Summary
The collagen domain, which is defined by the presence of the Gly-X-Y triplet repeats, is amongst the most versatile and widespread known structures found in proteins from organisms representing all three domains of life. The streptococcal collagen-like (Scl) proteins are widely present in pathogenic streptococci, including Streptococcus pyogenes, S. agalactiae, S. pneumoniae, and S. equi. Experiments and bioinformatic analyses support the hypothesis that all Scl proteins are homotrimeric and cell wall-anchored. These proteins contain the rod-shaped collagenous domain proximal to cell surface, as well as a variety of outermost non-collagenous domains that generally lack predicted functions but can be grouped into one of six clusters based on sequence similarity. The well-characterized Scl1 proteins of S. pyogenes show a dichotomous switch in ligand binding between human tissue and blood environments. In tissue, Scl1 adhesin specifically recognizes the wound microenvironment, promotes adhesion and biofilm formation, decreases bacterial killing by neutrophil extracellular traps, and modulates S. pyogenes virulence. In blood, ligands include components of complement and coagulation-fibrinolytic systems, as well as plasma lipoproteins. In all, the Scl proteins signify a large family of structurally related surface proteins, which contribute to the ability of streptococci to colonize and cause diseases in humans and animals.
Keywords: collagen-like proteins, streptococci, biofilm, fibronectin, immune evasion
Graphical abstract
Overview of collagens and collagen-like proteins
A common structure among diverse proteins
Collagens are ubiquitous in nature. The common feature of the collagen module is a triple-helical structure consisting of three polyproline-II-like helices supercoiled in a right-handed direction around a central axis (Berisio et al., 2002). The tight packing of the polypeptide chains requires a glycine every third residue, defining the Gly-X-Y repeat motif, where proline and hydroxyproline often occupy the X and Y positions of human collagen, respectively. Current knowledge on collagen structure is a culmination of fiber diffraction studies, modeling, and crystallographic studies on collagen mimetic peptides. Since a history of the structural dissection of collagen is out of the scope of this review, we direct the reader to several excellent reviews (Ramachandran, 1988, Brodsky & Persikov, 2005, Bella, 2016). The collagens emerged as an essential group of modular proteins of metazoans (Exposito et al., 2002). It is a versatile structure, appearing in human extracellular matrix proteins, host defense proteins, and anchoring fibrils (Brodsky & Persikov, 2005). The collagen domain is also present in important proteins in invertebrates, such as the exoskeleton collagens of sponges (Engel, 1997), holdfast structure of the mussel byssus (Suhre et al., 2014), and basement membrane and cuticle collagens of the nematode Caenorhabditis elegans (Kramer et al., 1982). There has been a large number of 18,874 collagen-like proteins (CLPs) annotated in bacteria, 695 in viruses, and 157 in archaea (search conducted on 9/24/16 in Uniprot database). The name streptococcal collagen-like proteins, Scl, in S. pyogenes was coined (Lukomski et al., 2000), which was followed by Bacillus proteins Bcl (Sylvestre et al., 2002, Pizarro-Guajardo et al., 2014), pneumococcal protein Pcl (Paterson et al., 2008), Lcl of Legionella pneumophila (Duncan et al., 2011), and Bucl proteins of Burkholderia spp. (Bachert et al., 2015). However, only a small proportion of predicted bacterial CLPs have been investigated thus far.
Origin of the collagenous domain in bacteria
The origin of the collagenous domain in prokaryotes is still unknown. However, the composition of the collagen domain in human and bacterial collagens differs considerably. In human collagens, proline and hydroxyproline residues are found preferentially in the X and Y positions, respectively, with frequencies of 27% and 38% (Bella, 2016). The abundance of hydroxyprolines in mammalian collagens is a major contributor to thermal stability of the triple helix (Chopra & Ananthanarayanan, 1982, Vitagliano et al., 2001) but prokaryotes lack the prolyl hydroxylase enzyme to perform this post-translational modification. Therefore, in bacteria more than 30% of proline residues are found in the X position but only 5% in the Y position (Berisio & Vitagliano, 2012). Despite the lack of hydroxyproline residues, bacterial CLPs have been shown to form stable triple helices, with thermal stabilities similar to human collagens, in the range of 35°–39°C, (Chan et al., 1997, Leikina et al., 2002, Xu et al., 2002, Han et al., 2006b, Xu et al., 2010). These CLPs rely on other mechanisms of helix stabilization, including hydration-mediated hydrogen bonding networks, electrostatic interactions between side chains, and the presence of specific stabilizing tripeptide repeats (Mohs et al., 2007, Xu et al., 2010). Proper folding of the triple helix is necessary for collagen functionality.
Horizontal transfer of collagenous sequences from eukaryotes to prokaryotes has been proposed (Rasmussen et al., 2003). However, recent studies have shown that collagen-like repeats likely arose in prokaryotes independently (Doxey & McConkey, 2013). Low-complexity repeats of the collagen triple helix could emerge by spontaneous mutations and amplify via simple repeat expansion, supporting the hypothesis that eukaryotic and prokaryotic collagens may have emerged by convergent evolution. Here, we present clustering analysis, which identified common domains in Scl proteins from different streptococcal species and subspecies, suggesting that horizontal scl-gene transfer is possible between bacteria that share the same human and animal hosts.
Classification of Scl proteins
CLuster ANalysis of Sequences (CLANS)
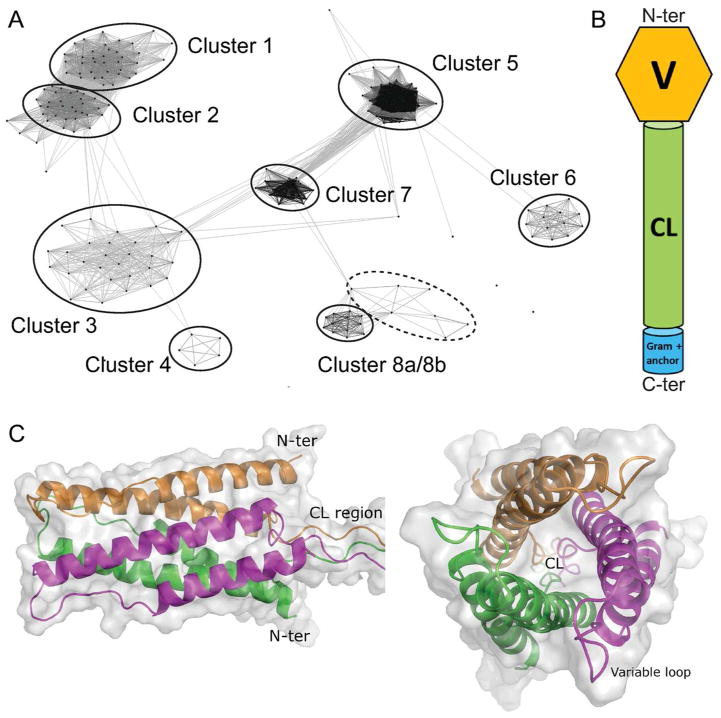
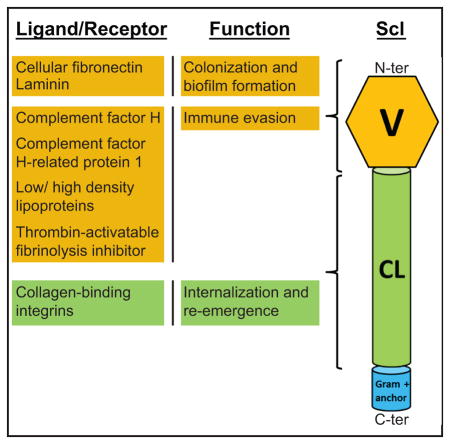
CLANS analysis was performed to obtain a sequence-based classification of Scl proteins. This analysis groups protein sequences based on all-against-all pairwise sequence similarities without phylogenetic reconstruction (Frickey & Lupas, 2004). CLANS identified six distinct clusters 1–6 of Scl proteins across pathogenic streptococci and two clusters 7–8 with phage proteins that will not be discussed here (Fig. 1A). Scl proteins harbored by streptococci infecting humans or animal species were found in separate clusters, suggesting co-evolution of Scl proteins with respective hosts. All Scl proteins share a distinct set of conserved features and a similar domain organization (Fig. 1B). They contain a signal peptide (not shown), an N-terminal non-collagenous sequence-variable (V) domain, a central collagen-like (CL) domain, and a cell-wall associated domain containing the LPXTG anchor motif (Gram-positive anchor). The CL domains of different Scl proteins are comprised of varying types of Gly-X-Y triplet repeats, and exhibit significant length variation due to the expansion and contraction of these repeats.
Fig. 1. Sequence similarity and structure of Scl proteins in streptococci.
A. Clustering of Scl proteins using CLuster Analysis of Sequences (CLANS). CLANS clusters and visualizes groups of protein sequences based on all-against-all pairwise sequence similarities (e-value cutoff for analysis was 10−13) without phylogenetic reconstruction. Each point on the plot represents a Scl amino acid sequence, while the connecting lines represent BLAST high-scoring segment pairs. High confidence clusters are indicated by solid circles, while dashed circle indicates a single low confidence cluster. Scl proteins identified in each cluster are listed in Table 1. Clusters 7 and 8a/8b consist of phage-associated proteins.
B. Cartoon representation of domain organization shared by mature Scl proteins. The Scl proteins consist of an N-terminal non-collagenous variable domain (V), a collagen-like domain (CL), and a C-terminally located cell wall-associated Gram-positive anchor with LPXTG motif.
C. Ribbon and surface models of the homotrimeric Scl2-protein globular domain. Left. Side view of the six-helix bundle, containing three pairs of antiparallel helices, colored green, orange and magenta, and the point of CL-region attachment. Right. Top view, showing the location of exposed variable loops between each alpha-helix pair and the arrangement of the three external helices wrapped around three internal helices.
Scl proteins of S. pyogenes (clusters 1,2)
The Scl1 and Scl2 proteins (also known as SclA and SclB) of S. pyogenes were the first bacterial CLPs to be reported and studied (Lukomski et al., 2000, Rasmussen et al., 2000, Lukomski et al., 2001, Rasmussen & Björck, 2001, Whatmore, 2001). Both proteins are homotrimeric and contain the globular V domain projected away from the cell surface by the rod-shaped CL domain. The V-region sequences differ significantly both between and within Scl1 and Scl2 variants; however, Scl variants are conserved in strains of the same M-type. The scl1 and scl2 genes have been found in all S. pyogenes strains tested and are co-expressed in the exponential phase of growth, although their expression is regulated differently. Transcription of the scl1 gene is positively regulated by the multiple gene regulator of S. pyogenes, Mga (Rasmussen et al., 2000, Lukomski et al., 2001, Almengor & McIver, 2004, Almengor et al., 2006). Scl2 expression depends on phase variation associated with CAAAA repeats located downstream of the start codon (Lukomski et al., 2001, Rasmussen & Björck, 2001, Whatmore, 2001).
Scl proteins in streptococci pathogenic to animals (clusters 3, 4 and 6)
The Scl3, 4 and 6 proteins are found in S. zooepidemicus, a commensal organism found in domesticated animals rarely transmitting to humans, and S. equi, a causative agent of the serious disease strangles in horses (Timoney, 2004). CLANS classified several Scl3 proteins, originally denoted SclC for group C Streptococcus (Karlström et al., 2004), as well as the related proteins SclD-I and SclZ.1–5, 7 and 12 (Beres et al., 2008), as belonging to the same cluster 3, whereas SclF (Karlström et al., 2006) formed an independent cluster 4. SclC was shown to be expressed during strangles infection, and immunization with the recombinant SclC protein partially protected against infection in both mouse and horse models (Flock et al., 2006, Waller et al., 2007). An effective multi-component vaccine for strangles was developed that include recombinant SclC (Guss et al., 2009). Several cluster 3 proteins share signature sequences with cluster 2 proteins of S. pyogenes (Karlström et al., 2004), suggesting an inter-species horizontal gene transfer. Some Scl3 variants contain CAAAA repeats in the 5′ translated region, indicating this protein may be regulated by phase variation similarly to Scl2 (Karlström et al., 2004). A second group of proteins found in both S. equi and S. zooepidemicus, classified in cluster 6, included the SclZ.6, 9, and 10 proteins (Beres et al., 2008), as well as FneC, E, and F that were annotated as fibronectin-binding proteins (Lannergård, 2006).
Scl proteins in S. pneumoniae and S. agalactiae (cluster 5)
The vast majority of Scl5 proteins were annotated in S. pneumoniae and S. agalactiae, but also in viridans species associated with endocarditis, S. mitis and S. tigurinus (Zbinden et al., 2015), as well as in fish pathogen S. ictaluri (Shewmaker et al., 2007). The clustering of Scl5 proteins from several related species provides evidence for horizontal gene transfer. The pneumococcal PclA (Paterson et al., 2008), is a large surface protein (265 kDa) that contains predicted G5 and FIVAR domains, though FIVAR was predicted with low confidence. The Scl5 variants in S. agalactiae contain the G5 domain, but lack the FIVAR domain.
The non-collagenous domain in streptococcal collagen-like proteins
The best structurally characterized streptococcal collagens are Scl1 and Scl2 of S. pyogenes. The proteins form stable triple-helical structures when expressed as recombinant (rScl) polypeptides (Xu et al., 2002, Mohs et al., 2007). The non-collagenous V region constitutes a trimerization domain that augments proper collagen assembly to avoid the misfolding of the triple helix due to its repeating structure. However, the V domain of Scls is not necessary for triple helix formation in vivo, since the CL region of Scl1 can be expressed in E. coli without the V region as recombinant protein in a folded triple-helical state; still, this rScl-CL construct could not re-fold after thermal denaturation in vitro (Xu et al., 2002, Yu et al., 2010). These results suggest that V domains of Scls present both structural and ligand-binding functions reviewed below.
Recently, the crystal structure of the V domain of Scl2 from M3-type S. pyogenes was reported (Squeglia et al., 2013, Squeglia et al., 2014). It folds into a six-helical bundle, with three pairs of antiparallel alpha helices, each connected by a variable loop region. Three of the helices are wound in a left-handed super-helix forming the inner core, which is further wrapped by three antiparallel external alpha helices (Fig. 1C). The six-helix bundle forms an elongated cylinder measuring about 30 Å in diameter and 60 Å in height. This fold is consistent with previous secondary structure predictions that deduced a helix-loop-helix motif in the primary amino acid sequence (Rasmussen et al., 2000, Han et al., 2006a) and is predicted to be conserved among Scl1 and Scl2 proteins. Indeed, hydrophobic residues located at regular positions of Scl sequences were shown to play a central role in the stabilization of the inner core of the 6-helix bundle fold. In addition, the variable loops adopt a well-defined polyproline-II-type conformation (Squeglia et al., 2014).
Scl proteins in pathogenesis: ligand-binding specificities in human tissue and blood
The Scl1 variants of S. pyogenes bind a wide range of host ligands. The switch of Scl1 ligands between tissue and blood environments is highly significant, as described below (Fig. 2). In contrast, the Scl2 proteins failed to bind the majority of ligands and their role in pathogenesis is less understood.
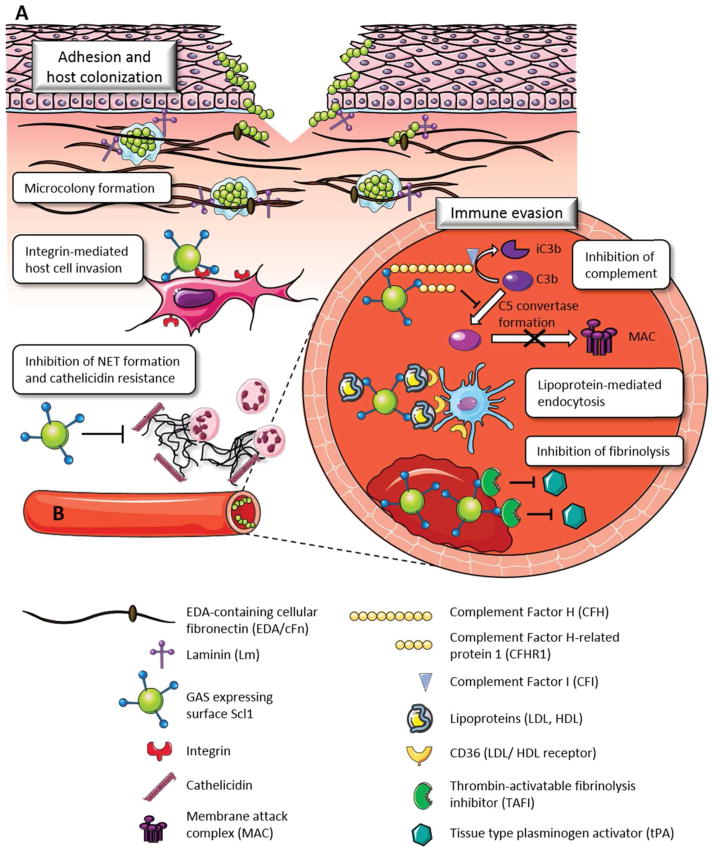
Fig. 2. Dichotomous nature of ligand binding by Scl1 in human tissue and blood.
A. Ligand binding in tissue. S. pyogenes accesses human tissue via portal of entry (skin, pharyngeal mucosa) represented here as a breach of skin epidermis extending into the underlying dermis. The globular domain of Scl1 surface adhesin selectively recognizes the EDA-containing isoforms of cellular fibronectin (EDA/cFn) and laminin (Lm). Scl1-mediated tissue microcolonies, embedded in glycocalyx, are formed and stabilized by interactions between Scl1 and the surrounding wound microenvironment. The collagenous domain of Scl1 directly binds human collagen receptors, integrins α2β1 and α11β1 on the host cell surface, promoting pathogen internalization and reemergence. Scl1 decreases the formation of extracellular traps (NETs) produced by infiltrating neutrophils and killing by NET-associated cathelicidin.
B. Ligand binding in blood. During dissemination to the blood and deeper tissue, Scl1 contributes to immune evasion and survival of S. pyogenes via several mechanisms. Binding to complement factor H (CFH) mediates cleavage of C3b by factor I, thus, preventing S. pyogenes opsonization, while binding to factor H-related protein 1 (CFHR1) prevents the formation of the C5 convertase and assembly of the membrane attack complex (MAC). Scl1-mediated adsorption of plasma lipoproteins LDL/HDL on the S. pyogenes surface may prevent immune recognition and/or promote LDL/HDL receptor-mediated endocytosis, intracellular survival and reemergence. Scl1 binding to and activation of the thrombin-activatable fibrinolysis inhibitor (TAFI) prevents clot breakdown, providing a protective niche for streptococci.
Binding specificities in tissue to extracellular matrix and cellular receptors
Scl1 selectively binds cellular, but not plasma, fibronectin and laminin (Caswell et al., 2010). Both main forms of fibronectin are encoded by a single gene and contain a conserved structure, consisting of three regions of repeats, type I, II, and III (Ffrench-Constant, 1995, To & Midwood, 2011). However, cellular fibronectin (cFn) isoforms may include extra domains A (EDA) and B (EDB), as well as varying numbers of the variable V domain (Ffrench-Constant, 1995). Scl1 proteins specifically bind cFn via recognition of the type III repeat, EDA (Oliver-Kozup et al., 2013). EDA/cFn isoforms are found in low levels in normal adult tissue but are upregulated in wounded tissue (Ffrench-Constant et al., 1989, Jarnagin et al., 1994), where the EDA domain interacts with keratinocyte integrin receptors (Shinde et al., 2008) and is important in the wound healing processes (Singh et al., 2004). S. pyogenes strains may express multiple fibronectin-binding proteins, including SfbI/PrtF1, PrtF2, SOF, FbaB, SfbX, and Shr (reviewed in Yamaguchi et al., 2013) that bind the type I and type II repeats; thus, Scl1 binds cFn via unique mechanism different from other Fn-binding proteins of S. pyogenes.
The putative fibronectin-binding Scl6 proteins of S. equi and S. zooepidemicus, including FneC, E, and F, likely contribute to colonization of animal hosts. These three proteins contain N-terminal regions similar to the fibronectin-binding domains of the proteins FNE and FNEB of S. equi, previously shown to bind fibronectin from horse serum (Lannergård et al., 2005, Tiouajni et al., 2014). Interestingly, FNE and FNEB lack the collagen-like domain found in FneC, E, and F. It is possible the Scl6 proteins diverged from FNE/FNEB and acquired the collagenous domain for stability or to project the N-terminal ligand-binding domains in a favorable position for ligand interaction. Therefore, proteins FneC, E, and F that clustered together with the SclZ.6, 9, and 10 proteins are all predicted fibronectin-binding Scl6 proteins.
Laminin binding was observed for the same Scl1 variants that bind EDA/cFn isoforms (Caswell et al., 2010). The molecular basis for the recognition of both ligands is undetermined due to a lack of sequence similarity between EDA and laminin chains α, β, or γ. At least 15 laminin heterotrimers have been identified in human tissues and binding studies with individual laminins represent a technical challenge. Two laminin-binding proteins, Lbp and Shr, in S. pyogenes have been reported in addition to Scl1 (Terao et al., 2002, Fisher et al., 2008, Linke et al., 2009). Given the localization of laminins to basement membranes, Scl1 binding likely represents a relevant pathogenesis trait in S. pyogenes.
The Scl1.41 variant, expressed by M41-type strains, directly binds human collagen receptors α2β1 and α11β1 integrins through the GLPGER motif in the Scl1-collagenous domain (Humtsoe et al., 2005, Caswell et al., 2008a). A similar binding motif GF/LOGER (O represents hydroxyproline), as well as derived sequence motifs GR/AOGER and GASGER, were identified in human collagens as integrin-binding sites (Kim et al., 2005). In addition, the GAPGER and GKPGER motifs are found in SclZ/Scl3 proteins (Beres et al., 2008). The integrin binding motifs RGD and KGD that are found within the collagen triple helix, become active for binding during tissue remodeling (Pfaff et al., 1993). These integrin binding sites are also found in Scl2 and Scl3 proteins (Beres et al., 2008; Lukomski et al., unpublished data).
Significance of ligand-binding in tissue
S. pyogenes forms biofilms, or microcolonies, in infected tissue (Akiyama et al., 2003, Roberts et al., 2012). In vitro assays showed enhanced biofilms formed by strains of multiple M-types on extracellular matrix coatings, including fibronectin and collagens type I and IV (Lembke et al., 2006, Courtney et al., 2009). Scl1 plays an important role in biofilm formation, as isogenic scl1-inactivated mutants had significantly reduced overall biofilm biomass and decreased biofilm thickness (Oliver-Kozup et al., 2011). In addition, enhanced Scl1-mediated biofilms were observed on simple cFn and laminin coatings, as well as on complex extracellular matrix deposited by human dermal fibroblasts (Oliver-Kozup et al., 2013). Inhibition experiments employing EDA-derived peptide and anti-EDA mAb showed significantly reduced adherence of S. pyogenes cells to fibroblast-derived matrix, indicating the Scl1-EDA/cFn interaction supports bacterial adherence.
The M3-type S. pyogenes strains, which are particularly invasive to humans, lack Scl1 protein due to a null mutation within the CL-region coding sequence (Lukomski et al., 2000), and form insignificant biofilms (Oliver-Kozup et al., 2011). Restoration of full-length Scl1 on the surface of M3-type S. pyogenes restored biofilm formation on cFn and laminin coatings (Bachert et al., 2016). Also, biofilm-capable Scl1-expressing M41-type strain produced glycocalyx-embedded tissue microcolonies in an in vitro skin equivalent infection model, while biofilm-poor Scl1-lacking M3 strain did not. Rare M3 strains containing the “scl1.3 carrier allele”, with restored open reading frame, were less invasive in a mouse model of necrotizing fasciitis than typical M3 strains lacking Scl1 (Flores et al., 2015). Scl1-deficient mutants of M28- and M41-type S. pyogenes, displayed increased lesion size in mice (Bachert et al., 2016). Altogether, these reports support the concept that S. pyogenes adherence to extracellular matrix and stable biofilm formation, conferred by surface-attached Scl1, promote localized infection over invasive spread.
The fibronectin-binding Scl6 proteins of S. equi with similarity to FNE and FNEB likely exhibit a different mechanism of fibronectin binding than Scl1 of S. pyogenes. The fibronectin-binding domain of Scl6 in S. equi was similar to that found in prtF1 of S. pyogenes and FnBPA of S. aureus (Lannergård et al., 2005). This mechanism of binding, which involves the N-terminal type I repeats of fibronectin, may also mediate molecular bridging to integrins on host cells, thus promoting bacterial internalization, as demonstrated for streptococci and staphylococci (Ozeri et al., 1998, Sinha et al., 1999). We suggest that Scl6 proteins mediate the adherence and cellular invasion of the zoonotic S. equi and S. zooepidemicus pathogens.
The Scl5 proteins of S. pneumoniae and S. agalactiae, harboring the G5 domain are likely involved in adherence and biofilm formation. The G5 domain is associated with binding to N-acetylglucosamine and biofilm formation for a variety of proteins found in streptococcal and staphylococcal species (Bateman et al., 2005), as well as the mycobacterial protein, RpfB, which also contains a triple helix motif (Ruggiero et al., 2009, Ruggiero et al., 2016). Additionally, the FIVAR domain in PclA of S. pneumoniae, is also found in surface-associated proteins SasC of Staphylococcus aureus and Embp of S. epidermidis that promote fibronectin binding and biofilm formation (Schroeder et al., 2009, Christner et al., 2010). Indeed, PclA was demonstrated to contribute to adherence and invasion of nasopharyngeal and epithelial cells by S. pneumoniae (Paterson et al., 2008). Scl5-harboring viridans streptococci are major etiological agents of infective endocarditis, and fibronectin binding within endothelial sites or deposited on prosthetic valves represents an important pathogenicity factor in these organisms (Munro & Macrina, 1994).
Scl1 binding to the α2β1 integrin via GLPGER motif promoted fibroblast adhesion and spreading, and induced intracellular signaling typical of integrin pathway, thus, mimicking the functional role of human collagen. Direct binding of Scl1 to the α2β1 integrins promotes S. pyogenes internalization by epithelial cells, resulting in an increased pool of intracellular bacteria and increased level of pathogen re-emergence (Caswell et al., 2007). The α2β1 and α11β1 integrins are expressed by fibroblasts, endothelial, and epithelial cells (Zutter & Santoro, 1990, Popova et al., 2007), indicating Scl1 has the potential to adhere to and invade a variety of cell types in the human host. The GAPGER and GKPGER, and possibly RGD and KGD, motifs identified in Scl2, Scl3 and Scl6 proteins of S. pyogenes and S. zooepidemicus may be similarly involved in host cell signaling and internalization upon infecting human and animal hosts.
A recent study reported a novel role for Scl1 of M1 S. pyogenes in resistance to neutrophil extracellular traps (Dohrmann et al., 2014). The Scl1-deficient isogenic mutant produced smaller skin lesions in mice, compared to the parental wild-type strain. Neutrophils incubated with this mutant displayed increased extracellular trap formation, compared to wild-type strain, which was associated with increased killing by the phagocytic cells, implying Scl1 expression impaired neutrophil function in vitro. The mutant strain also showed increased sensitivity to mouse cathelicidin killing. However, scl1 of M1-type S. pyogenes was absent from the list of genes found to be required for survival in human blood, as assessed by functional screening of a transposon library (Le Breton et al., 2013). This indicates that function(s) conferred by Scl1 in the M1-type strains, may not be significant for immune evasion in the blood but play a more important role during tissue infection.
Ligand binding in blood
The Scl1 proteins from M6- and M55-type S. pyogenes bind the complement regulatory proteins factor H and factor H-related protein 1 via Scl1’s globular V domain (Caswell et al., 2008b). Factor H is composed of 20 short consensus repeats (SCRs) that facilitate ligand binding and co-factor function for complement factor I-mediated C3b degradation; the C-terminal SCR 18–20 are involved in binding to cell surfaces, while SCR 1–4 harbor the co-factor function (Rodríguez de Córdoba et al., 2004). Factor H-related protein 1 comprises five SCRs, of which the C-terminal SCR 3–5 share 99% sequence identity, and therefore surface-binding function, with the carboxyl-terminus of factor H. It inhibits the C5 convertase and the formation of the membrane attack complex (Józsi & Zipfel, 2008). The Scl1 binding site was mapped in the C-terminal SCR 19–20 of factor H and SCR 4 of factor H-related protein 1 (Reuter et al., 2010). Borrelia burgdorferi also expresses CRASP surface proteins with similar binding characteristics to Scl1 (Kraiczy et al., 2001a, Kraiczy et al., 2001b). Notably, several other known factor H-binding proteins of S. pyogenes, such as certain M and M-like proteins and Fba, bind to SCR7, as well as complement factor H-like protein (Reuter et al., 2010).
Recombinant Scl1 proteins derived from diverse M-types bind the apolipoprotein ApoB100 associated with low density lipoprotein (LDL) (Han et al., 2006a). Binding of rScl1 constructs to LDL was facilitated by the Scl1-globular V domain with binding affinities of KD values in the nanomolar range. Importantly, LDL from human plasma was absorbed by the wild-type cells of S. pyogenes but not by the scl1-inactivated mutant cells (Han et al., 2006a). A similar binding with ApoAI apolipoprotein associated with high density lipoprotein (HDL) was demonstrated for rScl1 construct derived from M41-type S. pyogenes (Gao et al., 2010). HDL binding was inhibited by low concentration of the nonionic detergent Tween20, suggesting hydrophobic interaction to both rScl1 protein and S. pyogenes cells.
Some recombinant Scl proteins also bind thrombin-activatable fibrinolysis inhibitor (TAFI) with KD values in the nanomolar range (Påhlman et al., 2007) and the binding site was mapped to residues 205–232 within the TAFI protein (Valls Seron et al., 2011). TAFI is a zinc-dependent procarboxypeptidase, which acts as an important fibrinolysis regulator and inflammatory mediator upon activation by thrombin, thrombin-thrombomodulin complex, and plasmin.
Significance of Scl1 ligand-binding in blood
Diverse pathogens express surface proteins that bind complement regulatory proteins, including Neisseria gonorrhoeae, Borrelia burgdorferi, Yersinia enterocolitica, and several streptococcal species (Zipfel et al., 2002). Both factor H and factor H-related protein 1 bound to M6-S. pyogenes cells. Factor H binding to the cell surface prevents C3b deposition and phagocytosis, as well as downstream complement-mediated cell lysis. In vitro, rScl1-bound factor H retained its co-factor function by mediating the proteolytic breakdown of C3b by complement factor I (Caswell et al., 2008b). Similarly, factor H-related protein 1 binding to rScl1 inhibited the formation of the membrane attack complex in vitro (Reuter et al., 2010). However, factor H binding by M5 S. pyogenes did not contribute significantly to phagocytosis resistance or virulence (Gustafsson et al., 2013). Since M6 protein was the first bacterial factor H-binding protein reported (Horstmann et al., 1988), the combined factor H binding by Scl1 and M6 proteins might be necessary for the optimal anti-phagocytic phenotype of M6 S. pyogenes.
Plasma lipoproteins have been increasingly recognized as innate immune components. For example, HDL and LDL neutralize LPS endotoxin (Wurfel et al., 1994) and Staphylococcus aureus α-toxin (Bhakdi et al., 1983), whereas HDL is known to downregulate host adhesion molecules and inflammatory cytokines (Murch et al., 2007). Elevated lipoprotein levels may be protective against bacterial infections and sepsis in humans (Ravnskov, 2003), and LDL-deficient mice showed increased susceptibility to infections with Gram-negative bacteria and Candida albicans (Netea et al., 1996, Netea et al., 1997). Interaction between Yersinia pestis and ApoB-containing lipoproteins prevented binding of the bacteria to macrophages in vitro (Makoveichuk et al., 2003). In contrast, LDL and HDL could act as opsonins to increase S. pyogenes phagocytosis and killing via a CD36-mediated endocytosis (Liu et al., 2015, Zhou et al., 2016). Notably, about half of S. pyogenes strains express serum opacity factor disrupting the HDL structure (Courtney et al., 2006), which could protect S. pyogenes from lipoprotein-mediated opsonization. Clearly, the Scl1-lipoprotein interactions may have multiple functions during infection and in vivo studies are required to determine the effects of these interactions on the host.
Thrombin-activatable fibrinolysis inhibitor recruited by Scl proteins to the S. pyogenes cell surface was cleaved and activated by plasmin and thrombin-thrombomodulin (Påhlman et al., 2007). TAFI regulates inflammation by cleaving the C-terminal residues of bradykinin, osteopontin, and the chemoattractants C3a, C5a, and chemerin (Plug & Meijers, 2016). It functions by removing exposed C-terminal lysine residues from fibrin during blood clot formation, thereby preventing recognition and cleavage of these residues by the tissue-type plasminogen activator, ultimately inhibiting fibrinolysis or the breakdown of clots (Bajzar, 2000). Therefore, Scl-TAFI interaction may represent a mechanism for bacteria to remain associated with the fibrin clot and evade recognition by immune defenses. Additionally, activation of TAFI on the S. pyogenes cell surface induced inflammation via modulation of the kallikrein/kinin system (Bengtson et al., 2009).
Final Remarks
Modular collagens evolved in higher eukaryotes as members of extracellular matrix to support tissue structure and provide an essential network for cell function. Bacterial collagens likely emerged by convergent evolution via simple Gly-X-Y-repeat amplification.
Separate clustering of Scl proteins in streptococci infecting humans or animal species suggests co-evolution of Scl proteins with respective hosts, while sequence similarity amongst Scl proteins suggests horizontal gene transfer between closely related species.
All Scl proteins are predicted to be homotrimeric and surface attached. The amino-terminal globular “sensing” domain is variable in primary sequence but displays a conserved structure, while the C-terminal rod-shaped collagenous domain is variable in length and Gly-X-Y composition.
The Scl1 proteins sense distinct host environments, and have adapted to bind ligands in both tissue and blood to mediate S. pyogenes colonization and immune evasion.
Multiple Scl proteins, found in human pathogens S. pyogenes, S. pneumoniae, and S. agalactiae as well as horse pathogens S. equi and S. zooepidemicus, have adapted fibronectin-binding domains and integrin-binding motifs for augmenting colonization of human and animal hosts. Several of these proteins also mediate biofilm formation.
Table 1.
Scl classification by CLANS analysis
| Cluster No. | Proteins identified | Organisms* |
|---|---|---|
| 1 | Scl1 (SclA) | S. pyogenes (GAS) |
| 2 | Scl2 (SclB) | S. pyogenes (GAS) |
| 3 | SclC, D, E, G, H, I SclZ.1–5, 7, 12 |
S. equi, S. zooepidemicus (GCS) |
| 4 | SclF | S. equi, S. zooepidemicus (GCS) |
| 5 | PclA | S. pneumoniae, S. agalactiae (GBS) |
| 6 | SclZ.6, 9, 10 FneC, E, F |
S. equi, S. zooepidemicus (GCS) |
| 7 | Phage minor structural protein | S. pneumoniae, S. agalactiae (GBS) |
| 8a/b | Phage-associated hyaluronidase | S. pyogenes (GAS), S. equi and S. dysgalactiae (GCS) |
Abbreviations in parentheses refer to group A, B, and C Streptococcus
Acknowledgments
We are indebted to Drs. Janusz M. Bujnicki and Marcin Pawlowski for performing CLANS analysis and helpful discussions. We also thank Dr. Nyles Charon and members of Lukomski’s laboratory for comments on the manuscript. The authors declare no conflict of interest. This work was supported by National Institutes of Health Grants AI50666 and AI083683 (to S.L.). B.B. was supported by the NSF-EPSCoR Graduate Fellowship Program under the Research Infrastructure Improvement (RII) Track-1 award, Cooperative agreement 1003907 and was also a recipient of the Dr. Jennifer Gossling Scholarship in Microbiology.
References
- Akiyama H, Morizane S, Yamasaki O, Oono T, Iwatsuki K. Assessment of Streptococcus pyogenes microcolony formation in infected skin by confocal laser scanning microscopy. J Dermatol Sci. 2003;32:193–199. doi: 10.1016/s0923-1811(03)00096-3. [DOI] [PubMed] [Google Scholar]
- Almengor AC, McIver KS. Transcriptional activation of sclA by Mga requires a distal binding site in Streptococcus pyogenes. J Bacteriol. 2004;186:7847–7857. doi: 10.1128/JB.186.23.7847-7857.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almengor AC, Walters MS, McIver KS. Mga is sufficient to activate transcription in vitro of sof-sfbX and other Mga-regulated virulence genes in the group A Streptococcus. J Bacteriol. 2006;188:2038–2047. doi: 10.1128/JB.188.6.2038-2047.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachert BA, Choi SJ, LaSala PR, Harper TL, McNitt DH, Boehm DT, Caswell CC, Ciborowski P, Keene DR, Flores AR, Musser JM, Squeglia F, Marasco D, Berisio R, Lukomski S. Unique footprint in the scl1.3 locus affects adhesion and biofilm formation of the invasive M3-type group A Streptococcus. Front Cell Infect Microbiol. 2016;6:90. doi: 10.3389/fcimb.2016.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachert BA, Choi SJ, Snyder AK, Rio RVM, Durney BC, Holland LA, Amemiya K, Welkos SL, Bozue JA, Cote CK, Berisio R, Lukomski S. A unique set of the Burkholderia collagen-like proteins provides insight into pathogenesis, genome evolution and niche adaptation, and infection detection. PLoS ONE. 2015;10:e0137578. doi: 10.1371/journal.pone.0137578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajzar L. Thrombin activatable fibrinolysis inhibitor and an antifibrinolytic pathway. Arterioscler Thromb Vasc Biol. 2000;20:2511–2518. doi: 10.1161/01.atv.20.12.2511. [DOI] [PubMed] [Google Scholar]
- Bateman A, Holden MT, Yeats C. The G5 domain: a potential N-acetylglucosamine recognition domain involved in biofilm formation. Bioinformatics. 2005;21:1301–1303. doi: 10.1093/bioinformatics/bti206. [DOI] [PubMed] [Google Scholar]
- Bella J. Collagen structure: new tricks from a very old dog. Biochem J. 2016;473:1001–1025. doi: 10.1042/BJ20151169. [DOI] [PubMed] [Google Scholar]
- Bengtson SH, Sanden C, Mörgelin M, Marx PF, Olin AI, Leeb-Lundberg LM, Meijers JC, Herwald H. Activation of TAFI on the surface of Streptococcus pyogenes evokes inflammatory reactions by modulating the kallikrein/kinin system. J Innate Immun. 2009;1:18–28. doi: 10.1159/000145543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beres S, Sesso R, Pinto S, Hoe N, Porcella S, Deleo F, Musser J. Genome sequence of a lancefield group C Streptococcus zooepidemicus strain causing epidemic nephritis: new information about an old disease. PLoS ONE. 2008;3:e3026. doi: 10.1371/journal.pone.0003026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berisio R, Vitagliano L. Polyproline and triple helix motifs in host-pathogen recognition. Curr Protein Pept Sci. 2012;13:855–865. doi: 10.2174/138920312804871157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berisio R, Vitagliano L, Mazzarella L, Zagari A. Crystal structure of the collagen triple helix model [(Pro-Pro-Gly)10]3. Protein Sci. 2002;11:262–270. doi: 10.1110/ps.32602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhakdi S, Tranum-Jensen J, Utermann G, Fussle R. Binding and partial inactivation of Staphylococcus aureus alpha-toxin by human plasma low density lipoprotein. J Biol Chem. 1983;258:5899–5904. [PubMed] [Google Scholar]
- Brodsky B, Persikov AV. Molecular structure of the collagen triple helix. Adv Protein Chem. 2005;70:301–339. doi: 10.1016/S0065-3233(05)70009-7. [DOI] [PubMed] [Google Scholar]
- Caswell CC, Barczyk M, Keene DR, Lukomska E, Gullberg DE, Lukomski S. Identification of the first prokaryotic collagen sequence motif that mediates binding to human collagen receptors, integrins α2β1 and α11β1. J Biol Chem. 2008a;283:36168–36175. doi: 10.1074/jbc.M806865200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell CC, Han R, Hovis KM, Ciborowski P, Keene DR, Marconi RT, Lukomski S. The Scl1 protein of M6-type group A Streptococcus binds the human complement regulatory protein, factor H, and inhibits the alternative pathway of complement. Mol Microbiol. 2008b;67:584–596. doi: 10.1111/j.1365-2958.2007.06067.x. [DOI] [PubMed] [Google Scholar]
- Caswell CC, Lukomska E, Seo NS, Höök M, Lukomski S. Scl1-dependent internalization of group A Streptococcus via direct interactions with the α2β1 integrin enhances pathogen survival and re-emergence. Mol Microbiol. 2007;64:1319–1331. doi: 10.1111/j.1365-2958.2007.05741.x. [DOI] [PubMed] [Google Scholar]
- Caswell CC, Oliver-Kozup H, Han R, Lukomska E, Lukomski S. Scl1, the multifunctional adhesin of group A Streptococcus, selectively binds cellular fibronectin and laminin, and mediates pathogen internalization by human cells. FEMS Microbiol Lett. 2010;303:61–68. doi: 10.1111/j.1574-6968.2009.01864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan VC, Ramshaw JA, Kirkpatrick A, Beck K, Brodsky B. Positional preferences of ionizable residues in Gly-X-Y triplets of the collagen triple-helix. J Biol Chem. 1997;272:31441–31446. doi: 10.1074/jbc.272.50.31441. [DOI] [PubMed] [Google Scholar]
- Chopra RK, Ananthanarayanan VS. Conformational implications of enzymatic proline hydroxylation in collagen. Proc Natl Acad Sci U S A. 1982;79:7180–7184. doi: 10.1073/pnas.79.23.7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christner M, Franke GC, Schommer NN, Wendt U, Wegert K, Pehle P, Kroll G, Schulze C, Buck F, Mack D, Aepfelbacher M, Rohde H. The giant extracellular matrix-binding protein of Staphylococcus epidermidis mediates biofilm accumulation and attachment to fibronectin. Mol Microbiol. 2010;75:187–207. doi: 10.1111/j.1365-2958.2009.06981.x. [DOI] [PubMed] [Google Scholar]
- Courtney HS, Ofek I, Penfound T, Nizet V, Pence MA, Kreikemeyer B, Podbielski A, Hasty DL, Dale JB. Relationship between expression of the family of M proteins and lipoteichoic acid to hydrophobicity and biofilm formation in Streptococcus pyogenes. PLoS ONE. 2009;4:e4166. doi: 10.1371/journal.pone.0004166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney HS, Zhang YM, Frank MW, Rock CO. Serum opacity factor, a streptococcal virulence factor that binds to apolipoproteins A-I and A-II and disrupts high density lipoprotein structure. J Biol Chem. 2006;281:5515–5521. doi: 10.1074/jbc.M512538200. [DOI] [PubMed] [Google Scholar]
- Dohrmann S, Anik S, Olson J, Anderson EL, Etesami N, No H, Snipper J, Nizet V, Okumura CY. Role for streptococcal collagen-like protein 1 in M1T1 group A Streptococcus resistance to neutrophil extracellular traps. Infect Immun. 2014;82:4011–4020. doi: 10.1128/IAI.01921-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxey AC, McConkey BJ. Prediction of molecular mimicry candidates in human pathogenic bacteria. Virulence. 2013;4:453–466. doi: 10.4161/viru.25180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan C, Prashar A, So J, Tang P, Low DE, Terebiznik M, Guyard C. Lcl of Legionella pneumophila is an immunogenic GAG binding adhesin that promotes interactions with lung epithelial cells and plays a crucial role in biofilm formation. Infect Immun. 2011;79:2168–2181. doi: 10.1128/IAI.01304-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel J. Versatile collagens in invertebrates. Science. 1997;277:1785–1786. doi: 10.1126/science.277.5333.1785. [DOI] [PubMed] [Google Scholar]
- Exposito JY, Cluzel C, Garrone R, Lethias C. Evolution of collagens. Anat Rec. 2002;268:302–316. doi: 10.1002/ar.10162. [DOI] [PubMed] [Google Scholar]
- Ffrench-Constant C. Alternative splicing of fibronectin--many different proteins but few different functions. Exp Cell Res. 1995;221:261–271. doi: 10.1006/excr.1995.1374. [DOI] [PubMed] [Google Scholar]
- Ffrench-Constant C, Van De Water L, Dvorak HF, Hynes RO. Reappearance of an embryonic pattern of fibronectin splicing during wound healing in the adult rat. J Cell Biol. 1989;109:903–914. doi: 10.1083/jcb.109.2.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher M, Huang YS, Li X, McIver KS, Toukoki C, Eichenbaum Z. Shr is a broad-spectrum surface receptor that contributes to adherence and virulence in group A Streptococcus. Infect Immun. 2008;76:5006–5015. doi: 10.1128/IAI.00300-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flock M, Karlström Å, Lannergård J, Guss B, Flock JI. Protective effect of vaccination with recombinant proteins from Streptococcus equi subspecies equi in a strangles model in the mouse. Vaccine. 2006;24:4144–4151. doi: 10.1016/j.vaccine.2006.02.016. [DOI] [PubMed] [Google Scholar]
- Flores AR, Jewell BE, Versalovic EM, Olsen RJ, Bachert BA, Lukomski S, Musser JM. Natural variant of collagen-like protein A in serotype M3 group A Streptococcus increases adherence and decreases invasive potential. Infect Immun. 2015;83:1122–1129. doi: 10.1128/IAI.02860-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frickey T, Lupas A. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics. 2004;20:3702–3704. doi: 10.1093/bioinformatics/bth444. [DOI] [PubMed] [Google Scholar]
- Gao Y, Liang C, Zhao R, Lukomski S, Han R. The Scl1 of M41-type group A Streptococcus binds the high-density lipoprotein. FEMS Microbiol Lett. 2010;309:55–61. doi: 10.1111/j.1574-6968.2010.02013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guss B, Flock M, Frykberg L, Waller AS, Robinson C, Smith KC, Flock JI. Getting to grips with strangles: an effective multi-component recombinant vaccine for the protection of horses from Streptococcus equi infection. PLoS Pathog. 2009;5:e1000584. doi: 10.1371/journal.ppat.1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson MC, Lannergård J, Nilsson OR, Kristensen BM, Olsen JE, Harris CL, Ufret-Vincenty RL, Stalhammar-Carlemalm M, Lindahl G. Factor H binds to the hypervariable region of many Streptococcus pyogenes M proteins but does not promote phagocytosis resistance or acute virulence. PLoS Pathog. 2013;9:e1003323. doi: 10.1371/journal.ppat.1003323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han R, Caswell CC, Lukomska E, Keene DR, Pawlowski M, Bujnicki JM, Kim JK, Lukomski S. Binding of the low-density lipoprotein by streptococcal collagen-like protein Scl1 of Streptococcus pyogenes. Mol Microbiol. 2006a;61:351–367. doi: 10.1111/j.1365-2958.2006.05237.x. [DOI] [PubMed] [Google Scholar]
- Han R, Zwiefka A, Caswell CC, Xu Y, Keene DR, Lukomska E, Zhao Z, Höök M, Lukomski S. Assessment of prokaryotic collagen-like sequences derived from streptococcal Scl1 and Scl2 proteins as a source of recombinant GXY polymers. Appl Microbiol Biotechnol. 2006b;72:109–115. doi: 10.1007/s00253-006-0387-5. [DOI] [PubMed] [Google Scholar]
- Horstmann RD, Sievertsen HJ, Knobloch J, Fischetti VA. Antiphagocytic activity of streptococcal M protein: selective binding of complement control protein factor H. Proc Natl Acad Sci U S A. 1988;85:1657–1661. doi: 10.1073/pnas.85.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humtsoe JO, Kim JK, Xu Y, Keene DR, Höök M, Lukomski S, Wary KK. A streptococcal collagen-like protein interacts with the α2β1 integrin and induces intracellular signaling. J Biol Chem. 2005;280:13848–13857. doi: 10.1074/jbc.M410605200. [DOI] [PubMed] [Google Scholar]
- Jarnagin WR, Rockey DC, Koteliansky VE, Wang SS, Bissell DM. Expression of variant fibronectins in wound healing: cellular source and biological activity of the EIIIA segment in rat hepatic fibrogenesis. J Cell Biol. 1994;127:2037–2048. doi: 10.1083/jcb.127.6.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Józsi M, Zipfel PF. Factor H family proteins and human diseases. Trends Immunol. 2008;29:380–387. doi: 10.1016/j.it.2008.04.008. [DOI] [PubMed] [Google Scholar]
- Karlström Å, Jacobsson K, Flock M, Flock J, Guss B. Identification of a novel collagen-like protein, SclC, in Streptococcus equi using signal sequence phage display. Vet Microbiol. 2004;104:179–188. doi: 10.1016/j.vetmic.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Karlström Å, Jacobsson K, Guss B. SclC is a member of a novel family of collagen-like proteins in Streptococcus equi subspecies equi that are recognised by antibodies against SclC. Vet Microbiol. 2006;114:72–81. doi: 10.1016/j.vetmic.2005.10.036. [DOI] [PubMed] [Google Scholar]
- Kim J, Xu Y, Xu X, Keene D, Gurusiddappa S, Liang X, Wary K, Höök M. A novel binding site in collagen type III for the integrins, α1β1 and α2β1. J Biol Chem. 2005;280:32512–32520. doi: 10.1074/jbc.M502431200. [DOI] [PubMed] [Google Scholar]
- Kraiczy P, Skerka C, Brade V, Zipfel PF. Further characterization of complement regulator-acquiring surface proteins of Borrelia burgdorferi. Infect Immun. 2001a;69:7800–7809. doi: 10.1128/IAI.69.12.7800-7809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraiczy P, Skerka C, Kirschfink M, Brade V, Zipfel PF. Immune evasion of Borrelia burgdorferi by acquisition of human complement regulators FHL-1/reconectin and Factor H. Eur J Immunol. 2001b;31:1674–1684. doi: 10.1002/1521-4141(200106)31:6<1674::aid-immu1674>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Kramer JM, Cox GN, Hirsh D. Comparisons of the complete sequences of two collagen genes from Caenorhabditis elegans. Cell. 1982;30:599–606. doi: 10.1016/0092-8674(82)90256-2. [DOI] [PubMed] [Google Scholar]
- Lannergård J. Microbiology. Uppsala: Swedish University of Agricultural Sciences; 2006. Potentially virulence-related extracellular proteins of Streptococcus equi; pp. 32–34. [Google Scholar]
- Lannergård J, Flock M, Johansson S, Flock JI, Guss B. Studies of fibronectin-binding proteins of Streptococcus equi. Infect Immun. 2005;73:7243–7251. doi: 10.1128/IAI.73.11.7243-7251.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Breton Y, Mistry P, Valdes KM, Quigley J, Kumar N, Tettelin H, McIver KS. Genome-wide identification of genes required for fitness of group A Streptococcus in human blood. Infect Immun. 2013;81:862–875. doi: 10.1128/IAI.00837-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leikina E, Mertts MV, Kuznetsova N, Leikin S. Type I collagen is thermally unstable at body temperature. Proc Natl Acad Sci U S A. 2002;99:1314–1318. doi: 10.1073/pnas.032307099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lembke C, Podbielski A, Hidalgo-Grass C, Jonas L, Hanski E, Kreikemeyer B. Characterization of biofilm formation by clinically relevant serotypes of group A streptococci. Appl Environ Microbiol. 2006;72:2864–2875. doi: 10.1128/AEM.72.4.2864-2875.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke C, Caradoc-Davies TT, Young PG, Proft T, Baker EN. The laminin-binding protein Lbp from Streptococcus pyogenes is a zinc receptor. J Bacteriol. 2009;191:5814–5823. doi: 10.1128/JB.00485-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Zhou L, Li Y, Bai W, Liu N, Li W, Gao Y, Liu Z, Han R. High-density lipoprotein acts as an opsonin to enhance phagocytosis of group A Streptococcus by U937 cells. Microbiol Immunol. 2015;59:419–425. doi: 10.1111/1348-0421.12270. [DOI] [PubMed] [Google Scholar]
- Lukomski S, Nakashima K, Abdi I, Cipriano VJ, Ireland RM, Reid SD, Adams GG, Musser JM. Identification and characterization of the scl gene encoding a group A Streptococcus extracellular protein virulence factor with similarity to human collagen. Infect Immun. 2000;68:6542–6553. doi: 10.1128/iai.68.12.6542-6553.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukomski S, Nakashima K, Abdi I, Cipriano VJ, Shelvin BJ, Graviss EA, Musser JM. Identification and characterization of a second extracellular collagen-like protein made by group A Streptococcus: control of production at the level of translation. Infect Immun. 2001;69:1729–1738. doi: 10.1128/IAI.69.3.1729-1738.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makoveichuk E, Cherepanov P, Lundberg P, Lundberg S, Forsberg A, Olivecrona G. pH6 antigen of Yersinia pestis interacts with plasma lipoproteins and cell membranes. J Lipid Res. 2003;44:320–330. doi: 10.1194/jlr.M200182-JLR200. [DOI] [PubMed] [Google Scholar]
- Mohs A, Silva T, Yoshida T, Amin R, Lukomski S, Inouye M, Brodsky B. Mechanism of stabilization of a bacterial collagen triple helix in the absence of hydroxyproline. J Biol Chem. 2007;282:29757–29765. doi: 10.1074/jbc.M703991200. [DOI] [PubMed] [Google Scholar]
- Munro C, Macrina FL. Molecular pathogenesis of viridans streptococcal endocarditis. In: Kado CI, Crosa JH, editors. Molecular Mechanisms of Bacterial Virulence. Dordrecht: Springer Netherlands; 1994. pp. 249–265. [Google Scholar]
- Murch O, Collin M, Hinds CJ, Thiemermann C. Lipoproteins in inflammation and sepsis. I. Basic science. Intensive Care Med. 2007;33:13–24. doi: 10.1007/s00134-006-0432-y. [DOI] [PubMed] [Google Scholar]
- Netea MG, Demacker PN, de Bont N, Boerman OC, Stalenhoef AF, van der Meer JW, Kullberg BJ. Hyperlipoproteinemia enhances susceptibility to acute disseminated Candida albicans infection in low-density-lipoprotein-receptor-deficient mice. Infect Immun. 1997;65:2663–2667. doi: 10.1128/iai.65.7.2663-2667.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea MG, Demacker PN, Kullberg BJ, Boerman OC, Verschueren I, Stalenhoef AF, van der Meer JW. Low-density lipoprotein receptor-deficient mice are protected against lethal endotoxemia and severe gram-negative infections. J Clin Invest. 1996;97:1366–1372. doi: 10.1172/JCI118556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver-Kozup H, Martin KH, Schwegler-Berry D, Green BJ, Betts C, Shinde AV, Van De Water L, Lukomski S. The group A streptococcal collagen-like protein-1, Scl1, mediates biofilm formation by targeting the extra domain A-containing variant of cellular fibronectin expressed in wounded tissue. Mol Microbiol. 2013;87:672–689. doi: 10.1111/mmi.12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver-Kozup HA, Elliott M, Bachert BA, Martin KH, Reid SD, Schwegler-Berry DE, Green BJ, Lukomski S. The streptococcal collagen-like protein-1 (Scl1) is a significant determinant for biofilm formation by group A Streptococcus. BMC Microbiol. 2011;11:262. doi: 10.1186/1471-2180-11-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozeri V, Rosenshine I, Mosher DF, Fassler R, Hanski E. Roles of integrins and fibronectin in the entry of Streptococcus pyogenes into cells via protein F1. Mol Microbiol. 1998;30:625–637. doi: 10.1046/j.1365-2958.1998.01097.x. [DOI] [PubMed] [Google Scholar]
- Påhlman LI, Marx PF, Mörgelin M, Lukomski S, Meijers JC, Herwald H. Thrombin-activatable fibrinolysis inhibitor binds to Streptococcus pyogenes by interacting with collagen-like proteins A and B. J Biol Chem. 2007;282:24873–24881. doi: 10.1074/jbc.M610015200. [DOI] [PubMed] [Google Scholar]
- Paterson GK, Nieminen L, Jefferies JMC, Mitchell TJ. PclA, a pneumococcal collagen-like protein with selected strain distribution, contributes to adherence and invasion of host cells. FEMS Microbiol Lett. 2008;285:170–176. doi: 10.1111/j.1574-6968.2008.01217.x. [DOI] [PubMed] [Google Scholar]
- Pfaff M, Aumailley M, Specks U, Knolle J, Zerwes HG, Timpl R. Integrin and Arg-Gly-Asp dependence of cell adhesion to the native and unfolded triple helix of collagen type VI. Exp Cell Res. 1993;206:167–176. doi: 10.1006/excr.1993.1134. [DOI] [PubMed] [Google Scholar]
- Pizarro-Guajardo M, Olguin-Araneda V, Barra-Carrasco J, Brito-Silva C, Sarker MR, Paredes-Sabja D. Characterization of the collagen-like exosporium protein, BclA1, of Clostridium difficile spores. Anaerobe. 2014;25:18–30. doi: 10.1016/j.anaerobe.2013.11.003. [DOI] [PubMed] [Google Scholar]
- Plug T, Meijers JC. Structure-function relationships in thrombin-activatable fibrinolysis inhibitor. J Thromb Haemost. 2016;14:633–644. doi: 10.1111/jth.13261. [DOI] [PubMed] [Google Scholar]
- Popova SN, Lundgren-Akerlund E, Wiig H, Gullberg D. Physiology and pathology of collagen receptors. Acta Physiol (Oxf) 2007;190:179–187. doi: 10.1111/j.1748-1716.2007.01718.x. [DOI] [PubMed] [Google Scholar]
- Ramachandran GN. Stereochemistry of collagen. Int J Pept Protein Res. 1988;31:1–16. [PubMed] [Google Scholar]
- Rasmussen M, Björck L. Unique regulation of SclB - a novel collagen-like surface protein of Streptococcus pyogenes. Mol Microbiol. 2001;40:1427–1438. doi: 10.1046/j.1365-2958.2001.02493.x. [DOI] [PubMed] [Google Scholar]
- Rasmussen M, Eden A, Björck L. SclA, a novel collagen-like surface protein of Streptococcus pyogenes. Infect Immun. 2000;68:6370–6377. doi: 10.1128/iai.68.11.6370-6377.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen M, Jacobsson M, Björck L. Genome-based identification and analysis of collagen-related structural motifs in bacterial and viral proteins. J Biol Chem. 2003;278:32313–32316. doi: 10.1074/jbc.M304709200. [DOI] [PubMed] [Google Scholar]
- Ravnskov U. High cholesterol may protect against infections and atherosclerosis. QJM Mon J Assoc Phys. 2003;96:927–934. doi: 10.1093/qjmed/hcg150. [DOI] [PubMed] [Google Scholar]
- Reuter M, Caswell CC, Lukomski S, Zipfel PF. Binding of the human complement regulators CFHR1 and factor H by streptococcal collagen-like protein 1 (Scl1) via their conserved C termini allows control of the complement cascade at multiple levels. J Biol Chem. 2010;285:38473–38485. doi: 10.1074/jbc.M110.143727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AL, Connolly KL, Kirse DJ, Evans AK, Poehling KA, Peters TR, Reid SD. Detection of group A Streptococcus in tonsils from pediatric patients reveals high rate of asymptomatic streptococcal carriage. BMC Pediatr. 2012;12:3. doi: 10.1186/1471-2431-12-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez de Córdoba S, Esparza-Gordillo J, Goicoechea de Jorge E, Lopez-Trascasa M, Sanchez-Corral P. The human complement factor H: functional roles, genetic variations and disease associations. Mol Immunol. 2004;41:355–367. doi: 10.1016/j.molimm.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Ruggiero A, Squeglia F, Romano M, Vitagliano L, De Simone A, Berisio R. Structure and dynamics of the multi-domain resuscitation promoting factor RpfB from Mycobacterium tuberculosis. J Biomol Struct Dyn. 2016:1–9. doi: 10.1080/07391102.2016.1182947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiero A, Tizzano B, Pedone E, Pedone C, Wilmanns M, Berisio R. Crystal structure of the resuscitation-promoting factor (DeltaDUF)RpfB from M. tuberculosis. J Mol Biol. 2009;385:153–162. doi: 10.1016/j.jmb.2008.10.042. [DOI] [PubMed] [Google Scholar]
- Schroeder K, Jularic M, Horsburgh SM, Hirschhausen N, Neumann C, Bertling A, Schulte A, Foster S, Kehrel BE, Peters G, Heilmann C. Molecular characterization of a novel Staphylococcus aureus surface protein (SasC) involved in cell aggregation and biofilm accumulation. PLoS ONE. 2009;4:e7567. doi: 10.1371/journal.pone.0007567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewmaker PL, Camus AC, Bailiff T, Steigerwalt AG, Morey RE, da Carvalho MG. Streptococcus ictaluri sp. nov., isolated from Channel Catfish Ictalurus punctatus broodstock. Int J Syst Evol Microbiol. 2007;57:1603–1606. doi: 10.1099/ijs.0.64810-0. [DOI] [PubMed] [Google Scholar]
- Shinde AV, Bystroff C, Wang C, Vogelezang MG, Vincent PA, Hynes RO, Van De Water L. Identification of the peptide sequences within the EIIIA (EDA) segment of fibronectin that mediate integrin α9β1-dependent cellular activities. J Biol Chem. 2008;283:2858–2870. doi: 10.1074/jbc.M708306200. [DOI] [PubMed] [Google Scholar]
- Singh P, Reimer CL, Peters JH, Stepp MA, Hynes RO, Van De Water L. The spatial and temporal expression patterns of integrin α9β1 and one of its ligands, the EIIIA segment of fibronectin, in cutaneous wound healing. J Invest Dermatol. 2004;123:1176–1181. doi: 10.1111/j.0022-202X.2004.23485.x. [DOI] [PubMed] [Google Scholar]
- Sinha B, Francois PP, Nusse O, Foti M, Hartford OM, Vaudaux P, Foster TJ, Lew DP, Herrmann M, Krause KH. Fibronectin-binding protein acts as Staphylococcus aureus invasin via fibronectin bridging to integrin α5β1. Cell Microbiol. 1999;1:101–117. doi: 10.1046/j.1462-5822.1999.00011.x. [DOI] [PubMed] [Google Scholar]
- Squeglia F, Bachert B, De Simone A, Lukomski S, Berisio R. The crystal structure of the streptococcal collagen-like protein 2 globular domain from invasive M3-type group A Streptococcus shows significant similarity to immunomodulatory HIV protein gp41. J Biol Chem. 2014;289:5122–5133. doi: 10.1074/jbc.M113.523597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squeglia F, Bachert B, Romano M, Lukomski S, Berisio R. Crystallization and preliminary X-ray crystallographic analysis of the variable domain of Scl2.3, a streptococcal collagen-like protein from invasive M3-type Streptococcus pyogenes. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013;69:1023–1025. doi: 10.1107/S174430911302068X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhre MH, Gertz M, Steegborn C, Scheibel T. Structural and functional features of a collagen-binding matrix protein from the mussel byssus. Nat Commun. 2014;5:3392. doi: 10.1038/ncomms4392. [DOI] [PubMed] [Google Scholar]
- Sylvestre P, Couture-Tosi E, Mock M. A collagen-like surface glycoprotein is a structural component of the Bacillus anthracis exosporium. Mol Microbiol. 2002;45:169–178. doi: 10.1046/j.1365-2958.2000.03000.x. [DOI] [PubMed] [Google Scholar]
- Terao Y, Kawabata S, Kunitomo E, Nakagawa I, Hamada S. Novel laminin-binding protein of Streptococcus pyogenes, Lbp, is involved in adhesion to epithelial cells. Infect Immun. 2002;70:993–997. doi: 10.1128/iai.70.2.993-997.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timoney JF. The pathogenic equine streptococci. Vet Res. 2004;35:397–409. doi: 10.1051/vetres:2004025. [DOI] [PubMed] [Google Scholar]
- Tiouajni M, Durand D, Blondeau K, Graille M, Urvoas A, Valerio-Lepiniec M, Guellouz A, Aumont-Nicaise M, Minard P, van Tilbeurgh H. Structural and functional analysis of the fibronectin-binding protein FNE from Streptococcus equi spp. equi. FEBS J. 2014;281:5513–5531. doi: 10.1111/febs.13092. [DOI] [PubMed] [Google Scholar]
- To WS, Midwood KS. Plasma and cellular fibronectin: distinct and independent functions during tissue repair. Fibrogenesis Tissue Repair. 2011;4:21. doi: 10.1186/1755-1536-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valls Seron M, Plug T, Marquart JA, Marx PF, Herwald H, de Groot PG, Meijers JC. Binding characteristics of thrombin-activatable fibrinolysis inhibitor to streptococcal surface collagen-like proteins A and B. J Thromb Haemost. 2011;106:609–616. doi: 10.1160/TH11-03-0204. [DOI] [PubMed] [Google Scholar]
- Vitagliano L, Berisio R, Mazzarella L, Zagari A. Structural bases of collagen stabilization induced by proline hydroxylation. Biopolymers. 2001;58:459–464. doi: 10.1002/1097-0282(20010415)58:5<459::AID-BIP1021>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Waller A, Flock M, Smith K, Robinson C, Mitchell Z, Karlström Å, Lannergård J, Bergman R, Guss B, Flock JI. Vaccination of horses against strangles using recombinant antigens from Streptococcus equi. Vaccine. 2007;25:3629–3635. doi: 10.1016/j.vaccine.2007.01.060. [DOI] [PubMed] [Google Scholar]
- Whatmore AM. Streptococcus pyogenes sclB encodes a putative hypervariable surface protein with a collagen-like repetitive structure. Microbiology. 2001;147:419–429. doi: 10.1099/00221287-147-2-419. [DOI] [PubMed] [Google Scholar]
- Wurfel MM, Kunitake ST, Lichenstein H, Kane JP, Wright SD. Lipopolysaccharide (LPS)-binding protein is carried on lipoproteins and acts as a cofactor in the neutralization of LPS. J Exp Med. 1994;180:1025–1035. doi: 10.1084/jem.180.3.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Yu Z, Inouye M, Brodsky B, Mirochnitchenko O. Expanding the family of collagen proteins: recombinant bacterial collagens of varying composition form triple-helices of similar stability. Biomacromolecules. 2010;11:348–356. doi: 10.1021/bm900894b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Keene DR, Bujnicki JM, Höök M, Lukomski S. Streptococcal Scl1 and Scl2 proteins form collagen-like triple helices. J Biol Chem. 2002;277:27312–27318. doi: 10.1074/jbc.M201163200. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Terao Y, Kawabata S. Pleiotropic virulence factor - Streptococcus pyogenes fibronectin-binding proteins. Cell Microbiol. 2013;15:503–511. doi: 10.1111/cmi.12083. [DOI] [PubMed] [Google Scholar]
- Yu Z, Mirochnitchenko O, Xu C, Yoshizumi A, Brodsky B, Inouye M. Noncollagenous region of the streptococcal collagen-like protein is a trimerization domain that supports refolding of adjacent homologous and heterologous collagenous domains. Protein Sci. 2010;19:775–785. doi: 10.1002/pro.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zbinden A, Bostanci N, Belibasakis GN. The novel species Streptococcus tigurinus and its association with oral infection. Virulence. 2015;6:177–182. doi: 10.4161/21505594.2014.970472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Liu L, Yang J, Li Y, Bai W, Liu N, Li W, Gao Y, Xu L, Liu Z, Han R. LDL acts as an opsonin enhancing the phagocytosis of group A Streptococcus by monocyte and whole human blood. Med Microbiol Immunol. 2016;205:155–162. doi: 10.1007/s00430-015-0436-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel PF, Skerka C, Hellwage J, Jokiranta ST, Meri S, Brade V, Kraiczy P, Noris M, Remuzzi G. Factor H family proteins: on complement, microbes and human diseases. Biochem Soc Trans. 2002;30:971–978. doi: 10.1042/bst0300971. [DOI] [PubMed] [Google Scholar]
- Zutter MM, Santoro SA. Widespread histologic distribution of the α2β1 integrin cell-surface collagen receptor. Am J Pathol. 1990;137:113–120. [PMC free article] [PubMed] [Google Scholar]