Abstract
The chipmunk hibernation-related protein 25 (HP-25) is involved in the circannual control of hibernation in the brain. The liver-specific expression of the HP-25 gene is repressed in hibernating chipmunks under the control of endogenous circannual rhythms. However, the molecular mechanisms that differentially regulate the HP-25 gene during the nonhibernation and hibernation seasons are unknown. Here, we show that the hibernation-associated HP-25 expression is regulated epigenetically. Chromatin immunoprecipitation analyses revealed that significantly less hepatocyte nuclear receptor HNF-4 bound to the HP-25 gene promoter in the liver of hibernating chipmunks compared to nonhibernating chipmunks. Concurrently in the hibernating chipmunks, coactivators were dissociated from the promoter, and active transcription histone marks on the HP-25 gene promoter were lost. On the other hand, small heterodimer partner (SHP) expression was upregulated in the liver of hibernating chipmunks. Overexpressing SHP in primary hepatocytes prepared from nonhibernating chipmunks caused HNF-4 to dissociate from the HP-25 gene promoter, and reduced the HP-25 mRNA level. These results suggest that hibernation-related HP-25 expression is epigenetically regulated by the binding of HNF-4 to the HP-25 promoter, and that this binding might be modulated by SHP in hibernating chipmunks.
Of the many physiological seasonal adaptations that occur in animals in response to the physical world, hibernation is one of the most extreme. Mammalian hibernation is a unique circannual physiological adaptation that allows life to be sustained at extremely low body temperatures, which can fall below 10 °C or even 5 °C. During hibernation, heart and breathing rates drop, and the metabolic rate falls to only a few percent of the euthermic level, resulting in a considerable conservation of energy1. The mechanistic basis of these dynamic physiological changes in hibernation is not well understood.
The Tamias asiaticus (chipmunk) hibernation-related protein 25 is a component in a 140-kDa complex, consisting of HP-20, HP-25, HP-27, and HP-55, that decreases drastically in the blood during hibernation2. HP-20, HP-25, and HP-27 contain collagen-like Gly-X-Y repeats near the N terminus and a globular domain in the C terminus, and are highly homologous to each other2,3. The 140-kDa complex starts decreasing before the onset of hibernation, stays at reduced levels during hibernation, and starts increasing before hibernation ends2. Kondo et al. showed that the onset of hibernation is marked by an increase in the HP-20c protein complex in the brain; this complex is composed of HP-20, HP-25, and HP-274. Furthermore, hibernation is shortened when an antibody against HP-20c is administered via the lateral ventricles4, indicating that HP-20c has a critical role in hibernation.
The HP-25 gene, which is expressed specifically in the liver, is downregulated in hibernating chipmunks3. The HP-25 mRNA level in the liver fluctuates in parallel with levels of the 140-kDa complex in the blood. Since the levels of the 140-kDa complex fluctuate independently of changes in body temperature4, the HP-25 mRNA expression in the liver is thought to be controlled by endogenous circannual rhythms, rather than by changes in body temperature or by metabolic inhibition at low body temperatures. Thus, elucidating the mechanism of such hibernation-associated endogenous circannual rhythms will shed light on the molecular mechanisms of mammalian hibernation.
Our previous in vitro studies revealed that hepatocyte nuclear factor 4 (HNF-4) activates HP-25 transcription5. HNF-4 is a member of the nuclear receptor family6. Fatty acids and linoleic acids are assumed to be the endogenous ligands of HNF-47,8. HNF-4 is mainly expressed in the liver and binds to direct repeat 1 like motifs as a homodimer6,9. HNF-4 is a critical transcriptional regulator for many genes specifically expressed in the liver10. In addition, HNF-4 interacts with various partner proteins that change its transcriptional activity11,12. HNF-4 can also recruit several coactivators that modulate chromatin13.
In this study, we investigated the mechanism of the hibernation-associated loss of HP-25 expression in chipmunks. We first analyzed whether the hibernation-associated loss of HP-25 expression is regulated at the transcriptional level. Subsequently, we focused on the HNF-4-regulated transcriptional activity and on hibernation-associated changes in epigenetic marks on the HP-25 gene promoter region. Our results suggest that HNF-4 and its coactivators bind the HP-25 promoter region to epigenetically regulate the HP-25 transcription changes associated with circannual hibernation.
Results
HP-25 gene transcription is regulated in association with hibernation
We previously showed that the chipmunk HP-25 gene is downregulated in the liver of hibernating chipmunks, based on Northern analysis3. However, Northern analysis only indirectly measures gene transcription, since mRNA levels represent the sum of both transcriptional and post-transcriptional events. Here, we verified our initial finding by analyzing the levels of HP-25 primary transcripts; this approach to determining a gene’s transcriptional state is an alternative to the nuclear run-on assay14. Total RNA was isolated from the liver of six nonhibernating chipmunks and six hibernating chipmunks, and the level of HP-25 primary transcripts was compared by RT-PCR using a pair of primers, one located in exon 1 and the other in intron 1 (Fig. 1 and Supplementary Fig. S1). The PCR amplimers were confirmed to be a single band of the correct size by Southern blot analysis (Supplementary Fig. S1). The HP-25 mRNA level was also analyzed by RT-PCR using a pair of primers, one located in exon 2 and the other in exon 3. Since the albumin mRNA level remains constant regardless of hibernation state3, we used the level of albumin primary transcripts as a control. The HP-25 mRNA level was reduced as previously reported3, and similarly, the level of HP-25 primary transcripts was greatly reduced in the hibernating chipmunks compared with nonhibernating chipmunks. Although the albumin primary transcript levels differed slightly between individuals, the level was almost constant in nonhibernating and hibernating chipmunks. These results indicate that the HP-25 gene is in a transcriptionally active state in the liver of nonhibernating chipmunks and shifts to a transcriptionally repressed state in the liver of hibernating chipmunks. This observation was in clear contrast to the constitutively active albumin gene.
Figure 1. HP-25 is regulated at the transcriptional level in association with hibernation.
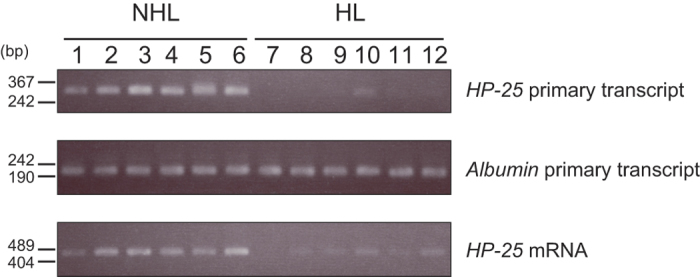
HP-25 primary transcript, HP-25 mRNA and albumin primary transcript were amplified by RT-PCR using total RNA extracted from the liver of nonhibernating (NHL; lanes 1–6) and hibernating chipmunks (HL; lanes 7–12). The PCR products were separated by electrophoresis on a 2% agarose gel.
Amount of HNF-4 bound to the HP-25 gene promoter decreased during hibernation
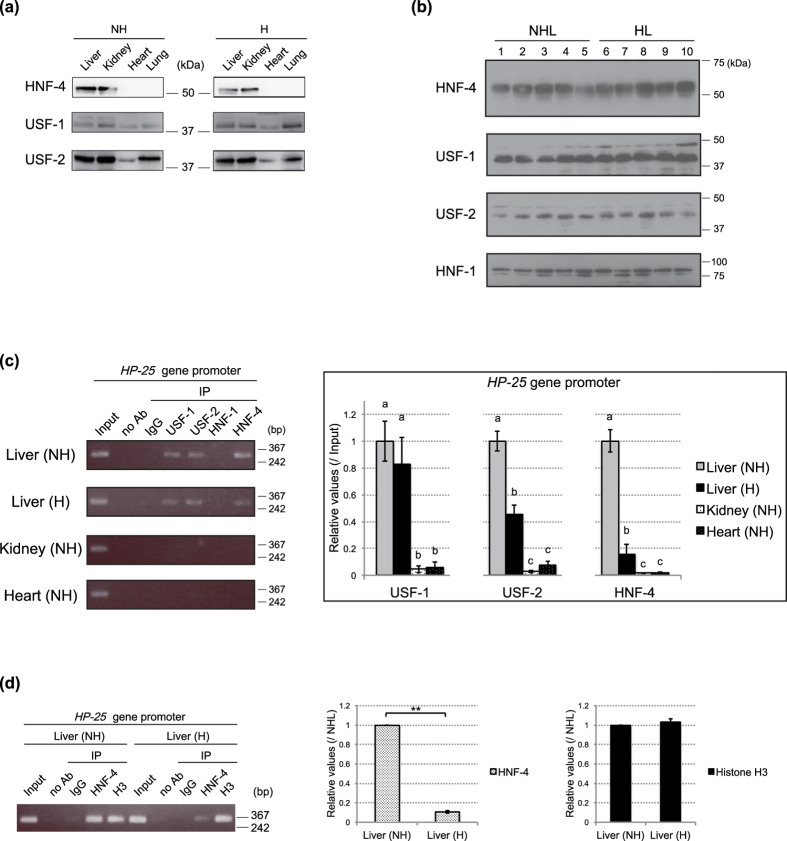
Our previous in vitro studies revealed that the HP-25 gene transcription is activated by HNF-4, which is a hepatocyte-enriched transcription factor belonging to the nuclear receptor superfamily, and by upstream stimulatory factor (USF), which is a helix-loop-helix transcription factor5,15. Here, immunoblotting analyses of nuclear extracts from several tissues of nonhibernating and hibernating chipmunks using antibodies validated for corresponding chipmunk proteins (Supplementary Fig. S2) revealed that HNF-4, USF-1, and USF-2 were present in the kidney, where the HP-25 gene is not expressed [HP-25 mRNA was detected only in the liver, but not in the kidney, by northern analysis3], as well as in the liver (Fig. 2a). Furthermore, although the amounts of HNF-4 and USF in the liver differed between individuals to some extent, the average amounts were comparable in nonhibernating and hibernating chipmunks (Fig. 2b and Supplementary Fig. S3).
Figure 2. Amount of HNF-4 bound to the HP-25 promoter increased in the nonhibernation season.
(a) Immunoblot analyses of HNF-4, USF-1, and USF-2 using nuclear extracts prepared from the liver, kidney, heart, and lung of a nonhibernating (NH) and a hibernating (H) chipmunk. (b) Immunoblot analyses of HNF-4, USF-1, USF-2, and HNF-1 were performed as in (a) using the liver of five nonhibernating (NHL; lanes 1–5) and five hibernating chipmunks (HL; lanes 6–10). (c) Binding of transcription factors to the HP-25 gene promoter was analyzed by ChIP (left panel) and ChIP-qPCR (right panel). ChIP and ChIP-qPCR were performed with chromatin from the liver, kidney, and heart of a nonhibernating chipmunk (NH) and the liver of a hibernating chipmunk (H) using the indicated antibodies, normal rabbit IgG (IgG), or no antibody (no Ab). Following DNA purification, the samples were analyzed by PCR using primer set specific for the HP-25 gene promoter region. One hundredth of four percent of the total input sample (Input) was also examined by PCR. Results are representative of more than three each of nonhibernating and hibernating chipmunks. In ChIP-qPCR, the values were normalized to the total input values, and the results are shown as the fold increase over the values for the liver of the nonhibernating chipmunk (NH). Results are means ± SEM for two independent experiments using the samples prepared from different individuals. Different letters (a–c) are significantly different at p < 0.05; one-way ANOVA with Tukey-Kramer post test. (d) ChIP (left panel) and ChIP-qPCR (middle and right panels) were performed with the indicated antibodies using chromatin from the liver of a nonhibernating chipmunk (NH) and a hibernating chipmunk (H) different from those used in (c). In ChIP-qPCR, the values were normalized as in (c). Results are means ± SEM for three independent experiments. **p < 0.01; Student’s t-test.
We next used chromatin immunoprecipitation (ChIP) to determine whether HNF-4, USF-1, and USF-2 were involved in the liver-specific transcription of the HP-25 gene (Fig. 2c). In the kidney, these transcription factors were hardly present on the promoter region of the HP-25 gene (Fig. 2c). In the liver of nonhibernating chipmunks, USF-1, USF-2, and HNF-4 bound to the HP-25 promoter region, as expected from our previous results5,15. To assess whether HNF-4 binds to the HP-25 gene promoter liver-specifically, we compared HNF-4-binding to the promoter regions of the HP-25 and Pklr genes by ChIP (Supplementary Fig. S4). While the HP-25 gene is expressed in liver but not in kidney3, the Pklr gene is expressed in both liver and kidney (Supplementary Fig. S4a). The results showed that HNF-4 bound to the Pklr gene promoter in both liver and kidney, but to the HP-25 gene promoter liver-specifically (Fig. 2c and Supplementary Fig. S4d), consistent with its liver-specific transcription. Of note, USF-1, USF-2, and HNF-4 bound to the HP-25 gene promoter region even in the liver of hibernating chipmunks (Fig. 2c), where this gene is transcriptionally repressed. However, the amount of HNF-4 bound to the HP-25 gene promoter region was drastically decreased in hibernating chipmunks (Fig. 2c). In the case of the albumin gene, which is transcriptionally upregulated by HNF-116, HNF-1 and Pol II bound the promoter region in the liver of both nonhibernating and hibernating chipmunks (Supplementary Fig. S5), consistent with its constitutively active transcription. ChIP-qPCR showed that about 10 times more HNF-4 was bound to the HP-25 gene promoter in the liver of nonhibernating compared to hibernating chipmunks (Fig. 2d). On the other hand, the amount of histone H3 on the HP-25 gene promoter region was almost the same in the liver of both nonhibernating and hibernating chipmunks (Fig. 2d). These findings suggested that the dissociation of HNF-4 from the HP-25 gene promoter region is one of the major causes of the repression of HP-25 gene transcription in hibernating chipmunks.
Histone modifications on the HP-25 gene promoter region change in association with hibernation
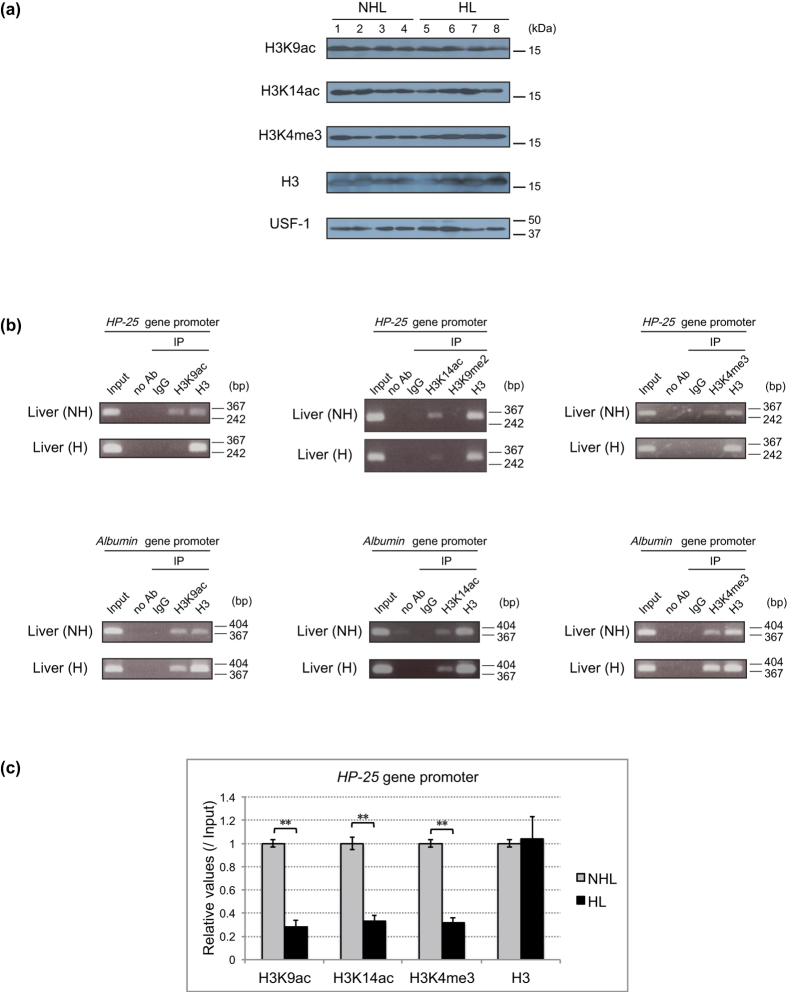
The acetylation and methylation of histone at certain residues, such as the trimethylation of histone H3 at lysine (K) 4 and the acetylation at K9 and K14, are generally involved in gene activation, whereas the lack of histone acetylation is a hallmark of gene repression17,18,19,20. Histone modifications for active transcription destabilize higher-order chromatin structures and render nucleosomal DNA more accessible to transcription factors21. To examine the involvement of epigenetic regulation in HP-25 gene transcription, we first compared the total amount of histone H3 acetylated at K9 and K14 and trimethylated at K4 in the liver between nonhibernating and hibernating chipmunks by immunoblotting (Fig. 3a). The total amounts of the modified histones in the liver were almost equal between nonhibernating and hibernating chipmunks. We then analyzed the modification states of the histone H3 specifically bound to the HP-25 gene promoter region by ChIP (Fig. 3b,c). Histone H3 bound to the HP-25 promoter region was significantly more acetylated at K9 and K14 and more trimethylated at K4 in the liver of nonhibernating chipmunks than in hibernating chipmunks (Fig. 3b upper part and 3c), but H3K9 dimethylation, one of the repressive histone modifications, was not detected even in hibernating chipmunks (Fig. 3b middle upper part). On the other hand, the histone H3 bound to the albumin promoter region was almost equally acetylated at K9 and K14 and trimethylated at K4 in the liver of nonhibernating and hibernating chipmunks (Fig. 3b lower part). These results were consistent with our findings on the transcriptional states of the HP-25 and albumin genes (Fig. 1). Considering that the total amounts of the modified histones were similar between nonhibernating and hibernating chipmunks (Fig. 3a), these histone modifications are probably regulated at the promoter region of specific genes involved in hibernation, including at least the HP-25 gene.
Figure 3. Increased active transcription histone markers on the HP-25 promoter in the liver of nonhibernating chipmunks.
(a) Comparison of the amounts of modified histones in the liver of nonhibernating and hibernating chipmunks. Immunoblotting was performed as in Fig. 2b using nuclear extracts from the liver of four nonhibernating chipmunks (NHL; lanes 1–4) and four hibernating chipmunks (HL; lanes 5–8). USF-1 was used as a loading control. (b) ChIP was performed as in Fig. 2c with chromatin from the liver of a nonhibernating chipmunk (NH) and a hibernating chipmunk (H) using the indicated antibodies and primer sets specific for the indicated gene promoter regions. (c) ChIP-qPCR was performed as in Fig. 2d with chromatin from the liver of a nonhibernating (NHL) and a hibernating chipmunk (HL) using the indicated antibodies. Results are means ± SEM for three independent experiments performed in technical triplicate. **p < 0.01. The chipmunks used in (b) and (c) are different individuals.
HNF-4 and coactivators activate HP-25 transcription in the liver of nonhibernating chipmunks
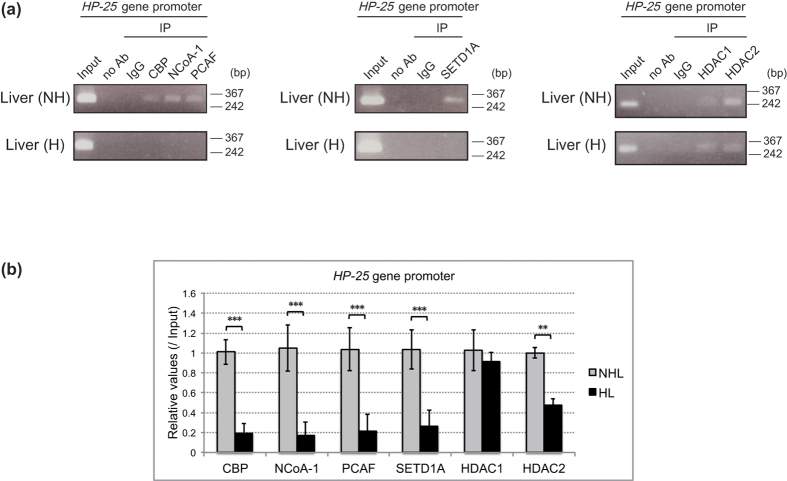
To elucidate the mechanism of the hibernation-associated changes in histone modifications on the HP-25 gene promoter, we used ChIP to analyze the histone acetyltransferases (HATs), histone methyltransferases (HMTs), and histone deacetylases (HDACs) bound to the HP-25 promoter region (Fig. 4a,b). Histone H3 can be acetylated at K9 and K14 by HATs such as CBP, NCoA-1, PCAF, p300, GCN5, MOZ, MORF, and TFIIIC, and can be methylated at K4 by HMTs such as SETD1A, SETD1B, MLL1, MLL2, and ASH1L17,19,22,23. HDACs are classified into groups I–IV, and the class I HDACs HDAC1, HDAC2, HDAC3, and HDAC8 generally function as subunits in multiprotein complexes that are crucial for repressing transcription24. ChIP analyses revealed that HDAC1 and HDAC2 bound the HP-25 promoter region in the liver of both nonhibernating and hibernating chipmunks (Fig. 4a right panel and 4b). In contrast, CBP, NCoA-1, PCAF, and SETD1A were found on the HP-25 gene promoter mainly in the liver of nonhibernating chipmunks (Fig. 4a left and middle panels and 4b). Thus, hibernation-associated histone modifications are likely to be modulated by HAT and HMT, which are associated with the HP-25 gene promoter in nonhibernating chipmunks and dissociated from the promoter region in hibernating chipmunks.
Figure 4. Coactivators enhance the HP-25 transcriptional activation in nonhibernating chipmunks.
(a) ChIP was performed as in Fig. 2c using the indicated antibodies. (b) ChIP-qPCR was performed as in Fig. 2d with chromatin from the liver of two nonhibernating (NHL) and two hibernating chipmunks (HL) using the indicated antibodies. Results are means ± SEM for two independent experiments performed in technical triplicate. **p < 0.01, ***p < 0.001. The chipmunks used in (a) and (b) are different individuals.
To investigate the involvement of HATs and HMT(s) in activating the HP-25 gene transcription, we transiently transfected HeLa cells with an HP-25 gene promoter-reporter plasmid and expression plasmids for CBP, SETD1A, and HNF-4 singly (Supplementary Fig. S6a) or in combination (Supplementary Fig. S6b). Although neither CBP nor SETD1A alone could activate the HP-25 transcription, both could facilitate its transcriptional activation by HNF-4, and the combination of CBP and SETD1A further enhanced the HP-25 transcription (Supplementary Fig. S6a,b). CBP’s enhancement of the HP-25 transcriptional activation was repressed by curcumin, a specific inhibitor of CBP HAT activity25, in a dose-dependent manner (Supplementary Fig. S6c). These results indicate that the HP-25 gene transcription is activated by HNF-4 in combination with HATs and HMT(s) in the liver of nonhibernating chipmunks.
SHP upregulation represses HP-25 transcription by suppressing HNF-4 activity during hibernation
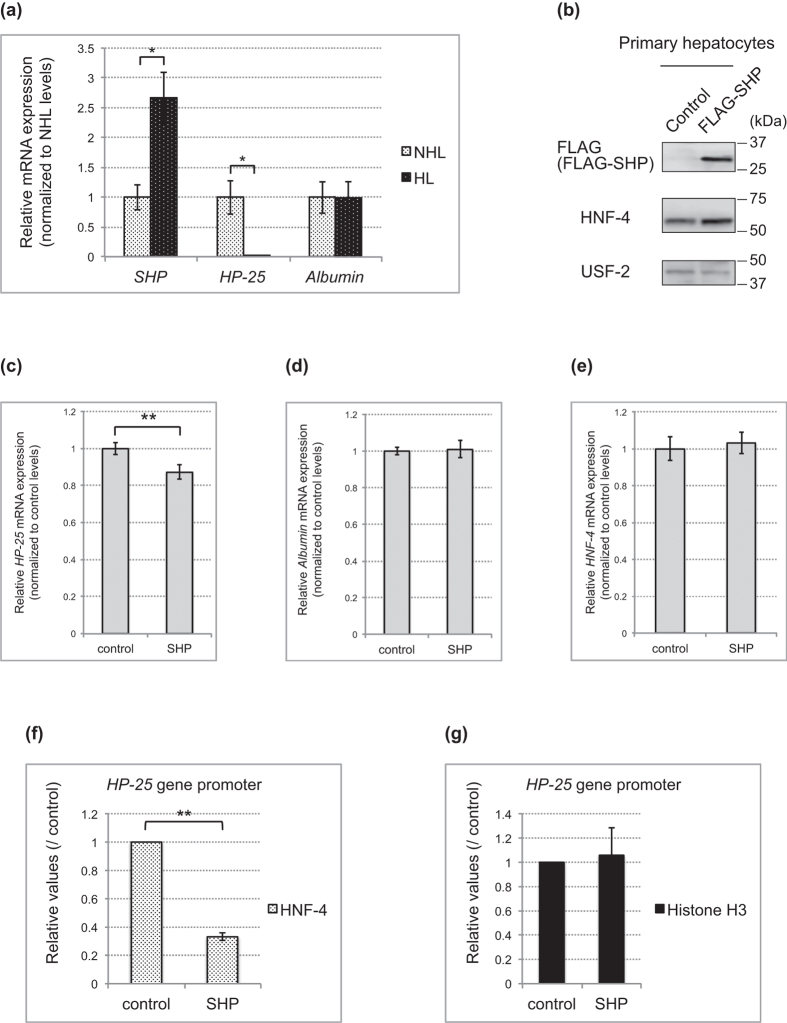
Our results thus far indicated that the HP-25 transcription was probably repressed in hibernating chipmunks due to reduced levels of HNF-4 bound to the promoter (Fig. 2d) and the dissociation of coactivators from the promoter (Fig. 4a,b). Although several microRNAs have been implicated in the translational repression of HNF-4 expression26,27, the observation that the HNF-4 levels were unchanged in nonhibernating and hibernating chipmunks would exclude microRNAs from the cause of this repression. Small heterodimer partner (SHP), which belongs to a group of atypical nuclear receptors that lack a classical DNA-binding domain and are corepressors of a number of nuclear receptors28, represses HNF-4 activity by competing with the coactivator SRC-329. We could not detect endogenous SHP protein expression in the liver with any of several commercial anti-SHP antibodies, so we used RT-qPCR to compare SHP expression in the liver of nonhibernating and hibernating chipmunks (Fig. 5a). Notably, unlike the HP-25 mRNA levels, the amount of SHP mRNA was about 2.5-fold higher in hibernating than in nonhibernating chipmunks. To examine whether chipmunk SHP affects the transcriptional activity of HNF-4, we conducted a transient transfection assay using an HP-25 gene promoter-reporter plasmid and expression plasmids for HNF-4, CBP, and SHP singly (Supplementary Fig. S6d) or in combination (Supplementary Fig. S6e), in HeLa cells. We found that SHP repressed HP-25 gene transcription activated by HNF-4 alone or in combination with CBP (Supplementary Fig. S6d,e). We next examined interactions between chipmunk HNF-4 and chipmunk SHP in cultured cells. In HeLa cells transfected with chipmunk HNF-4 and FLAG-tagged chipmunk SHP, immunocytochemistry using fluorescent and confocal microscopies showed that significant portions of HNF-4 and SHP were colocalized in nucleus (Supplementary Fig. S6f), and HNF-4 and SHP were co-precipitated in a co-immunoprecipitation assay (Supplementary Fig. S6g). Taken together, these results indicate that chipmunk SHP interacts with chipmunk HNF-4 in the nucleus and represses the HP-25 gene transcription activated by HNF-4.
Figure 5. SHP represses HP-25 gene transcription by inhibiting HNF-4 binding to the promoter.
(a) SHP, HP-25, and albumin mRNA levels in the liver of four nonhibernating chipmunks (NHL) and four hibernating chipmunks (HL), measured by RT-qPCR. The results were normalized to the amount of mRNA of the respective genes in the NHL. The results are shown as means ± SEM. *p < 0.05. (b–g) Primary hepatocytes prepared from a nonhibernating chipmunk were transfected with a FLAG-SHP expression plasmid or an empty vector plasmid (control). In (b), whole-cell extracts from the primary hepatocytes were immunoblotted with the indicated antibodies. USF-2 was used as a loading control. In (c–e), total RNA was prepared from the primary hepatocytes (SHP or control), and the endogenous HP-25 (c), albumin (d), and HNF-4 (e) expression was assessed by RT-qPCR. The results were normalized to the amount of mRNA for each gene in control cells. Results show means ± SEM for three independent experiments. **p < 0.01. In (f and g), ChIP-qPCR was performed with chromatin from the primary hepatocytes using anti-HNF-4 (f) or anti-histone H3 (g) antibodies, and the values were normalized to the total input value. Results are shown as the fold increase over the control value. Results show means ± SEM for three independent experiments. **p < 0.01.
Finally, to examine the effect of ectopic SHP expression on endogenous HP-25 gene expression, we transfected a FLAG-tagged SHP-expression construct into primary hepatocytes prepared from a nonhibernating chipmunk. The expression of FLAG-tagged SHP was verified by immunoblotting (Fig. 5b). RT-qPCR analysis revealed that although SHP overexpression did not affect the albumin mRNA levels, the HP-25 mRNA was reduced by 15% (Fig. 5c,d). Ectopic SHP expression did not affect the amount of HNF-4 protein or HNF-4 mRNA (Fig. 5b,e). ChIP-qPCR revealed that SHP overexpression decreased the amount of HNF-4 bound to the HP-25 gene promoter by about 70% (Fig. 5f), although the amount of histone H3 on the HP-25 gene promoter region was almost the same with or without SHP transfection (Fig. 5g). Taken together, these results indicate that SHP expression is upregulated in hibernating chipmunks and that SHP represses HP-25 expression by removing HNF-4 from the promoter.
Discussion
Mammalian hibernation is accompanied by various physiological changes1 that are assumed to be under genetic control. Although there is growing evidence for differential gene expression during hibernation30,31,32,33,34,35,36,37,38, the molecular mechanisms underlying hibernation-associated gene regulation are unknown. In this study, we found that differential HP-25 transcription in the liver between nonhibernating and hibernating chipmunks is epigenetically regulated: H3K9 and K14 were highly acetylated and H3K4 was highly trimethylated in the HP-25 promoter region in nonhibernating chipmunks, but not in hibernating chipmunks. There was a positive correlation among HNF-4 binding, HAT/HMT binding, and histone acetylation/trimethylation levels. Notably, SHP, which negatively regulates HNF-4, was upregulated in the liver of hibernating chipmunks, and ectopically expressing SHP in primary hepatocytes prepared from nonhibernating chipmunks resulted in the dissociation of HNF-4 from the HP-25 promoter and the repression of HP-25 gene transcription.
Our ChIP analyses revealed that in tissues like the kidney, where HP-25 is not expressed, the transcription factors HNF-4, USF-1, and USF-2 were present but hardly bound to the HP-25 promoter region (Fig. 2). These results indicate that the liver-specific transcription of the HP-25 gene is established by assembling a higher order chromatin structure around the promoter region, which is inaccessible by transcription-factors, in nonexpressing tissues. Our previous finding that USF binding to the CACGTG sequence in the HP-27 promoter region is inhibited by CpG methylation in nonexpressing tissues supports this idea39. On the other hand, in the liver of hibernating chipmunks, where HP-25 is transcriptionally repressed, the transcription factors HNF-4, USF-1, and USF-2 still bound the HP-25 gene promoter (Fig. 2), but a repressive histone mark was not observed (Fig. 3b), indicating that the chromatin structures surrounding the HP-25 gene do not change so drastically that they prevent transcription factors from binding, unlike in nonexpressing tissues like the kidney.
HDACs are traditionally considered to be transcriptional corepressors24. However, HDAC1 and HDAC2 were found on the HP-25 promoter region in the liver of nonhibernating chipmunks (Fig. 4a,b). Recent genome-wide mapping revealed that HDACs localize to many active genes40,41. These observations support the idea that HDACs are recruited to active genes to reset the chromatin modification states and maintain an adequate level of histone acetylation40,42, and our result is consistent with this idea. On the other hand, HDACs are also thought to enforce the hypo-acetylated chromatin state of the HP-25 gene in hibernating chipmunks (Fig. 4a,b).
In this study, we showed that the transcriptional repression of the HP-25 gene in hibernating chipmunks results mainly from the dissociation of HNF-4 from the promoter, possibly due to SHP, which is upregulated during hibernation. SHP is proposed to repress gene transcription by interacting with other nuclear receptors via two dimerization LXXLL-related motifs in a ligand-binding domain43, or by binding to the C-terminal activation function-2 helix of other nuclear receptors through these motifs28. Since chipmunk SHP also possesses both LXXLL-related motifs in a ligand-binding domain, chipmunk SHP probably interacts with chipmunk HNF-4 through these two functional motifs. SHP is also reported to interact with lysine-specific histone demethylase 1 (LSD1)44,45, which is associated with a CoREST–SIRT1 repressor complex that removes H3K4 mono-, di- and trimethylation marks45. These reports imply that SHP might also directly repress HP-25 transcription by recruiting histone modification enzymes to the HP-25 gene promoter via an interaction with HNF-4 bound to the promoter.
The HP-25 gene expression in the liver is thought to be regulated by the endogenous circannual rhythm responsible for hibernation. Our results suggest that this hibernation-associated regulation of HP-25 transcription occurs as follows: in the liver of nonhibernating chipmunks, HNF-4 binds the HP-25 promoter region to activate HP-25 transcription in combination with coactivators and active histone modification marks; in hibernating chipmunks, HNF-4 and its coactivators dissociate from the HP-25 gene promoter, possibly by an interaction with SHP (which is now upregulated), and active transcription histone marks are removed from the promoter region (Fig. 6). Since transcription and translation are drastically decreased at low body temperatures46,47,48, these repression processes are likely to occur before or during the entry into hibernation. On the other hand, a reduction in SHP during the nonhibernation season would allow HNF-4 to bind to the HP-25 promoter, which could facilitate H3K4 trimethylation and histone acetylation on the promoter region by recruiting HMTs and HATs, leading to HP-25 transcriptional activation. Together, controlled HNF-4 binding to the HP-25 gene promoter is likely to play a key role in the differential regulation of HP-25 transcription between the nonhibernating and hibernating chipmunks. Recently, it has been reported that not only hibernating mammals but also non-hibernating mammals contain HP-20, HP-25 and HP-27 genes, and in cow, whose hibernation-related protein (HP) genes are expressed specifically in the liver, circulating levels of HP-25 and HP-27 in the blood and cerebrospinal fluid change seasonally as is the case with the chipmunk HPs49,50. Even when chipmunks were kept under conditions of constant warmth (23 °C) with a 12 h:12 h light:dark period cycle, the HP level in the blood and their gene expression in the liver still showed seasonal oscillations similar to those in animals with a low body temperature (Tb) during hibernation season, indicating that the gene expression of HPs is regulated by endogenous circannual rhythms, and independent of Tb changes4. Since there is a HNF-4 binding site in the cow HP-25 gene promoter (data not shown), transcription of the cow HP-25 gene may be regulated by a general nuclear receptor HNF-4 as the chipmunk HP-25 gene. Although it has not been established that cow HPs oscillate in an endogenously generated circannual manner49, the gene regulation of HPs is likely to be under the control of a circannual clock common to chipmunk and cow. In recent years, the presence of a circannual pacemaker in the pars tuberalis of the pituitary gland has been suggested51,52. Identifying the repressors that regulate HP-25 transcription and HNF-4’s binding to the HP-25 promoter poses an interesting challenge. Based on our results, elucidating the upstream factors that control the epigenetic regulation of HP-25 transcription will help to identify the circannual signals from the circannual pacemaker common to hibernators and nonhibernators. Also, comparative analyses of the pathways affecting the HP-25 gene transcriptional regulation between hibernators and nonhibernators, together with functional analysis of the HP-25 protein, may lead to identification of hibernation signals.
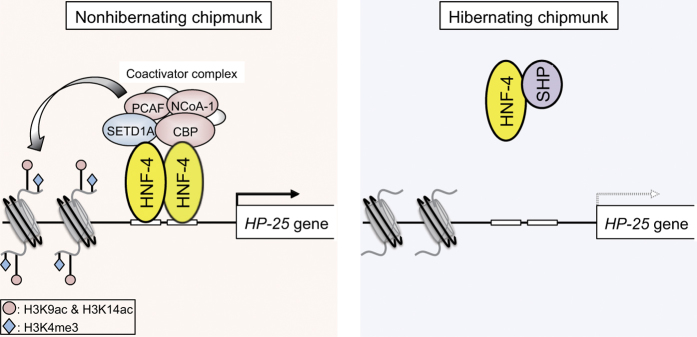
Figure 6. A schematic model of the hibernation-associated transcriptional regulation of the HP-25 gene based on this study.
In nonhibernating chipmunks, HNF-4 binds the HP-25 gene promoter region to activate HP-25 transcription in combination with coactivators and active histone modification marks (left side part). On the other hand, in hibernating chipmunks, HNF-4 and its coactivators dissociate from the promoter, possibly by an interaction of HNF-4 with SHP whose expression is upregulated during hibernation, and active transcription histone marks are removed, resulting in transcriptional repression of the HP-25 gene (right side part).
Material and Methods
Ethics statement
All recombinant DNA experiments in this study strictly followed a protocol and guidelines approved by the Institutional Recombinant DNA Experiment Safety Committee of Kitasato University.
Animals
Male chipmunks (Tamias asiaticus) purchased from Pet Easy Space (Osaka) were individually housed and offered standard rodent chow and water ad libitum. The chipmunks were kept at 23 °C with a 12 h:12 h light:dark period during the nonhibernation season (April–September) and at 5 °C in darkness during the hibernation season (October–March). Nonhibernating chipmunks were summer-active animals sampled in June or July. Hibernating chipmunks were sampled approximately three months after first immergence into torpor. The topor bouts usually last 5–6 days. At the sampling time, these hibernating chipmunks had been hibernating for 3–4 days: they were torpid, and their body surface temperatures were under 10 °C measured by an infrared thermometer. All animals were deeply anesthetized with isoflurane prior to sacrifice. Tissues were immediately excised, frozen in liquid nitrogen, and stored at −80 °C until use. All of the protocols were in accordance with the guidelines of the Institutional Animal Care and Use Committee of Kitasato University, and all experimental procedures were approved by the same committee.
RT-PCR of primary transcripts
Total RNA prepared from the liver using the RNeasy Mini Kit (Qiagen) was treated with DNase I using the TURBO DNA-free kit (Ambion). The reverse transcription of total RNA and subsequent PCR were performed using the mRNA Selective PCR Kit (Takara) to amplify the HP-25 primary transcript, HP-25 mRNA, and albumin primary transcript. The PCR products were fractionated using a 2% agarose gel. The following primer sets were used for PCR: CM25 + 114F_exon1, 5′-ATGCCTGCACAAAGAGG-3′, and CM25 + 409R_intron1, 5′-GGCCTGATATCATTCCTCC-3′; cmHP25 + 216F_exon2, 5′-AATTCTGAACCCTGTGGACCT-3′, and cmHP25 + 683R_exon3, 5′-TTCCAGCCAGACTTTGTCCC-3′; CMALB + 868F_exon2, 5′-GGGAGAGCAAAATTTTAAAGG-3′, and CMALB + 1066R_intron2, 5′-ACCAGTGGCTACCATATTAGA-3′.
Immunoblot analysis
Nuclear extracts were prepared from the liver, kidney, heart, and lung of nonhibernating and hibernating chipmunks39. Immunoblotting was performed as described previously39. The anti-USF-1 (sc-229) and anti-USF-2 (sc-862) antibodies were purchased from Santa Cruz Biotechnology. The anti-HNF-4 antibodies (sc-8987 and H1415) were purchased from Santa Cruz Biotechnology and Perseus Proteomics, respectively. The anti-HNF-1 antibody (H69220) was purchased from Transduction Laboratories. The anti-acetyl-histone H3 (Lys9) (07-352) and anti-acetyl-histone H3 (Lys14) (07-353) antibodies were purchased from Millipore. The anti-histone H3 (tri methyl K4) (ab8580) and anti-histone H3 (ab1791) antibodies were purchased from Abcam. The anti-DYKDDDDK antibody (KO602) was purchased from TransGenic Inc.
ChIP analysis
Chromatin immunoprecipitation (ChIP) was performed as described previously15. All ChIP experiments were performed three or more times using independent chromatin preparations. The normal rabbit IgG (sc-2027), and antibodies against HNF-4 (sc-8987), HNF-1 (sc-8986), Pol II (sc-9001), NCoA-1 (sc-8995), and PCAF (sc-8999) were purchased from Santa Cruz Biotechnology. The anti-dimethyl-histone H3 (Lys9) antibody (07-441) was purchased from Millipore. The anti-HDAC1 (ab7028), anti-HDAC2 (ab7029), and anti-CBP (ab2832) antibodies were purchased from Abcam. The anti-SETD1A antibody (NB100–559A) was purchased from Novus Biologicals. PCR was carried out with the following primer sets: CM25-230F, 5′-TCTTGCTCTGATGGTTTGTTGACAC-3′, and CM25 + 79 R, 5′-GCCTCTCTCCTCTTTAGTTTTCTTC-3′, which amplicon size is 310 bp; CMALB-380F, 5′-ATTTCCAAGCAGAATCATTGG-3′, and CMALB-11R, 5′-ATAGAAAAGGTAGGATGAATG-3′, which amplicon size is 392 bp. ChIP quantitative PCR (ChIP-qPCR) was performed with the indicated number of samples in technical triplicate using SsoFast EvaGreen Supermix (Bio-Rad) with the following primer set: CM25-166F, 5′-GTGACCACGTAAGGGGCAGG-3′, and CM25-24R, 5′-CTGATCTTTCCTGAGGTCAGTGTC-3′, which primer set was more efficient and selective in SsoFast PCR amplification. Both HP-25 gene primer sets, CM25-230F and CM25 + 79 R, and CM25-166F and CM25-24R, can amplify the regions encompassing the USF-1, USF-2, and HNF-4 binding sites. The PCR amplimers were confirmed to be a single band of the correct size by agarose gel electrophoresis. ChIP-qPCR data were normalized by the % input method and were shown as the fold increase over the value in the nonhibernating chipmunk liver. Results are expressed as means ± SEM of the indicated number of independent determinations.
Transfection and luciferase assays
The promoter-reporter plasmid construction of pCM25G-260/luc was described previously5. HeLa cells were cultured as described previously5. Cells were plated at 4 × 104 cells in a 24-well plate. After 24 h, the cells were transfected with 400 ng of a firefly luciferase promoter-reporter plasmid, and with 10 ng of the Renilla luciferase internal control plasmid pRL-SV40 (Promega), using TransIT-LT1 reagent (Mirus). Where denoted, the cells were cotransfected with 100 ng of the mouse CBP expression construct pcDNA3/mCBP, 100 ng of the mouse SETD1A expression construct pcDNA3/mSETD1A, 5 ng of the chipmunk HNF-4 expression construct pcDNA3/cmHNF-4, and/or 100 ng of the chipmunk SHP expression construct pcDNA3/FLAG-cmSHP. After 24 h, the luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega). Where denoted, curcumin (Sigma) was added at the indicated concentrations 12 h after transfection, and the luciferase activity was measured after another 12 h.
Immunocytochemistry
HeLa cells were grown on uncoated glass coverslips and transfected with pcDNA3/cmHNF-4 and pcDNA3/FLAG-cmSHP using Polyethylenimine Max (Polysciences, Inc.). At 24 h after transfection, the cells were fixed with 10% Formaldehyde Neutral Buffer Solution (Nacalai Tesque) for 10 min, mounted on glass slides, and observed with a BZ-8100 fluorescence microscope (Keyence) or an LSM510 confocal laser-scanning microscope (Zeiss). To detect HNF-4 and FLAG-SHP, the cells were incubated with an anti-HNF-4 antibody and anti-DYKDDDDK antibody overnight at 4 °C, washed three times with 0.5% BSA in PBS, and incubated with Alexa Fluor 488 goat anti-rabbit IgG and Alexa Fluor 594 goat anti-mouse IgG (Thermo Scientific) for 1 h at room temperature. To detect nuclei using a fluorescence microscope, the cells were washed twice with PBS and stained with 1 μg/ml Hoechst 33258 (Thermo Scientific) solution for 10 min at room temperature. The cells were then washed with H2O, mounted on a slide, and observed under the microscope.
Co-immunoprecipitation Assays
HeLa cells were plated on a 10-cm dish, and transfected with pcDNA3/cmHNF-4 and pcDNA3/FLAG-cmSHP using Polyethylenimine Max (Polysciences, Inc.). At 48 h after transfection, the cells were lysed in RIPA buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, and protease inhibitor cocktails) for 30 min at 4 °C. The lysates were centrifuged at 12,000 x g for 30 min at 4 °C. The supernatants were collected and incubated with Ab-Capcher (ProteNova) and IgG. The pre-cleared samples were centrifuged, and the supernatants were collected. The pre-cleared supernatants were incubated with an anti-HNF-4 antibody, anti-FLAG M5 antibody (Sigma Aldrich), or pre-immune IgG (used as a negative control) at 4 °C overnight. Immunoprecipitated samples were then captured by Ab-Capcher. The Ab-Capcher beads were washed three times with wash buffer (10 mM Tris-HCl pH 7.5, 1 mM EDTA pH 8.0, 150 mM NaCl, 1% Triton X-100, and protease inhibitor cocktail), and the immunoprecipitated proteins were eluted with SDS-PAGE loading buffer. The eluates were analyzed by immunoblotting using an anti-HNF-4 or anti-DYKDDDDK antibody.
Reverse transcription qPCR (RT-qPCR)
Total RNA was obtained from chipmunk livers and primary hepatocytes using ISOGEN II (Nippon Gene). First-strand cDNA was synthesized using the PrimeScript 1st Strand cDNA Synthesis Kit (Takara) with random primers following the manufacturer’s instructions; qPCR was performed with four samples in technical triplicate using the SYBR Green Realtime PCR Master Mix (Toyobo) and a Rotor-Gene Q apparatus (Qiagen). The following PCR primers were used: cmSHP + 490 F, 5′-CTGGGCCCCAAAGAGTATGC-3′, and cmSHP + 572 R, 5′-ATGTGGGAGGAGGCATGAAG-3′; cmHP25 + 607 F, 5′-AAGCCAATGACAGCTACAAACAT-3′, and cmHP25 + 799 R, 5′-TTCAGAATTACAAGAGGGATGGA-3′; cmALB + 502 F, 5′-AGAAAATAACCTGAGGCTTTTGG-3′, and cmALB + 675 R, 5′-TTCATAGCCTCAATCTTTGGTGT-3′; cmACTB + 226 F, 5′-GTCACCAACTGGGACGACAT-3′, and cmACTB + 401 R, 5′-ACATACATGGCTGGGGTGT-3′. Beta-actin (ACTB) was used as an endogenous control to normalize gene expression. Relative mRNA expression levels were presented as means ± SEM. Statistical differences were analyzed by Student’s t-test.
Primary hepatocyte cultures and transfection
Primary hepatocytes of nonhibernating chipmunks were obtained by the conventional collagenase method with minor modifications. Following liver perfusion with collagenase type IV (Worthington), the dispersed cells were filtered successively through 500-μm and 125-μm stainless steel sieves, and washed with DMEM four times. In all experiments, the hepatocytes were more than 80% viable as judged by trypan blue exclusion. After centrifugation, the cell pellet was resuspended in Williams’ medium E supplemented with 10% FCS, 1 mM insulin, 1 mM dexamethasone, 100 U/ml penicillin, 0.1 mg/ml streptomycin, 0.5 mg/ml fungizone, and 0.1 mg/ml kanamycin, and the hepatocytes were plated on collagen-coated BioCoat dishes (Corning). On the next day, pcDNA3/FLAG-cmSHP or pcDNA3/FLAG were transfected into the hepatocytes using Lipofectamine 2000 (Invitrogen) following the manufacturer’s instructions. The hepatocytes were cultured for 2 additional days, and then whole-cell extracts and total RNA were prepared.
Statistical analysis
Student’s t-test (2-tailed 2, type 2), Tukey-Kramer Multiple Comparison test, one-way ANOVA, or two-way ANOVA were used to compare the difference from the control group. When the interaction was significant at p < 0.05 by one-way ANOVA or two-way ANOVA, post hoc multiple comparison was made using Tukey-Kramer test. Two-way ANOVA was also used to analyze the interaction between treatments.
Additional Information
Accession codes: The accession number for the chipmunk SHP sequence reported in this paper is Genbank: LC144633.
How to cite this article: Tsukamoto, D. et al. HNF-4 participates in the hibernation-associated transcriptional regulation of the chipmunk hibernation-related protein gene. Sci. Rep. 7, 44279; doi: 10.1038/srep44279 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported in part by grants-in-aid from the Ministry of Education, Science, Sports and Culture in Japan, and by the Grant for Joint Research Project of the Institute of Medical Science, University of Tokyo.
Footnotes
The authors declare no competing financial interests.
Author Contributions D.T. and N.T. designed this experiments, wrote the manuscript; D.T. conducted the experiments; D.T., M.I., and N.T. discussed the data. All authors gave final approval for publication.
References
- Wang L. C. H. & Lee T. F. Topor and Hibernation in Mammals: Metabolic, Physiological, and Biochemical Adaptations. In Comprehensive Physiology 507–532 (American Physiological Society, 2011). [Google Scholar]
- Kondo N. & Kondo J. Identification of novel blood proteins specific for mammalian hibernation. J. Biol. Chem. 267, 473–478 (1992). [PubMed] [Google Scholar]
- Takamatsu N., Ohba K., Kondo J., Kondo N. & Shiba T. Hibernation-associated gene-regulation of plasma-proteins with a collagen-like domain in mammalian hibernators. Mol. Cell. Biol. 13, 1516–1521 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo N. et al. Circannual control of hibernation by HP complex in the brain. Cell 125, 161–172, doi: 10.1016/j.cell.2006.03.017 (2006). [DOI] [PubMed] [Google Scholar]
- Kojima M., Takamatsu N., Ishii T., Kondo N. & Shiba T. HNF-4 plays a pivotal role in the liver-specific transcription of thechipmunk HP-25 gene. Eur. J. Biochem. 267, 4635–4641, doi: 10.1046/j.1432-1327.2000.01499.x (2000). [DOI] [PubMed] [Google Scholar]
- Sladek F. M., Zhong W. M., Lai E. & Darnell J. E. Liver-enriched transcription factor HNF-4 is a novel member of the steroid-hormone receptor superfamily. Genes. Dev. 4, 2353–2365, doi: 10.1101/gad.4.12b.2353 (1990). [DOI] [PubMed] [Google Scholar]
- Wisely G. B. et al. Hepatocyte nuclear factor 4 is a transcription factor that constitutively binds fatty acids. Structure 10, 1225–1234 (2002). [DOI] [PubMed] [Google Scholar]
- Yuan X. H. et al. Identification of an Endogenous Ligand Bound to a Native Orphan Nuclear Receptor. Plos One 4, e5609, doi: 10.1371/journal.pone.0005609 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang G. Q., Nepomuceno L., Hopkins K. & Sladek F. M. Exclusive homodimerization of the orphan receptor hepatocyte nuclear factor-4 defines a new subclass of nuclear receptors. Mol. Cell. Biol. 15, 5131–5143 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt A. J., Garrison W. D. & Duncan S. A. HNF4: A central regulator of hepatocyte differentiation and function. Hepatology 37, 1249–1253, doi: 10.1053/jhep.2003.50273 (2003). [DOI] [PubMed] [Google Scholar]
- Peng H., Zhu Q. S., Zhong S. P. & Levy D. Transcription of the human microsomal epoxide hydrolase gene (EPHX1) is regulated by an HNF-4 alpha/CAR/RXR/PSF complex. Biochim. Biophys. Acta 1829, 1000–1009, doi: 10.1016/j.bbagrm.2013.05.003 (2013). [DOI] [PubMed] [Google Scholar]
- Zhang Y. X., Bonzo J. A., Gonzalez F. J. & Wang L. Diurnal Regulation of the Early Growth Response 1 (Egr-1) Protein Expression by Hepatocyte Nuclear Factor 4 alpha (HNF4 alpha) and Small Heterodimer Partner (SHP) Cross-talk in Liver Fibrosis. J. Biol. Chem. 286, 29635–29643, doi: 10.1074/jbc.M111.253039 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida E. et al. Functional association between CBP and HNF4 in trans-activation. Biochem. Biophys. Res. Commun. 241, 664–669, doi: 10.1006/bbrc.1997.7871 (1997). [DOI] [PubMed] [Google Scholar]
- Elferink C. J. & Reiners J. J. Quantitative RT-PCR on CYP1A1 heterogeneous nuclear RNA: A surrogate for the in vitro transcription run-on assay. Biotechniques 20, 470–477 (1996). [DOI] [PubMed] [Google Scholar]
- Tsukamoto D. et al. USF is involved in the transcriptional regulation of the chipmunk HP-25 gene. Gene 396, 268–272, doi: 10.1016/j.gene.2007.03.014 (2007). [DOI] [PubMed] [Google Scholar]
- Maire P., Wuarin J. & Schibler U. The role of cis-acting promoter elements in tissue-specific albumin gene expression. Science 244, 343–346 (1989). [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell 128, 693–705, doi: 10.1016/j.cell.2007.02.005 (2007). [DOI] [PubMed] [Google Scholar]
- Cheung P. & Lau P. Epigenetic regulation by histone methylation and histone variants. Mol. Endocrinol. 19, 563–573, doi: 10.1210/me.2004-0496 (2005). [DOI] [PubMed] [Google Scholar]
- Sterner D. E. & Berger S. L. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64, 435–459, doi: 10.1128/mmbr.64.2.435-459.2000 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak G. G. & Strahl B. D. Hitting the ‘mark’: Interpreting lysine methylation in the context of active transcription. Biochim. Biophys. Acta 1839, 1353–1361, doi: 10.1016/j.bbagrm.2014.03.002 (2014). [DOI] [PubMed] [Google Scholar]
- Wolffe A. P. & Pruss D. Targeting chromatin disruption: Transcription regulators that acetylate histones. Cell 84, 817–819, doi: 10.1016/s0092-8674(00)81059-4 (1996). [DOI] [PubMed] [Google Scholar]
- Spencer T. E. et al. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature 389, 194–198 (1997). [DOI] [PubMed] [Google Scholar]
- Zhang Y. & Reinberg D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes. Dev. 15, 2343–2360, doi: 10.1101/gad.927301 (2001). [DOI] [PubMed] [Google Scholar]
- Yang X. J. & Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell. Biol. 9, 206–218, doi: 10.1038/nrm2346 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanyam K. et al. Curcumin, a novel p300/CREB-binding protein-specific inhibitor of acetyltransferase, represses the acetylation of histone/nonhistone proteins and histone acetyltransferase-dependent chromatin transcription. J. Biol. Chem. 279, 51163–51171, doi: 10.1074/jbc.M409024200 (2004). [DOI] [PubMed] [Google Scholar]
- Hatziapostolou M. et al. An HNF4 alpha-miRNA Inflammatory Feedback Circuit Regulates Hepatocellular Oncogenesis. Cell 147, 1233–1247, doi: 10.1016/j.cell.2011.10.043 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. Y. & Burke P. A. The role of microRNAs in hepatocyte nuclear factor-4alpha expression and transactivation. Biochim. Biophys. Acta 1829, 436–442, doi: 10.1016/j.bbagrm.2012.12.009 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. X., Hagedorn C. H. & Wang L. Role of nuclear receptor SHP in metabolism and cancer. Biochim. Biophys. Acta 1812, 893–908, doi: 10.1016/j.bbadis.2010.10.006 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. K., Dell H., Dowhan D. H., Hadzopoulou-Cladaras M. & Moore D. D. The orphan nuclear receptor SHP inhibits hepatocyte nuclear factor 4 and retinoid X receptor transactivation: Two mechanisms for repression. Mol. Cell. Biol. 20, 187–195 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin P. J. & Storey K. B. Evidence for a reduced transcriptional state during hibernation in ground squirrels. Cryobiology 53, 310–318, doi: 10.1016/j.cryobiol.2006.08.002 (2006). [DOI] [PubMed] [Google Scholar]
- Hittel D. & Storey K. B. Differential expression of adipose- and heart-type fatty acid binding proteins in hibernating ground squirrels. Biochim. Biophys. Acta 1522, 238–243, doi: 10.1016/s0167-4781(01)00338-4 (2001). [DOI] [PubMed] [Google Scholar]
- Yan J., Barnes B. M., Kohl F. & Marr T. G. Modulation of gene expression in hibernating arctic ground squirrels. Physiol. Genomics 32, 170–181, doi: 10.1152/physiolgenomics.00075.2007 (2008). [DOI] [PubMed] [Google Scholar]
- Williams D. R. et al. Seasonally hibernating phenotype assessed through transcript screening. Physiol. Genomics 24, 13–22, doi: 10.1152/physiolgenomics.00301.2004 (2005). [DOI] [PubMed] [Google Scholar]
- Srere H. K., Wang L. C. H. & Martin S. L. Central role for differential gene-expression in mammalian hibernation. Proc. Natl. Acad. Sci. USA 89, 7119–7123, doi: 10.1073/pnas.89.15.7119 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton M. et al. Deep Sequencing the Transcriptome Reveals Seasonal Adaptive Mechanisms in a Hibernating Mammal. Plos One 6, 13, doi: 10.1371/journal.pone.0027021 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer B. B., Barnes B. M., Lowell B. B. & Grujic D. Differential regulation of uncoupling protein gene homologues in multiple tissues of hibernating ground squirrels. Am. J. Physiol. 275, R1232–R1238 (1998). [DOI] [PubMed] [Google Scholar]
- O’Hara B. F. et al. Gene expression in the brain across the hibernation cycle. Am. J. Physiol. 19, 3781–3790 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epperson L. E. & Martin S. L. Quantitative assessment of ground squirrel mRNA levels in multiple stages of hibernation. Physiol. Genomics 10, 93–102, doi: 10.1152/physiolgenomics.00004.2002 (2002). [DOI] [PubMed] [Google Scholar]
- Fujii G. et al. CpG methylation at the USF-binding site is important for the liver-specific transcription of the chipmunk HP-27 gene. Biochem. J. 395, 203–209, doi: 10.1042/bj20051802 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. B. et al. Genome-wide Mapping of HATs and HDACs Reveals Distinct Functions in Active and Inactive Genes. Cell 138, 1019–1031, doi: 10.1016/j.cell.2009.06.049 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram O. et al. Combinatorial Patterning of Chromatin Regulators Uncovered by Genome-wide Location Analysis in Human Cells. Cell 147, 1628–1639, doi: 10.1016/j.cell.2011.09.057 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S. L. The complex language of chromatin regulation during transcription. Nature 447, 407–412, doi: 10.1038/nature05915 (2007). [DOI] [PubMed] [Google Scholar]
- Johansson L. et al. The orphan nuclear receptor SHP utilizes conserved LXXLL-related motifs for interactions with ligand-activated estrogen receptors. Mol. Cell. Biol. 20, 1124–1133, doi: 10.1128/mcb.20.4.1124-1133.2000 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. C. et al. Farnesoid X receptor-induced lysine-specific histone demethylase reduces hepatic bile acid levels and protects the liver against bile acid toxicity. Hepatology 62, 220–231, doi: 10.1002/hep.27677 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan P. et al. A SIRT1-LSD1 Corepressor Complex Regulates Notch Target Gene Expression and Development. Mol. Cell 42, 689–699, doi: 10.1016/j.molcel.2011.04.020 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Breukelen F. & Martin S. L. Reversible depression of transcription during hibernation. J. Comp. Physiol. B. 172, 355–361, doi: 10.1007/s00360-002-0256-1 (2002). [DOI] [PubMed] [Google Scholar]
- Van Breukelen F. & Martin S. L. Translational initiation is uncoupled from elongation at 18 degrees C during mammalian hibernation. Am. J. Physiol. Regul. Integr. Comp. Physiol. 281, R1374–R1379 (2001). [DOI] [PubMed] [Google Scholar]
- Osborne P. G., Gao B. & Hashimoto M. Determination in vivo of newly synthesized gene expression in hamsters during phases of the hibernation cycle. Jpn. J. Physiol. 54, 295–305, doi: 10.2170/jjphysiol.54.295 (2004). [DOI] [PubMed] [Google Scholar]
- Seldin M. M. et al. Seasonal oscillation of liver-derived hibernation protein complex in the central nervous system of non-hibernating mammals. J. Exp. Biol. 217, 2667–2679, doi: 10.1242/jeb.095976 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita S. et al. Identification of bovine hibernation-specific protein complex and evidence of its regulation in fasting and aging. J. Biochem. 153, 453–461, doi: 10.1093/jb/mvt008 (2013). [DOI] [PubMed] [Google Scholar]
- Hut R. A., Dardente H. & Riede S. J. Seasonal Timing: How Does a Hibernator Know When to Stop Hibernating? Curr. Biol. 24, R602–R605, doi: 10.1016/j.cub.2014.05.061 (2014). [DOI] [PubMed] [Google Scholar]
- de Miera C. S. et al. A Circannual Clock Drives Expression of Genes Central for Seasonal Reproduction. Curr. Biol. 24, 1500–1506, doi: 10.1016/j.cub.2014.05.024 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.