis buffer (Thermo Fisher Scientific) supplemented with Halt Protease Inhibitor Cocktail (Thermo Fisher Scientific). The resultant lysates were subjected to western blot analysis in order to examine expression levels of various AR-V constructs. Androgen-depleted M12 cells were transfected with the expression vectors for FLAG-tagged AR-FL or AR-V. At 24 h after transfection, cells were lysed in the IP buffer (150 mm NaCl, 20 mm Tris, pH 7.8 and 0.5% Nonidet P-40, 20 mm sodium molybdate). Subsequently, anti-FLAG M2 agarose beads (Sigma) were incubated with cleared lysates, washed four times with the IP buffer and boiled with 2 × SDS–PAGE sample buffer. The eluted proteins were resolved by SDS–PAGE gels, followed by western blot analysis using the specified antibodies.
Immunofluorescence
Androgen-depleted M12 were transfected with the expression vectors for FLAG-tagged AR-FL or AR-V. The cells were separately transfected with the expression construct for GFP-2xNES.PKI The cells were treated 24 h later with DHT (1 nm), leptomycin B (10 ng/ml) or vehicle control. Cells were 4 h later fixed in 4% paraformaldehyde, permeabilized with 0.2% Triton X-100 and processed for immunostaining for AR-FL and AR-Vs with FLAG antibody. Fluorescent microscopic images were captured on a DeltaVision Elite Microscopy System. Images were processed with Adobe Photoshop CS5 (Adobe Systems, Seattle, WA, USA). Subcellular localization of AR-FL and AR-V was determined in approximately 200 cells per each condition by visualizing FLAG immunofluorescence with Zeiss Axiovert 40 CFL microscope (Carl Zeiss Microscopy, LLC, Thornwood, NY, USA).
Table 1. DNA oligonucleotides used in this study.
| Name | Sequence (5′ to 3′) and Notes |
|---|---|
| NES1 | GAC TCA GAT CTC GAG CTC AAG CTT CGA ATT C-EcoRI site is in bold |
| NES2 | CAT TGC TAC CAC CAC CTT CTG TCT TGT TGA TAT CAA GAC CTG CTA ATT TCA AGG CTA ATT CAT TGC TCT TGT ACA GCT CGT CCA TGC CGA GAG |
| NES3 | CCT GGC GGC CGC TCA ACC TTC TGT CTT GTT GAT ATC AAG ACC TGC TAA TTTC AAG GCT AAT TCAT TGC TAC CAC CAC CTT CTG TCT TGT TGA TATC |
| NotI site is single underlined | |
| g4A gDNA sense | CACCGTCTCCTCTTTGTTAGTTCGG |
| g4A gDNA antisense | AAACCCGAACTAACAAAGAGGAGAC |
| g4B gDNA sense | CACCGATGCCACCGAACTAACAAAG |
| g4B gDNA antisense | AAACCTTTGTTAGTTCGGTGGCATC |
| g5 gDNA sense | CACCGTGCTGTATGAGGTCCCTCGA |
| g5 gDNA antisense | AAACTCGAGGGACCTCATACAGCAC |
| P1 | AGGCTACTTCAGAGATTGGGC |
| P2 | CTGGAAGGATTCTCTGTGGTGAGA |
| P3 | GGAATGTGTGAATGTGAAGGCAC |
| P4 | GAAGGCTATGAATGTCAGCCCATC |
| P5 | CAGCCCATCCACTGGAATAATGC |
| P6 | CAGGTCAAAAGTGAACTGATGCAGCTCTCTC |
| qPCR primers | |
| RPL13A forward | CCTGGAGGAGAAGAGGAAAGAGA |
| RPL13A reverse | TTGAGGACCTCTGTGTATTTGTCAA |
| ATF1 forward | GCCATTGGAGAGCTGTCTTC |
| ATF1 reverse | GGGCCATCTGGAACATAAGA |
Acknowledgments
This work was supported by the Department of Defense (W81XWH-12-PCRP-TIA), Veterans Affairs Research Program (to SRP) and National Cancer of Institute (P01CA163227, P50 CA 097186) (to SRP and PSN).
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
The authors declare no conflict of interest.
Supplementary Material
References
- Attard G, Parker C, Eeles RA, Schröder F, Tomlins SA, Tannock I et al. Prostate cancer. Lancet 2016; 387: 70–82. [DOI] [PubMed] [Google Scholar]
- Ferraldeschi R, Welti J, Luo J, Attard G, de Bono JS. Targeting the androgen receptor pathway in castration-resistant prostate cancer: progresses and prospects. Oncogene 2015; 34: 1745–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centenera MM, Harris JM, Tilley WD, Butler LM. The contribution of different androgen receptor domains to receptor dimerization and signaling. Mol Endocrinol 2008; 22: 2373–2382. [DOI] [PubMed] [Google Scholar]
- Claessens F, Denayer S, Van Tilborgh N, Kerkhofs S, Helsen C, Haelens A. Diverse roles of androgen receptor (AR) domains in AR-mediated signaling. Nucl Recept Signal 2008; 6: e008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan MH, Li J, Xu HE, Melcher K, Yong EL. Androgen receptor: structure, role in prostate cancer and drug discovery. Acta Pharmacol Sin 2015; 36: 3–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman BJ, Feldman D. The development of androgen-independent prostate cancer. Nat Rev Cancer 2001; 1: 34–45. [DOI] [PubMed] [Google Scholar]
- Mostaghel EA, Plymate SR, Montgomery B. Molecular pathways: targeting resistance in the androgen receptor for therapeutic benefit. Clin Cancer Res 2014; 20: 791–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehm SM, Schmidt LJ, Heemers HV, Vessella RL, Tindall DJ. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res 2008; 68: 5469–5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Guo Z, Sun F, Li W, Alfano A, Shimelis H et al. Novel membrane-associated androgen receptor splice variant potentiates proliferative and survival responses in prostate cancer cells. J Biol Chem 2011; 286: 36152–36160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R, Dunn TA, Wei S, Isharwal S, Veltri RW, Humphreys E et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res 2009; 69: 16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Sprenger CC, Vessella RL, Haugk K, Soriano K, Mostaghel EA et al. Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. J Clin Invest 2010; 120: 2715–2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehm SM, Tindall DJ. Alternatively spliced androgen receptor variants. Endocr Relat Cancer 2011; 18: R183–R196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Luo J. Decoding the androgen receptor splice variants. Transl Androl Urol 2013; 2: 178–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprenger CC, Plymate SR. The link between androgen receptor splice variants and castration-resistant prostate cancer. Horm Cancer 2014; 5: 207–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med 2011; 364: 1995–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med 2012; 367: 1187–1197. [DOI] [PubMed] [Google Scholar]
- Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med 2014; 371: 1028–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörnberg E, Ylitalo EB, Crnalic S, Antti H, Stattin P, Widmark A et al. Expression of androgen receptor splice variants in prostate cancer bone metastases is associated with castration-resistance and short survival. PLoS One 2011; 6: e19059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R, Isaacs WB, Luo J. A snapshot of the expression signature of androgen receptor splicing variants and their distinctive transcriptional activities. Prostate 2011; 71: 1656–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015; 161: 1215–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Network CGAR. The Molecular Taxonomy of Primary Prostate Cancer. Cell 2015; 163: 1011–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson PA, Arora VK, Sawyers CL. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer 2015; 15: 701–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Thomas TZ, Kasper S, Matusik RJ. A small composite probasin promoter confers high levels of prostate-specific gene expression through regulation by androgens and glucocorticoids in vitro and in vivo. Endocrinology 2000; 141: 4698–4710. [DOI] [PubMed] [Google Scholar]
- Cao B, Qi Y, Zhang G, Xu D, Zhan Y, Alvarez X et al. Androgen receptor splice variants activating the full-length receptor in mediating resistance to androgen-directed therapy. Oncotarget 2014; 5: 1646–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Zhan Y, Qi Y, Cao B, Bai S, Xu W et al. Androgen receptor splice variants dimerize to transactivate target genes. Cancer Res 2015; 75: 3663–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung JK, Banuelos CA, Fernandez JG, Mawji NR, Wang J, Tien AH et al. An androgen receptor N-terminal domain antagonist for treating prostate cancer. J Clin Invest 2013; 123: 2948–2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marivoet S, Van Dijck P, Verhoeven G, Heyns W. Interaction of the 90-kDa heat shock protein with native and in vitro translated androgen receptor and receptor fragments. Mol Cell Endocrinol 1992; 88: 165–174. [DOI] [PubMed] [Google Scholar]
- Gillis JL, Selth LA, Centenera MM, Townley SL, Sun S, Plymate SR et al. Constitutively-active androgen receptor variants function independently of the HSP90 chaperone but do not confer resistance to HSP90 inhibitors. Oncotarget 2013; 4: 691–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saporita AJ, Zhang Q, Navai N, Dincer Z, Hahn J, Cai X et al. Identification and characterization of a ligand-regulated nuclear export signal in androgen receptor. J Biol Chem 2003; 278: 41998–42005. [DOI] [PubMed] [Google Scholar]
- Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M et al. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 1997; 390: 308–311. [DOI] [PubMed] [Google Scholar]
- Wen W, Harootunian AT, Adams SR, Feramisco J, Tsien RY, Meinkoth JL et al. Heat-stable inhibitors of cAMP-dependent protein kinase carry a nuclear export signal. J Biol Chem 1994; 269: 32214–32220. [PubMed] [Google Scholar]
- Maquat LE. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol 2004; 5: 89–99. [DOI] [PubMed] [Google Scholar]
- Li Y, Chan SC, Brand LJ, Hwang TH, Silverstein KA, Dehm SM. Androgen receptor splice variants mediate enzalutamide resistance in castration-resistant prostate cancer cell lines. Cancer Res 2013; 73: 483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sack JS, Kish KF, Wang C, Attar RM, Kiefer SE, An Y et al. Crystallographic structures of the ligand-binding domains of the androgen receptor and its T877A mutant complexed with the natural agonist dihydrotestosterone. Proc Natl Acad Sci USA 2001; 98: 4904–4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SC, Li Y, Dehm SM. Androgen receptor splice variants activate androgen receptor target genes and support aberrant prostate cancer cell growth independent of canonical androgen receptor nuclear localization signal. J Biol Chem 2012; 287: 19736–19749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helsen C, Dubois V, Verfaillie A, Young J, Trekels M, Vancraenenbroeck R et al. Evidence for DNA-binding domain–ligand-binding domain communications in the androgen receptor. Mol Cell Biol 2012; 32: 3033–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Kemppainen JA, Voegel JJ, Gronemeyer H, Wilson EM. Activation function 2 in the human androgen receptor ligand binding domain mediates interdomain communication with the NH(2)-terminal domain. J Biol Chem 1999; 274: 37219–37225. [DOI] [PubMed] [Google Scholar]
- Dehm SM, Tindall DJ. Ligand-independent androgen receptor activity is activation function-2-independent and resistant to antiandrogens in androgen refractory prostate cancer cells. J Biol Chem 2006; 281: 27882–27893. [DOI] [PubMed] [Google Scholar]
- Gardner LB. Nonsense-mediated RNA decay regulation by cellular stress: implications for tumorigenesis. Mol Cancer Res 2010; 8: 295–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Alsagabi M, Fan D, Bova GS, Tewfik AH, Dehm SM. Intragenic rearrangement and altered RNA splicing of the androgen receptor in a cell-based model of prostate cancer progression. Cancer Res 2011; 71: 2108–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Hwang TH, Oseth LA, Hauge A, Vessella RL, Schmechel SC et al. AR intragenic deletions linked to androgen receptor splice variant expression and activity in models of prostate cancer progression. Oncogene 2012; 31: 4759–4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LL, Xie N, Sun S, Plymate S, Mostaghel E, Dong X. Mechanisms of the androgen receptor splicing in prostate cancer cells. Oncogene 2014; 33: 3140–3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An J, Wang C, Deng Y, Yu L, Huang H. Destruction of full-length androgen receptor by wild-type SPOP, but not prostate-cancer-associated mutants. Cell Rep 2014; 6: 657–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thadani-Mulero M, Portella L, Sun S, Sung M, Matov A, Vessella RL et al. Androgen receptor splice variants determine taxane sensitivity in prostate cancer. Cancer Res 2014; 74: 2270–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darshan MS, Loftus MS, Thadani-Mulero M, Levy BP, Escuin D, Zhou XK et al. Taxane-induced blockade to nuclear accumulation of the androgen receptor predicts clinical responses in metastatic prostate cancer. Cancer Res 2011; 71: 6019–6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonarakis ES, Lu C, Luber B, Wang H, Chen Y, Nakazawa M et al. Androgen receptor splice variant 7 and efficacy of taxane chemotherapy in patients with metastatic castration-resistant prostate cancer. JAMA Oncol 2015; 1: 582–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Soest RJ, van Royen ME, de Morrée ES, Moll JM, Teubel W, Wiemer EA et al. Cross-resistance between taxanes and new hormonal agents abiraterone and enzalutamide may affect drug sequence choices in metastatic castration-resistant prostate cancer. Eur J Cancer 2013; 49: 3821–3830. [DOI] [PubMed] [Google Scholar]
- Zhang G, Liu X, Li J, Ledet E, Alvarez X, Qi Y et al. Androgen receptor splice variants circumvent AR blockade by microtubule-targeting agents. Oncotarget 2015; 6: 23358–23371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalman FC, Scherrer LC, Taylor LP, Akil H, Pratt WB. Localization of the 90-kDa heat shock protein-binding site within the hormone-binding domain of the glucocorticoid receptor by peptide competition. J Biol Chem 1991; 266: 3482–3490. [PubMed] [Google Scholar]
- Godowski PJ, Rusconi S, Miesfeld R, Yamamoto KR. Glucocorticoid receptor mutants that are constitutive activators of transcriptional enhancement. Nature 1987; 325: 365–368. [DOI] [PubMed] [Google Scholar]
- Picard D, Yamamoto KR. Two signals mediate hormone-dependent nuclear localization of the glucocorticoid receptor. EMBO J 1987; 6: 3333–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storer CL, Dickey CA, Galigniana MD, Rein T, Cox MB. FKBP51 and FKBP52 in signaling and disease. Trends Endocrinol Metab 2011; 22: 481–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Nguyen MM, Masoodi KZ, Singh P, Jing Y, O'Malley K et al. Splicing factor Prp8 interacts with NES(AR) and regulates androgen receptor in prostate cancer cells. Mol Endocrinol 2015; 29: 1731–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Céraline J, Cruchant MD, Erdmann E, Erbs P, Kurtz JE, Duclos B et al. Constitutive activation of the androgen receptor by a point mutation in the hinge region: a new mechanism for androgen-independent growth in prostate cancer. Int J Cancer 2004; 108: 152–157. [DOI] [PubMed] [Google Scholar]
- Steinkamp MP, O'Mahony OA, Brogley M, Rehman H, Lapensee EW, Dhanasekaran S et al. Treatment-dependent androgen receptor mutations in prostate cancer exploit multiple mechanisms to evade therapy. Cancer Res 2009; 69: 4434–4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Furusato M, Allsbrook WC, Nishii H, Wakui S, Barrett JC et al. Prevalence of androgen receptor gene mutations in latent prostatic carcinomas from Japanese men. Cancer Res 1995; 55: 1621–1624. [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 2014; 11: 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Li W, Zhang Y, Yuan X, Xu K, Yu J et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell 2009; 138: 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.