Abstract
Oral diseases, including dental caries and periodontitis, are among the most prevalent diseases worldwide and develop as a consequence of a microbial dysbiosis. Several bacterial strains are being tested as potential oral health-promoting organisms, but usually they are species isolated from niches other than the site where they must exert its probiotic action, typically from fecal samples. We hypothesize that oral inhabitants associated to health conditions will be more effective than traditional, gut-associated probiotic species in key aspects such as colonization of the oral site where disease takes place or the possession of oral health promoting functions, as well as more practical issues like safety and toxicity, and establishing proper doses for administration. As an example of these active colonizers, we describe the case of Streptococcus dentisani, a new streptococcal species isolated from dental plaque of caries-free individuals. We have detected it in 98% of dental plaque samples from healthy individuals and, as expected, it does not produce any toxic secondary metabolite and does not survive a simulated stomach digestion, preventing potential secondary effects. Besides, this species has a double probiotic action, as it inhibits the growth of major oral pathogens through the production of bacteriocins, and also buffers acidic pH (the primary cause of dental caries) through an arginolytic pathway. We propose the use of S. dentisani as a promising probiotic against tooth decay.
Keywords: probiotics, dental caries, pH buffering, arginolytic pathway, bacteriocins, Streptococcus dentisani
Introduction
Oral diseases such as dental caries (tooth decay) and periodontitis (gum disease) are caused by microorganisms. However, they are currently not considered infectious diseases in classical terms because their etiology is clearly polymicrobial (Fejerskov, 2004; Simón-Soro and Mira, 2015), and because the pathogenic bacteria involved are also found at lower proportions in healthy individuals (Hajishengallis and Lamont, 2012; Camelo-Castillo et al., 2015). Thus, it has been pointed out that antimicrobial strategies may not be effective against oral diseases, and new preventive or therapeutic approaches based on restoring the microbial ecological balance in the oral cavity have been proposed (Marsh, 2015; Marsh et al., 2015). Those new preventive measures could include the use of prebiotic compounds to promote the growth of health-associated bacteria (Santarpia et al., 2014), or the application of probiotic bacteria with beneficial features (Saha et al., 2012). In the case of dental caries, health-associated microbial communities have been identified using omics approaches (Zaura et al., 2009; Belda-Ferre et al., 2011; Alcaraz et al., 2012), and therefore culturing those microorganisms could provide potential beneficial bacteria to prevent oral diseases.
However, due to the greater development of probiotics in gut pathologies, and the strong safety evidence accumulated for gut bacteria, many microorganisms isolated from human or animal fecal samples with beneficial properties are being developed as potential probiotics to promote oral health. These often include strains of lactobacilli and bifidobacteria, which had previously been shown to have inmunomodulatory, anti-inflammatory and anti-bacterial properties in different in vitro studies and also in clinical trials (see Table 1; Reid et al., 2011; Cagetti et al., 2013). However, the use of many of these strains in oral diseases like dental caries can be problematic because both Lactobacillus and Bifidobacterium species have been shown to be acidogenic and to be involved in tooth decay (Badet and Thebaud, 2008; Mantzourani et al., 2009a). Even if weak acidogenic strains are selected, the capacity of gut bacteria to colonize the oral niche and to produce anti-caries effects has still to be demonstrated, and the use of gut probiotics in the oral cavity has been criticized. For instance, when the strain Lactobacillus salivarius W24 was tested in an in vitro oral biofilm model, it was shown that this strain further lowered the pH and affected the compositional stability of oral communities (Pham et al., 2009). Thus, the identification of novel strains isolated from the oral cavity itself could be instrumental for the development of efficient probiotics applied to oral health.
Table 1.
Bacteria tested in clinical trials as oral care probiotics.
| Strain | Origin | Brand | Disease | Reference1 |
|---|---|---|---|---|
| Lactobacillus rhamnosus GG | Human intestinal tract | Caries | Aminabadi et al., 2011 | |
| Lactobacillus rhamnosus LB21 | Human intestinal tract | Caries | Stecksén-Blicks et al., 2009 | |
| Lactobacillus reuteri 55730 + PTA 5289 | Woman breast milk | ProdentisTM | Ginvival health, plaque reduction and caries | Krasse et al., 2006; Caglar et al., 2008a |
| Lactobacillus brevis CD2 | Dairy products | Inersan® | Periodontal disease and caries | Riccia et al., 2007; Campus et al., 2014 |
| Lactobacillus paracasei SD1 | Human oral cavity | Caries | Ritthagol et al., 2014 | |
| Bifidobacterium animalis DN-173 010 | Dairy products | ACTIVIA® | Caries | Caglar et al., 2005 |
| Bifidobacterium animalis subsp. lactis BB-12® | Dairy cultures | BB-12® | Caries | Caglar et al., 2008b |
| Streptococcus salivarius M18 | Saliva of a healthy child | BLIS M18® | Plaque reduction | Burton et al., 2013 |
| Lactobacillus salivarius WB21 | Mammalian digestive tract | Gingival health, periodontal disease, halitosis and caries | Shimauchi et al., 2008; Mayanagi et al., 2009; Iwamoto et al., 2010; Suzuki et al., 2014 | |
| Streptococcus oralis KJ3®, Streptococcus uberis KJ2® and Streptococcus rattus JH145® | Healthy mouths | ProBiora(3)® | Caries, periodontal disease and halitosis | Zahradnik et al., 2009 |
| Weissella cibaria CMU, CMS-1, CMS-2 and CMS-3 | Children’s saliva | Halitosis and plaque reduction | Kang et al., 2006a,b | |
| Lactobacillus reuteri DSM 17938 + PTA 5289 | Human saliva | BioGaia ProDentis | Halitosis | Keller et al., 2012 |
| Lactobacillus plantarum 7481 and Lactobacillus brevis 7480 | Children’s saliva | AB:Dentis® | Halitosis and gingivitis | WO 2012022773 A1∗ |
| Streptococcus salivarius K12 | Children’s saliva | BLIS K12® | Halitosis | Burton et al., 2006 |
| Lactobacillus acidophilus NK1 + 100ash + K3III24 | Human intestinal tract | Acilact (solely Russia) | Periodontal disease | Pozharitskaia et al., 1994 |
| Bifidobacterium bifidum N1 and Lactobacillus acidophilus NK1 | Human intestinal tract | Bifidumbacterin + Acilact (solely Russia) | Periodontal disease | Grudianov et al., 2002 |
| Lactobacillus casei 37 | Human intestinal tract | Periodontal disease | Volozhin et al., 2004 | |
| Lactobacillus paracasei GMNL-33 (ADP-1) | Human oral cavity | Dental-LacTM | Periodontal disease | Chuang et al., 2011 |
| Lactobacillus salivarius TI 2711 (LS 1) | Human saliva | Periodontal disease | Matsuoka et al., 2006 | |
| Lactobacillus rhamnosus GG (53103) + LC705 | Human intestinal tract | Oral candidasis | Hatakka et al., 2007 |
1Reference = Publication with results of clinical trial; ∗Patent number.
In the current manuscript, we describe the potential probiotic features of the new species Streptococcus dentisani that we recently isolated from the dental plaque of caries-free individuals (Camelo-Castillo et al., 2014). There are currently six isolates for which the genome sequence is available and that robust phylogenomic analysis include within the dentisani cluster (Jensen et al., 2016). This cluster forms part of the S. oralis clade and differs from the S. oralis and S. tigurinus clusters mainly in their ability to hydrolyze arginine (Jensen et al., 2016). In this manuscript, we study strains 7746 and 7747T, which were isolated from two different individuals and that were shown to be different by comparison of their genomes (Camelo-Castillo et al., 2014). We describe their ability to inhibit the growth of caries-producing bacteria, as Streptococcus mutans and Streptococcus sobrinus, and to buffer extracellular acidic pH, which is the underlying cause of tooth decay. In addition, we show that, being a commensal species in the oral cavity of healthy individuals, their safety features are robust and the appropriate dose for probiotic treatment can be easily determined experimentally.
Materials and Methods
Inhibition Assays with S. dentisani Supernatants
Inhibition experiments against cariogenic bacteria were carried out with concentrated supernatants of well grown cultures of S. dentisani 7746 and 7747 (stationary phase). In order to obtain 5 ml of the concentrated supernatants, single colonies of each S. dentisani strain were inoculated into 50 ml of brain hearth infusion broth (BHI, Biolife) and incubated aerobically at 37°C without agitation during 48 h, or until they reached an optical density of around 1.5 at 610 nm (O.D. 610). After the incubation period, the cultures were centrifuged at 4000 rpm 10 min and the pellets discarded. The obtained supernatants were filtered throughout 0.2 μm pore-size filters (Millipore) and ten-fold concentrated by rotary evaporation on a RV 10 digital device (VWR) with the following settings: heating bath at 40°C, 70 rpm of rotation, 100 mbar of pressure, and 30 min of operating time. The resulting 5 ml of concentrated supernatants were filtered throughout 0.2 μm pore-size filters (Millipore) and stored at –20°C until use.
The inhibitory activity of the supernatants was determined by monitoring the growth of S. mutans ATCC 25175 and S. sobrinus CECT 4034 in the presence/absence of the potential inhibitor by means of optical density measurements. Pre-inocula of S. mutans and S. sobrinus were obtained by inoculating a single colony of each strain in 10 ml of BHI liquid medium and incubated aerobically overnight at 37°C without agitation. The O.D. 610 of the pre-inocula was measured in a spectrophotometer (BioPhotometer, Eppendorf) and diluted with BHI liquid medium to obtain an optical density of 0.1. To assess the inhibitory effect of S. dentisani on the growth of S. mutans and S. sobrinus, 160 μl of the bacterial suspensions were mixed with 40 μl of the concentrated supernatants and loaded by triplicate into a Nunc Microwell 96-well microplate by triplicate. As controls, 160 μl of the bacterial suspensions were mixed with 40 μl of 10-fold concentrated BHI and loaded by triplicate into the 96-well microplate. The microplate was loaded into a microplate reader (Infinite 200 PRO, Tecan) and incubated at 37°C during 24 h. The O.D. 610 of each inoculated well was measured every 30 min during the incubation time.
To determine the active size fraction, the concentrated supernatant was size-fractionated by sequential filtering throughout 10 KDa and 3 KDa filters (Amicon), following the manufacturer’s recommendations. By this way, we obtained three size fractions (>10 KDa, 3–10 KDa, and <3KDa) that were tested by triplicate against S. mutans cultures by the same methodology explained above. To confirm the peptidic nature of the inhibitory compounds and discard that the inhibition was produced by hydrogen peroxide we proceed as described in Zhu and Kreth (2012). Briefly, S. dentisani was grown on a BHI plate for 24 h and peroxidase (40 μg), peptidase (64 μg), or phosphate-buffered saline were applied beside each colony for 10 min before the other species were inoculated at the same spot. S. mutans and S. sanguinis were used as controls of proteinaceus inhibitory substance and H2O2, respectively.
Besides, we used scanning electron microscopy (SEM) to directly observe the effect of the S. dentisani supernatants on the three type strains S. mutans ATCC 25175, Prevotella intermedia NCTC 13070, and Fusobacterium nucleatum DSMZ 20482 cells. Briefly, 160 μl of the bacterial cultures in exponential phase (O.D. 610 = 0.6) were mixed with 40 μl of the concentrated supernatant and incubated for 30, 60, and 90 min at 37°C. After the incubation period, the suspensions were centrifuged at 4000 rpm 10 min and the supernatant discarded. The pellets were fixed in 2.5% paraformaldehyde and 0.5% glutaraldehyde, washed three times with PBS buffer and exposed to 1% osmium tetroxide in PBS buffer for 1 h. The samples were rinsed with PBS buffer three more times and then moved through a gradual process of dehydration, starting with 30% ethanol and ending with absolute ethanol (multiple rinse steps at each 30, 50, 70, 80, 90, and 100% ethanol). Finally, the samples were mounted on scanning electron micrograph stubs, sputter coated with gold, and viewed on a JEOL JSM 840 scanning electron microscope.
pH Range of Growth and Resistance of S. dentisani to the Digestive Process
To know the optimal and pH growth range of S. dentisani strains 7746 and 7747 we prepared BHI broth at different pHs (4.7, 5.5, 6, 6.5, 7, and 7.5) by adding NaOH 10 N or HCl 10 N to the commercial medium (Biolife). Pre-inocula were obtained as explained above. Two-hundred microliters of the BHI broths at different pHs were inoculated by triplicate with 20 μl of the inocula into a Nunc Microwell 96-well microplate. The O.D. 610 of each inoculated well was measured every 2 h and plotted versus the time of incubation to obtain the growth curves.
To evaluate the resistance of S. dentisani to the digestive process, the viability of the strains 7746 and 7747 was checked after a treatment with the salivary enzyme alpha amylase, followed by digestion with gastric enzymes under progressive acidic conditions to simulate digestion, following the protocol described in Khalf et al. (2010) and Martínez et al. (2011). The assays were performed in an in vitro model stomach (or dynamic digester) by the external services of the Food Industry Research Association AINIA (Valencia, Spain). Each strain was tested by duplicate whereas the control was assayed once. Colony forming units (CFU) counts were obtained in BHI agar before any treatment (reference cultures), after the chewing step, and during the gastric digestion at three different times (30, 60, and 120 min). The plates were incubated at 37°C during 24–48 h. As the medium used for CFU counts was not selective for S. dentisani, the same assay was made without inoculation and used as blank to discount any contamination, obtaining 0 CFUs in this control.
Growth of S. dentisani in an Arginine Enriched Medium
Streptococcus dentisani 7746 was grown in 300 ml of BHI medium amended with 5 g/l of L-arginine monohydrochloride 98% (Alfa-Aesar). The pre-inoculum was obtained by inoculating a single colony of the strain in 10 ml of BHI broth and incubating overnight at 37°C without agitation. Inoculation was made with the volume required to obtain an initial O.D. 610 of 0.02. For comparison, the same volume was used to inoculate 300 ml of BHI without arginine. In both conditions the initial pH was set at 7.3. The cultures were incubated at 37°C during 24 h and aliquots of 1 ml were taken every hour for measuring the pH and the O.D. 610. Both conditions were assayed by triplicate.
Prevalence of S. dentisani and Other Oral Probiotics in the Dental Plaque of Healthy Individuals
To assess the prevalence of S. dentisani in the dental plaque of healthy individuals we compared the genomic sequences of the S. dentisani strains 7746 and 7747 (1.98 and 1.74 Mb, respectively) against 118 metagenomes of the dental plaque of healthy individuals available at The Human Microbiome Project Consortium (HMP, Turnbaugh et al., 2007). The metagenomic reads were mapped against the sequenced reference genomes using the NUCmer and PROmer v3.06 alignment algorithms (Kurtz et al., 2004) with the default parameters, following the methodology of Belda-Ferre et al. (2011). We considered that S. dentisani was present in a sample if at least 20 sequences >100 bp of the metagenome showed a similarity equal to or higher than 99% with the sequenced genomes. For comparison, the same approach was used to analyze the prevalence of S. salivarius in dental plaque.
The presence of the genus Lactobacillus in different parts of the oral cavity was performed by the analysis of the oral 16S rDNA sequences deposited in the Human Microbiome Project database in year 20141. The niches analyzed were the keratinized gingiva, buccal mucosa, hard palate, palatine tonsils, saliva, supra- and subgingival plaque, and tongue dorsum. The analyses were carried out with a total of 4.3 × 108 sequences. The taxonomic affiliation was performed using the Megablast tool implemented in the NCBI against the SILVA ribosomal database2, with the following parameters: e value <1e–10, percent identity >97%, alignment length >350 bp.
Quantification of S. dentisani in the Dental Plaque of Healthy Individuals
Two healthy volunteers, named MG01 and MG02, were selected for sampling. They were males aged 20–30 years, non-smokers, with 28 teeth excluding third molars, with good dental and periodontal health: in both, absence of caries (non-cavitated level), DMF = 0, OHI = 0, GI = 1, and CPI = 1 (following nomenclature by World Health Organization [WHO], 1997). They had not been treated with antibiotics in the 6 months prior to the study nor presented antecedents of routine use of oral antiseptics. The two donors signed a written informed consent and the sampling procedure was approved by the Ethics Committee from the DGSP-CSISP (Valencian Health Authority), with reference 10/11/2009. Supragingival and subgingival dental plaque samples were taken from vestibular (buccal) and lingual (palatine) surfaces of 28 teeth in each volunteer. Streptococcus dentisani was quantified in individual free surfaces of each tooth type (incisor, canine, premolar, and molar) from one quadrant and the absolute numbers calculated by multiplying the obtained value by the number of each tooth type in the mouth. The same procedure was followed for the total bacterial content quantification.
Dental plaque samples were resuspended in 100 μl of PBS buffer and DNA was extracted with the MagnaPure LC JE379 instrument and the MagnaPure LC DNA Isolation Kit (Roche). The quantification of the DNA was done with the Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen), and real-time PCR was performed in a LightCycler 480 System with the LightCycler 480 SYBR Green I Master Mix (Roche). In every step, we followed the manufacturer’s recommendations.
The specific primers used for the quantification of S. dentisani were designed for this study and targeted the genes for the carbamate kinase (arcC): CkSdF 5′-GTAACCAACCGCCCAGAAGG-3′ and CkSdR 5′-CCGCTTTCGGACTCGATCA-3′; and the ORF540: Orf540F (5′-ATGTTCATCGGCTTGACAGGCTT-3′) and Orf540R (5′- TAAGCAAGCATAGAACCGCGCC-3′). Primers specificity was predicted in silico by the primerBLAST tool implemented in the NCBI3 and confirmed by absence of amplification by qPCR with 5 ng/μl DNA from S. mutans, S. sobrinus, S. sanguinis, S. salivarius, S. mitis, S. pneumoniae, S. infantis, and S. oralis. The specificity of the primers was also checked by scrutinizing the melting profiles after every assay. Primers for the Ck gene did not amplify any of the tested streptococcal species. Primers for the ORF540 amplified only the DNA of S. pneumoniae, which is a rare inhabitant of dental plaque. Amplification was performed in a 20 μl final volume containing 1 μl of template DNA (at concentrations 5–22 ng/μl), 10 μl of the LightCycler 480 SYBR Green I Master Mix, 0.4 μl of each primer, and 7.2 μl of nuclease-free water. The thermocycling protocol used was as follows: an initial step of 95°C for 5 min, and 40 cycles of 10 s at 95°C, 20 s at 65°C, and 25 s at 72°C. All the quantifications were made by duplicate.
The concentration of S. dentisani in each sample was calculated by comparison with the Cq values obtained from a standard curve. This was generated using serial 10-fold dilutions of DNA extracted from 2 × 107 CFUs/ml (counted by serial dilutions in agar plates). Finally, the Cq values obtained with the plaque samples were replaced in the standard equation and expressed in absolute numbers per tooth type analyzed.
Results
Inhibitory Activity of the S. dentisani Supernatants
As shown in Figure 1A, the addition of the S. dentisani 7746 concentrated supernatant produced a marked inhibitory effect on the growth of both S. mutans and S. sobrinus, as compared to the curves obtained when only concentrated BHI was added. Similarly, the concentrated supernatant completely inhibited the growth of other S. mutans strains (strains OMZ175 and ATCC 700610, data not shown). Regarding the curves obtained with the different size fractions, the <3 KDa fraction retained the inhibitory activity when tested against S. mutans ATCC 25175, while the 3–10 KDa and >10 KDa fractions did not (Figure 1B). The last two even enhanced the growth of S. mutans, probably due to a higher availability of nutrients coming from the concentrated culture medium and/or the S. dentisani metabolism. The same results were obtained with strain 7747 (data not shown).
FIGURE 1.
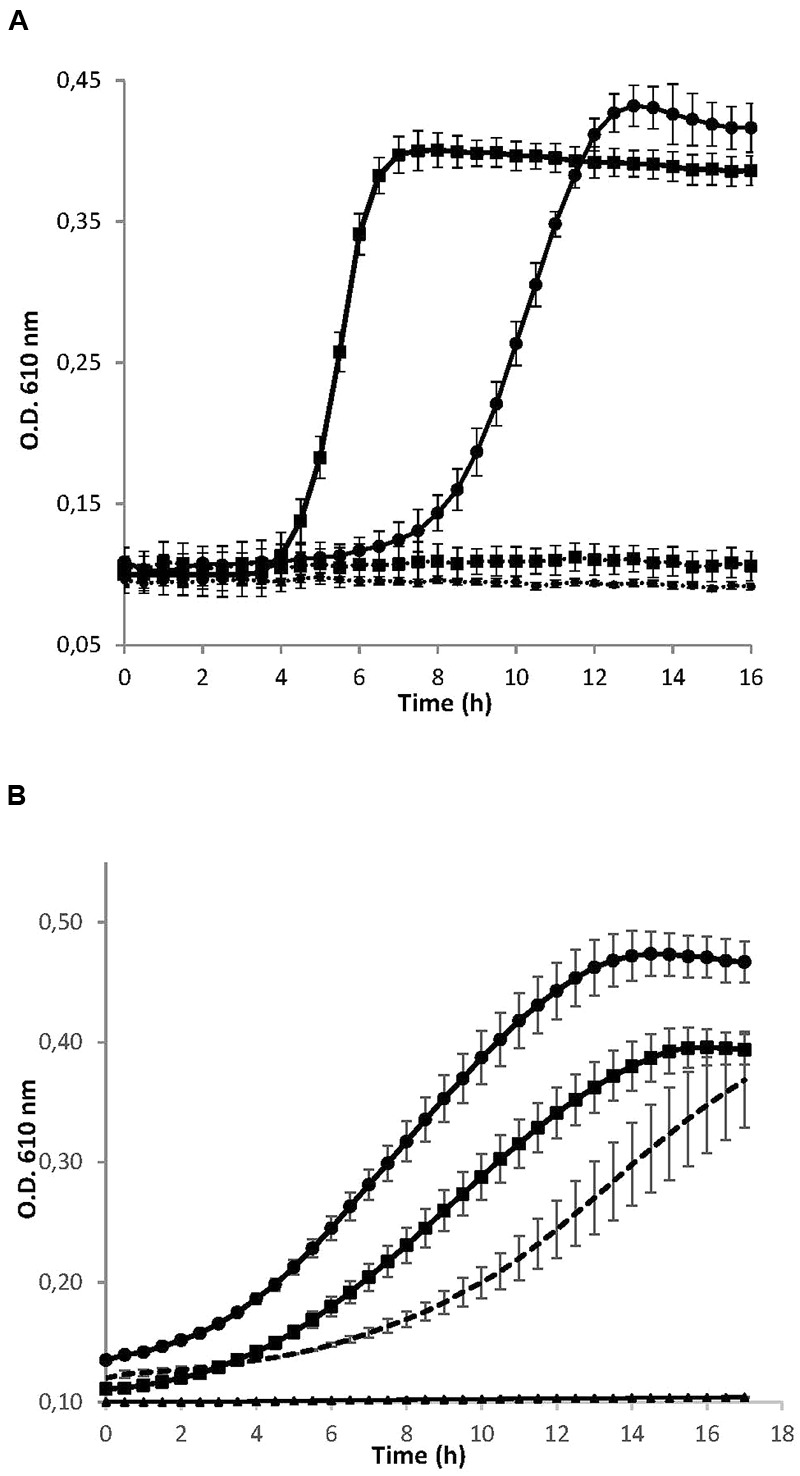
(A) Growth curves of the cariogenic bacteria Streptococcus mutans ATCC 25175 (squares) and S. sobrinus CECT 4034 (circles) in the presence (dotted lines) and absence (solid lines) of concentrated supernatant of S. dentisani strain 7746. (B) Growth curves of S. mutans ATCC 25175 in the presence of different size fractions of the 10× concentrated supernatant of S. dentisani 7746. Circles correspond to the fraction >10 KDa, squares to the 3–10 KDa fraction, and triangles to the fraction <3 KDa. For comparisons, S. mutans was grown in the presence of 10x concentrated BHI medium (dotted line). Means ± SD from three independent replicates are plotted.
Microscopy visualizations revealed that after 30 min of incubation with concentrated supernatants of S. dentisani, the cells of S. mutans, F. nucleatum, and P. intermedia were clearly affected, showing pores in the surface of the cells (S. mutans), changes in the cell walls’ structure (P. intermedia) and cell lysis (F. nucleatum) (Figures 2A–C).
FIGURE 2.
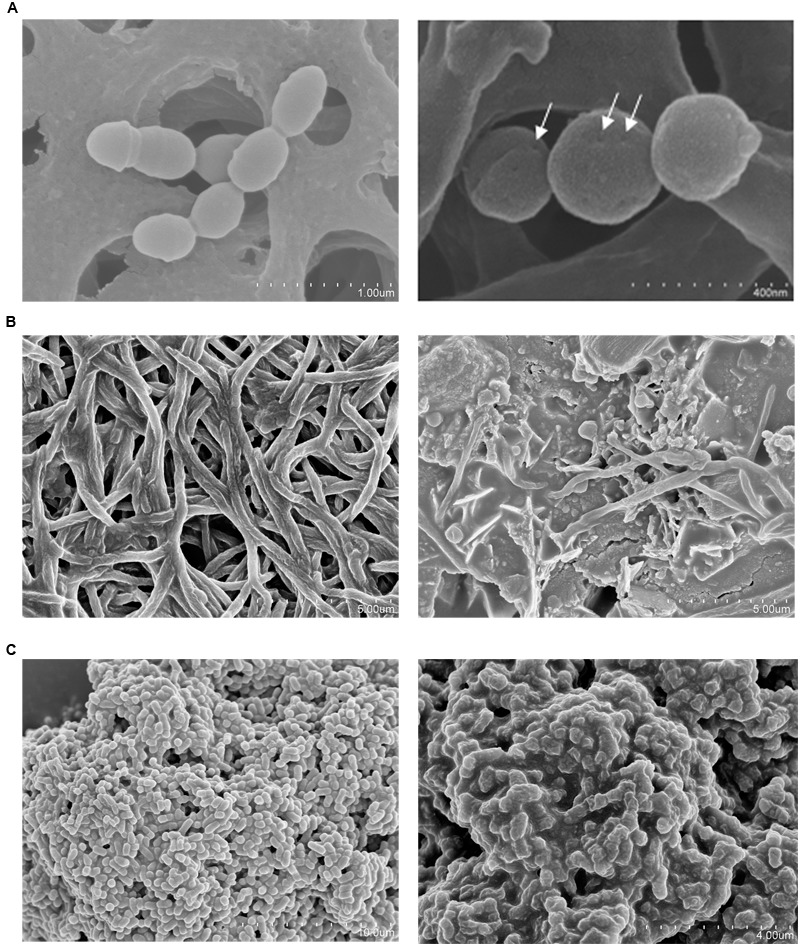
Scanning electron microscopy (SEM) of three oral pathogens before (left) and after (right) a 30-min treatment with 10-fold concentrated supernatant of S. dentisani strain 7746. (A) Streptococcus mutans (pores are pointed with arrows). (B) Fusobacterium nucleatum. (C) Prevotella intermedia.
Proteinase treatment of the supernatant provoked the absence of growth inhibition when tested against S. mutans, while the peroxidase treatment did not affect the inhibitory activity (Figure 3). Taken together, the results indicated that the inhibition is not due to the production of hydrogen peroxide, and supports the idea that S. dentisani inhibit oral pathogenic bacteria by the production of inhibitors of peptidic nature, such as bacteriocin-like peptides.
FIGURE 3.
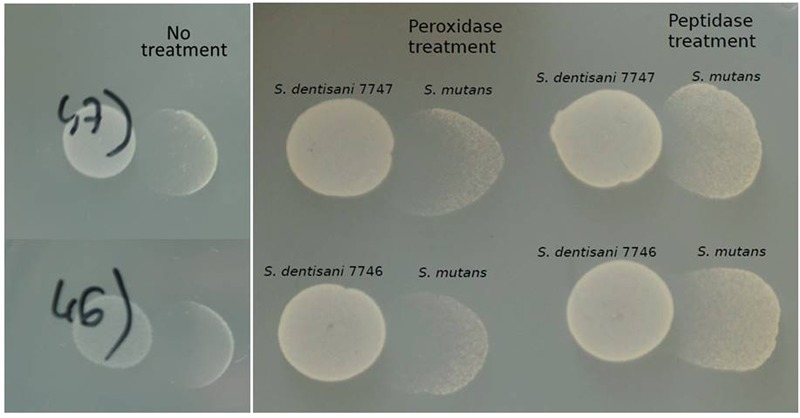
Inhibition of S. mutans by S. dentisani in the presence of peroxidase and proteinase. The tests were performed with S. dentisani strains 7746 and 7747, treating with the enzymes the spots where S. mutans would later be grown. The controls on the left panel show the inhibition without treatment.
pH Tolerance and Resistance of S. dentisani to the Digestive Process
Streptococcus dentisani was tested for its ability to grow at different pHs. The growth curves obtained showed that both strains displayed a very similar behavior, with an optimal growth pH close to 7 (Supplementary Figures S1A,B). It is noteworthy that they were able to grow at pH values between 6 and 7.5 but not between 4.7 and 5.5, indicating that S. dentisani can endure moderately acidic conditions but is not an acidophilic organism. Surprisingly, the growth of both strains at pH 6 was better than at 6.5, suggesting the activation of a buffering metabolic pathway at pH values around 6 (see Section “pH Buffering Capacity of S. dentisani” below).
Regarding the simulated digestive process, and as expected from the pH curves obtained before, S. dentisani was not able to maintain the initial viability during digestion simulations. As starting values of viability we obtained 6.5 × 108 and 3.9 × 108 CFUs/ml for the strains 7746 and 7747, respectively. After the chewing simulation with salivary enzymes these values decreased in only two to three orders of magnitude, showing that both strains could remain in the mouth at high levels after chewing, whereas the gastric digestion strongly inhibited their growth, even after the first step of the simulation (30 min, see Table 2). As neither strain of S. dentisani survived the gastric process, the simulation of an intestinal digestion was not performed.
Table 2.
Viability, expressed in CFU/ml, of the two strains of S. dentisani after the chewing and gastric digestion processes (G.D.) at three different times.
| Strain | Starting culture | Chewing pH 6.9 | G.D. 30 min pH 3 | G.D. 60 min pH 2.1 | G.D. 120 min pH 1.7 |
|---|---|---|---|---|---|
| S. dentisani 7746 | 6.5 × 108± 3.05 × 108 | 5.3 × 106± 2.8 × 108 | <10 | <10 | <10 |
| S. dentisani 7747 | 3.9 × 108± 4 × 107 | 1 × 106± 9.8 × 105 | <10 | <10 | <10 |
The values shown are the mean of two independent replicates and the standard deviation is specified.
pH Buffering Capacity of S. dentisani
Figure 4 depict the growth curves obtained by triplicate when S. dentisani 7746 was grown in presence/absence of arginine. During the initial 6 h of incubation, the growth seemed to be fueled by the sugars present in the BHI medium, as the growth curves were identical in both conditions (with and without arginine), and the culture pH dropped from 7.3 to about 6.2. Soon after completion of this first phase, in the cultures containing arginine the pH starts to rise, reaching almost the initial pH value 12 h after inoculation. Contrarily, in the medium without arginine the pH continuously decreased to reach a value of about 6.2. In both conditions the O.D. 610 measurements were similar, with a maximum value of around 1.3 after 12 h of incubation.
FIGURE 4.
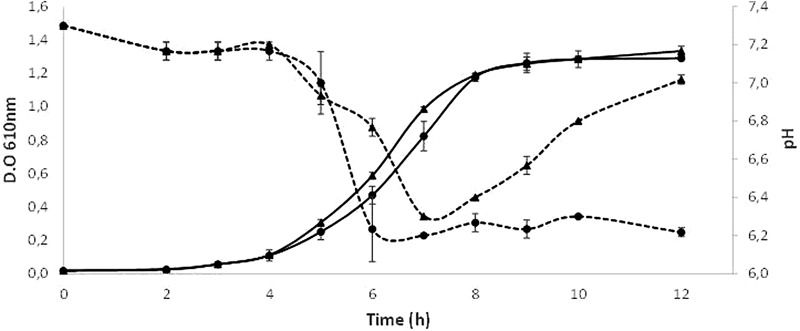
Growth curves of S. dentisani strain 7746 in a medium with (triangles) and without (circles) the addition of 5 g/l of arginine. The mean ± SD of O.D. 610 (solid lines) and pH of the culture (dotted lines) from three replicates are depicted.
The analysis of the genome of S. dentisani showed that strains 7746 and 7747T contain the key genes of the arginine deiminase system: arcA (arginine deiminase, locus tag HK29_RS02275), arcC (carbamate kinase, locus tag HK29_RS02285), and arcB (ornithine carbamoyltransferase, locus tag HK29_RS02280), involved in the ammonia generation through arginine metabolism, a mechanism that releases ammonnia extracellularly producing the alkalinization of the environment (Liu et al., 2008, 2012).
Prevalence of S. dentisani in the Dental Plaque of Healthy Individuals
The recruitment analyses showed that sequences of S. dentisani were present in most of the analyzed metagenomes (direct, whole-sequenced DNA coming from dental plaques of healthy individuals at the Human Microbiome Project). At least 20 metagenomic sequences >100 bp were found to have a similarity ≥99% compared with the 7746 genome in 116 out of 118 individuals, and in the strain 7747 this threshold was fulfilled in all 118 individuals. Contrarily, the species S. salivarius that has been proposed as a probiotic agent against caries was only detected in three of these metagenomes, in agreement with this species inhabiting mucosal surfaces and not the hard tissues. In addition, we have analyzed the Lactobacillus 16S rRNA gene sequences from oral samples contained in the HMP database, as this genus contains species commonly used as oral probiotics (namely L. acidophilus, L. reuteri, and L. rhamnosus). The results showed that Lactobacillus spp. is present in very low numbers in the oral cavity, accounting for less than 0.05% of the bacterial genera in the buccal mucosa, hard palate, palatine tonsils, saliva, tongue, and supragingival plaque. The highest numbers of Lactobacillus 16S rDNA sequences were obtained in the subgingival plaque, but even here this genus accounted for only 0.94% of the total bacterial population. We analyzed in the same oral niches the prevalence of the genus Streptococcus, which was found to be present at very high numbers in the keratinized gingiva, buccal mucosa, hard palatine, palatine tonsil, tongue dorsum, and saliva (59.5, 58.8, 54, 27.1, 26.8, and 19.7 per cent of the total bacterial members, respectively). Furthermore, the percentage of the genus Streptococcus in the subgingival and supragingival plaques was high (18.8 and 20.2%, respectively). Overall, the results showed that the genus Streptococcus is much more abundant than Lactobacillus in every analyzed oral niche, and particularly in those directly affected by the cariogenic processes, being Streptococcus between 20 and 600 times more abundant than Lactobacillus in the subgingival and supragingival plaque, respectively.
Quantification of S. dentisani in the Dental Plaque of Healthy Individuals by q-PCR
The absolute numbers of S. dentisani cells were obtained for the teeth’s free surfaces of two healthy volunteers by the use of two set of S. dentisani-specific primers, which provided similar estimates (Table 3). The results showed that both individuals contain very similar values of abundance of S. dentisani (1.04 × 107 and 3.14 × 107 cells for MG01 and MG02, respectively, as estimated by the ORF540 primers; 4.46 × 107 and 6.94 × 107 cells for MG01 and MG02, as estimated by the carbamate kinase primers). However, although the calculated S. dentisani numbers in the mouth of both individuals was highly similar, its distribution was very different. As shown in Table 3, MG01 did not show important differences in the amounts of S. dentisani between tooth types nor between the lingual and vestibular surfaces. Patient MG02 however, had a more heterogeneous distribution, with higher proportions of S. dentisani in premolars and molars, and on the lingual surfaces of every tooth type.
Table 3.
Total cell counts of S. dentisani on supragingival dental plaque in the vestibular (V) and lingual (L) parts of different tooth types in two caries-free individuals (MG01 and MG02).
| Incisor | Canine | Premolar | Molar | |
|---|---|---|---|---|
|
MG01 (V) (L) |
2.1 × 105/2.8 × 104 2.8 × 105/2.7 × 104 |
4.7 × 105/7.9 × 104 1.4 × 105/4.1 × 105 |
2.9 × 105/3.46 × 104 4.6 × 105/9.8 × 106 |
3.1 × 105/3.1 × 104 4.3 × 105/6.9 × 104 |
|
MG02 (V) (L) |
5.5 × 102/7.5 × 103 3.4 × 104/5.1 × 104 |
2.7 × 103/4.4 × 103 2.8 × 104/5.9 × 103 |
1.2 × 103/1.3 × 104 7.1 × 104/5.0 × 105 |
2.5 × 104/4.5 × 104 4.7 × 104/1.7 × 107 |
Data show the estimates of bacterial numbers obtained by qPCR with two different sets of S. dentisani-specific primers (orf540/CK).
Discussion
Dental caries is considered the most prevalent disease worldwide, with up to 80% of the human population being affected at some point during their lives (Petersen and Lennon, 2004). There are however no efficient means to prevent it, as the polymicrobial nature of the infection and its complex etiology make passive and active immunization strategies (e.g., a caries vaccine) ineffective (Fejerskov, 2004; Simón-Soro and Mira, 2015). Thus, new strategies directed towards the re-establishment of the natural balance in the oral microbiome like the use of pre- and probiotics have been proposed (Marsh et al., 2015). However, as shown in Table 1, most of the probiotics proposed to promote oral health are bacteria isolated from environments other than the oral cavity, usually the human gut (Cagetti et al., 2013). In addition, the most commonly used probiotic bacteria to treat dental caries are lactobacilli, due to their well-proven safety characteristics. However, the use of non-oral bacteria may prevent efficient colonization of the oral niche, which is a vital and desirable feature of probiotics. This is supported by the low frequencies of lactobacilli detected in tooth tissues by molecular methods, as reported in the current manuscript. In addition, lactobacilli are important acid-producers and long known to be associated to dental caries lesions (Badet and Thebaud, 2008; Plonka et al., 2012), as well as bifidobacteria (Mantzourani et al., 2009b; Beighton et al., 2010), which are also common oral probiotics. This can be the reason why in vitro experiments with these bacteria, even if low-acid producing strains are selected, may result in pH acidification (Pham et al., 2009). These results underline the need to use potential probiotics from the oral cavity, and therefore the isolation of a bacteriocin-producing probiotic Streptococcus salivarius which inhibited the cariogenic agent S. mutans was promising (Wu et al., 2015). However, the comparison of the S. salivarius genome against a high number of metagenomes from supragingival plaque presented in the current manuscript reveals its absence in the tooth, in agreement with its soft-tissues-associated nature. Streptococcus salivarius is a typical inhabitant of the buccal epithelium, tongue, and dorsal epithelium (Bowden et al., 1979; Power et al., 2008) and comprises an important part of the total cultivable flora on the soft tissues of the mouth (Wilson, 2005). Probably for this reason, it has been proposed as a probiotic for the pharyngeal mucosa (Guglielmetti et al., 2010) but its inability to colonize the tooth surface may hamper its potential as an anti-caries probiotic. The high heterogeneity of environments in the mouth and the resulting adaptation of microorganisms to those specific microniches (Simon-Soro et al., 2013) underscores the importance of selecting oral bacteria adapted to live in hard tissues as probiotics against dental caries to guarantee a proper colonization. In agreement with this view, our data show that Streptococcus dentisani, which was isolated from supragingival dental plaque of caries-free individuals (Belda-Ferre et al., 2011; Camelo-Castillo et al., 2014), was widespread among dental plaque samples from healthy subjects.
Apart from dental colonization, we show that an important probiotic feature of S. dentisani is its anti-microbial properties, inhibiting the growth of important oral pathogens like S. mutans, S. sobrinus, or Prevotella intermedia. In addition, the killing of Fusobacterium nucleatum is unusual among oral probiotics and may provide an important beneficial effect, as this organism is responsible for a large number of co-aggregation patterns with many oral species and is considered the main “bridge” bacteria between early and late colonizers of dental plaque (Kolenbrander et al., 2002). Thus, its inhibition could contribute to impede the adhesion of pathogenic organisms that co-aggregate with this bacterium and by doing so hamper dental plaque development, whose maturity has been shown to considerably increase the drop in pH after a meal (Firestone et al., 1987). In addition, F. nucleatum itself has been associated with halitosis due to the production of volatile sulfur compounds (Krespi et al., 2006), and therefore its growth inhibition should be investigated in the future as a way to diminish the severity of this condition.
The different experiments performed in the current work suggest a peptidic nature of the inhibitory molecules, as the inhibitory effect was significantly reduced by proteinase treatment but was unaffected by peroxidase, indicating that the inhibition was not due to the production of hydrogen peroxide as it is common in other streptococci (Zhu and Kreth, 2012). This, together with the small size of the molecules responsible for the inhibition (<3 KDa) and the appearance of pores in the membrane of sensitive bacteria as identified by SEM, strongly suggest that the inhibition is caused by bacteriocins, and future work should be directed towards identifying and characterizing such antimicrobial peptides.
Dental caries, as well as other oral diseases like periodontitis or halitosis are not considered typical infectious diseases in classical terms, as there is more than one species responsible for their etiology, the microbial consortia causing the disease varies considerably between individuals and even between lesions of the same patient, and pathogenic organisms normally can be isolated also from healthy individuals (Hajishengallis and Lamont, 2012; Simon-Soro et al., 2014; Camelo-Castillo et al., 2015; Marsh et al., 2015; Simón-Soro and Mira, 2015). For these reasons, antimicrobial properties of probiotics may not be sufficient for effectively preventing dental caries (ten Cate and Zaura, 2012) and the ability to restore the microbial ecological balance after pH acidification, for instance by alkali production, is a promising probiotic feature (Huang et al., 2015). Production of ammonia from urea or arginine by several oral bacteria has been shown to efficiently buffer salivary pH and has been associated to a reduction in caries risk (Reyes et al., 2014; Moncada et al., 2015). The analysis of the genome of Streptococcus dentisani revealed all genes from the arginolytic pathway, including the arginine deiminase and the carbamate kinase, a feature which appears to be common to all members of the cluster (Jensen et al., 2016). In addition, the agmatine deiminase gene, involved in the production of ammonia (Liu et al., 2012) was also present in both analyzed strains. In agreement with this, our data show that S. dentisani was able to efficiently buffer extracellular pH in the presence of arginine. Arginine is present in saliva in variable concentrations and it has also been added as a prebiotic to toothpaste, with strong clinical evidence for reducing enamel demineralization and buffering acidic pH (Yin et al., 2013; Santarpia et al., 2014). In addition, we have found in the genome of S. dentisani genes encoding for different aminopeptidases that can liberate arginine from peptides and proteins (Gonzales and Robert-Baudouy, 1996), which could then enter the arginolytic pathway. This is supported by the higher growth of the bacterium at pH 6 than at pH 6.5 in standard BHI growth medium (Supplementary Figure S1). Given that its optimal growth pH is neutral, and that the arginolytic pathway has been shown to be activated by low pH in other species (Liu et al., 2008), the higher growth rates at pH 6 are probably the result of protein degradation liberating arginine, which would allow the production of NH4, as well as the production of ATP derived from this metabolic route. These results also suggest that the full activation of the arginine degradation genes occurs at pH < 6.5, and the regulation of this pathway should be studied in the future. In Figure 5, we have illustrated the buffering effect that could take place in the oral cavity due to the enrichment of the dental plaque bacterial community with S. dentisani. When acidification occurs as a consequence of the consumption of dietary carbohydrates, and the pH drops to values around 6, S. dentisani would be able to activate the ADS system producing ammonia and, consequently, buffering the pH of its close environment.
FIGURE 5.
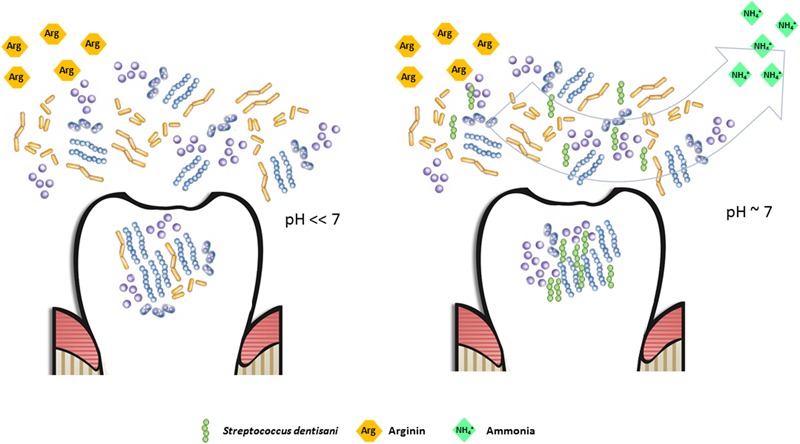
Potential buffering effect in the oral cavity due to the enrichment of the dental plaque bacterial community with Streptococcus dentisani. When arginine is available (either as an added prebiotic or as a product of peptide degradation) and S. dentisani is established on the tooth surfaces, the pH drop caused by the accumulation of organic acids can be buffered by the production of NH4+ through the arginolytic pathway, maintaining the pH close to neutrality (right panel). In the absence of S. dentisani, or if present at low proportions, the buffering effect would be diminished, with the consequent acidification and enamel damage. This would be the perfect scenario for the proliferation of acidogenic/cariogenic microorganisms (left panel).
The double beneficial action of S. dentisani (i.e., anti-microbial and anti-acid) makes it a promising probiotic bacterium. These benefits, together with its dental colonization capacity, derive to a big extent from its oral inhabitance. In addition, the choice of a dental plaque species as probiotic allows the quantification of the species in caries-free individuals, in order to use those levels as the appropriate administration dose, instead of using the dosages normally delivered for gut probiotics. In the current manuscript, we have quantified S. dentisani amounts in dental plaque for just two individuals and similar estimates for larger sample sizes could serve to accurately determine the dosage for an appropriate treatment.
For all these reasons, we propose the use of probiotics which are active colonizers, that is microorganisms which inhabit the site where the disease takes place, and that are isolated from healthy individuals. We believe that the administration of these organisms will maximize the chance of colonization and the potential beneficial effects for human and animal health. In the case of dental caries, we encourage the search of probiotic bacteria that are normal inhabitants of the human supragingival plaque in healthy individuals and propose the use of S. dentisani in clinical trials to test its potential in promoting oral health.
Author Contributions
AM conceived the work, was implied in the analyses and interpretation of data and wrote the manuscript together with AL-L. AL-L, AC-C, and MF worked in the designing and performance of the experiments, data acquisition, analyses, and interpretation of the results. AS-S collected the plaque samples and made the bioinformatic analyses.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The research leading to these results has received funding from the Spanish Government under grant agreements BIO2012-40007 and CSD2009-0006.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00379/full#supplementary-material
Growth curves of Streptococcus dentisani strains 7746 (A) and 7747 (B) in BHI medium at different starting pHs: 4.7 (red), 5.5 (orange), 6 (yellow), 6.5 (light blue), 7 (dark blue), and 7.5 (purple).
References
- Alcaraz L. D., Belda-Ferre P., Cabrera-Rubio R., Romero H., Simón-Soro A., Pignatelli M., et al. (2012). Identifying a healthy oral microbiome through metagenomics. Clin. Microbiol. Infect. 18 54–57. 10.1111/j.1469-0691.2012.03857.x [DOI] [PubMed] [Google Scholar]
- Aminabadi N. A., Erfanparast L., Ebrahimi A., Oskouei S. G. (2011). Effect of chlorhexidine pretreatment on the stability of salivary lactobacilli probiotic in six- to twelve-year-old children: a randomized controlled trial. Caries Res. 45 148–154. 10.1159/000325741 [DOI] [PubMed] [Google Scholar]
- Badet C., Thebaud N. B. (2008). Ecology of Lactobacilli in the oral cavity: a review of literature. Open Microbiol. J. 2 38–48. 10.2174/1874285800802010038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beighton D., Al-Haboubi M., Mantzourani M., Gilbert S. C., Clark D., Zoitopoulos L., et al. (2010). Oral Bifidobacteria: caries-associated bacteria in older adults. J. Dent. Res. 89 970–974. 10.1177/0022034510369319 [DOI] [PubMed] [Google Scholar]
- Belda-Ferre P., Alcaraz L. D., Cabrera R., Simon-Soro A., Romero H., Pignatelli M., et al. (2011). The oral metagenome in health and disease. ISME J. 6 46–56. 10.1038/ismej.2011.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden G. H. W., Ellwood D. C., Hamilton I. R. (1979). “Microbial ecology of the oral cavity,” in Advances in Microbial Ecology ed. Alexander M. (New York, NY: Plenum Press; ) 135–217. 10.1007/978-1-4615-8279-3_4 [DOI] [Google Scholar]
- Burton J. P., Chilcott C. N., Moore C. J., Speiser G., Tagg J. R. (2006). A preliminary study of the effect of probiotic Streptococcus salivarius K12 on oral malodour parameters. J. Appl. Microbiol. 100 754–764. 10.1111/j.1365-2672.2006.02837.x [DOI] [PubMed] [Google Scholar]
- Burton J. P., Drummond B. K., Chilcott C. N., Tagg J. R., Thomson W. M., Hale J. D., et al. (2013). Influence of the probiotic Streptococcus salivarius strain M18 on indices of dental health in children: a randomized double-blind, placebo-controlled trial. J. Med. Microbiol. 62 875–884. 10.1099/jmm.0.056663-0 [DOI] [PubMed] [Google Scholar]
- Cagetti M. G., Marstroberardino S., Milia E., Cocco F., Lingström P., Campus G. (2013). The use of probiotic strains in caries prevention: a systematic review. Nutrients 5 2530–2550. 10.3390/nu5072530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caglar E., Kuscu O. O., Cildir S. K., Kuvvetli S. S., Sandalli N. (2008a). A probiotic lozenge administered medical device and its effect on salivary mutans streptococci and lactobacilli. Int. J. Paedr. Dent. 18 35–39. [DOI] [PubMed] [Google Scholar]
- Caglar E., Kuscu O. O., Selvi Kuvvetli S., Kavaloglu Cildir S., Sandalli N., Twetman S. (2008b). Short-term effect of ice-cream containing Bifidobacterium lactis Bb-12 on the number of salivary mutans streptococci and lactobacilli. Acta Odontol. Scand. 66 154–158. 10.1080/00016350802089467 [DOI] [PubMed] [Google Scholar]
- Caglar E., Sandalli N., Twetman S., Kavaloglu S., Ergeneli S., Selvi S. (2005). Effect of yogurt with Bifidobacterium DN-173 010 on salivary mutans streptococci and lactobacilli in young adults. Acta Odontol. Scand. 63 317–320. 10.1080/00016350510020070 [DOI] [PubMed] [Google Scholar]
- Camelo-Castillo A., Benítez-Páez A., Belda-Ferré P., Cabrera-Rubio R., Mira A. (2014). Streptococcus dentisani sp. nov., a novel member of the Mitis group. Int. J. Syst. Appl. Microbiol. 64 60–65. 10.1099/ijs.0.054098-0 [DOI] [PubMed] [Google Scholar]
- Camelo-Castillo A., Mira A., Pico A., Nibali L., Henderson B., Tomás I. (2015). Subgingival microbiota in health compared to periodontitis and the influence of smoking. Front. Microbiol. 6:119 10.3389/fmicb.2015.00119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campus G., Cocco F., Carta G., Cagetti M. G., Simark-Mattson C., Strohmenger L., et al. (2014). Effect of a daily dose of Lactobacillus brevis CD2 lozenges in high caries risk schoolchildren. Clin. Oral Investig. 18 555–561. 10.1007/s00784-013-0980-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang L. C., Huang C. S., Ou-Yang L. W., Lin S. Y. (2011). Probiotic Lactobacillus paracasei effect on cariogenic bacterial flora. Clin. Oral Investig. 15 471–476. 10.1007/s00784-010-0423-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fejerskov O. (2004). Changing paradigms in concepts on dental caries: consequences for oral health care. Caries Res. 38 182–191. 10.1159/000077753 [DOI] [PubMed] [Google Scholar]
- Firestone A. R., Imfeld T., Schiffer S., Lutz F. (1987). Measurement of interdental plaque pH in humans with an indwelling glass pH electrode following a sucrose rinse: a long-term retrospective study. Caries Res. 21 555–558. 10.1159/000261066 [DOI] [PubMed] [Google Scholar]
- Gonzales T., Robert-Baudouy J. (1996). Bacterial aminopeptidases: properties and functions. FEMS Microbiol. Rev. 18 319–344. 10.1111/j.1574-6976.1996.tb00247.x [DOI] [PubMed] [Google Scholar]
- Grudianov A. I., Dmitrieva N. A., Fomenko E. V. (2002). Use of probiotics Bifidumbacterin and Acilact in tablets in therapy of periodontal inflammations [Article in Russian]. Stomatologiia (Mosk) 81 39–43. [PubMed] [Google Scholar]
- Guglielmetti S., Taverniti V., Minuzzo M., Arioli S., Stuknyte M., Karp M., et al. (2010). Oral bacteria as potential probiotics for the pharyngeal mucosa. Appl. Environ. Microbiol. 76 3948–3958. 10.1128/AEM.00109-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G., Lamont R. J. (2012). Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol. Oral Microbiol. 6 409–419. 10.1111/j.2041-1014.2012.00663.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakka K., Ahola A. J., Yli-Knuuttila H., Richardson M., Poussa T., Meurman J. H., et al. (2007). Probiotics reduce the prevalence of oral candida in the elderly–A randomized controlled trial. J. Dent. Res. 86 125–130. 10.1177/154405910708600204 [DOI] [PubMed] [Google Scholar]
- Huang X., Schulte R. M., Burne R. A., Nascimento M. M. (2015). Characterization of the arginolytic microflora provides insights into pH homeostasis in human oral biofilms. Caries Res. 49 165–176. 10.1159/000365296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto T., Suzuki N., Tanabe T., Takeshita T., Hirofuji T. (2010). Effects of probiotic Lactobacillus salivarius WB21 on halitosis and oral health: an open-labeled pilot trial. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 110 201–208. 10.1016/j.tripleo.2010.03.032 [DOI] [PubMed] [Google Scholar]
- Jensen A., Scholz C. F. P., Kilian M. (2016). Re-evaluation of the taxonomy of the Mitis group of the genus Streptococcus based on whole genome phylogenetic analyses, and proposed reclassification of Streptococcus dentisani as Streptococcus oralis subsp. dentisani comb. nov., Streptococcus tigurinus as Streptococcus oralis subsp. tigurinus comb. nov., and Streptococcus oligofermentans as a later synonym of Streptococcus cristatus. Int. J. Syst. Evol. Microbiol. 66 4803–4820. 10.1099/ijsem.0.001433 [DOI] [PubMed] [Google Scholar]
- Kang M. S., Chung J., Kim S. M., Yang K. H., Oh J. S. (2006a). Effect of Weissella cibaria isolates on the formation of Streptococcus mutans biofilm. Caries Res. 40 418–425. [DOI] [PubMed] [Google Scholar]
- Kang M. S., Kim B. G., Chung J., Lee H. C., Oh J. S. (2006b). Inhibitory effect of Weissella cibaria isolates on the production of volatile sulphur compounds. J. Clin. Periodontol. 33 226–232. [DOI] [PubMed] [Google Scholar]
- Keller M. K., Bardow A., Jensdottir T., Lykkeaa J., Twetman S. (2012). Effect of chewing gums containing the probiotic bacterium Lactobacillus reuteri on oral malodour. Acta Odontol. Scand. 70 246–250. 10.3109/00016357.2011.640281 [DOI] [PubMed] [Google Scholar]
- Khalf M., Dabour N., Kheadr E., Fliss I. (2010). Viability of probiotic bacteria in maple sap products under storage and gastrointestinal conditions. Bioresour. Technol. 101 7966–7972. 10.1016/j.biortech.2010.05.053 [DOI] [PubMed] [Google Scholar]
- Kolenbrander P. E., Andersen R. N., Blehert D. S., Egland P. G., Foster J. S., Palmer R. J., Jr. (2002). Communication among oral bacteria. Microbiol. Mol. Biol. 66 3486–3505. 10.1128/MMBR.66.3.486-505.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasse P., Carlsson B., Dahl C., Paulsson A., Nilsson A., Sinkiewicz G. (2006). Decreased gum bleeding and reduced gingivitis by the probiotic Lactobacillus reuteri. Swed. Dent. J. 30 55–60. [PubMed] [Google Scholar]
- Krespi Y. P., Shrime M. G., Kacker A. (2006). The relationship between oral malodor and volatile sulfur compound-producing bacteria. Otolaryngol. Head Neck Surg. 135 671–676. 10.1016/j.otohns.2005.09.036 [DOI] [PubMed] [Google Scholar]
- Kurtz S., Phillippy A., Delcher A. L., Smoot M., Shumway M., Antonescu C., et al. (2004). Versatile and open software for comparing large genomes. Genome Biol. 5:R12 10.1186/gb-2004-5-2-r12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Dong Y., Chen Y. Y., Burne R. A. (2008). Arginine deiminase gene regulation in Streptococcus gordonii. J. Dent. Res. 87 714–721. [Google Scholar]
- Liu Y. L., Nascimento M., Burne R. A. (2012). Progress toward understanding the contribution of alkali generation in dental biofilms to inhibition of dental caries. Int. J. Oral Sci. 4 135–140. 10.1038/ijos.2012.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantzourani M., Fenlon M., Beighton D. (2009a). Association between Bifidobacteriaceae and the clinical severity of root caries lesions. Mol. Oral Microbiol. 24 32–37. 10.1111/j.1399-302X.2008.00470.x [DOI] [PubMed] [Google Scholar]
- Mantzourani M., Gilbert S. C., Sulong H. N., Sheehy E. C., Tank S., Fenlon M., et al. (2009b). The isolation of bifidobacteria from occlusal carious lesions in children and adults. Caries Res. 43 308–313. 10.1159/000222659 [DOI] [PubMed] [Google Scholar]
- Marsh P. D. (2015). The commensal microbiota and the development of human disease–an introduction. J. Oral Microbiol. 7:29128 10.3402/jom.v7.29128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh P. D., Head D. A., Devine D. A. (2015). Ecological approaches to oral biofilms: control without killing. Caries Res. 49 46–54. 10.1159/000377732 [DOI] [PubMed] [Google Scholar]
- Martínez R. C. R., Aynaou A., Albrecht S., Schols H. A., De Martinis E. C. P., Zoetendal E. G., et al. (2011). In vitro evaluation of gastrointestinal survival of Lactobacillus amylovorus DSM 16698 alone and combined with galactooligosaccharides, milk and/or Bifidobacterium animalis subsp. lactis Bb-12. Int. J. Food Microbiol. 149 152–158. 10.1016/j.ijfoodmicro.2011.06.010 [DOI] [PubMed] [Google Scholar]
- Matsuoka T., Sugano N., Takigawa S., Takane M., Yoshimura N., Ito K., et al. (2006). Effect of oral Lactobacillus salivarius TI 2711 (LS1) administration on periodontopathic bacteria in subgingival plaque. J. Jpn. Soc. Periodontol. 48 315–324. 10.2329/perio.48.315 [DOI] [Google Scholar]
- Mayanagi G., Kimura M., Nakaya S., Hirata H., Sakamoto M., Benno Y., et al. (2009). Probiotic effects of orally administered Lactobacillus salivarius WB21-containing tablets on periodontopathic bacteria. J. Clin. Periodontol. 36 506–513. 10.1111/j.1600-051X.2009.01392.x [DOI] [PubMed] [Google Scholar]
- Moncada G., Maureira J., Neira M., Reyes E., Oliveira Junior O. B., Faleiros S., et al. (2015). Salivary urease and ADS enzymatic activity as endogenous protection against dental caries in children. J. Clin. Pediatr. Dent. 39 358–363. 10.17796/1053-4628-39.4.358 [DOI] [PubMed] [Google Scholar]
- Petersen P. E., Lennon M. A. (2004). Effective use of fluorides for the prevention of dental caries in the 21st century: the WHO approach. Community Dent. Oral Epidemiol. 32 319–321. 10.1111/j.1600-0528.2004.00175.x [DOI] [PubMed] [Google Scholar]
- Pham L. C., van Spanning R. J. M., Roling W. F. M., Prosperi A. C., Terefework Z., ten Cate J. M., et al. (2009). Effects of probiotic Lactobacillus salivarius W24 on the compositional stability of oral microbial communities. Arch. Oral Microbiol. 54 132–137. 10.1016/j.archoralbio.2008.09.007 [DOI] [PubMed] [Google Scholar]
- Plonka K. A., Pukallus M. L., Barnett A. G., Walsh L. J., Holcombe T. F., Seow W. K. (2012). A longitudinal study comparing mutans streptococci and lactobacilli colonisation in dentate children aged 6 to 24 months. Caries Res. 46 385–393. 10.1159/000339089 [DOI] [PubMed] [Google Scholar]
- Power D. A., Burton J. P., Chilcott C. N., Dawes P. J., Tagg J. R. (2008). Preliminary investigations of the colonisation of upper respiratory tract tissues of infants using a paediatric formulation of the oral probiotic Streptococcus salivarius K12. Eur. J. Clin. Microbiol. Infect. Dis. 27 1261–1263. 10.1007/s10096-008-0569-4 [DOI] [PubMed] [Google Scholar]
- Pozharitskaia M. M., Morozova L. V., Mel’nichuk G. M., Mel’nichuk S. S. (1994). The use of the new bacterial biopreparation Acilact in the combined treatment of periodontitis [Article in Russian]. Stomatologiia (Mosk) 73 17–20. [PubMed] [Google Scholar]
- Reid G., Younes J. A., Van der Mei H. C., Gloor G. B., Knight R., Busscher H. J. (2011). Microbiota restoration: natural and supplemented recovery of human microbial communities. Nat. Rev. Microbiol. 9 27–38. 10.1038/nrmicro2473 [DOI] [PubMed] [Google Scholar]
- Reyes E., Martin J., Moncada G., Neira M., Palma P., Gordan V., et al. (2014). Caries-free subjects have high levels of urease and arginine deiminase activity. J. Appl. Oral Sci. 22 235–240. 10.1590/1678-775720130591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccia D. D., Bizzini F., Perilli M., Polimeni A., Trinchieri V., Amicosante G., et al. (2007). Anti-inflammatory effects of Lactobacillus brevis (CD2) on periodontal disease. Oral Dis. 13 376–385. 10.1111/j.1601-0825.2006.01291.x [DOI] [PubMed] [Google Scholar]
- Ritthagol W., Saetang C., Teanpaisan R. (2014). Effect of probiotics containing Lactobacillus paracasei SD1 on salivary mutans Streptococci and Lactobacilli in orthodontic cleft patients: a double-blinded, randomized, placebo-controlled study. Cleft Palate Craniofac. J. 51 257–263. 10.1597/12-243 [DOI] [PubMed] [Google Scholar]
- Saha S., Tomaro-Duchesneau C., Tabrizian M., Prakash S. (2012). Probiotics as oral health biotherapeutics. Exp. Opin. Biol. Ther. 12 1207–1220. 10.1517/14712598.2012.693474 [DOI] [PubMed] [Google Scholar]
- Santarpia P., Lavender S., Gittins E., Vandeven M., Cummins D., Sullivan R. (2014). A 12-week clinical study assessing the clinical effects on plaque metabolism of a dentifrice containing 1.5% arginine, an insoluble calcium compound and 1,450 ppm fluoride. Am. J. Dent. 27 100–105. [PubMed] [Google Scholar]
- Shimauchi H., Mayanagi G., Nakaya S., Minamibuchi M., Ito Y., Yamaki K., et al. (2008). Improvement of periodontal condition by probiotics with Lactobacillus salivarius WB21: a randomized, double-blind, placebo-controlled study. J. Clin. Periodontol. 35 897–905. 10.1111/j.1600-051X.2008.01306.x [DOI] [PubMed] [Google Scholar]
- Simon-Soro A., Guillen-Navarro M., Mira A. (2014). Metatranscriptomics reveals overall active bacterial composition in caries lesions. J. Oral Microbiol. 6:25443 10.3402/jom.v6.25443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simón-Soro A., Mira A. (2015). Solving the etiology of dental caries. Trends Microbiol. 23 76–82. 10.1016/j.tim.2014.10.010 [DOI] [PubMed] [Google Scholar]
- Simon-Soro A., Tomás I., Cabrera-Rubio R., Catalan M. D., Nyvad B., Mira A. (2013). Microbial geography of the oral cavity. J. Dent. Res. 92 616–621. 10.1177/0022034513488119 [DOI] [PubMed] [Google Scholar]
- Stecksén-Blicks C., Sjöström I., Twetman S. (2009). Effect of long-term consumption of milk supplemented with probiotic lactobacilli and fluoride on dental caries and general health in preschool children: a cluster-randomized study. Caries Res. 43 374–381. 10.1159/000235581 [DOI] [PubMed] [Google Scholar]
- Suzuki N., Yoneda M., Tanabe K., Fujimoto A., Iha K., Seno K., et al. (2014). Lactobcaillus salivarius WB21-containing tablets for the treatment of oral malodor: a double blind, randomized, placebo-controlled crossover trial. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 117 462–470. 10.1016/j.oooo.2013.12.400 [DOI] [PubMed] [Google Scholar]
- ten Cate J. M., Zaura E. (2012). The numerous microbial species in oral biofilms: how could antibacterial therapy be effective? Adv. Dental Res. 24 108–111. 10.1177/0022034512450028 [DOI] [PubMed] [Google Scholar]
- Turnbaugh P. J., Ley R. E., Hamady M., Fraser-Liggett C., Knight R., Gordon J. I. (2007). The human microbiome project: exploring the microbial part of ourselves in a changing world. Nature 18 804–810. 10.1038/nature06244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volozhin A. I, Il’in V. K., Maksimovskiĭ IuM, Sidorenko A. B., Istranov L. P., Tsarev V. N., et al. (2004). Development and use of periodontal dressing of collagen and Lactobacillus casei 37 cell suspension in combined treatment of periodontal disease of inflammatory origin (a microbiological study). Stomatologiia (Mosk) 83 6–8. [PubMed] [Google Scholar]
- World Health Organization [WHO] (1997). Oral Health Surveys: Basic Methods. Geneva: World Health Organization. [Google Scholar]
- Wilson M. (2005). Microbial Inhabitants of Humans: Their Ecology and Role in Health and Disease. Cambridge: Cambridge University Press. [Google Scholar]
- Wu C. C., Lin C. T., Wu C. Y., Peng W. S., Lee M. J., Tsai Y. C. (2015). Inhibitory effect of Lactobacillus salivarius on Streptococcus mutans biofilm formation. Mol. Oral Microbiol. 30 16–26. 10.1111/omi.12063 [DOI] [PubMed] [Google Scholar]
- Yin W., Hu D. Y., Li X., Fan X., Zhang Y. P., Pretty I. A., et al. (2013). The anti-caries efficacy of a dentifrice containing 1.5% arginine and 1450 ppm fluoride as sodium monofluorophosphate assessed using Quantitative Light-induced Fluorescence (QLF). J. Dent. 41(Suppl. 2) S22–S28. 10.1016/j.jdent.2010.04.004 [DOI] [PubMed] [Google Scholar]
- Zahradnik R. T., Magnusson I., Walker C., McDonell E., Hillman C. H., Hillman J. D. (2009). Preliminary assessment of safety and effectiveness in humans of ProBiora3, a probiotic mouthwash. J. Appl. Microbiol. 107 682–690. 10.1111/j.1365-2672.2009.04243.x [DOI] [PubMed] [Google Scholar]
- Zaura E., Keijser B. J. F., Huse S., Crielaard W. (2009). Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 9:259 10.1186/1471-2180-9-259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Kreth J. (2012). The role of hydrogen peroxide in environmental adaptation of oral microbial communities. Oxid. Med. Cell. Longev. 2012:717843 10.1155/2012/717843 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Growth curves of Streptococcus dentisani strains 7746 (A) and 7747 (B) in BHI medium at different starting pHs: 4.7 (red), 5.5 (orange), 6 (yellow), 6.5 (light blue), 7 (dark blue), and 7.5 (purple).


