Abstract
Little is known about how chromatin folds in its native state. Using optimized in situ hybridization and live imaging techniques have determined compaction ratios and fiber flexibility for interphase chromatin in budding yeast. Unlike previous studies, ours examines nonrepetitive chromatin at intervals short enough to be meaningful for yeast chromosomes and functional domains in higher eukaryotes. We reconcile high-resolution fluorescence in situ hybridization data from intervals of 14–100 kb along single chromatids with measurements of whole chromosome arms (122–623 kb in length), monitored in intact cells through the targeted binding of bacterial repressors fused to GFP derivatives. The results are interpreted with a flexible polymer model and suggest that interphase chromatin exists in a compact higher-order conformation with a persistence length of 170–220 nm and a mass density of ≈110–150 bp/nm. These values are equivalent to 7–10 nucleosomes per 11-nm turn within a 30-nm-like fiber structure. Comparison of long and short chromatid arm measurements demonstrates that chromatin fiber extension is also influenced by nuclear geometry. The observation of this surprisingly compact chromatin structure for transcriptionally competent chromatin in living yeast cells suggests that the passage of RNA polymerase II requires a very transient unfolding of higher-order chromatin structure.
Keywords: higher-order structure, 30-nm fiber, nucleosomes
Genetic studies indicate that the spatial positioning of the genome in interphase contributes to the regulation of nuclear functions, yet the principles that govern the organization of interphase chromosomes (Chrs) are largely unknown. At the simplest level, DNA is folded through interaction with histones forming the nucleosome core particle (NCP), which yields a 6:1 or 7:1 compaction ratio depending on linker length. Arrays of nucleosomes are further condensed by ≈6-fold into a higher-order structure, the so-called 30-nm fiber, whose in vivo architecture is unresolved. Several models have been proposed for this structure (ref. 1 and reviewed in ref. 2), yet species' specific variation in linker histones and nucleosome repeat lengths may lead to variation in fiber characteristics. How interphase Chrs fold beyond this level of organization is even less well understood, although in addition to fiber compaction, local looping and/or anchoring to subnuclear elements may influence chromatin conformation (3, 4).
To analyze chromatin compaction ratios in interphase nuclei, laboratories have generally applied fluorescence in situ hybridization (FISH), using differentially derivatized probes. These data were interpreted as identifying mega bp-sized loops (averaging 3,000 kb) with the bases of the loops being distributed in a random walk throughout the nucleoplasm (5–7) or as a chain of chromosomal subcompartments each comprising ≈10–20 loops of ≈120 kb (8). The random distribution of the chain may reflect local chromatin dynamics, which have been recently well documented in living cells by rapid time-lapse microscopy of GFP-tagged loci (4, 9–11). Because the FISH probes previously used to study compaction were cosmids of 30–40 kb spaced at intervals ranging from 150 kb to 190 Mb (5–7), such studies could not address the local folding characteristics of the nucleosomal fiber, nor the organization within transcription units or replicons, which generally range in size from 50 to 200 kb. This problem of scale is even more pronounced with respect to the yeast genome, where entire Chr arms encompass only 120–800 kb. An earlier attempt to determine chromatin compaction rates in yeast made use of FISH probes ranging from 35 to 70 kb in length over intervals of 145 and 255 kb in swollen or lysed nuclei (12). This led, not surprisingly, to large errors in estimated compaction ratios.
Improved imaging techniques now permit one to monitor chromatin compaction in living cells. Time-lapse studies show that the interphase chromatin fiber is quite flexible, and a given locus can be observed to move in a nearly random fashion, within a spatially constrained volume (4, 9–11). Fiber flexibility can be described mathematically by polymer chain models, where its stiffness is quantified as the persistence length (Lp; refs. 13–15). For genomic separations of more than a few Lp (e.g., distances of >150–200 nm), the fiber between the two markers cannot be considered a rod, and local compaction ratios (i.e., the linear mass density expressed in bp/nm of chromatin fiber) are not accurately calculated by dividing the end-to-end distances that separate two probes by the number of bp. However, to determine both the compaction ratio and Lp of the chromatin fiber independently, one needs end-to-end distance values for genomic separations on the order of 1 to 3 times the Lp of the fiber. In previous studies that determined chromatin flexibility from cross-linking (16) or recombination probabilities between specific sites (17), it was necessary to assume a certain value for the compaction ratio to allow calculation of the Lp.
Here, we determine end-to-end distances for a range of genomic intervals by using unique techniques for high-resolution FISH (18, 19) and live GFP-fusion imaging based on repressor binding to chromosomally integrated, nonamplified lac or tet operator arrays (20). This process, combined with immunofluorescence (IF), allows us to examine chromatin folding over small distances in intact yeast cells. We also compare arm length measurements with distances separating different repressor array insertions. The polymer chain model then allows us to determine both the persistence length and mass density of chromatin from these end-to-end distance values. Chromatin dynamics (10, 11) suggest that a population of conformations exist, further justifying the use of flexible polymer modeling to analyze chromatin compaction. The compaction value we obtain for G1-phase yeast chromatin is remarkably high and remains fairly uniform over intervals ranging from 14 to 623 kb. From this finding we predict a steady-state interphase chromatin structure equivalent to a 30-nm fiber with 7–10 nucleosomes per 11-nm superhelical turn and a Lp of 170–210 nm. Variations in whole Chr arm extension suggest further that the path of long Chr arms may be influenced by nuclear geometry.
Materials and Methods
Plasmids and Strains. Plasmid pAFS59T was created by inserting a BamHI–SacI fragment from pRS306tetO2×112 (21) containing 224 tet operators into the backbone of pAFS59 (20). PCR-amplified genomic fragments (Saccharomyces Genome Database coordinates) were inserted into this and pAFS52LO (22): 15160–15773 for Tel3L, 294892–295241 for Tel3R, 9645–11059 for Tel5R, 558701–559863 for Tel5L, 16431–17993 for Tel6L, 256581–256893 for Tel6R, 18832–19853 for Tel14L, 68363–68569 for ARS603, and 256581–256893 for ARS607, or into the TRP1 gene (17 kb left of CEN4) for ARS1/CEN4. Genomic integration was verified by colony PCR and pulsed-field electrophoresis. pGVH29, a plasmid containing the Tetr–GFP fusion, was cloned by inserting an EcoRI–SapI fragment from p128tetR–GFP (21) into the same sites of pCJ097 (23). pGVH30, a plasmid containing both the Tetr–yellow fluorescent protein (YFP) and the laci–cyan fluorescent protein (CFP) fusions, was cloned by inserting a KpnI–SacI blunt fragment of CFP–laci from pDH3 (gift of M. Peter, Eidgenössische Technische Hochschule, Zurich) into the XhoI site of pCJ097. Genotypes of strains and parental backgrounds are in Table 3, which is published as supporting information on the PNAS web site.
Combined IF/FISH on Yeast Cells. For two-color and centromere FISH, diploid GA-1190 and haploid GA-1188 strains were grown in rich medium and synchronized by the temperature-sensitive mutation (cdc4-3; refs. 18 and 19). GA-180 was synchronized in G1 by pheromone for localization of Cen3, Cen5, and Cen6. All antibodies have been described (18, 19) and were preadsorbed against yeast spheroplasts before use. Cell integrity was monitored by the equatorial pore-to-pore distance (haploid maxima, 2.0 μm; diploid maxima, 2.4 μm). FISH distances were measured in single confocal sections, i.e., not projections, by using the Zeiss LSM510 2.0 line tool or version 2.5 for 3D epifluorescence signals. Experiments with GFP derivatives and spindle pole body (SPB) IF were performed on α-factor-synchronized (G1) or random cultures in which G1- and S-phase cells were distinguished by bud morphology, although both populations yielded similar values (<10% mean difference).
Microscopy. Live imaging was performed as described (10, 22) on rich media. Because Tel3L and Tel3R tagging with CFP–laci/YFP–tetr was too weak, Tel3L and Tel3R were visualized by GFP–laci and GFP–tetr bound to the same insertions, distinguishable by spot size. IF and FISH imaging were performed in multitracking mode on the Zeiss LSM510 (or LSM410) with a ×63 or ×100 Plan-Apochromat objective (numerical aperture, 1.4; zoom, 3 or 1.8, such that pixel size was 88 or 100 nm). For IF, 3D stacks on fixed cells were typically 16 steps of 0.2 μm or 10 steps of 0.25 μm. Chromatic aberration was corrected before image capture by alignment of 0.1- and 0.2-μm Tetraspeck Microsphere signals (Molecular Probes). Live imaging was performed in single tracking mode with closed pinhole (1–1.2 Airy units; GFP at 488 nm with 0.1–1.0% transmission; CFP–YFP at 458 and 514 nm, with 1–25% transmission; four averages/region of interest 30 × 30).
Brownian Dynamics Modeling. The end-to-end distance distribution of a chromatin chain confined in a spherical nucleus was simulated by using the program corchy++ (24). Brownian dynamics trajectories were calculated for a 3-μm long chain with a Lp of 190 nm and a diameter of 30 nm, in a sphere of 300–4,000 nm diameter. The first 10 ms of the trajectory was used for equilibration; the end-to-end distance was averaged over 100–850 ms thereafter.
Results
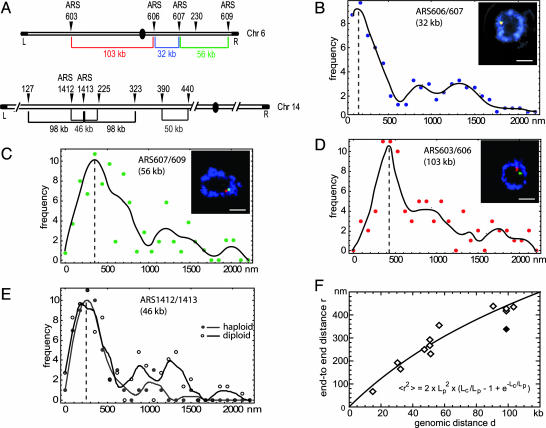
To achieve high-resolution labeling of short DNA intervals in intact nuclei, we have improved FISH techniques for budding yeast such that short probes (derivatized 200-nt fragments spanning 5–8 kb) could be detected efficiently in intact cells. Nuclear integrity was monitored by 3D confocal imaging and measurement of nuclear diameter by means of antipore staining (18). Probes were designed to detect pairs of unique sites on individual arms of Chrs 5, 6, and 14 at intervals ranging from 14 to 100 kb (Fig. 1A). Analysis was performed on rapidly growing cultures synchronized in late G1 phase and fixed with formaldehyde in rich medium. Confocal images were captured on 31–127 cells for each probe pair, often in more than one independent experiment, and the separation between the centers of gravity for the two fluorochromes was monitored within a single focal plane. Examples of the end-to-end distance distribution are presented in Fig. 1 B–E for four pairs of probes. The vertical dotted lines represent the maxima of the fitted curves, which can be taken as a measure of the most probable distance between probes.
Fig. 1.
Compaction ratios determined by high-resolution two-color FISH. (A) Schematic representation of the location and spacing of the Chr6 and Chr14 FISH probes. (B–E) Combined IF/FISH on diploid cdc4-3 cells synchronized in late G1. Representative Zeiss LSM410 confocal images of nuclear midsections show nuclear pores (mAb414; blue) and the two specified FISH probes (red/green; see Insets). Distances between the centers of gravity of the two probes were scored on similar single confocal planes. A cubic spline fit of the frequencies (black line) defines the peak distance as the plot maximum. Results for haploid (gray curve) and diploid (black curve) cells are compared in E. (Bars, 1 μm.) (F) Worm-like chain fit (Eq. 1; black line) of the peak distances versus genomic separation.
Initially we used diploid cells for this study because their larger nuclear diameter facilitated measurement. In most cases the fluorescent signals for the two contiguous loci were found in one focal section, whereas the equivalent pair of signals for the other homologue was not. Nonetheless, the occasional presence of three or four signals in the plane of focus led to a background of dispersed values much larger than the peaks (note that these larger separation values occur less frequently in haploid cells; Fig. 1 B–D). However, if we compare the end-to-end distance peak values for a given chromosomal interval in both haploid and diploid cells (probe pair ARS1412/ARS1413 in Fig. 1E), we find that the peak distances are identical in diploid and haploid cells. We therefore used the peak values as the average end-to-end distances for the 12 probe pairs analyzed in our additional calculations (Table 1). The true means for all distances measured (rms values) have large SD values because of diploid scatter. When we use rms values for haploid samples alone (see Table 4, which is published as supporting information on the PNAS web site), we obtain comparable parameters for higher-order chromatin folding although the calculations are not statistically robust.
Table 1. Genomic and physical distances between FISH probes.
| FISH probe name | Chr arm | Genomic distance, kb | Average peak distance, nm |
|---|---|---|---|
| ARS607 + C6-230 | Chr6R (2n) | 30 | 187.0 |
| ARS606 + ARS607 | Chr6R (2n) | 32 | 158.4 |
| ARS607 + 609 | Chr6R (2n) | 56 | 353.0 |
| ARS603 + 606 | Chr6R (2n) | 103 | 436.0 |
| ARS1412 + 1413 | Chr14L (2n) | 46 | 246.4 |
| ARS1412 + 1413 | Chr14L (1n) | 46 | 250.0 |
| C14-210-257 | Chr14L (2n) | 47 | 231.0 |
| C14-127-225 | Chr14L (1n) | 98 | 330.0 |
| C14-225-323 | Chr14L (1n) | 98 | 417.0 |
| C14-390-440 | Chr14L (1n) | 50 | 265.0 |
| C14-300-ARS1413 | Chr14L (1n) | 50 | 290.4 |
| C14-300-390 | Chr14L (1n) | 90 | 440.0 |
| ARS501/ChrV-562 | Chr5R (1n) | 14 | 60.0 |
In situ hybridization of probes located on the right arm of Chr 6 or 5 or the left arm of Chr14 in either haploid (1n) or diploid (2n) cells was performed on G1-arrested cells.
To determine both compaction and Lp simultaneously, we modeled the stiffness of chromatin as a flexible polymer through a nonlinear fit by using the known relationship for the Porod–Kratky chain (Eq. 1) (13):
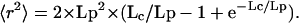 |
[1] |
In Eq. 1, the contour length Lc (in nm) is the ratio of the genomic distance d (in bp) divided by the linear mass density of the chromatin chain c (in bp/nm) or Lc = d/c. Fig.1F shows the spatial distances r as a function of the genomic separation d for all of the peak end-to-end distances and the corresponding fit to Eq. 1. This process yields values for the Lp = 197 ± 62 nm and mass density c = 144 ± 17 bp/nm. Intriguingly, one peak value (Chr14, 127–225 kb) appeared different from all other data points, yielding significantly different parameters. This value, which results from two probes on Chr14 flanking a cluster of late-replicating origins (25), was therefore omitted from the fit. The chromatin in this region is able to impose late replication on an early firing origin inserted into it, presumably reflecting either a unique chromatin structure or subnuclear context (16, 25). Given its functional characteristics, we find it noteworthy that this domain appears to be organized either as unusually compact or unusually flexible chromatin.
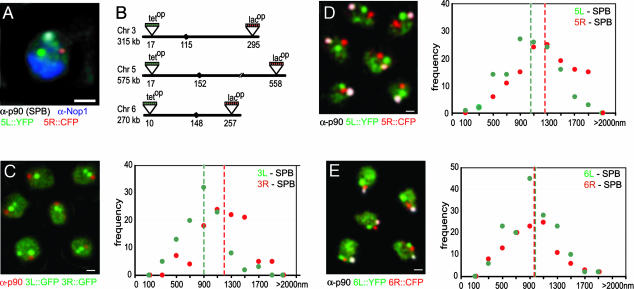
We next verified whether the chromatin compaction parameters determined for the FISH probe pairs spanning relatively short distances would accurately reflect the organization of entire Chr arms monitored in cells that are not subjected to the fairly harsh conditions necessary for FISH. To this end, we inserted arrays of either lac or tet operator sites (lacop, ref. 20; and tetop, ref. 12) into the most distal unique sequence on Chrs 3, 5, 6, and 14, such that the natural organization of subtelomeric repeats is maintained (see Fig. 3). This process is coupled with staining of the SPB, which is situated near centromeres (18, 26, 27).
Fig. 3.
Quantitative fluorescence analysis of Chr arm length. (A) Four-color epifluorescence and IF (full 3D rendering, imaris) of a G1-arrested haploid GA-2201 cell probed with anti-Nop1-CY5 (blue), anti-p90 (SPB)-CY3 (white), Tel5R::CFP (red), and Tel5L::YFP (green). (B) Schematic representation of tet or lac operator insertion sites near the telomeres (distances indicated in kb from left telomere; black sphere, centromere). (C–E) As in A except that the Nop1 signal is not shown. The Tetr–YFP background indicated the nucleoplasm in these maximal projections of 10 0.25-μm z sections of GA-2195 (SPB, red; 3L::GFP, green; 3R::GFP, green) (C), GA-2201 (SPB, white; 5L::YFP, green; 5R::CFP, red) (D), and GA-2199 (SPB, white; 6L::YFP, green; 6R::CFP, red) (E). Frequencies of 3D telomere-SPB distances are plotted by 0.2-μm categories (median value ± 0.1). The summary of the average distances is given in Table 2. (Bars, 1 μm.)
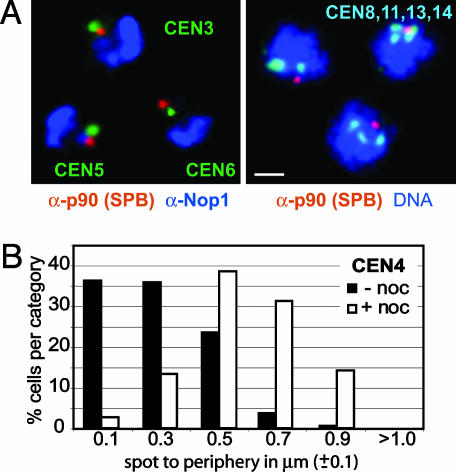
In Fig. 2 we show by centromere FISH coupled with immunostaining of the SPB that centromeres in G1-phase cells cluster around this membrane-embedded structure, yet are generally not coincident with it. Measurements taken from 3D stacks for centromere-SPB distances (n = 20) ranged from 250 to 400 nm. In 2D time-lapse movies of living cells (10), distances ranging from 100 to 300 nm were shown to separate a lacop-tagged Cen4 from the nuclear envelope, labeled with Nup49–GFP. This interphase centromere–nuclear envelope distance increases significantly when microtubules are disrupted with nocodazole (Fig. 2B), suggesting that short microtubules hold interphase centromeres near the SPB. Finally, measurement of the distances separating a CFP-tagged SPB component (CFP–Spc42) and a GFP-tagged centromere factor (Cse4–GFP) in living cells also yielded values that peak at 300 nm (K.B., unpublished work). These measurements justify use of the immunostained SPB, coupled with a correction of 300 nm, as a centromere marker for Chr arm length measurements, thus avoiding perturbation that might arise from FISH or inserts of repeats at centromeres.
Fig. 2.
Centromere clustering around the SPB is microtubule-dependent. (A) Combined IF/FISH images of G1-arrested cells probed with anti-p90 (SPB)-CY3 (red), anti-Nop1 (nucleolus)-CY5 (blue), and CEN3, CEN5, or CEN6 (green, as indicated). Full 3D rendering was performed with imaris (Bitplane, Zurich). Combined IF/FISH of diploid cells synchronized in G1 with combined probes to CEN8, CEN11, CEN13, and CEN14 (light blue), DNA stain (TOTO-3; blue), and anti-p90 (red). (Bar, 1 μm.) (B) Localization of GFP-tagged CEN4 relative to the nuclear periphery in G1-phase cells exposed to 1% DMSO with (filled bars) or without (open bars) 10 μg/ml nocodazole (noc). Time-lapse movies were acquired as in ref. 10. Relative 2D spot-periphery distances were averaged over six to nine movies (1,200–1,800 frames) and plotted in intervals of 0.2 μm (median value ± 0.1).
To perform 3D measurements on telomere-SPB distances, we captured four-color through-focus stacks of confocal images on fixed cells bearing lacop and tetop arrays in the indicated subtelomeric regions (Fig. 3 A and B). These cells express CFP–laci and YFP–tetr and were further stained for both the SPB and a nucleolar marker (Nop1, Fig. 3A), with which we monitored nuclear integrity and cell-cycle stage. Representative images are shown as projections in Fig. 3 C–E, from which we omit the Nop1 channel for clarity. Telomere-SPB distances scored in 3D in 48–160 G1- or S-phase cells are plotted in Fig. 3 C–E, and the arithmetic means are summarized in Table 2. The spacing frequencies show Gaussian distributions, which may stem in part from an inherent, random “Brownian-like” chromatin movement (10, 11). In this assay, all chromatic aberration was removed before image capture and optical resolution was limited to ≈1 voxel per signal (pixel size of 100 nm for x and y axis and 200–250 nm for the z axis).
Table 2. 3D telomere-SPB distances from combined IF and epifluorescence labels.
| Chr arm | Genomic distance, kb | Measured Tel-SPB distance, nm | SD | n |
|---|---|---|---|---|
| 3R | 200 | 1,202 | 343 | 160 |
| 3L | 115 | 898 | 295 | 160 |
| 5R | 423 | 1,277 | 425 | 133 |
| 5L | 152 | 1,071 | 373 | 133 |
| 6R | 122 | 942 | 346 | 111 |
| 6L | 148 | 960 | 342 | 159 |
| 14L | 623 | 1,393 | 467 | 48 |
Mean distances in nm measured between each telomere and the SPB are given with SD, without correction for the SPB-centromere distance.
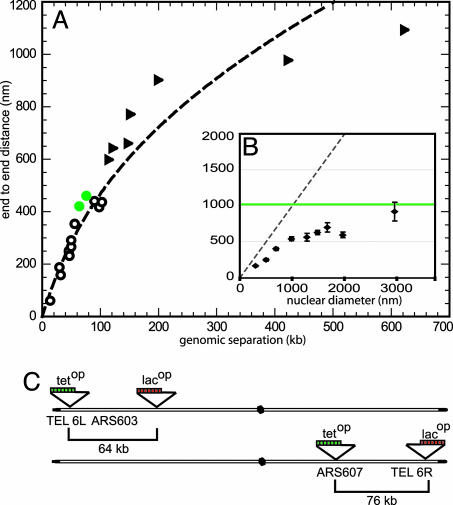
These epifluorescence/IF data yield average measured lengths for whole Chr arms that range from 122 to 623 kb. The mean end-to-end values were corrected for a SPB-centromere distance of 300 nm and are plotted together with the FISH data and the curve determined from those results in Fig. 4A (see triangles). Although most IF data agree well with the FISH-determined curve, we note that the two longest Chr arm measurements are systematically shorter than that predicted by the flexible polymer theory. One explanation for this finding would be that the chain segment behaves like a free worm-like chain only at shorter genomic intervals. For longer separations (>200 kb), which approach the length of the nuclear radius, the confinement of the polymer within the nuclear volume is predicted to reduce end-to-end distances below those expected for a worm-like chain (28). Indeed, if we use the values for Lp and mass density obtained for yeast chromatin (see above), we predict that the squared end-to-end length of the DNA chain that is 3 μm in length unfolded (corresponding to ≈430 kb), should be 1.03 μm when folded as chromatin in vivo. However, simulations of Brownian dynamics show that for nuclear radii of <2 μm the chain's end-to-end distance becomes significantly smaller than the unconstrained value (Fig. 4B Inset). As a result, the deduced Lp from the simulated end-to-end distance yields a lower apparent Lp value (120 nm rather than 197 nm). This finding could account for our observation that end-to-end values for the longest Chr arms are slightly below what is predicted from the curve based on shorter genomic intervals. We argue from this that nuclear geometry itself can influence long-range chromatin folding in yeast, where the nuclear diameter is 2 μm or less.
Fig. 4.
Compaction ratio of yeast interphase chromatin. (A) SPB-telomere distances corrected for the SPB-centromere distance of 300 nm (▴) are plotted on the polymer fit from Eq. 1 by using the peak FISH values (○). The equivalent plot with adjusted rms values for the FISH data is in Fig. 5. (B) Simulated rms end-to-end distances of a 3-μm chromatin chain with a Lp of 190 nm and a diameter of 30 nm confined into a spherical nucleus of varying diameter. The green line indicates the theoretical rms end-to-end distance of 1.03 μm for the same chain without confinement. The dashed line indicates the maximum possible extension of the chain (end-to-end distance is equivalent to nuclear diameter). (C) Schematic representation of tet or lac operator insertion sites for in vivo distance measures that are plotted in green in A.
Because the above measurements required the fixation of cells, we proceeded to confirm the validity of our curve by monitoring in living cells distances that separate lacop and tetop arrays integrated along the same chromatid arm. By combining differentially derivatised laci and tetr fusions (i.e., to CFP, YFP, or GFP) we can distinguish the two insertions as discrete sites. This cannot be done accurately if the second repeat array contains the same operators, because of artifactual interactions between identical simple repeats in trans (29). Two intervals of 64 and 76 kb in length, respectively, on the left and right arms of Chr6 (Tel6L–ARS603 and ARS607–Tel6R) were measured (Fig. 4C). Distances determined in 3D were 420 ± 128 nm (n = 75) for the 64-kb interval and 460 ± 150 nm (n = 48) for the 76-kb interval, which both coincide well with the fit obtained from the FISH data (green circles in Fig. 4A). Other independent studies estimated the 100-kb distance between HMR and MAT on Chr3 to be ≈600–650 nm (30, 31), which is larger than we would predict. Those studies, however, used GFP–lacr bound to two integrations of lacop repeats, and because two GFP spots cannot be resolved below distances of 300 nm, the average separation based on two distinct spots would naturally slightly overestimate the average distance (single spots were not counted). This underscores the importance of using two-color, chromatic aberration-corrected imaging for high-resolution distance measurements. We further exploited live two-color measurements to examine whether compaction levels change as cells progress into G2/M. This does not appear to be the case; cells blocked in nocodazole have an identical distance separation for Tel6L and ARS603 (412 ± 195 nm for 64 kb; n = 111), although a 2-fold increase in rRNA-encoding DNA compaction was previously proposed (12).
Finally, we recalculated the fit of all data to Eq. 1, combining the FISH data and in vivo distances and Chr arm lengths. We excluded the largest Chr arm values because of the influence that nuclear geometry has on these end-to-end distances. The combined data sets yielded a mass density value of c = 142 ± 21 bp/nm and Lp = 211 ± 68 nm, which are very similar to the parameters determined with the FISH data alone. Given the diversity of measurements used for the determination of the curve, we propose that these parameters reliably describe the steady-state level of compaction and flexibility of yeast interphase chromatin. Calculations using adjusted rms values (Table 4) for the FISH data alone (c = 110 ± 32 bp/nm and Lp = 167 ± 95 nm), or together with the shorter Chr arms and the in vivo distance measurements (c = 118 ± 17 bp/nm and Lp = 218 ± 63 nm), yield a slightly less compact fiber, although the curve is not statistically distinct from that determined from peak values (Fig. 5, which is published as supporting information on the PNAS web site).
Discussion
Using end-to-end distances derived from high-resolution microscopy, we model interphase yeast chromatin as a flexible polymer chain and determine a mass density between 110 and 150 bp/nm and a Lp of 170–220 nm. These results correspond to an average compaction ratio of ≈40-fold. The Lp value suggests that the chromosomal fiber is relatively stiff over intervals of 10–20 kb, with an Lp much higher than that predicted from single-molecule stretching experiments (30–50 nm; ref. 32). These latter determinations are performed at low ionic strengths in vitro, which are conditions that tend to unfold chromatin. Although our value is somewhat higher than that estimated from formaldehyde cross-linking studies of Chr3 (16), it is consistent with the values predicted from simulations and FISH measurements in human fibroblast nuclei (5, 14, 33). Our study predicts Lp based on measurements over small intervals monitored under physiological conditions. The results support computer modeling of the chromatin fiber, based on conclusions from assumptions of chain geometry, DNA elasticity, and nucleosome interaction potentials. For nucleosome repeat lengths found in yeast, such models also suggest a stiff 30-nm-like structure with an Lp of 150–250 nm (ref. 15 and J.L., unpublished work).
Given a nucleosomal repeat length in yeast of 165 bp (34), the estimated mass density c of 110–150 bp/nm corresponds to a surprisingly compact structure, equivalent to 7–10 yeast NCPs per 11-nm turn of a 30-nm-like fiber. Chicken erythrocyte chromatin was found to have six or seven NCPs per 11 nm at nearly physiological salt concentrations (80 mM, ref. 35). Thoma et al. (36) determined a density of six to eight NCPs per 11 nm at 100 mM NaCl in chromatin extracted from rat liver nuclei, whereas electron microscopy and mass density analyses on isolated chromatin suggested a zigzag helical ribbon model that allowed linker-length independent folding into a 30-nm helical structure consistent with 11.6 NCPs per 11 nm at 150 mm NaCl (37). Our data are thus compatible with the proposed zigzag nucleosomal array.
Perhaps the most remarkable aspect of our measurements, and the compact fiber they predict, is that they reflect measurements in growing budding yeast cells, over intervals that contain almost exclusively actively transcribed genes or genes poised for transcription. The average gene density in budding yeast is one ORF per 2 kb. Because our measurements were performed either in living cells exposed to rich media or cells fixed while growing in rich medium, normal transcriptional rates should not have been significantly altered by the conditions of visualization. Moreover, the agreement we detect among data obtained from FISH and in vivo imaging renders it unlikely that we are observing a methodological artifact. We therefore propose that the unfolding of the compact fiber into a 10-nm array, or even the displacement or shifting of nucleosomes, occurs very transiently in living cells, just long enough to allow an engaged polymerase to traverse a given site. We predict that the nucleosomal fiber refolds immediately after passage of the polymerase to account for the global compaction rates we have detected. This idea is consistent with observations from Grunstein and colleagues (38) who found that histone deacetylation follows immediately after RNA polymerase II as it traverses chromatin in yeast, potentially facilitating rapid refolding by histone tail deacetylation once the polymerase passes through. Rapid refolding of a transcribed chromatin fiber was also observed by Daneholt (39) in electron micrographs of the Chironomus Balbiani ring.
More recent microscopy studies have monitored bulk chromatin volume changes during transcriptional activation in mammalian cells and are consistent with this model. For instance, Tumbar et al. (40) proposed the existence of ≈100-nm chromonema fibers, consisting of compact 30-nm loops in transcriptionally active Chinese hamster ovary nuclei. Transient, partial unfolding of such fibers could be induced by the activator VP16 (41) without unfolding the entire domain. Similarly, analyses of mouse mammary tumor virus promoter arrays suggest that transcription from a natural promoter occurs at a chromatin compaction ratio of ≈50-fold in vivo (42).
With what frequency does one expect yeast chromatin to unfold for transcription? Given a RNA polymerase II rate of 20 nt/s (43), a gene density of 1 per 2 kb, a mean transcript abundance of 0.1–1 per cell (44), and mRNA half-life of 19 min (http://web.wi.mit.edu/young/CTD_phosphatase/home.html), one predicts at most five to eight engaged polymerases at any one moment along a typical 150-kb stretch of a yeast Chr. This low frequency is consistent with the rare visualization of mRNA transcripts reported for live imaging of transcription reported for living mammalian cells (45) and calculations for tissue-specific gene expression in mammals, which suggest an average transcript, like that encoding the TATA box-binding protein, is synthesized once per hour (46). Thus we conclude that most chromatin is compacted into folded fiber most of the time, as suggested by our analysis of living yeast cells.
Is histone H1 necessary for this folded state? Although yeast nucleosomes have short linker DNA, a protein related to the H1 linker histone called Hho1p is encoded in the yeast genome and is present in roughly one copy per four nucleosomes (47). Deletion analysis suggests that Hho1p does not affect chromatin structure per se, but influences recombination rates and may occlude sites of recognition for DNA binding proteins or chromatin remodelers (47). Thus although linker histones may well serve to stabilize a folded 30-nm structure, we suggest that folding into a higher-order fiber like that described here could also occur by default. We propose that “open” or transcriptionally competent chromatin, rather than switching to a different higher-order structure, simply has the propensity to unfold rapidly and transiently in the presence of the transcription machinery, maintaining its steady-state, 30-nm fiber organization.
Supplementary Material
Acknowledgments
We thank G. Van Houwe (University of Geneva) for creating strains for CFP/YFP double tagging, M. Gartenberg (University of Medicine and Dentistry, Piscataway, NJ) for tagging Tel3R, and C. Janke (Centre National de la Recherche Scientifique, Montpellier, France), L. Hartwell (Fred Hutchinson Cancer Center, Seattle), E. Hurt (University of Heidelberg, Heidelberg), J. Kilmartin (Medical Research Council, Cambridge), K. Nasmyth (Institute of Molecular Pathology, Vienna), P. Philippsen (University of Basel, Basel), M. Peter (Eidgenossiche Technische Hochschule, Zurich), and H. Scherthan (Max Planck Institute for Molecular Genetics, Berlin) for plasmids, strains, or antibodies. The Gasser laboratory is supported by the Swiss National Science Foundation and National Center of Competence in Research Frontiers in Genetics program.
Author contributions: K.B., P.H., and S.M.G. designed research; K.B., P.H., and L.G. performed research; K.B., L.G., J.L., and S.M.G. analyzed data; K.B., P.H., and S.M.G. contributed new reagents/analytic tools; and K.B., J.L., and S.M.G. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: FISH, fluorescence in situ hybridization; Lp, persistence length; IF, immunofluorescence; YFP, yellow fluorescent protein; CFP, cyan fluorescent protein; SPB, spindle pole body; Chr, chromosome; NCP, nucleosome core particle.
References
- 1.Woodcock, C. L. & Dimitrov, S. (2001) Curr. Opin. Genet. Dev. 11, 130–135. [DOI] [PubMed] [Google Scholar]
- 2.Hansen, J. C. (2002) Annu. Rev. Biophys. Biomol. Struct. 31, 361–392. [DOI] [PubMed] [Google Scholar]
- 3.Gasser, S. M. (2002) Science 296, 1412–1416. [DOI] [PubMed] [Google Scholar]
- 4.Marshall, W. F. (2002) Curr. Biol. 12, R185–R192. [DOI] [PubMed] [Google Scholar]
- 5.van den Engh, G., Sachs, R. & Trask, B. J. (1992) Science 257, 1410–1412. [DOI] [PubMed] [Google Scholar]
- 6.Yokota, H., van den Engh, G., Hearst, J. E., Sachs, R. K. & Trask, B. J. (1995) J. Cell Biol. 130, 1239–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sachs, R. K., van den Engh, G., Trask, B., Yokota, H. & Hearst, J. E. (1995) Proc. Natl. Acad. Sci. USA 92, 2710–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Munkel, C., Eils, R., Dietzel, S., Zink, D., Mehring, C., Wedemann, G., Cremer, T. & Langowski, J. (1999) J. Mol. Biol. 285, 1053–1065. [DOI] [PubMed] [Google Scholar]
- 9.Chubb, J. R. & Bickmore, W. A. (2003) Cell 112, 403–406. [DOI] [PubMed] [Google Scholar]
- 10.Heun, P., Laroche, T., Shimada, K., Furrer, P. & Gasser, S. M. (2001) Science 294, 2181–2186. [DOI] [PubMed] [Google Scholar]
- 11.Marshall, W. F., Straight, A., Marko, J. F., Swedlow, J., Dernburg, A., Belmont, A., Murray, A. W., Agard, D. A. & Sedat, J. W. (1997) Curr. Biol. 7, 930–939. [DOI] [PubMed] [Google Scholar]
- 12.Guacci, V., Hogan, E. & Koshland, D. (1994) J. Cell Biol. 125, 517–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kratky, O. & Porod, G. (1949) Rec. Trav. Chim. Pays-Bas 68, 1106–1123. [Google Scholar]
- 14.Ostashevsky, J. Y. & Lange, C. S. (1994) J. Biomol. Struct. Dyn. 11, 813–820. [DOI] [PubMed] [Google Scholar]
- 15.Wedemann, G. & Langowski, J. (2002) Biophys. J. 82, 2847–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dekker, J., Rippe, K., Dekker, M. & Kleckner, N. (2002) Science 295, 1306–1311. [DOI] [PubMed] [Google Scholar]
- 17.Ringrose, L., Chabanis, S., Angrand, P. O., Woodroofe, C. & Stewart, A. F. (1999) EMBO J. 18, 6630–6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heun, P., Laroche, T., Raghuraman, M. K. & Gasser, S. M. (2001) J. Cell Biol. 152, 385–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gotta, M., Laroche, T., Formenton, A., Maillet, L., Scherthan, H. & Gasser, S. M. (1996) J. Cell Biol. 134, 1349–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Straight, A. F., Belmont, A. S., Robinett, C. C. & Murray, A. W. (1996) Curr. Biol. 6, 1599–1608. [DOI] [PubMed] [Google Scholar]
- 21.Michaelis, C., Ciosk, R. & Nasmyth, K. (1997) Cell 91, 35–45. [DOI] [PubMed] [Google Scholar]
- 22.Hediger, F., Neumann, F. R., Van Houwe, G., Dubrana, K. & Gasser, S. M. (2002) Curr. Biol. 12, 2076–2089. [DOI] [PubMed] [Google Scholar]
- 23.Janke, C., Ortiz, J., Tanaka, T. U., Lechner, J. & Schiebel, E. (2002) EMBO J. 21, 181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klenin, K., Merlitz, H. & Langowski, J. (1998) Biophys. J. 74, 780–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedman, K. L., Diller, J. D., Ferguson, B. M., Nyland, S. V., Brewer, B. J. & Fangman, W. L. (1996) Genes Dev. 10, 1595–1607. [DOI] [PubMed] [Google Scholar]
- 26.Jin, Q., Trelles-Sticken, E., Scherthan, H. & Loidl, J. (1998) J. Cell Biol. 141, 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guacci, V., Hogan, E. & Koshland, D. (1997) Mol. Biol. Cell 8, 957–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langowski, J. & Schiessel, H. (2004) in Chromatin Structure: State of the Art, eds. Zlatanova, S. & Leuba, S. H. (Elsevier, New York), pp. 397–400.
- 29.Aragon-Alcaide, L. & Strunnikov, A. V. (2000) Nat. Cell Biol. 2, 812–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simon, P., Houston, P. & Broach, J. (2002) EMBO J. 21, 2282–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bressan, D. A., Vazquez, J. & Haber, J. E. (2004) J. Cell Biol. 164, 361–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cui, Y. & Bustamante, C. (2000) Proc. Natl. Acad. Sci. USA 97, 127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trask, B. J., Allen, S., Massa, H., Fertitta, A., Sachs, R., van den Engh, G. & Wu, M. (1993) Cold Spring Harbor Symp. Quant. Biol. 58, 767–775. [DOI] [PubMed] [Google Scholar]
- 34.Holde, K. E. V. (1989) Chromatin (Springer, New York).
- 35.Bednar, J., Horowitz, R. A., Grigoryev, S. A., Carruthers, L. M., Hansen, J. C., Koster, A. J. & Woodcock, C. L. (1998) Proc. Natl. Acad. Sci. USA 95, 14173–14178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thoma, F., Koller, T. & Klug, A. (1979) J. Cell Biol. 83, 403–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woodcock, C. L., Frado, L. L. & Rattner, J. B. (1984) J. Cell Biol. 99, 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kurdistani, S. K., Robyr, D., Tavazoie, S. & Grunstein, M. (2002) Nat. Genet. 31, 248–254. [DOI] [PubMed] [Google Scholar]
- 39.Daneholt, B. (1992) Cell Biol. Int. Rep. 16, 709–715. [DOI] [PubMed] [Google Scholar]
- 40.Tumbar, T., Sudlow, G. & Belmont, A. S. (1999) J. Cell Biol. 145, 1341–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tumbar, T. & Belmont, A. S. (2001) Nat. Cell Biol. 3, 134–139. [DOI] [PubMed] [Google Scholar]
- 42.Muller, W. G., Walker, D., Hager, G. L. & McNally, J. G. (2001) J. Cell Biol. 154, 33–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edwards, A. M., Kane, C. M., Young, R. A. & Kornberg, R. D. (1991) J. Biol. Chem. 266, 71–75. [PubMed] [Google Scholar]
- 44.Wodicka, L., Dong, H., Mittmann, M., Ho, M. H. & Lockhart, D. J. (1997) Nat. Biotechnol. 15, 1359–1367. [DOI] [PubMed] [Google Scholar]
- 45.Janicki, S. M., Tsukamoto, T., Salghetti, S. E., Tansey, W. P., Sachidanandam, R., Prasanth, K. V., Ried, T., Shav-Tal, Y., Bertrand, E., Singer, R. H. & Spector, D. L. (2004) Cell 116, 683–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmidt, E. E. & Schibler, U. (1995) Development (Cambridge, U.K.) 121, 2373–2383. [DOI] [PubMed] [Google Scholar]
- 47.Downs, J. A., Kosmidou, E., Morgan, A. & Jackson, S. P. (2003) Mol. Cell 11, 1685–1689. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.