Abstract
Alternative splicing (AS) of pre-mRNAs is one of the most important post-transcriptional regulations that enable a single gene to code for multiple proteins resulting in the biodiversity of proteins in eukaryotes. Recently, we have shown that an Arabidopsis thaliana RNA recognition motif-containing protein RBM25 is a novel splicing factor to modulate plant response to ABA during seed germination and post-germination through regulating HAB1 pre-mRNA AS. Here, we show that RBM25 is preferentially expressed in stomata and vascular tissues in Arabidopsis and is induced by ABA and abiotic stresses. Loss-of-function mutant is highly tolerant to drought and sensitive to salt stress. Bioinformatic analysis and expression assays reveal that Arabidopsis RBM25 is induced by multiple abiotic stresses, suggesting a crucial role of RBM25 in Arabidopsis responses to adverse environmental conditions. Furthermore, we provide a comprehensive characterization of the homologous genes of Arabidopsis RBM25 based on the latest plant genome sequences and public microarray databases. Fourteen homologous genes are identified in different plant species which show similar structure in gene and protein. Notably, the promoter analysis reveals that RBM25 homologs are likely controlled by the regulators involved in multiple plant growth and abiotic stresses, such as drought and unfavorable temperature. The comparative analysis of general and unique cis regulatory elements of the RBM25 homologs highlights the conserved and unique molecular processes that modulate plant response to abiotic stresses through RBM25-mediated alternative splicing.
Keywords: RBM25, splicing factor, Arabidopsis, drought, abiotic stresses
Introduction
As plants are essentially sessile in nature, they have to face various environmental stresses during life time. Drought and high salinity are two of the most important factors that greatly affect plant growth, yield, and distribution. Global warming increases the frequency and severity of extreme weather that exacerbate the effects of abiotic stresses (i.e., drought, cold, and heat stresses) on plants. To deal with these challenges, plants have evolved numerous adaptive strategies at physiological, biochemical and molecular levels in order to survive better (Broun, 2004; Bohnert et al., 2006). Plant stress responses are regulated by numerous gene regulatory networks at multiple levels. In the past decades, the extensive efforts have been made to understand how plants respond to abiotic stresses at transcriptional level. However, how plants respond to abiotic stresses at post-transcriptional level is less clear.
Pre-mRNA splicing is a process that removes the introns and joins exons to produce mature mRNA. It is one of the important post-transcriptional regulation events that control gene expression and plant response to developmental and environmental stresses (Reddy, 2007). Alternative splicing (AS) produces transcript isoforms that encode proteins with different subcellular localization, stability and functions (Chung et al., 2010; Kriechbaumer et al., 2012; Remy et al., 2013). It has been shown that more than 61% genes in Arabidopsis undergo AS in normal conditions and the percentage increases greatly under stress conditions (Marquez et al., 2012). AS is carried out in a large complex called spliceosome, where ribonucleoprotein particles (snRNPs) and numerous other snRNP-associated proteins are assembled precisely and orderly (Makarova et al., 2001). A large number of splicing factors are involved in pre-mRNA splicing and are necessary for generating mature mRNAs. Recent research has discovered that some plant splicing factors play important roles in plant development and stress adaptation. The plant specific splicing factor SR45 functions in various developmental processes. The sr45 mutant shows defects in flowering, leaf and root development (Ali et al., 2007). SUPPRESSOR OF abi3-5 (SUA) affects seed maturation by regulating AS of ABSCISIC ACID INSENSITIVE3 (ABI3) (Sugliani et al., 2010). Loss-of-function in STA1, a pre-mRNA splicing factor, caused defects in the splicing of the cold-induced COR15A (COLD-REGULATED 15A) and subsequent altered stress response of plants (Lee et al., 2006). Mutations in SR34B affect splicing of Iron-regulated Transporter 1 (IRT1) pre-mRNAs under Cd stress to reduce the IRT1 mRNA accumulation resulting in increased sensitivity of plants to Cd stress (Zhang et al., 2014). Furthermore, it has been shown that AS of some stress-responsive genes is crucial for their functions in plant stress response to abiotic stress. For example, rice DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN2 (OsDREB2B), which is temperature- and drought-responsive, undergoes pre-mRNA AS that is required for its function in plant response to low temperature and osmotic stress (Matsukura et al., 2010). Thus, splicing factors and AS of pre-mRNAs play a fundamental role in plant development and plant stress adaptation.
Recently, we have shown that RBM25, an RNA-recognition motif (RRM) containing protein, acts as a key regulator of HAB1 pre-mRNA AS in Arabidopsis (Wang et al., 2015). RBM25 can bind to the intron of HAB1 to yield the opposite functional isoforms HAB1.2 and HAB1.1, and the ratio of HAB1.2/HAB1.1 regulates the switching off and on of the ABA signaling pathway that modulates seed germination and post-germinative growth in response to ABA treatment (Wang et al., 2015). However, many questions still remain unknown, such as whether RBM25 is involved in other biological progresses, how the RBM25 gene(s) is likely regulated and whether the RBM25 homologes genes/proteins share similar functions. Here, we show that RBM25 is preferentially expressed in vascular tissues in Arabidopsis and is induced by ABA, salt, drought, and other abiotic stresses. Intriguingly, mutation in RBM25 dramatically enhances plant tolerance to drought and salt stress. Moreover, we show that Arabidopsis RBM25 and its homologous genes share the highly conserved gene structures and protein motifs, indicating that they are involved in similar molecular processes and biological functions in plant growth and response to abiotic stresses. Notably, our data reveal that RBM25 is likely to mediate plant response to more abiotic stresses and to be regulated by manifold regulators that are involved in plant responsiveness to multiple plant growth regulators. Our results provide important and novel insights into the regulation of the Arabidopsis RBM25 and its homologous genes and the potential roles in regulating abiotic stress responses in plants.
Materials and Methods
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Columbia-0 (Col-0), T-DNA insertion mutant rbm25-1 and the complimented lines RBM25-9, RBM25-10 were used in this study (Wang et al., 2015). Seeds were sterilized with 50% bleach, washed five times using sterile water, and then sowed on MS medium containing 1% sucrose and 0.8% agar. The plates were stratified in darkness at 4°C for 2 days and then transferred to culture room at 22°C under a 16 h of light/8 h of dark photoperiod. After 7 days, the seedlings were planted in soil.
Gene Expression Analysis
For the expression assay of HAB1.2/HAB1.1, total RNA was extracted using Trizol reagent (Life Technologies, Carlsbad, CA, USA) from 3-week-old seedlings exposed in air for different time. Equal amounts of RNA samples were used for reverse transcription with RevertAidTM First Strand cDNA Synthesis Kit (Invitrogen) according to the manufacturer’s instructions. ACTIN2 was used as the internal control for quantitative RT-PCR (qRT-PCR). The relative expression levels of the target genes were calculated by the equation Y = 2Ct (Ct is the difference in Ct between the target and control products, i.e., Ct = CtHAB1.1-CtACTIN).
For the expression assay of RBM25, total RNA was extracted from 10-day-old Col-0 seedlings treated with different abiotic stresses. qRT-PCR was performed as described above, primers used are listed in Supplementary Table S1.
Phenotypic Analysis Assay
For phenotypic analysis in the presence of NaCl and Mannitol, MS medium was prepared supplemented with 1% sucrose and different concentrations of NaCl and Mannitol. Seeds were surface-sterilized and sowed on the MS medium with NaCl or Mannitol (three plates per treatment). The plates were stratified at 4°C for 2 days and then transferred to chamber at 22°C. Seedlings with elongated radicles or with green cotyledons were counted at the indicated time points.
GUS Histochemical Analysis
For the GUS histochemical analysis, a 3-kb DNA fragment upstream of RBM25 ATG start codon was cloned into pCAMBIA1391 to get PRBM25::GUS, using primers listed in Supplementary Table S1. The PRBM25::GUS construct was introduced into Agrobacterium tumefaciens strain GV3101 and then transformed into Col-0. The GUS assay was performed as previously described (Zhao et al., 2011). The seeds or seedlings were harvested and treated with MS or MS containing 50 uM ABA for 3 h or treated with different abiotic stresses. The histochemical staining was conducted by incubating those materials in freshly prepared buffer containing 5-bromo-4-chloro-3-indolyl-b-D-glucuronic acid for specified time at 37°C in the dark followed by clearing with 75% ethanol. The GUS-staining image was taken by optical microscope.
Drought Treatment and Stomatal Aperture Measurement
For the drought tolerance assay, 2-week-old seedlings were withheld from water for 21 days, and then survival rates were determined and the plants were re-watered. To analyze stomatal function, rosette leaves were incubated under light in the solution containing 50 mM KCl, 10 mM CaCl2, and 10 mM MES (pH 6.15) for at least 2 h to keep the stoma open totally. Then ABA was added to a final concentration of 10 uM. After ABA treatment for 3 h, stomatal apertures were measured as described previously (Ren et al., 2010).
Phylogenetic Tree and Gene Structure Analysis
We obtained the sequences of the identified Arabidopsis RBM25 and its homologous genes from a published database phytozome1, including genomic DNA sequences, coding sequences, and amino acid sequences. A phylogenetic tree was constructed based on the amino acid sequences of Arabidopsis RBM25 and its homologous proteins using MEGA6. The gene structures of Arabidopsis RBM25 and its homologous genes were determined using the Gene Structure Display Server (GSDS) website2.
Promoter Analysis
Sequences of a 3-kb fragments upstream of Arabidopsis RBM25 and its homologous genes were downloaded from website. The regulatory cis-elements of Arabidopsis RBM25 and its homologous genes were then analyzed using PlantCARE3.
Statistical Analysis
All data were analyzed using SigmaPlot 10.0 (Systat Software, Inc., Chicago, IL, USA). The averages and standard deviations of all results were calculated, and Student’s t-test were performed to generate P-values. And the statically significant treatments were marked with (∗∗P < 0.01). ns, no significant (P > 0.05).
Results
RBM25 Is Expressed Ubiquitously in All Tissues
Our previous results have shown that RBM25 was expressed in all organs during plant growth and development using qRT-PCR (Wang et al., 2015). To further determine the tissue and cell-specific pattern of RBM25 expression, we analyzed PRBM25::GUS transgenic plants. Histochemical staining of the PRBM25::GUS plants showed that RBM25 was expressed at all developmental stages from seed germination to reproductive growth, which is consistent with the previous result (Wang et al., 2015). High levels of RBM25 transcripts were observed in imbibed seed (Figure 1A), young seedling (Figures 1B–D), mature leaf (Figure 1E), and inflorescence (Figure 1F). In the germinating seedlings, RBM25 was mainly expressed in cotyledons and vascular of hypocotyl; in young seedlings, RBM25 was preferentially expressed in leaf vascular and in the vascular tissues at the shoot-root junction site (Figures 1B–E).
FIGURE 1.
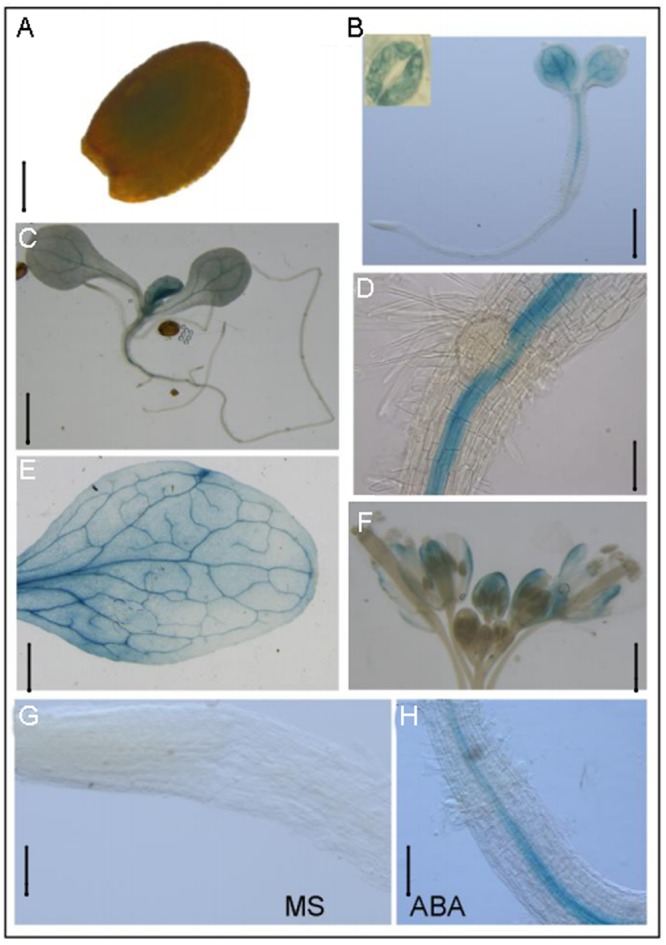
RBM25 is ubiquitously expressed. Histochemical analysis of RBM25. The GUS gene driven by the RBM25 promoter was expressed in (A) imbibed seed (Bar = 0.16 mm), (B) 4-day-old seedling (Bar = 0.5 mm), (C) 8-day-old seedling (Bar = 0.5 mm), (D) root (Bar = 0.5 mm), (E) mature leaf (Bar = 2.5 mm), (F) inflorescence (Bar = 1 mm), (G) root tip in normal condition (Bar = 0.3 mm), and (H) root tip with ABA treatment (Bar = 0.3 mm).
Previously, our qPCR results showed that RBM25 is responsive to ABA in young seedlings (Wang et al., 2015). To further check the expression pattern of RBM25, we treated 8-day-old seedlings with or without 50 μM ABA treatment for 3 h for GUS staining. GUS staining showed that RBM25 expression was substantially increased in vascular of root tip (Figures 1G,H). The result confirms that RBM25 is induced by ABA and reveals that RBM25 mainly functions at the vascular tissues of plants. Together, these results suggest that RBM25 plays crucial roles in ABA-mediated responses.
RBM25 Mutation Improves Plant Tolerance to Salt, Osmotic and Drought Stresses
Previously we have shown that RBM25 mediates plant response to ABA and the loss-of-function mutant rbm25 showed enhanced sensitivity to ABA during seed germination and cotyledon greening (Wang et al., 2015). Since RBM25 was ubiquitously expressed in plants and was induced by ABA, we questioned whether RBM25 mediates plant response to abiotic stress, such as drought, osmotic and high salinity. To this end, we first germinated the seeds of rbm25-1, the complimented lines RBM25-9/RBM25-10 and the wild type (WT) Col-0 on the MS medium with or without 100 or 150 mM NaCl, and evaluated the plant response to NaCl during germination and post-germination stages. In the absence of NaCl, the germination and cotyledon greening rates of the rbm25-1 mutant were comparable to the complementary lines and the WT (Figures 2A–C). By contrast, in the presence of NaCl, the germination and greening rates of rbm25-1 was significantly lower than that of the WT; while the complemented lines exhibited similar germination and greening rates to the WT (Figures 2A–C). We then germinated the seeds of these genotypes on MS medium supplemented with different concentration of mannitol to analyze the plant response to osmotic stress. As shown in Figure 2, rbm25-1 mutant also displayed increased sensitivity to mannitol compared with the WT, whereas the complementary lines showed similar phenotypes during seed germination and cotyledon greening (Figures 2A–C). The results suggest RBM25 is involved in plant responses to salt and osmotic stress during early development.
FIGURE 2.
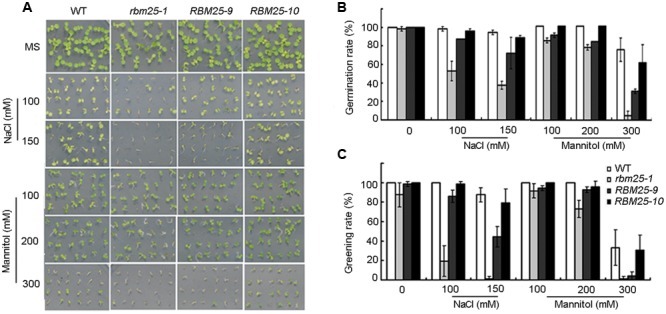
RBM25 improves plant tolerance to salt and osmotic stress. (A) Phenotypic analyses of wild type (WT), rbm25-1 and the complementation lines RBM25-9 and RBM25-10 treated with different concentration of NaCl or mannitol. The images were taken after 4 days after germination. (B) The germination rates and (C) greening rates of WT, rbm25-1 and the complementation lines RBM25-9 and RBM25-10 plants. The data were given as means plus the standard deviation of three independent replicates.
Next, we analyzed the phenotype of rbm25-1 and the complementary lines under drought conditions. We transplanted 8 days-old seedlings of rbm25-1, RBM25-9, RBM25-10 and the WT to the soil. One week after transplanting, the seedlings were subjected to drought stress by withholding water for 21 days. As shown in the Figure 3, all the plants of the WT and the complementary lines were severely wilted, while the rbm25-1 plants were more vigorous. Intriguingly, when the plants were rewatered, rbm25-1 plants exhibited a high survival rate (70%), in sharp contrast, none of the WT and the complementary lines survived (Figures 3A,B). This result suggests that RBM25 plays an important role in plant drought tolerance.
FIGURE 3.
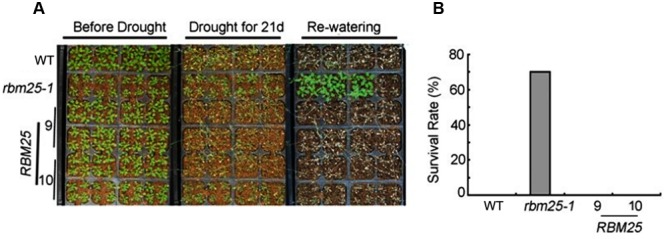
RBM25 functions in drought tolerance. (A) Drought tolerance assay of the 2-week-old seedlings of WT Col-0, rbm25-1, and the complementation lines RBM25-9 and RBM25-10. (B) Survival rate of the Col-0, rbm25-1, RBM25-9, and RBM25-10 under water stress. Three experiments were repeated with similar results, and one representative result was shown.
RBM25 Modulates ABA-Mediated Stomatal Closure
Stomata closure controls water evaporation and drought tolerance in plants in response to water stress. The fact that RBM25 was highly expressed in stomata and mature leaves (Figures 1B,E) made us curious about whether RBM25 regulates plant drought tolerance through stomata regulation. To test the possibility, we first examined the stomatal density of the mutant. The result showed that there was no significant difference between Col-0, rbm25-1 and the complementary line RBM25-9 (Supplementary Figure S1). We then examined the stomatal apertures of WT, rbm25-1 and the complementary line RBM25-9 plants. In the absence of ABA, the stomata opening of rbm25-1 mutant was comparable to that of WT and the complementary line RBM25-9 plants when they were treated with KCl (Figures 4A,B). However, when treated with 10 μM ABA, the guard cells of rbm25-1 mutant showed substantially enhanced sensitivity to ABA and the stomatal aperture was smaller than that of the WT (Figures 4A,B); while the stomatal aperture of the RBM25-9 plants was similar to that of the WT (Figures 4A,B). Taken together, these results suggest that RBM25 regulates drought tolerance by modulating ABA-mediated stomatal closure.
FIGURE 4.
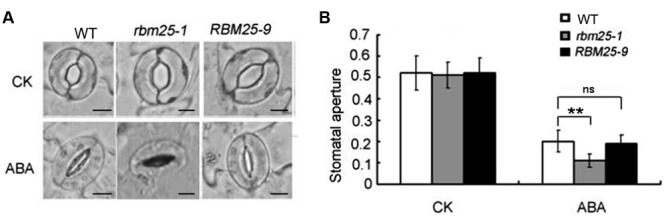
RBM25 modulates ABA-mediated stomatal closure. (A) Stomatal apertures of the Col-0, rbm25-1 and RBM25-9. Leaves of the Col-0, rbm25-1 and RBM25-9 were treated with 10 μM ABA for 3 h. CK represents leaves without ABA treatment. Bar = 0.1 mM. (B) Stomatal apertures of the Col-0, rbm25-1 and RBM25-9 corresponding to (A). Values are mean ratios of width to length ± standard deviations of three independent experiments (n = 30). The Student’s t-test was performed and the statically significant treatments were marked with (∗∗P < 0.01). ns, no significant (P > 0.05).
RBM25 Affects the Ratio of HAB1.2/HAB1.1 under Drought Treatment
As rbm25-1 plants were sensitive to ABA and tolerant to drought, and our previous report found that RBM25 affects the ratio of HAB1.2/HAB1.1 to modulate ABA signaling pathway (Wang et al., 2015), we hypothesized that RBM25 may regulate drought tolerance through the same mechanism. To test this hypothesis, we analyzed the expression of HAB1.1 and HAB1.2 and the ratio of HAB1.2/HAB1.1 in the WT, rbm25-1 and the complementary line RBM25-9 plants. Three-week-old seedlings were taken out from the soil and exposed to air for indicated time to simulate drought. Total RNA was then extracted and qPCR was used to check the expression levels of HAB1.1 and HAB1.2. As shown in the Figure 5, under normal conditions, the ratio of HAB1.2/HAB1.1 in rbm25-1 was up to about 2.2, whereas the ratios of HAB1.2/HAB1.1 in both Col-0 and complementary line RBM25-9 were lower than 0.5. Under drought treatment, the ratios of HAB1.2/HAB1.1 increased at 0.5 h and then decreased as time goes on in the WT, rbm25-1 and the complementary line RBM25-9 plants. However, the ratio of HAB1.2/HAB1.1 in rbm25-1 was significantly higher than that in Col-0 and RBM25-9 at all the time points (Figure 5). This result indicates that under drought stress RBM25 affects the ratio of HAB1.2/HAB1.1 that may modulate plant response to drought stress.
FIGURE 5.
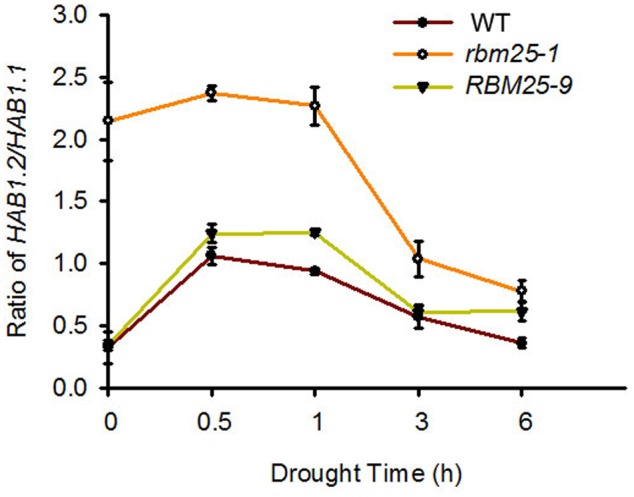
The ratio of HAB1.2/HAB1.1 in Col-0, rbm25-1, and RBM25-9. RNA was extracted from 3-week-old seedlings exposed to air for indicated time, and qRT-PCR was performed. Three independent experiments were performed with similar results, each with three replicates. Values are mean ± SE.
RBM25 is Responsive to Multiple Abiotic Stresses
The RNA-seq results of RBM25 mutant reveal that in addition to HAB1, RBM25 also affects expression of other genes (Zhan et al., 2015), indicating that RBM25 may also participate in other biological processes in Arabidopsis. As plants are sessile organisms, we speculated that RBM25 may play crucial roles in Arabidopsis adaptation to constantly changing environmental conditions. To test the possibility, we first analyzed the expression patterns of RBM25 in Arabidopsis treated with various abiotic stresses by collecting HiSeq data from the eFP website4. As shown in Supplementary Figure S2, RBM25 was induced by almost all of the abiotic stresses including osmotic, drought, genotoxic, oxidative, wounding and salt stresses, although there were some variations in its expression patterns (Supplementary Figure S2). To further test the result, we first detected the expression of RBM25 in response to different abiotic stresses. The result showed that RBM25 expression was up-regulated under drought, cold, heat, salt and osmotic stresses (Figure 6A). We then performed GUS staining for the PRBM25::GUS seedlings treated with drought, cold, heat, salt, and osmotic stresses. The result confirmed that RBM25 is induced by multiple abiotic stresses (Figure 6B). Interestingly, the HiSeq data indicates that RBM25 is induced significantly in root, while our GUS staining result showed that the induction of RBM25 expression preferentially occurred in shoots of the stressed plants (Figure 6B). This difference is likely due to the differences in seedling ages and stress treatments. Taken together, the results indicate that RBM25 may play a key role in plant responses to abiotic stresses.
FIGURE 6.
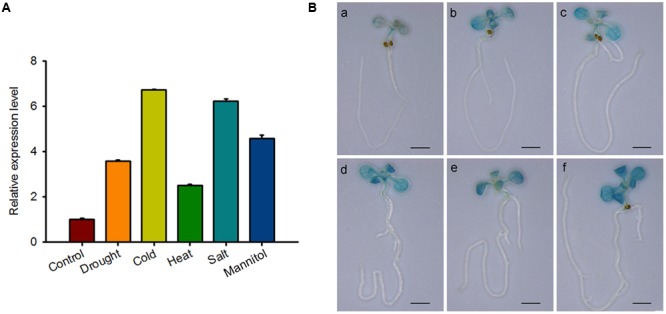
RBM25 is responsive to multiple abiotic stresses. (A) Ten-day-old seedlings were treated with drought (exposed to air for 5 min), cold (4°C for 6 h), heat (37°C for 6 h), salt (150 mM NaCl for 6 h) and osmotic stress (300 mM Mannitol for 6 h). RNA was extracted and qRT-PCR was performed. Three independent experiments were performed and each with three replicates. Values are mean ± SE. (B) Histochemical analysis of 10-day-old PRBM25::GUS seedlings treated with different stress as described in (A). Bar = 2 mm.
The Promoter of Arabidopsis RBM25 Contains Manifold cis Regulatory Elements
To further understand how RBM25 expression is regulated, we performed a promoter analysis to identify regulatory cis-elements using PlantCARE3. As shown in Figure 7, manifold cis regulat- ory elements were identified in a 3 kb DNA fragment upstream of the ATG start code of the RBM25 gene. As expected, there are two ABRE cis elements (CACGTG) which is involved in ABA response and one cis element (MBS: TAACTG) that is related to plant response to drought in the RBM25 promoter. It also contains several cis elements that are related to plant response to heat (HSE: AAAAAATTTC) and low temperature (LTR: CCGAAA). Unexpectedly, the RBM25 promoter contains cis elements involved in hormone signaling which functions in plant growth regulation and response to stresses, including auxin, gibberellin acid (GA), ethylene, and methyl Jasmonate acid (MeJA). Notably, the RBM25 promoter contains six cis regulatory elements involved in JA responsiveness. These observations implicate that RBM25 is an important player in plant responses to drought and unfavorable temperatures and it may modulate plant plastic development through hormonal regulation under stresses.
FIGURE 7.
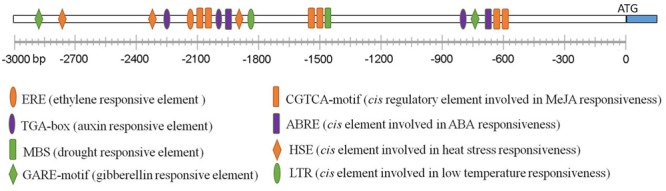
The promoter analysis of Arabidopsis RBM25. The 3-Kb DNA fragment upstream of the ATG staring code of the Arabidopsis RBM25 gene were analyzed using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Phylogenetic and Protein Structure Analyses of Arabidopsis RBM25 and its Homologous Genes in Plants
Previously we have shown that RBM25 is highly conserved to human RBM25 (Wang et al., 2015). To investigate whether RBM25 plays a conserved role in plants, we used Arabidopsis RBM25 as a query to against the genome and PGDD website to analyze the data collected from the Phytozome website1 and we found 14 homologous genes of Arabidopsis RBM25 including those from major crops, e.g., soybean, rice, maize, edible rape, and so on. Apparently, both monocots and dicots contain homologs of Arabidopsis RBM25, indicating that RBM25 is a conserved gene in plants.
To assess the phylogenetic relationships among the homologous genes, we constructed a phylogenetic tree using the neighbor-joining method in MEGA6.0. Among the homologs, 925031 from Arabidopsis lyrata and A02463 from Brassica rapa are closest to Arabidopsis RBM25 in the phylogenetic tree. Interestingly, we found two Arabidopsis RBM25 homolog genes in the soybean genome (Glyma.01G031400 and Glyma.02G034300) which show a close relationship to Arabidopsis RBM25. To gain better understanding of these RBM25 homolog genes, we downloaded the mRNA and genomic DNA sequences from the Phytozome database and analyzed the structures of these RBM25 and its homologous genes using GSDS 2.02. As shown in Figure 8A, the structures of the homologous genes are highly conserved and have a similar exon–intron pattern. All the RMB25 homologous genes contain seven exons and six introns, although there are variations in the lengths of exons or introns (Figure 8A).
FIGURE 8.
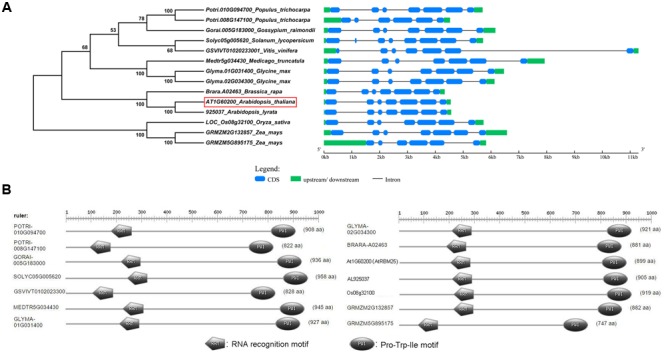
Phylogenetic relationships and conserved protein domains analyses of Arabidopsis RBM25 homologous proteins. (A) The phylogenetic tree (left panel) was constructed using MEGA6.0, and the gene structures were created using the GSDS website. (B) The conserved domains of the homologous proteins of Arabidopsis RBM25 in different species were analyzed using the PROSITE website (http://prosite.expasy.org/prosite.html).
Arabidopsis RBM25 is a RNA binding protein and functions as a splicing factor (Wang et al., 2015; Zhan et al., 2015). To obtain detailed information about these homologous proteins, deduced amino acid sequences of these RBM25 homolog proteins collected from the Phytozome database were aligned, and the protein structures were analyzed using PROSITE5. The result showed that the length of these proteins ranges from 747 aa to 958 aa and the structures are highly conserved with a RRM domain (an RRM) at its N terminal and a PWI domain (Pro-Trp-Ile) at its C terminal (Figure 8B). Both RRM and PWI motifs are capable of binding to DNAs and/or RNAs. The high degree of conservation among these homologous RBM25 proteins highlights that these RBM25 proteins may share highly conserved functions, including the RNA binding activity, AS of pre-mRNA and so on.
The Promoters of Soybean RBM25 Genes Contain both Common and Unique cis Regulatory Elements
The bioinformatics analysis data indicate that these RBM25 genes and the corresponding proteins are highly conserved in plants. The next question is whether these genes or proteins have their unique characteristics that can differentiate one from another. Then we attempted to see whether there are differences between these genes at transcriptional levels. To this end, we compared the promoters of soybean RBM25 homologs with Arabidopsis RBM25 promoter as an example. We found that both promoters of soybean RBM25 homologous genes contain cis regulatory elements related to auxin, GA, MeJA, ABA and the motifs involved in plant responses to drought and temperatures (Figure 9), which is similar to that of the promoter of Arabidopsis RBM25 (Figure 7). The observation suggests that these RBM25 genes are regulated by a common regulatory mechanism at the transcriptional level. However, we also found the differences between soybean and Arabidopsis RBM25 promoters. The major difference between Arabidopsis and soybean RBM25 promoters is that the former contains ethylene responsive cis element, while the latter does not have; however, the latter contains the cis element involved in SA responsiveness, whereas the former does not have. Notably, both soybean RBM25 promoters contain the cis regulatory element related to SA and lack the elements involved in ethylene. These results suggest that in addition to the conserved functions, the RBM25 homolog genes in different species may have their own unique features that are related to the species. The fact that two soybean RBM25 promoters are different in the numbers and the positions of each cis element indicates that the duplicates of the RBM25 homolog genes may function or may be regulated differently.
FIGURE 9.
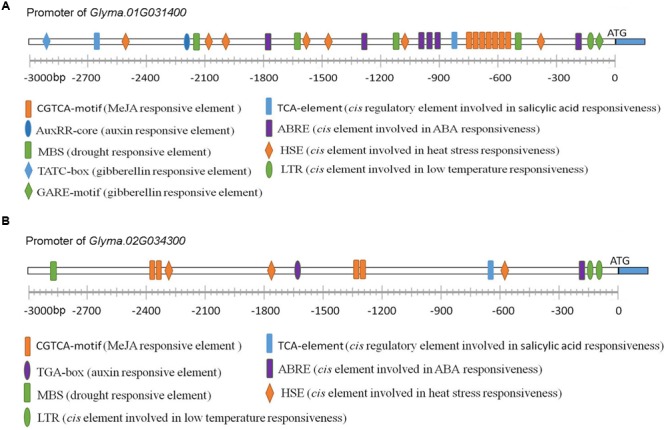
Promoter analysis of two RBM25 genes in soybean. The promoters of the genes (3,000 bp) were analyzed using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) to search for cis-elements. (A) The promoter of Glyma.01G031400. (B) The promoter of Glyma.02G034300.
Discussion
Pre-mRNA AS is an important process that regulates gene expression. It’s reported that about 61% intron-containing genes of Arabidopsis are subjected to AS under normal conditions (Marquez et al., 2012), suggesting the importance of AS in regulating gene expression during plant growth and development. When plants are exposed to abiotic stresses, the AS events of pre-mRNAs substantially increase (Filichkin and Mockler, 2012; Staiger and Brown, 2013). Thus, it has been proposed that pre-mRNA AS is an important post-transcriptional regulation of genes that meditate plant adaptation to changing growth conditions. RBM (RNA-binding motif) proteins are important regulators in the process of AS, and many of which are involved in the response of plant to growth and environmental stimuli (Reddy et al., 2013). Previously we and other labs have shown that RBM25 in Arabidopsis plays a crucial role in ABA signaling at the early developmental stage (Wang et al., 2015; Zhan et al., 2015). Here, we show that RBM25 is a positive regulator of plant tolerance to abiotic stresses, such as drought and salt stress. The RBM25 genes and proteins are highly conserved in the higher plants and their expression are possibly responsive to various hormones and abiotic stresses. Our results demonstrate that RBM25 play a conserved function in plant responses to abiotic stresses.
Previously, we have shown that RBM25 is responsive to ABA and modulates plant response to ABA during seed germination and post-germinative growth in Arabidopsis (Wang et al., 2015). Apparently, RBM25 is not only expressed in the early development of Arabidopsis. Our histochemical analysis results clearly show that RBM25 was expressed in various tissues and organs in the whole life cycle of Arabidopsis and was responsive to ABA (Figure 1), which is consistent with our qRT-PCR data (Wang et al., 2015). This expression patterns of RBM25 implicit its roles in plant responses to abiotic stresses during late developmental stages. In this study, we provide several lines of evidences to support the notion that RBM25 is a key regulator of plant responses to abiotic stresses. Firstly, RBM25 is required for plant responses to osmotic stresses during early development of Arabidopsis, because the rbm25 mutant plants were more sensitive to salt and mannitol during seed germination and post-germinative growth (Figure 2). Secondly, RBM25 is an important negative regulator of plant drought tolerance because loss of function in RBM25 results in greatly enhanced drought tolerance of the plants (Figures 3, 4). Thirdly, promoter analysis identifies several cis-elements related to abiotic stresses (dehydration, low temperature, etc.) in RBM25 promoter, including ABRE, MBS, HSE, LTR (Figure 7), suggesting that expression of RBM25 is responsive to the abiotic stresses. Finally, the expression data from the public website, our qRT-PCT and GUS staining assay results show that RBM25 was induced by abiotic stresses, including osmotic, drought, genotoxic, oxidative, wounding, and salt stresses (Figure 6 and Supplementary Figure S2). These results demonstrate that RBM25 play an important role in the response to abiotic stresses in Arabidopsis.
Previously, we have shown that RBM25 modulates seed germination and post-germinative growth in response to ABA through regulating AS of HAB1 pre-mRNA that produces two isoforms of HAB1, HAB1.1 and HAB1.2 with opposite functions in SnRK2.6 (SNF1-RELATED PROTEIN KINASE 2.6) regulation (Wang et al., 2015). HAB1.1 can interact with SnRK2.6 and inhibits the kinase activity of SnRK2.6 and by dephosphorylation of the protein, thereafter switching off the ABA signaling; while HAB1.2 interacts with SnRK2.6 but keeps SnRK2.6 in an active state and the ABA signaling on (Wang et al., 2015). Here, we show that RBM25-mediated drought tolerance is partially dependent on the alterative splicing of pre-mRNA of HAB1. The ratio of HAB1.2/HAB1.1 in the rbm25 mutant plants was much higher than that in the WT and complementary plants in response to drought treatment, and accordingly, the mutant plants exhibited dramatically smaller stomatal aperture and increased tolerance to drought (Figures 3, 4). As RBM25 affects AS of many genes based on the RNA-seq data (Zhan et al., 2015), we do not exclude the possibility that RBM25 also modulates plant drought tolerance via regulating AS of other genes that are responsible for the drought tolerance of plants. So far, we still do not know how RBM25 mediates plant responses to other abiotic stresses, such as low temperature. It is likely that RBM is in charge of AS of the genes involved in different abiotic stresses. Further identification and characterization of the target genes whose pre-mRNA AS are regulated by RBM25 in response to various stress conditions will help us to understand RBM25-mediated plant tolerance to various abiotic stresses in Arabidopsis.
It is worthy to note that RBM25 is highly conserved proteins in plants. These RBM25 proteins in different plant species share similar gene and protein structures. All the RBM25 proteins contain a RRM in the N-terminal ends and a PWI (Pro-Leu-Ile) motif in the C-terminal ends (Figure 8) which can bind to RNA or DNA. The high level of the protein identity of these RBM25 suggests a conserved role for these proteins in plant responses to various abiotic stresses. Comparison between Arabidopsis RBM25 and two soybean RBM25 homolog promoters favors idea that RBM25s are responsive to various abiotic stresses because both Arabidopsis and soybean RBM25 genes contain many cis-elements, including ABRE, MBS, HSE, and LTR in their promoters. Besides the stress-related cis-elements, there are also some elements involved in hormonal responses in plants, including CGTCA-motif responsive to methyl jasmonate, AuxRR-core to auxin, TATC-box and GARE-motif to gibberellin and TCA-element to salicylic acid. Multiple cis regulatory elements related to abiotic stress and hormonal responses in the promoters of RBM25 genes implicate that these RBM25 may function as a molecular node that integrates hormonal and abiotic signals to orchestrate plant adaptation to the changing environment. Interestingly, we also found some different cis elements (TCA-element to salicylic acid and ERE-element to ethylene), which are unique to Arabidopsis or soybean RBM25 genes (Figures 7, 9), suggesting that functional diversity of RBM25 also exists in different plant species. In summary, RBM25-mediated AS of pre-mRNAs plays a key role in plant response to abiotic stresses. Our results provide an overview of the main characteristics of these RBM25 genes and their potential functions in different plant species. Most importantly, these findings will facilitate our understanding of how plants tolerate abiotic stresses and provide novel insight into genetic improvement of plant stress tolerance.
Author Contributions
XL and ZW conceived the project and designed the experiments. ZW, CC, and BY performed the experiments, analyzed the data; ZW, CC, and XL prepared the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank ABRC (Arabidopsis Biological Resource Center) for providing T-DNA lines. We would like to acknowledge support from National Science Foundation (31230050 and 31200220), National Program on Key Basic Research Project (2012CB114300), the Ministry of Agriculture of the People’s Republic of China (2013ZX08009-02-03).
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00292/full#supplementary-material
References
- Ali G. S., Palusa S. G., Golovkin M., Prasad J., Manley J. L., Reddy A. S. (2007). Regulation of plant developmental processes by a novel splicing factor. PLoS ONE 2:e471 10.1371/journal.pone.0000471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert H. J., Gong Q., Li P., Ma S. (2006). Unraveling abiotic stress tolerance mechanisms–getting genomics going. Curr. Opin. Plant Biol. 9 180–188. 10.1016/j.pbi.2006.01.003 [DOI] [PubMed] [Google Scholar]
- Broun P. (2004). Transcription factors as tools for metabolic engineering in plants. Curr. Opin. Plant Biol. 7 202–209. 10.1016/j.pbi.2004.01.013 [DOI] [PubMed] [Google Scholar]
- Chung H. S., Cooke T. F., Depew C. L., Patel L. C., Ogawa N., Kobayashi Y., et al. (2010). Alternative splicing expands the repertoire of dominant JAZ repressors of jasmonate signaling. Plant J. 63 613–622. 10.1111/j.1365-313X.2010.04265.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filichkin S. A., Mockler T. C. (2012). Unproductive alternative splicing and nonsense mRNAs: a widespread phenomenon among plant circadian clock genes. Biol. Direct 2 7–20. 10.1186/1745-6150-7-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriechbaumer V., Wang P., Hawes C., Abell B. M. (2012). Alternative splicing of the auxin biosynthesis gene YUCCA4 determines its subcellular compartmentation. Plant J. 70 292–302. 10.1111/j.1365-313X.2011.04866.x [DOI] [PubMed] [Google Scholar]
- Lee B. H., Kapoor A., Zhu J., Zhu J. K. (2006). STABILIZED1, a stress-upregulated nuclear protein, is required for pre-mRNA splicing, mRNA turnover, and stress tolerance in Arabidopsis. Plant Cell 18 1736–1749. 10.1105/tpc.106.042184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova O. V., Makarov E. M., Luhrmann R. (2001). The 65 and 110 kDa SR-related proteins of the U4/U6.U5 tri-snRNP are essential for the assembly of mature spliceosomes. Embo J. 20 2553–2563. 10.1093/emboj/20.10.2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez Y., Brown J. W., Simpson C., Barta A., Kalyna M. (2012). Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 22 1184–1195. 10.1101/gr.134106.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsukura S., Mizoi J., Yoshida T., Todaka D., Ito Y., Maruyama K., et al. (2010). Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Mol. Genet. Genomics 283 185–196. 10.1007/s00438-009-0506-y [DOI] [PubMed] [Google Scholar]
- Reddy A. S. (2007). Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu. Rev. Plant Biol. 58 267–294. 10.1146/annurev.arplant.58.032806.103754 [DOI] [PubMed] [Google Scholar]
- Reddy A. S., Marquez Y., Kalyna M., Barta A. (2013). Complexity of the alternative splicing landscape in plants. Plant Cell 25 3657–3683. 10.1105/tpc.113.117523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remy E., Cabrito T. R., Baster P., Batista R. A., Teixeira M. C., Friml J., et al. (2013). A major facilitator superfamily transporter plays a dual role in polar auxin transport and drought stress tolerance in Arabidopsis. Plant Cell 25 901–926. 10.1105/tpc.113.110353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Chen Z., Liu Y., Zhang H., Zhang M., Liu Q., et al. (2010). ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 63 417–429. 10.1111/j.1365-313X.2010.04248.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger D., Brown J. W. (2013). Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 25 3640–3656. 10.1105/tpc.113.113803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugliani M., Brambilla V., Clerkx E. J., Koornneef M., Soppe W. J. (2010). The conserved splicing factor SUA controls alternative splicing of the developmental regulator ABI3 in Arabidopsis. Plant Cell 22 1936–1946. 10.1105/tpc.110.074674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Ji H., Yuan B., Wang S., Su C., Yao B., et al. (2015). ABA signalling is fine-tuned by antagonistic HAB1 variants. Nat. Commun. 6:8138 10.1038/ncomms9138 [DOI] [PubMed] [Google Scholar]
- Zhan X., Qian B., Cao F., Wu W., Yang L., Guan Q., et al. (2015). An Arabidopsis PWI and RRM motif-containing protein is critical for pre-mRNA splicing and ABA responses. Nat. Commun. 6:8139 10.1038/ncomms9139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Du B., Liu D., Qi X. (2014). Splicing factor SR34b mutation reduces cadmium tolerance in Arabidopsis by regulating iron-regulated transporter 1 gene. Biochem. Biophys. Res. Commun. 455 312–317. 10.1016/j.bbrc.2014.11.017 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Wang T., Zhang W., Li X. (2011). SOS3 mediates lateral root development under low salt stress through regulation of auxin redistribution and maxima in Arabidopsis. New Phytol. 189 1122–1134. 10.1111/j.1469-8137.2010.03545.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


