Abstract
Granulocyte colony-stimulating factor (GCSF) and its receptor (GCSFR), also known as CSF3 and CSF3R, are required to maintain normal neutrophil numbers during basal and emergency granulopoiesis in humans, mice and zebrafish. Previous studies identified two zebrafish CSF3 ligands and a single CSF3 receptor. Transient antisense morpholino oligonucleotide knockdown of both these ligands and receptor reduces neutrophil numbers in zebrafish embryos, a technique widely used to evaluate neutrophil contributions to models of infection, inflammation and regeneration. We created an allelic series of zebrafish csf3r mutants by CRISPR/Cas9 mutagenesis targeting csf3r exon 2. Biallelic csf3r mutant embryos are viable and have normal early survival, despite a substantial reduction of their neutrophil population size, and normal macrophage abundance. Heterozygotes have a haploinsufficiency phenotype with an intermediate reduction in neutrophil numbers. csf3r mutants are viable as adults, with a 50% reduction in tissue neutrophil density and a substantial reduction in the number of myeloid cells in the kidney marrow. These csf3r mutants are a new animal model of human CSF3R-dependent congenital neutropenia. Furthermore, they will be valuable for studying the impact of neutrophil loss in the context of other zebrafish disease models by providing a genetically stable, persistent, reproducible neutrophil deficiency state throughout life.
Granulocyte colony-stimulating factor (GCSF), also known as Colony-stimulating Factor 3 (CSF3), is a key regulator of neutrophil production and a wide range of neutrophil functions such as migration, antimicrobial activities and neutrophil survival1. These primary roles of GCSF in neutrophil cell biology are evolutionarily conserved between mammals such as humans and mice as well as fish including zebrafish2. GCSF signalling is initiated from the GCSF receptor (GCSFR), a class 1 cytokine receptor, and engages intracellular mediators, commonly the JAK/STAT/SOCS pathway.
The absolute requirement for GCSF signalling in granulopoiesis was first demonstrated by GCSF and GCSFR deficient mice, which have neutrophil and myeloid progenitor cell deficiencies, and exhibit vulnerability to infective challenges3,4. A rare form of human congenital neutropenia is due to biallelic GCSFR mutations5,6. Somatic GCSFR mutations are frequently acquired in long-standing GCSF-treated congenital neutropenia patients, and are associated with progression to acute myeloid leukaemia7.
Zebrafish granulopoiesis, at both primitive and definitive stages, is regulated through many cellular and molecular mechanisms that are largely conserved with mammalian granulopoiesis8,9. Hence, zebrafish models of myeloid development and neutrophil function have been exploited to gain new insights into the genetic and molecular regulation of neutrophil development, and the role of neutrophils in inflammatory and infective disease models. Specifically, GCSF/GCSFR signalling is conserved in zebrafish2,10 with two zebrafish GCSF/CSF3 ligands encoded by genes on chromosomes 12 (designated csf3a, ZFIN ID: ZDB-GENE-091229-1) and 19 (designated csf3b, ZFIN ID: ZDB-GENE-141212-221), which signal through a single receptor encoded by a gene located on chromosome 16 (csf3r, ZFIN ID: ZDB-GENE-080104-4). The ligands display different spatiotemporal expression patterns suggesting individualized function10. Indeed, a specific role for csf3b has been demonstrated in the later phase of neutrophil migration during the response to tissue injury11.
The requirement for both Csf3 ligands and Csf3r in zebrafish granulopoiesis has been demonstrated by transient loss-of-function studies employing antisense morpholino oligonucleotide knockdown strategies, which result in transient neutrophil depletion in zebrafish embryos2,10. The sufficiency of Csf3 signalling in adult zebrafish granulopoiesis is demonstrated by activity of the Csf3 ligands to support the in vitro development of myeloid-cell containing haemopoietic colonies10.
Here we describe the generation and characterisation of zebrafish gcsfr mutants using targeted CRISPR/Cas9 mutagenesis. Zebrafish csf3r mutants have a profound and stable neutrophil deficiency as embryos. The impairment of granulopoiesis persists into adulthood, manifesting as marked reduction of neutrophil abundance in kidney haematopoietic marrow and peripheral tissues. These studies confirm the primary role of Csf3/Csf3r signalling in granulopoiesis in zebrafish, and provide a new tool for assessing the contribution of neutrophils in embryonic and adult zebrafish disease models. Unlike transient knockdown approaches, which require potentially confounding experimental manipulations to induce neutrophil depletion, these csf3r mutants intrinsically provide a stable, basal neutrophil deficiency state in vivo.
Results
CRISPR/Cas9 mutagenesis of zebrafish csf3r
Three sgRNAs (C1, C2 and C3) designed to target exon 2 in the csf3r gene were injected (Fig. 1a,b, Supplementary Fig. S1). Only C3 sgRNA resulted in mutagenesis at the expected target site in F0 sgRNA-injected embryos. On-target mutagenesis in these F0 embryos was confirmed by sequencing the predicted target site in a cohort of embryos, which revealed corrupted sequence traces commencing in the vicinity of the sgRNA target sequence for the C3 sgRNA (Supplementary Fig. S2), but not for sgRNAs C1 or C2.
Figure 1. CRISPR/Cas9-induced mutant zebrafish csf3r alleles.
(a) Intron/exon structure of zebrafish csf3r locus. (b) Domain structure of zebrafish Csf3r protein. Ig = immunoglobulin, FBN = fibronectin. (c) Four CRISPR/Cas9-induced csf3r nonsense mutations identified in adult F1 DNA (designated alleles 1–4 for this report) aligned to WT sequence. The corresponding predicted truncated amino acid sequences are shown: blue = native Csf3r sequence, red = predicted non-native sequence downstream of the mutation site, *premature stop.
Adult F0 fish from sgRNA C3 injections were incrossed to enable immediate observation of the predicted phenotype of reduced neutrophil numbers in F1 embryos, although genetic complexity was anticipated due to a multiplicity of CRISPR/Cas9-induced mutations. Germline transmission of mutant alleles was confirmed by genotyping F1 embryos and observing duplex sequencing traces at the predicted target sites (Fig. 1). Genotyping of individual adult F1 fish revealed multiple csf3r alleles, four of which were selected for propagation and future study (designated alleles 1–4 for this report, Fig. 1c). These four csf3r mutant alleles in exon 2 were all predicted to result in premature stop codons leading to Csf3r proteins truncated within the extracellular immunoglobulin (Ig)-like domain (Fig. 1b,c) and are currently undergoing genetic segregation by outcrossing. The studies presented here are a combination of analyses of F1 embryos and adults arising from F0 incrosses, and of F2 embryos and adults arising from F1 incrosses, and hence include biallelic mutants of several compound heterozygous and homozygous genotypes that randomly resulted from mating the first-available cohort of genotyped fish. Specific csf3r−/− allelotypes contributing to each experiment are specified throughout.
csf3r mutant zebrafish embryos have a selective, persistent neutrophil deficiency
To facilitate recognition of a neutrophil-depletion phenotype, mutagenesis was performed on a Tg(mpx:EGFP) background where neutrophils are marked by EGFP fluorescence. Even as F0 embryos, populations of C3 sgRNA-injected F0 embryos had significantly fewer Tg(mpx:EGFP)-labelled neutrophils compared to uninjected controls and embryos injected with either C1 and C2 sgRNAs (Supplementary Fig. S2). The occurrence of a neutrophil-depletion phenotype only in C3-injected F0 embryos also correlated with C3 being the only sgRNA for which targeting of the csfr3r locus was successfully demonstrated by Sanger sequencing. Hence, despite their complex mosaic genotype and heterogeneous mix of numerous csf3r allelic variants (evidenced in individual F0 embryo sequences, Supplementary Fig. S2), the expected csf3r-null phenotype of neutrophil deficiency was clearly discernable in F0 embryos when csfr3r targeting occurred.
Adult F1 fish from F0 incrosses were genotyped and 6 fish carrying two csf3r null alleles were identified representing one homozygous and 3 compound heterozygous csf3r null genotypes (specific allelotypes: csf3r3/3, csf3r1/2, csf3r2/3, and csf3r1/3). Incrosses of these fish generated null mutant F2 embryos of multiple compound csf3r mutant allelotypes, all of which showed obvious neutrophil deficiency upon inspection, with neutrophil population sizes being 30%, 36%, 38% and 45% of normal at 2, 3, 4 and 5 dpf, respectively (p < 0.05) (Fig. 2a–e). The concordance of the phenotype across the multiple compound heterozygous and homozygous allelotypes represented in these crosses demonstrates non-complementation of the various alleles. It also provides genetic evidence for the specificity of the neutrophil depletion phenotype to on-target mutagenesis at the GCSFR locus and demonstrates the robustness of this genotype/phenotype association.
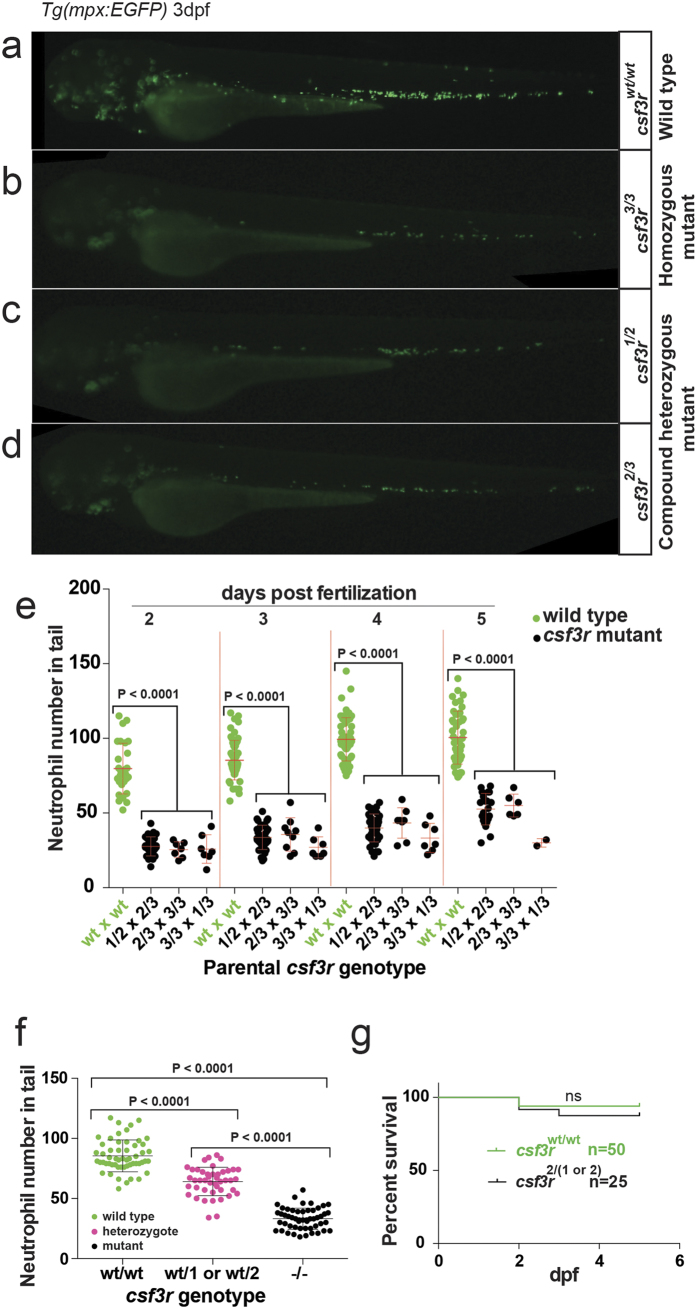
Figure 2. Neutrophil deficiency in csf3r null F2 embryos.
(a–d) Fluorescence micrographs of 3 dpf Tg(mpx:EGFP) embryos showing that several representative csf3r mutant null allelotypes (b–d) have substantially reduced numbers of fluorescent neutrophils compared to WT (a). Panel b is a representative homozygous csf3r3/3 null embryo, and panels c-d show representative compound heterozygous null embryos of the two allelotypes shown. (e) Quantification over 2–5 days post-fertilisation (dpf) of tail region neutrophil numbers in embryos of control (WT) matings and incrosses of 3 different pairs of csf3r null parents of the csf3r allelotypes shown. Data are mean ± SD. Within-day comparisons across genotypes are analysed by unpaired two-tailed t-tests, pooling data from the mutant compound heterozygous allelotypes shown, which are not significantly different to each other. These parental allelotype combinations were randomly selected, being those pairs that laid of those that were set up. They are not intended to be comprehensive, but they do demonstrate a consistent non-complementing neutrophil-depletion phenotype encompassing five embryonic allelotypes (1/2, 1/3, 2/2, 2/3 and 3/3). (f) Embryos carrying single csf3r null alleles have an intermediate neutrophil deficiency. A mix of heterozygous csf3rwt/(1or2) embryos in a 1:1 ratio were generated by outcrossing a parent of genotype csf3r1/2 to WT. Their neutrophil numbers at 3 dpf are compared with the non-contemporaneous csf3rWT/WT and pooled csf3r−/− mutant 3 dpf groups of panel (e). The mutant data were pooled as none of the mutant allelotype groups is significantly different to any other. p-values are from one-way ANOVA with Tukey’s multiple comparisons test. p < 0.0001. (g) csf3r null embryos have equivalent survival to WT embryos up to 5 days post-fertilisation (dpf). Kaplan-Meier plots compared by Wilcoxon rank sum test.
To test if there was a gene dosage-dependent neutrophil phenotype, a 1:1 mixture of heterozygous csf3rWT/1 or csf3rWT/2 F2 embryos was generated from an outcross of compound heterozygous fish A (csf3r1/2), and tail neut-ro-phil population sizes at 3 dpf compared with the pooled historical control data from 3 dpf csf3rWT/WT embryos and a heterogenous mix of csfr3r−/− allelotypes of Fig. 2e. WT (csf3rWT/WT), heterozygous carriers (csf3rWT/(1or2)) and csf3r−/− nulls had 85.4 ± 13.3, 64.1 ± 11.8 and 33.7 ± 8.2 neutrophils per tail region respectively, consistent with an intermediate haploinsufficiency phenotype (Fig. 2f). Also of note, the range of tail neutrophil numbers did not overlap between WT and mutant null embryos (WT:58–117 n = 55; null:18–57 neutrophils, n = 36). Furthermore, the upper 40% of WT and the lower 56% of csf3r1/2 mutant neutrophil counts fell outside the heterozygote range, these groups consistently reflecting their csf3r gene dosage.
Survival of csf3r mutant neutrophil-depleted embryos over this period was normal (Fig. 2g), indicating that neutrophil deficiency did not affect early embryo viability.
The myeloid deficiency in csf3r mutant embryos was neutrophil-specific and did not affect macrophage numbers. To quantify macrophage numbers, whole mount in situ hybridisation gene expression analysis for csfr1a/cfms was performed. There was no difference between the number of csfr1a/cfms-expressing macrophages located in the torso of csf3r mutant and WT embryos at 3 dpf (Fig. 3a,b).
Figure 3. Normal macrophage numbers in csf3r null F2 embryos.
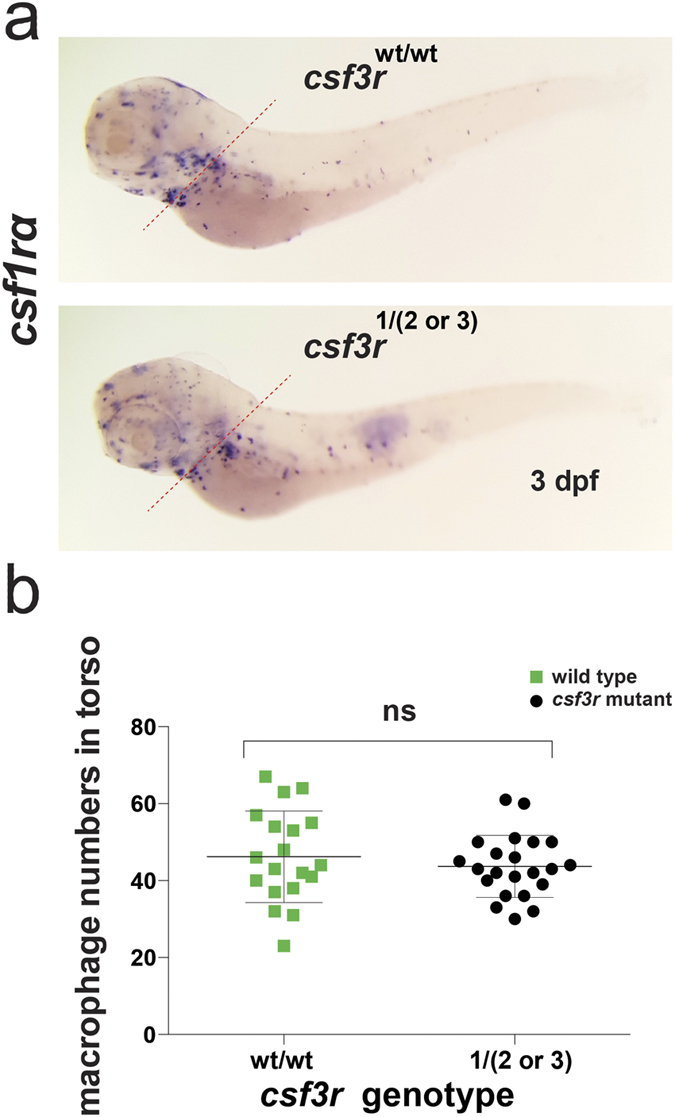
(a) Photomicrographs of representative 3 dpf WT and csf3r null embryos stained by whole mount in situ hybridisation (WISH) for the macrophage-specific marker csf1ra/cfms. (b) Quantification of torso-located macrophage numbers in csf1ra/cfms WISH embryos in shows no difference between genotypes (the torso being the region distal to the dotted red line in (a), selected for scoring for its lower density of macrophages and lower incidence of overlapping cells).
csf3r mutant zebrafish are adult viable despite neutrophil depletion in peripheral tissues
Csf3r−/− mutant zebrafish were viable as adults of both sexes. This is demonstrated by the six F1 adults incrossed for the analysis of Fig. 2e, which included parent fish with both homozygous and compound heterozygous mutant genotypes (specific allelotypes: csf3r3/3, csf3r1/2, csf3r2/3, and csf3r1/3). These fish retained a neutrophil-depleted phenotype into adulthood (Fig. 4a,b), demonstrated by quantification of neutrophil abundance and density in their tail fins. Csf3r mutant animals had a neutrophil density of only 41% of wildtype levels (Fig. 4c).
Figure 4. Neutrophil deficiency in csf3r null adults.
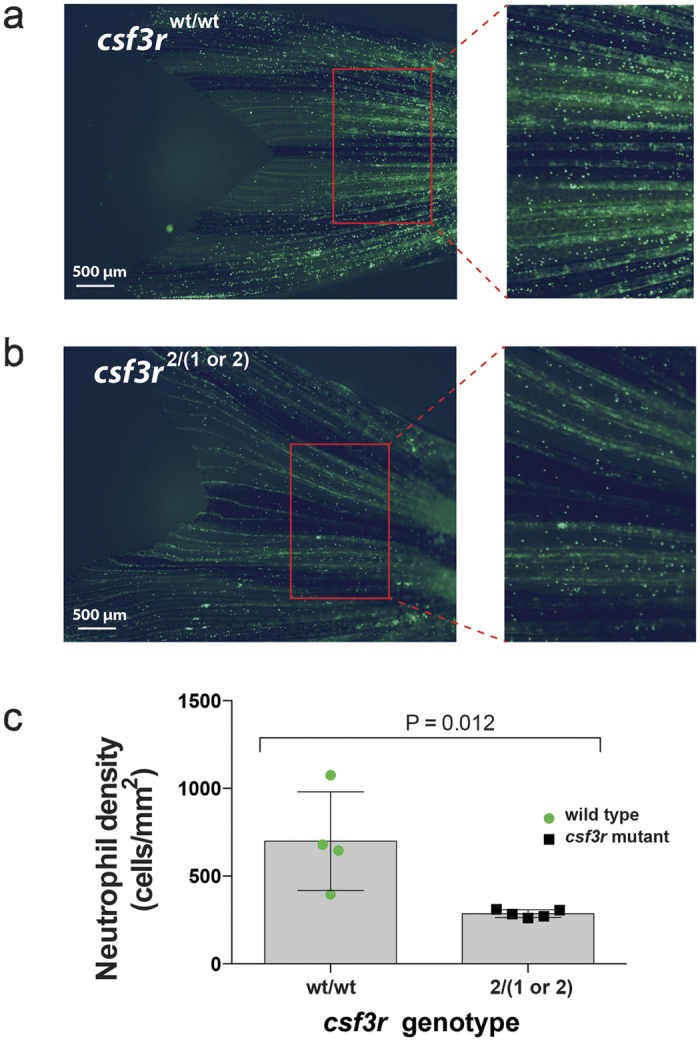
(a,b) Fluorescent photomicrographs of 5 month old adult Tg(mpx:EGFP) zebrafish tail fins showing reduced numbers of EGFP-positive neutrophils in csf3r null (b) vs WT (a) animals. (c) Quantification of tail fin neutrophil density. n = 4–5 animals/genotype as shown.
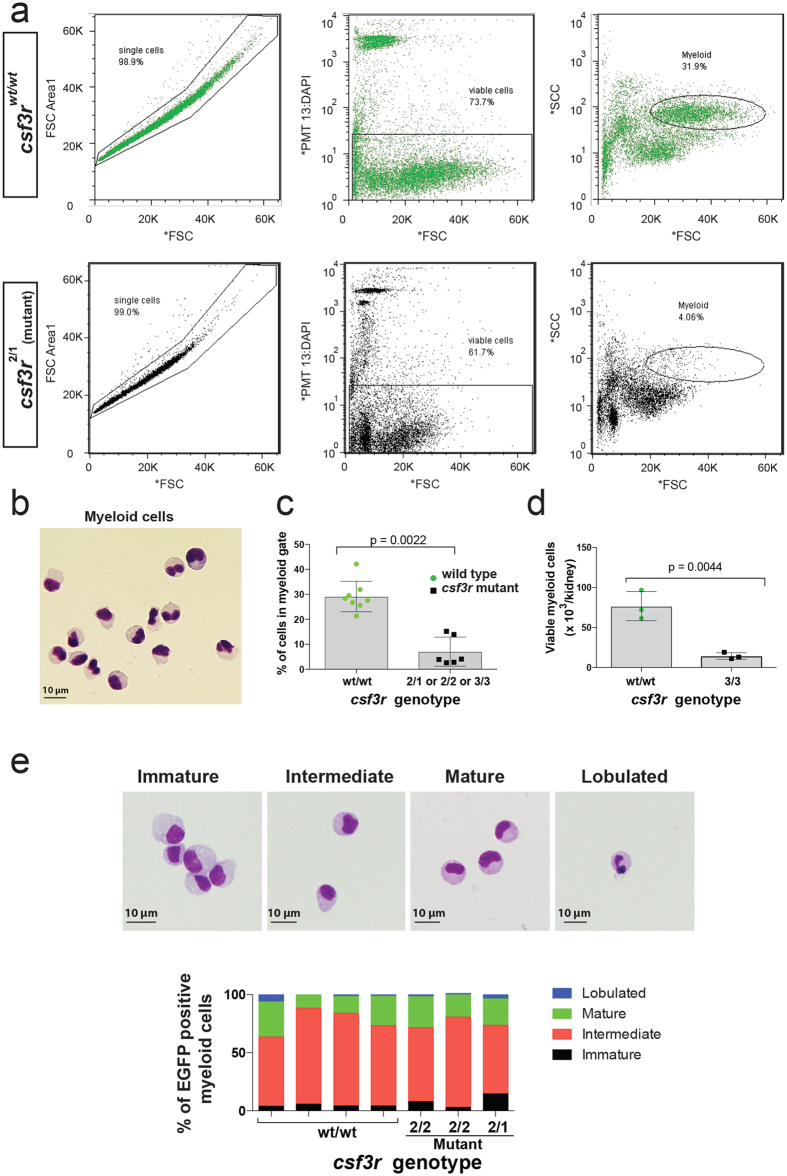
The kidney is the primary haematopoietic and granulopoietic organ in adult zebrafish8. Kidney marrow granulopoiesis in adult csf3r mutants was assessed by preparing single cell suspensions of adult kidney marrow followed by analysis of population sizes by FACS12 and cellular morphology by histological staining. The proportion of kidney marrow cells within the myelomonocytic gate was significantly reduced in csf3r mutant kidney marrow compared to wild type (7 ± 6% in csf3r−/− vs. 29 ± 6% in csf3rWT/WT, p = 0.0022) (Fig. 5a–c). This translated into a 5.3-fold reduction in the absolute number of myeloid cells per kidney (14 ± 4 vs. 77 ± 18 × 103 viable myeloid cells recovered/kidney from csf3r−/− and csf3rWT/WT respectively, p = 0.0044) (Fig. 5d).
Figure 5. Reduced granulopoiesis in csf3r null kidney marrow.
(a) Representative examples showing gating strategy on single and viable cells, and forward/side scatter profiles (FSC ad SSC) displaying reduced number of cells in the myeloid cell gate of csf3r-mutant kidney marrow cells compared to wildtype (WT). Supplementary Fig. S3 provides this information for all samples contributing to data in this figure. (b) May Grünwald-Giemsa stained cytospin of WT myeloid gate cells. (c) Percentage of viable cells falling within myeloid gate, which is significantly reduced in csf3r−/− mutant kidney marrows. Points represent different animals. Data are mean ± SD; p-value from Mann-Whitney test. n = 8 animals (WT) and 6 (mutant). (d) Number of viable myeloid cells/kidney, which is significantly reduced in csf3r−/− mutant kidney marrows. Data are mean ± SD; p-value from unpaired, 2-tailed t-test. n = 3 animals/group. (e) Four-category neutrophil differential counts based on the categories illustrated (top row of panels) in WT (n = 4) and csf3r−/− mutant (n = 3) kidney marrow cells, demonstrating no marked difference.
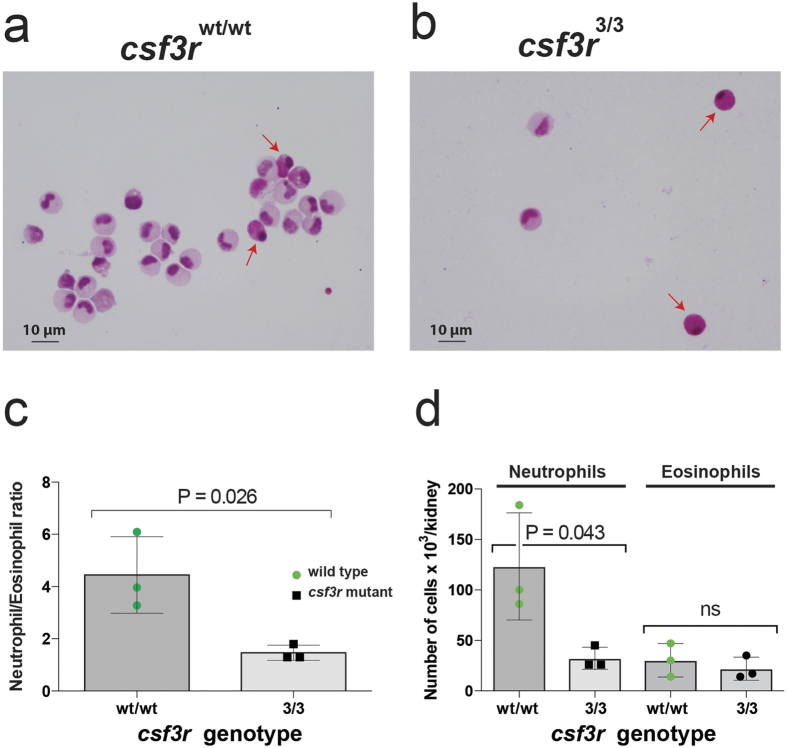
A 4-category differential count revealed that within the residual population of neutrophil lineage cells, csf3r-mutant kidney marrow granulopoiesis showed no marked maturational difference compared to WT (Fig. 5e). The morphologic assessment of myeloid cells in kidney marrow cytospin preparations also revealed an increased proportion of eosinophil granulocytes compared to neutrophil granulocytes (Fig. 6a,b). The neutrophil:eosinophil ratio in Periodic Acid-Schiff stained cytospins prepared from the myeloid gate was 1.5 ± 0.3 in csf3r−/− mutant vs. 4.4 ± 1.5 in csf3rWT/WT (p = 0.026) (Fig. 6c). A similar observation was made in May Grünwald-Giemsa stained preparations in an independent experiment (Supplementary Fig. S4). However, this change was relative, because the total numbers of eosinophils in kidney marrow suspensions was not different between wildtype and csf3r−/− mutants (Fig. 6d).
Figure 6. Relative preponderance of eosinophils in csf3r null kidney marrow.
(a,b) Periodic Acid-Schiff (PAS) stained cytospins of FACS-purified myeloid cells from WT (a) and csf3r3/3 mutant (b) kidney marrow. Red arrows indicate eosinophils, recognised by their PAS-positive strongly pink-staining cytoplasm and nuclear morphology and position. (c) Higher Neutrophil/Eosinophil ratio in WT vs csf3r3/3 mutant myeloid cells. Individual ratios determined from >150 cell differential counts. (d) Although neutrophil numbers are depleted in csf3r−/− kidney marrow, eosinophil numbers are not, indicating that the change in neutrophil:eosinophil ratio is due to relative change in the cell population sizes.
Discussion
These new csf3r−/− mutant zebrafish have a basal neutrophil deficiency that persists into adulthood. This phenotype is concordant with that of Csf3 and Csf3r knockout mice3,4. The reduced numbers of neutrophils in csf3r−/− zebrafish indicates that in zebrafish also, there is a requirement for Csf3r signalling to achieve a normal output from granulopoiesis. At the same time, the residual neutrophils present in csf3r−/− zebrafish indicate that, also in zebrafish, there are Csf3-independent mechanisms still capable of some neutrophil production.
These zebrafish csf3r−/− mutants also provide a new animal model of the rare form of autosomal recessive human severe congenital neutropenia due to biallelic CSF3R mutations (SCN7, OMIM #617014)5,6. The five mutant CSF3R alleles in the five patients described with SCN7 were nonsense mutations in either the extracellular cytokine homology domain (n = 1 allele), fibronectin-like domains (n = 3 alleles) or a p.Arg308Cys missense mutation in the cytokine homology domain (n = 1 allele)5,6. The more N-terminal csf3r mutations in this zebrafish allelic series occur in the immunoglobulin domain and best model the nonsense human SCN disease alleles. Furthermore, several of the human patients described were compound heterozygotes carrying two different CSF3R mutations5,6, a similar scenario to the compound heterozygotes resulting from our CRISPR/Cas9 mutagenesis and breeding strategy. The N-terminal nonsense mutations in the zebrafish mutants would not be expected to carry the leukaemogenic risk associated with the various C-terminal truncations that are prevalent as acquired somatic mutations in long-standing severe congenital neutropenia patients and some acute myeloid leukaemias, and which transmit hyperproliferative signals7. Interesting, the marrow of csf3r−/− mutant zebrafish had a proportional over-representation of eosinophils, a feature observed in some human cases of severe congenital neutropenia13,14.
Despite the lack of concordance reported between some zebrafish mutant phenotypes and that of their corresponding morphants15,16, for csf3r mutants and morphants, this aspect of the phenotype is concordant and confirms an absolute requirement for csf3r signalling in embryonic zebrafish granulopoiesis for which there are no compensatory genetic mechanisms.
We did not observe a reduction in macrophage numbers in csf3r−/− mutant embryos at 72 hpf, although at this time point, embryos with csf3a and csf3b overexpression from mRNA injections had increased macrophage numbers10, suggesting there are csf3-dependent signalling pathways in zebrafish that when driven strongly, are capable of impacting on macrophage abundance. In csf3r morphants, at 22 hpf macrophage numbers were normal, whereas at 96 hpf a ∼10% reduction was observed2. In both our mutants and these morphants, macrophages were scored as cells expressing csf1ra/cfms by whole mount in situ hybridisation. Whether or not there is a macrophage deficiency in older csf3r−/− mutant embryos or adults awaits further comprehensive studies; we suggest that this is best assessed by a range of macrophage markers such as crossing the mutant alleles onto mpeg1- or mfap4-driven fluorophore reporter lines17,18, which will take several generations.
There has been a considerable need in the field for methods to assess the specific contribution of neutrophils in zebrafish models of developmental processes, and in disease/biomedical models such as inflammation, infection and regeneration19. These neutrophil-depleted csf3r−/− mutant animals provide a new tool for doing this, with the advantages of specificity, reproducibility and persistence of the neutrophil-deficiency phenotype.
In particular, for studies that seek to assess the requirement or contribution of neutrophils to biological processes or disease models, this series of csf3r−/− mutants offers significant advantages over transient morphant knockdown approaches. These include lack of potential toxicity and no requirement for external manipulation, genetic stability, phenotype reproducibility and persistence, and concordance between different mutant allelotypes. There are 4 different csf3r morpholinos used in studies reported to date, targeting different regions of the Csf3r protein2,17,20,21. Although the csf3r morphant neutrophil deficiency phenotype is generally concordant for these different morphants, a direct comparison of all these reagents has not been conducted and the possibility remains of there being at least subtle differences between them in some or all of phenotype strength, persistence, and also potentially in confounding non-documented off-target effects. Groups of morphants also have heterogeneity reflecting the unavoidable variability inherent in dosing and delivery of morpholino oligonucleotides.
In other zebrafish mutants that have neutrophil deficiency, it occurs in the context of confounding multisystem phenotypes and often with accompanying macrophage deficiency. These mutants include, among many others: cloche/npas422,23, alk824, med1225, prpf826, plcg127 and runx128.
Experimentally, other transient genetic approaches have been used to deplete embryos of leukocytes including neutrophils. Normal lineage-balanced myelopoiesis requires appropriate levels of pu.1/spi1b, and pu.1/spi1b morphants have been used to induce a leukocyte-depleted state. However, the pu.1/spi1b morphant lacks specificity for neutrophil depletion and the outcome is highly dose-dependent: with modestly lowered Pu.1/Spi1b levels only macrophage development is impaired, and it is only with very substantive Pu.1/Spi1b depletion that neutrophil development is also impaired28,29. Imbalance of irf8 signalling also perturbs the balance of neutrophil/macrophage production in zebrafish embryos, and has often been used to do so, but in this case, the suppression of neutrophil production achieved by irf8 overexpression is accompanied by increased macrophage numbers, and so lacks specificity of effect30,31.
Although the genetically stable neutrophil deficiency of csf3r−/− mutants that persists through embryonic, larval and adult life is an advantage for some experimental questions, it also has its drawbacks. It provides a tool enabling the requirement for normal basal granulopoiesis to be evaluated in experimental models, but it complicates the assessment of emergency granulopoietic responses, including those driven by other haemopoietins, which in csf3r−/− mutants would be initiated from an already depleted basal state. It also does not permit the evaluation of the impact of acute, sudden neutrophil depletion on experimental models. A conditional system for achieving acute neutrophil ablation is provided by using our Tg(mpx:kalTA4)gl28Tg transgenic line to drive neutrophil-specific expression of the E. coli nitroreductase (NfsB) protein32. This approach has been employed in embryos (achieving up to 95% neutrophil depletion from 4 days of induction), but not yet in adults32.
In summary, we have built a new animal model of severe congenital neutropenia due to CSF3R mutation. These csf3r−/− mutant zebrafish will also be an extremely valuable resource for assessing the contribution of neutrophils in zebrafish developmental and disease models.
Methods
Zebrafish, husbandry and animal ethics
Fish were held in FishCore (Monash University) using standard practices. Embryos were incubated in egg water (0.06 g/L salt (Red Sea, Sydney, Australia)) or E3 medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4, equilibrated to pH 7.0). From 12 hpf, 0.003% (w/v) 1-phenyl-2-thiourea (Sigma-Aldrich) was used to inhibit pigmentation. Embryos were held at 28 °C in an incubator (Thermoline Scientific) following collection. The Tg(mpx:EGFP)il114Tg strain33 was used, which has been maintained on a majority Tübingen background by outcrossing to the Tübingen strain at least every second generation 1–2/yr for >10 years. The four new csf3r alleles called csf3r1,2,3,4 in this report (Fig. 1a) have been designated csf3rgl31,32,33,3431–34 respectively on the Zebrafish Information Network (ZFIN)34. Animal experiments followed National Health and Medical Research Council (NHMRC) guidelines (“Australian code of the care and use of animals for scientific purposes” 8th edition, NHMRC, 2013), were approved by the Monash University Animal Ethics Committees (protocol MAS-2010-18) and were conducted in accordance with these protocols.
CRISPR/Cas9 mutagenesis of zebrafish csf3r
CRISPR/Cas9 mutagenesis was based on the method of Gagnon et al.35. Briefly, the web tool “CHOPCHOP” was used to design a set of three sgRNA molecules (designated C1 to C3) to target the zebrafish csf3r gene. All three sgRNAs targeted exon 2. Each sgRNA contained a gene-specific spacer sequence (sequences in Fig. S1c) followed by a Cas9 enzyme binding sequence. These two sites were initially provided in different oligonucleotides called “site specific” and “constant” respectively. Oligonucleotides (Sigma) were annealed and single-stranded overhangs were filled in by T4 DNA polymerase (NEB) activity to form a double-stranded oligonucleotide, dsDNA (120 nucleotides after annealing and extension). All steps were performed in a 96-well T100™ Thermal Cycler (Bio-Rad). This was used as a template for in vitro transcription of sgRNA using mMESSAGE mMACHINE® T7 Transcription Kit (Ambion) followed by RNA clean up using Sephadex G-50 spin columns (Roche Diagnostics) according to the manufacturer’s instructions. synthesised sgRNA integrity was checked on a non-denaturing 1% TBE gel. For microinjection, individual sgRNAs (50–200 ng/μL) were mixed with Cas9 Nuclease 20 μM (New England Biolabs) at a 1:1 ratio and microinjected (500–1000 pg) directly into 1-cell Tg(mpx:EGFP) embryos.
Genotyping
DNA was extracted from single embryos or fin clips from adult fish using the HotSHOT protocol36 and purified PCR or gel extracted PCR products (AccuPrep® PCR/Gel purification kit, BIONEER) were sent for sequencing. 25 μL PCR reactions consisted of 0.5 μL Phusion High Fidelity DNA Polymerase (Thermo scientific), 5 μL 5X Phusion HF Buffer, 2 μL dNTP (2.5 mM), 0.5 μL forward primer (10 μM) (5′CCTTGCACATTTACTACCGACA3′), 0.5 μL reverse primer (10 μM) (5′GTCCTCCTGAACACACACAAGA3′), 5 μL genomic DNA and 11.5 μL of nuclease free water. Biorad T100 thermal cycler with the following program was used for amplification: 90 seconds at 95 °C as initial denaturation followed by 30 cycles of 30 sec at 95 °C for denaturation, 30 sec at 66 °C for annealing, 30 sec at 72 °C for extension, and final extension at 72 °C for 5 min. Sequencing was performed using the reverse primer (Micromon sequencing facility, Monash University). Sequencing traces were analysed using DNASTAR (Version 14) and ApE (A Plasmid Editor v.2.0.47). Mutations were identified manually by comparing mutant and wild-type traces.
Microscopy and neutrophil enumeration
An Olympus MVX10 microscope fitted with an Olympus DP72 camera and CellSens software version 1.11 was used for fluorescence and bright field imaging. EGFP-expressing neutrophil granulocyte numbers were quantified at various time points in the tail region caudal to the yolk extension, which includes the leukocyte-rich caudal haematopoietic tissue (CHT). Manual counting of neutrophil numbers was assisted by the brush tool in Paintbrush 2.1.2 (Soggy Waffles), which records clicks to avoid duplicate counting. For Supplementary Fig. S2 data, “Neutrophil Units” refer to a neutrophil number determined with assistance of the “Find maxima” function in Fiji (Version 1.47n).
Neutrophil density in adult tail fins was determined from Tg(mpx-EGFP) tail fin images of the appropriate csf3r genotype anaesthetised with 160 mg/L tricaine.
Whole-mount in situ hybridisation
Whole-mount in situ hybridisation was performed as previously described22,37 using a csf1ra digoxigenin-labelled riboprobe38. Cells were counted assisted by the Paintbrush 2.1.2 (Soggy Waffles), which records clicks to avoid duplicate counting, and by zooming to optimally resolve aggregates of cells into single cells.
Adult neutrophil isolation and evaluation
Kidneys were dissected from adult zebrafish euthanased by dense anaesthesia with 300 mg/L tricaine. A kidney single cell suspension was prepared for FACS in ice-cold 0.9X PBS containing 5% FBS12. Yield of viable cells per kidney was determined by trypan dye exclusion using a haemocytometer. FACS analysis was performed in Monash Flowcore using an Influx2 cell sorter (BD Biosciences) and data were analysed using FlowJo software (version 7.6.1). Gating strategies for all samples are displayed in Supplementary Fig. S3. For sorting cells to generate the histological images and differential counts of Fig. 5b and e, the myeloid gate shown was carefully reproduced by eye for each sorted sample. Cytospins of FACS-purified myeloid cell populations (gated as displayed in Fig.5a) were prepared at 800 r.p.m. for 5 minutes using a Shandon Elliott Cytospin machine and stained with May Grünwald-Giemsa22. Periodic Acid-Schiff staining used undiluted May Grünwald solution (VWR Biosciences 352622 M) as fixative followed by staining with Periodic Acid-Schiff according to the manufacturer’s instructions (Sigma 395B-1KT).
Statistics
Descriptive and analytical statistics were generated in Prism 6.0 f (GraphPad Software). Throughout this report, parametric data are presented as mean ± SD. n values are indicated by dots in histograms; every individual n value represents a different animal (i.e. there was no replicate scoring within a single animal). Statistical analysis used unpaired two-tailed t-tests, Mann-Whitney test, one-way ANOVA with Tukey’s multiple comparisons test, and the Wilcoxon rank sum test. p < 0.05 was taken to indicate a significant difference.
Additional Information
How to cite this article: Pazhakh, V. et al. A GCSFR / CSF3R zebrafish mutant models the persistent basal neutrophil deficiency of severe congenital neutropenia. Sci. Rep. 7, 44455; doi: 10.1038/srep44455 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank F. Ellett and J. Wittig for early contributions to the project, S. Varma for technical assistance, FishCore staff for excellent animal care, and FlowCore, Monash Micro Imaging and Micromon platform technologies for services and their staff for technical assistance. VP was supported by a Monash International Postgraduate Research Scholarship, a Monash Graduate Scholarship, and a Monash Postgraduate Publication Award. This work was supported by the NHMRC (1044754 [GL], 1069284 [GL], 1070687 [GL, MCK]). The Australian Regenerative Medicine Institute is supported by funds from the State Government of Victoria and the Australian Federal Government.
Footnotes
The authors declare no competing financial interests.
Author Contributions Author contributions (following the CRediT taxonomy of contributor roles). Conceptualization: V.P., G.J.L. Writing - Original Draft: V.P., G.J.L. Writing - Review and Editing: V.P., S.C., M.C.K., G.J.L. Investigation: V.P., M.C.K., S.C. Formal Analysis: V.P., G.J.L. Visualisation: V.P., G.J.L. Supervision: M.C.K., G.J.L. Funding Acquisition: G.J.L.
References
- Demetri G. D. & Griffin J. D. Granulocyte colony-stimulating factor and its receptor. Blood 78, 2791–2808 (1991). [PubMed] [Google Scholar]
- Liongue C., Hall C. J., O’Connell B. A., Crosier P. & Ward A. C. Zebrafish granulocyte colony-stimulating factor receptor signaling promotes myelopoiesis and myeloid cell migration. Blood 113, 2535–2546, doi: 10.1182/blood-2008-07-171967 (2009). [DOI] [PubMed] [Google Scholar]
- Lieschke G. J. et al. Mice lacking granulocyte colony-stimulating factor have chronic neutropenia, granulocyte and macrophage progenitor cell deficiency, and impaired neutrophil mobilization. Blood 84, 1737–1746 (1994). [PubMed] [Google Scholar]
- Liu F., Wu H. Y., Wesselschmidt R., Kornaga T. & Link D. C. Impaired production and increased apoptosis of neutrophils in granulocyte colony-stimulating factor receptor-deficient mice. Immunity 5, 491–501 (1996). [DOI] [PubMed] [Google Scholar]
- Klimiankou M. et al. GM-CSF stimulates granulopoiesis in a congenital neutropenia patient with loss-of-function biallelic heterozygous CSF3R mutations. Blood 126, 1865–1867, doi: 10.1182/blood-2015-07-661264 (2015). [DOI] [PubMed] [Google Scholar]
- Triot A. et al. Inherited biallelic CSF3R mutations in severe congenital neutropenia. Blood 123, 3811–3817, doi: 10.1182/blood-2013-11-535419 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touw I. P. Game of clones: the genomic evolution of severe congenital neutropenia. Hematology/the Education Program of the American Society of Hematology. American Society of Hematology. Education Program 2015, 1–7, doi: 10.1182/asheducation-2015.1.1 (2015). [DOI] [PubMed] [Google Scholar]
- Carradice D. & Lieschke G. J. Zebrafish in hematology: sushi or science? Blood 111, 3331–3342, doi: 10.1182/blood-2007-10-052761 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson A. J. & Zon L. I. The ‘definitive’ (and ‘primitive’) guide to zebrafish hematopoiesis. Oncogene 23, 7233–7246, doi: 10.1038/sj.onc.1207943 (2004). [DOI] [PubMed] [Google Scholar]
- Stachura D. L. et al. The zebrafish granulocyte colony-stimulating factors (Gcsfs): 2 paralogous cytokines and their roles in hematopoietic development and maintenance. Blood 122, 3918–3928, doi: 10.1182/blood-2012-12-475392 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galdames J. A., Zuniga-Traslavina C., Reyes A. E. & Feijoo C. G. Gcsf-Chr19 promotes neutrophil migration to damaged tissue through blood vessels in zebrafish. J Immunol 193, 372–378, doi: 10.4049/jimmunol.1303220 (2014). [DOI] [PubMed] [Google Scholar]
- Traver D. et al. Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nature immunology 4, 1238–1246, doi: 10.1038/ni1007 (2003). [DOI] [PubMed] [Google Scholar]
- Andrews J. P., Mc C. J. & Scott C. H. Lethal congenital neutropenia with eosinophilia occurring in two siblings. Am J Med 29, 358–362 (1960). [DOI] [PubMed] [Google Scholar]
- Zeidler C., Germeshausen M., Klein C. & Welte K. Clinical implications of ELA2-, HAX1-, and G-CSF-receptor (CSF3R) mutations in severe congenital neutropenia. British journal of haematology 144, 459–467, doi: 10.1111/j.1365-2141.2008.07425.x (2009). [DOI] [PubMed] [Google Scholar]
- Kok F. O. et al. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev Cell 32, 97–108, doi: 10.1016/j.devcel.2014.11.018 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi A. et al. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 524, 230–233, doi: 10.1038/nature14580 (2015). [DOI] [PubMed] [Google Scholar]
- Ellett F., Pase L., Hayman J. W., Andrianopoulos A. & Lieschke G. J. mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood 117, e49–56, doi: 10.1182/blood-2010-10-314120 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton E. M., Cronan M. R., Beerman R. W. & Tobin D. M. The Macrophage-Specific Promoter mfap4 Allows Live, Long-Term Analysis of Macrophage Behavior during Mycobacterial Infection in Zebrafish. PloS one 10, e0138949, doi: 10.1371/journal.pone.0138949 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley M. C., Wang C. H., Pazhakh V. & Lieschke G. J. Delineating the roles of neutrophils and macrophages in zebrafish regeneration models. The international journal of biochemistry & cell biology 56, 92–106, doi: 10.1016/j.biocel.2014.07.010 (2014). [DOI] [PubMed] [Google Scholar]
- Hall C., Flores M. V., Storm T., Crosier K. & Crosier P. The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. BMC developmental biology 7, 42, doi: 10.1186/1471-213X-7-42 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall C. J. et al. Immunoresponsive gene 1 augments bactericidal activity of macrophage-lineage cells by regulating beta-oxidation-dependent mitochondrial ROS production. Cell metabolism 18, 265–278, doi: 10.1016/j.cmet.2013.06.018 (2013). [DOI] [PubMed] [Google Scholar]
- Lieschke G. J., Oates A. C., Crowhurst M. O., Ward A. C. & Layton J. E. Morphologic and functional characterization of granulocytes and macrophages in embryonic and adult zebrafish. Blood 98, 3087–3096 (2001). [DOI] [PubMed] [Google Scholar]
- Reischauer S. et al. Cloche is a bHLH-PAS transcription factor that drives haemato-vascular specification. Nature 535, 294–298, doi: 10.1038/nature18614 (2016). [DOI] [PubMed] [Google Scholar]
- Hogan B. M. et al. Specification of the primitive myeloid precursor pool requires signaling through Alk8 in zebrafish. Current biology: CB 16, 506–511, doi: 10.1016/j.cub.2006.01.047 (2006). [DOI] [PubMed] [Google Scholar]
- Keightley M. C., Layton J. E., Hayman J. W., Heath J. K. & Lieschke G. J. Mediator subunit 12 is required for neutrophil development in zebrafish. PloS one 6, e23845, doi: 10.1371/journal.pone.0023845 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley M. C. et al. In vivo mutation of pre-mRNA processing factor 8 (Prpf8) affects transcript splicing, cell survival and myeloid differentiation. FEBS Lett 587, 2150–2157, doi: 10.1016/j.febslet.2013.05.030 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing C. B. et al. Phospholipase C gamma-1 is required for granulocyte maturation in zebrafish. Developmental biology 374, 24–31, doi: 10.1016/j.ydbio.2012.11.032 (2013). [DOI] [PubMed] [Google Scholar]
- Jin H. et al. Runx1 regulates embryonic myeloid fate choice in zebrafish through a negative feedback loop inhibiting Pu.1 expression. Blood 119, 5239–5249, doi: 10.1182/blood-2011-12-398362 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes J. et al. Interplay of pu.1 and gata1 determines myelo-erythroid progenitor cell fate in zebrafish. Dev Cell 8, 97–108, doi: 10.1016/j.devcel.2004.11.014 (2005). [DOI] [PubMed] [Google Scholar]
- Li L., Jin H., Xu J., Shi Y. Q. & Wen Z. L. Irf8 regulates macrophage versus neutrophil fate during zebrafish primitive myelopoiesis. Blood 117, 1359–1369, doi: 10.1182/blood-2010-06-290700 (2011). [DOI] [PubMed] [Google Scholar]
- Ellett F. & Lieschke G. J. Computational quantification of fluorescent leukocyte numbers in zebrafish embryos. Methods in enzymology 506, 425–435, doi: 10.1016/B978-0-12-391856-7.00046-9 (2012). [DOI] [PubMed] [Google Scholar]
- Okuda K. S. et al. A zebrafish model of inflammatory lymphangiogenesis. Biol Open 4, 1270–1280, doi: 10.1242/bio.013540 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renshaw S. A. et al. A transgenic zebrafish model of neutrophilic inflammation. Blood 108, 3976–3978, doi: 10.1182/blood-2006-05-024075 (2006). [DOI] [PubMed] [Google Scholar]
- Howe D. G. et al. ZFIN, the Zebrafish Model Organism Database: increased support for mutants and transgenics. Nucleic Acids Res 41, D854–860, doi: 10.1093/nar/gks938 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon J. A. et al. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PloS one 9, e98186, doi: 10.1371/journal.pone.0098186 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeker N. D., Hutchinson S. A., Ho L. & Trede N. S. Method for isolation of PCR-ready genomic DNA from zebrafish tissues. BioTechniques 43, 610, 612, 614 (2007). [DOI] [PubMed] [Google Scholar]
- Thisse C. & Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nature protocols 3, 59–69, doi: 10.1038/nprot.2007.514 (2008). [DOI] [PubMed] [Google Scholar]
- Parichy D. M., Ransom D. G., Paw B., Zon L. I. & Johnson S. L. An orthologue of the kit-related gene fms is required for development of neural crest-derived xanthophores and a subpopulation of adult melanocytes in the zebrafish, Danio rerio. Development 127, 3031–3044 (2000). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.