Abstract
Second messenger cAMP regulates many cellular functions through its effectors, such as cAMP-dependent protein kinase (PKA) and Epac (exchange proteins directly activated by cAMP). Spatial and temporal control of cAMP signaling is crucial to differential regulation of cellular targets involved in various signaling cascades. To investigate the compartmentalized cAMP signaling, we constructed fluorescent indicators that report intracellular cAMP dynamics and Epac activation by sandwiching the full-length Epac1 between cyan and yellow mutants of GFP. Elevations of cAMP decreased FRET and increased the ratio of cyan-to-yellow emissions by 10–30% in living mammalian cells. This response can be reversed by removing cAMP-elevating agents and abolished by mutating the critical residue responsible for cAMP binding. Targeting of the reporter to the plasma membrane, where cAMP is produced in response to the activation of β-adrenergic receptor, revealed a faster cAMP response at the membrane than in the cytoplasm and mitochondria. Simultaneous imaging with targeted cAMP indicator and PKA activity reporter allowed the detection of a much delayed PKA response in the nucleus after the rapid accumulation of cAMP at the plasma membrane of the same cell, despite the immediate presence of a pool of cAMP in the nucleus. Thus, cAMP dynamics and the activation of its effectors are precisely controlled spatiotemporally in vivo.
Keywords: FRET, cAMP-dependent protein kinase
Signaling through cAMP, the paradigm for the second messenger concept, governs many fundamental cellular functions, including metabolic, electrical, cytoskeletal, and transcriptional responses within the cell (1, 2). Many of these responses are mediated by cAMP-dependent protein kinase (PKA), which has served as a model of protein kinase structure and regulation (3). On the other hand, the exchange protein directly activated by cAMP (Epac) has been discovered as a cAMP receptor protein mediating cAMP signaling through its cAMP-dependent guanine nucleotide exchange activity toward a small guanosine triphosphatase, Rap1 (4, 5). Epac has been shown to be involved in cAMP-modulated insulin secretion and cell adhesion; however, the molecular mechanisms of Epac-mediated signaling remain to be unraveled (6).
Subcellular compartmentalization of cAMP action was suggested >20 years ago (7), and recent studies provide new evidence for spatial organization of the components that regulate cAMP signaling. In mammalian cells, cAMP is produced by two classes of adenylyl cyclases (AC). Transmembrane ACs are tethered to the plasma membrane and regulated by heterotrimeric G proteins in response to hormones. Soluble AC is localized to nucleus, mitochondria, microtubules, and centrioles and is regulated by bicarbonate and calcium (8, 9). The distinct territories of two classes of ACs led to the hypothesis that discrete cAMP microdomains exist and that different pools of cAMP can be generated in response to extracellular signals or intrinsic signals such as intracellular pH and the metabolite CO2. However, the diffusion of cAMP in the cytosol is rapid, and local control and restricted diffusion of cAMP would be essential for the establishment of cAMP signaling microdomains. This restriction is partly achieved by phosphodiesterase (PDE), the enzyme responsible for the degradation of cAMP (10).
Microdomains or distinct pools of cAMP formed at different subcellular sites appear to be crucial to specific cAMP actions. For instance, in cardiac myocytes, activation of β-adrenergic receptor (β-AR) correlated with activation of the particulate or membrane-bound PKA, whereas prostaglandin E1 (PGE1) elevated cAMP in the soluble fraction, and the subsequent activation of PKA in the soluble pool was without functional consequences (11). The availability of different pools of cAMP to the local effectors within the same compartments should enhance the specificity and efficiency of cAMP signaling. As shown in cardiac myocytes (12), the cAMP rise within restricted domains appears necessary for limiting the activation to a specific sub-population of PKA molecules that, anchored through A-kinase-anchoring proteins, AKAPs (13), lie in proximity to their targets. Specific subcellular localization of Epac has also been shown at the mitochondria and nuclear envelop and to mitotic spindle and centrosomes in the metaphase of cell cycle (14).
To analyze the spatiotemporal dynamics of cAMP signaling, a first generation of cAMP indicator was constructed based on cAMP-sensitive FRET between two fluorophores chemically attached to subunits of PKA (15). Replacing the fluorophores with variants of GFP produced a genetically encodable fluorescent indicator (16). In addition, cAMP levels can be monitored by measuring the current flowing through the expressed recombinant cAMP-gated channel (17, 18). Dynamic visualization of compartmentalized PKA activities downstream of cAMP production has been achieved by genetically encoded phosphorylation indicators for PKA (19, 20). In the case of A-kinase activity reporter (AKAR), phosphorylation-dependent conformational change of a tandem fusion domain, composed of a substrate peptide for PKA and a phosphorylation recognition domain, alters the distance and/or relative orientation between two GFP proteins fused to the N and C termini and generates a FRET change. AKAR shares a general design with many genetically encoded indicators containing conformationally responsive elements sandwiched between two mutants of GFP capable of FRET, which have successfully monitored Ca2+, cGMP fluctuations, and protein phosphorylation in live cells (21, 22). Here we present fluorescent reporters based on cAMP-dependent conformational change of Epac for monitoring cAMP dynamics and Epac activation in living mammalian cells. We use this readily targetable indicator of cAMP using Epac (ICUE) to analyze the spatiotemporal dynamics of cAMP signaling at different subcellular locations.
Experimental Procedures
FRET Indicator Construction. The full-length Epac1 (1–881) and truncated forms of Epac2 (T316-A501 and P350-A501) were created by PCR using Epac1 (4) or Epac2 (23) as the templates. Mutation R522E was incorporated by the QuikChange method (Stratagene). Enhanced cyan fluorescent protein (ECFP) and citrine were fused to the N and C termini of the individual gene constructs (Fig. 1). The constructs were first generated in pRSET B (Invitrogen) and subcloned into pcDNA3 (Invitrogen) behind a Kozak sequence for mammalian expression.
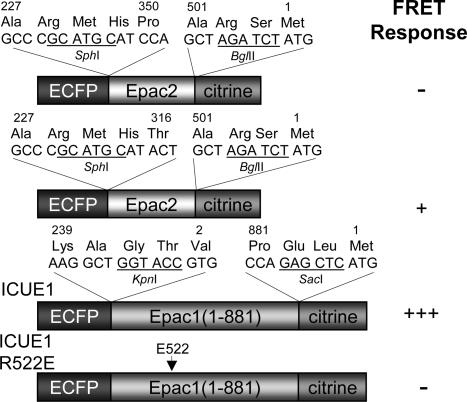
Fig. 1.
Domain structure and comparison of FRET responses for Epac-based cAMP reporters. Sandwiched between ECFP and citrine are truncated forms of Epac2, full-length Epac1 with or without R522E mutation, with R522 corresponding to R279 in Epac1. The construct that generated the biggest FRET response was designated as ICUE1.
For plasma membrane targeting of ICUE1, the sequence KKKKKSKTKCVIM was inserted at the C terminus. For nuclear targeting, the nuclear localization signal (NLS) PKKKRKVEDA was added to the C terminus. The signal sequence MAIQLRSLFPLALPGMLALLGWWWFFSRKK was inserted at the N terminus for targeting ICUE to mitochondria. Localization to the mitochondrial matrix was achieved by fusing the first 12 aa of subunit IV of human cytochrome oxidase c to the N terminus of the construct.
Cell Culture. HEK-293, HeLa, and PC12 cells were plated onto sterilized glass coverslips in 35-mm dishes and grown to 50–90% confluency in DMEM (10% FBS at 37°C with 5% CO2). Cells were then transfected with FuGENE-6 transfection reagent (Roche) or calcium phosphate and allowed to grow for 12–24 h before imaging. Colocalization studies were performed by incubating transfected HEK-293 cells with MitoTracker Red 580 or Hoechst 33342 cell-permeable dyes (Molecular Probes) for staining mitochondria or nucleic acids, respectively.
Imaging. Cells were washed twice with Hanks' balanced salt solution buffer after a 12- to 24-h incubation at 37°C in culture medium. Cells were maintained in buffer in the dark at room temperature with the addition of isoproterenol (ISO; Aldrich), forskolin (Calbiochem), PGE1 (Sigma), and 8-(4-chlorophenylthio)-2′-O-methyl adenosine 3′,5′-monophosphate (8-pCPT-2′-O-Me-cAMP) (Axxora Biolog, San Diego) as indicated. Cells were also treated with P-(4,5-dimethoxy-2-nitrobenzyl) cAMP (Molecular Probes). Uncaging of cAMP was performed as previously described (20).
Cells were imaged on a Zeiss Axiovert 200M microscope with a cooled charge-coupled device camera MicroMAX BFT512 (Roper Scientific, Trenton, NJ) controlled by metafluor 6.2 software (Universal Imaging, Downingtown, PA). Dualemission ratio imaging used a 420DF20 excitation filter, a 450DRLP dichroic mirror, and two emission filters (475DF40 for ECFP and 535DF25 for citrine) alternated by a filter-changer Lambda 10–2 (Sutter Instruments, Novato, CA). Exposure time was 100–500 ms, and images were taken every 8–30 s. Fluorescent images were background-corrected by subtracting autofluorescence intensities of untransfected cells (or background with no cells) from the emission intensities of fluorescent cells expressing reporters. The ratios of cyan-to-yellow emissions were then calculated at different time points and normalized by dividing all ratios by the emission ratio just before stimulation, thereby setting the basal emission ratio to 1. FRET efficiency was determined by acceptor photobleaching as reported (24). Briefly, citrine was photobleached at the end of the experiment by intense illumination with a 525DF40 filter. ECFP fluorescence intensities before (Fda) and after (Fd) citrine photobleaching and the equation E = 1 – (Fda/Fd) were then used to calculate the FRET efficiency.
Results
Development of Epac-Based Fluorescent Reporters. It has been suggested that cAMP binding to Epac induces a conformational change that liberates the catalytic domain of Epac from intrasubunit allosteric inhibition (25, 26). We hypothesize that sandwiching such Epac-based conformationally responsive elements between a FRET pair would allow the detection of cAMP rise and Epac activation by changes in FRET. Therefore, we generated a number of chimeric proteins by fusing the N terminus of various Epac truncations to ECFP and the C terminus to citrine, an improved version of yellow fluorescent protein (YFP) (Fig. 1).
In one construct, ECFP and citrine were fused together with a domain (P350-A501) containing the second cyclic nucleotide monophosphate-binding domain from Epac2 and a C-terminal lid (Fig. 1). When expressed in HEK-293 cells, this protein showed variable ratios of cyan-to-yellow emissions that are inversely correlated with expression level of the protein. This concentration dependence is indicative of intermolecular FRET between different reporter molecules that may occur because of oligomerization or aggregation (27). Upon cAMP elevations, this protein did not show a cAMP-dependent FRET change. We incorporated a larger portion of Epac2 sequence N-terminal to the binding domain (Fig. 1) (T316-A501) and obtained a construct that showed more homogeneous emission ratios and a 5% increase in emission ratio of cyan-to-yellow upon cAMP elevations (data not shown).
To improve the dynamic range of the response and to develop a reporter for Epac activation, we sandwiched the full-length Epac1 between ECFP and citrine (Fig. 1). When this reporter (designated as ICUE1) was transfected in HEK-293 cells, the fluorescence was uniformly distributed in the cytosolic compartment in 60% of the cells (Fig. 2A, first image). In the remaining 40% of the cells, the chimeric protein was localized to perinuclear region or mitochondria, consistent with our previous observation using full-length Epac1 fused to GFP (14). A similar expression pattern was also observed in HeLa and PC12 cells (data not shown).
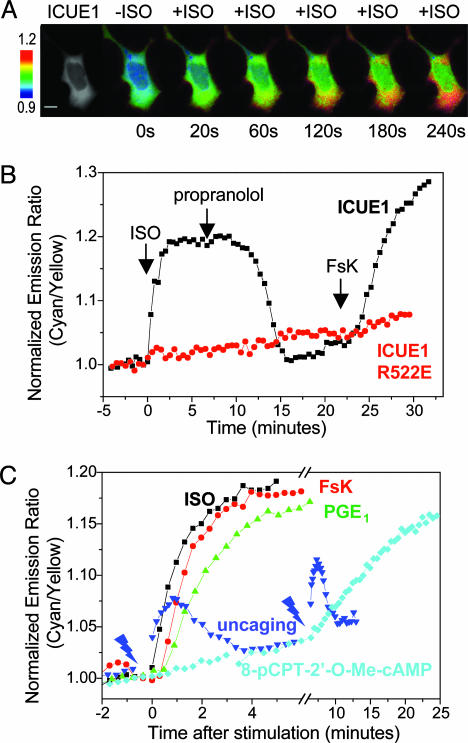
Fig. 2.
Responses of ICUE1 to changes in cellular cAMP levels. (A) FRET response of HEK-293 cells transfected with ICUE1. The first image is a YFP-only image. Pseudocolor images depict the FRET response of the reporter to ISO stimulation at various time points. (Scale bar, 10 μm.) (B) Representative emission ratio time courses of ICUE1 and the R522E mutant stimulated with 10 μM ISO followed by 10 μM propranolol and 50 μM forskolin (FsK). (C) Representative emission ratio time courses of ICUE1 stimulated with 10 μM ISO, 50 μM forskolin, 10 μM PGE1, 300 μM 8-pCPT-2′-O-Me-cAMP, and 100 μM P-(4,5-dimethoxy-2-nitrobenzyl) cAMP followed by UV uncaging. The flash signs indicate 5-s UV flashes at two different time points.
Stimulation of endogenous β-AR with ISO generated a FRET decrease in HEK-293 cells expressing ICUE1, resulting in an increase in the ratio of cyan-to-yellow emissions (Fig. 2 A and B). The change in emission ratios was detectable within several seconds and reached a plateau of 16.8 ± 1.0% (n = 8) [average ± SEM (n value)] signal increase within 1.5–3 min. This FRET change consisted of reciprocal decreases in yellow and increases in cyan emission, and the FRET efficiencies were measured by acceptor photobleaching to be 29 ± 3% and 21 ± 1% (n = 3), respectively, before and after ISO stimulation. The presence of propranolol, a β-AR antagonist, prevented the ISO-stimulated response (data not shown).
We next tested whether the FRET response was reversible. Addition of 10 μM propranolol after the ISO-stimulated response reached the plateau resulted in an initial decrease in emission ratio of cyan-to-yellow in 2–3 min and a full recovery over 6–8 min. Removal of ISO had the same effect (data not shown). Finally, a second rise in emission ratio was induced by addition of forskolin to activate AC and elevate cAMP. The change in emission ratio reached a plateau in 3–5 min (Fig. 2B).
To verify that the FRET response is caused by cAMP binding, we generated a variant of the reporter that carries a point mutation in the cAMP binding domain. Arginine 279 in Epac1 is a conserved residue that contributes to cAMP binding (26). EpacR279E has been shown to be defective in cAMP binding (4, 28), and mutation of the same arginine to glutamate (R522E) in the reporter completely abolished the FRET change induced by ISO and forskolin (Fig. 2B; n = 7).
Different means of elevating intracellular cAMP revealed different kinetics for the FRET response (Fig. 2C). As shown in Fig. 2 B and C, activation of β-AR with a selective agonist such as ISO (10 μM) induced decreases in FRET in 1.5–3 min, whereas stimulation of AC required slightly longer treatment with forskolin (3–5 min) to produce a maximal increase in emission ratios (19.6 ± 1.0% response; n = 8) (Fig. 2C). Addition of 10 μM PGE1 to increase cAMP level (17) induced a FRET response (13.3 ± 0.9%; n = 7) within 3–5 min, noticeably delayed compared with the response induced by ISO (Fig. 2C). A newly characterized analogue of cAMP, 8-pCPT-2′-O-Me-cAMP, specifically activates Epac but not PKA (29). This cAMP analog, when administrated at 300 μM, required 10–15 min to produce a half-maximal increase in emission ratios (t1/2) (Fig. 2C, 12.8 ± 0.9%; n = 6). The fastest intracellular responses were generated by photolytic release (“uncaging”) of cAMP from a membrane-permeant ester, P-(4,5-dimethoxy-2-nitrobenzyl) cAMP (30). Cells expressing ICUE1 were first incubated with 100 μM P-(4,5-dimethoxy-2-nitrobenzyl) cAMP for 3 min and were exposed to UV light to uncage the cAMP intracellularly. A flash of 5 s acutely increased the emission ratio by 4.7 ± 0.7% (n = 8) in just 15–30 s (Fig. 2C). The response was then quickly reversed because of the degradation of uncaged cAMP by PDE. The slower time courses of the other responses are presumably caused by rate-limiting steps in activating AC and accumulating sufficient cAMP rather than the kinetics of cAMP binding to Epac1 or the FRET response of the reporter.
cAMP Dynamics Within Subcellular Compartments. To directly monitor cAMP dynamics at different subcellular locations inside cells, we prepared several fusions of ICUE1 to various specific targeting motifs (Fig. 3A). To localize the reporter to the plasma membrane, we fused the plasma membrane-targeting signal of small guanosine triphosphatase K-ras4B (31) to the C terminus of ICUE1. This targeting motif combined a farnesylated cysteine residue with a strongly polybasic sequence and effectively targeted the reporter to the plasma membrane (Fig. 3B). As shown in Fig. 3C, plasma membrane-targeted ICUE1 generated a FRET response of 18.3 ± 1.2% (n = 8) upon stimulation with ISO. The response time (t1/2 = 24.9 ± 2.8 s; n = 8) was shortened by 40% compared with the time course for the cytoplasmically distributed ICUE1 (t1/2 = 40.5 ± 3.3 s; n = 8), whereas both plasma membrane-targeted and cytoplasmically distributed ICUE1 generated rapid responses upon whole-cell cAMP uncaging (data not shown). These results indicate that this delay in response of untargeted ICUE1 is not caused by the intrinsic kinetic properties of the localized reporters but is most likely caused by restricted release of cAMP from the plasma membrane to cytosol (18).
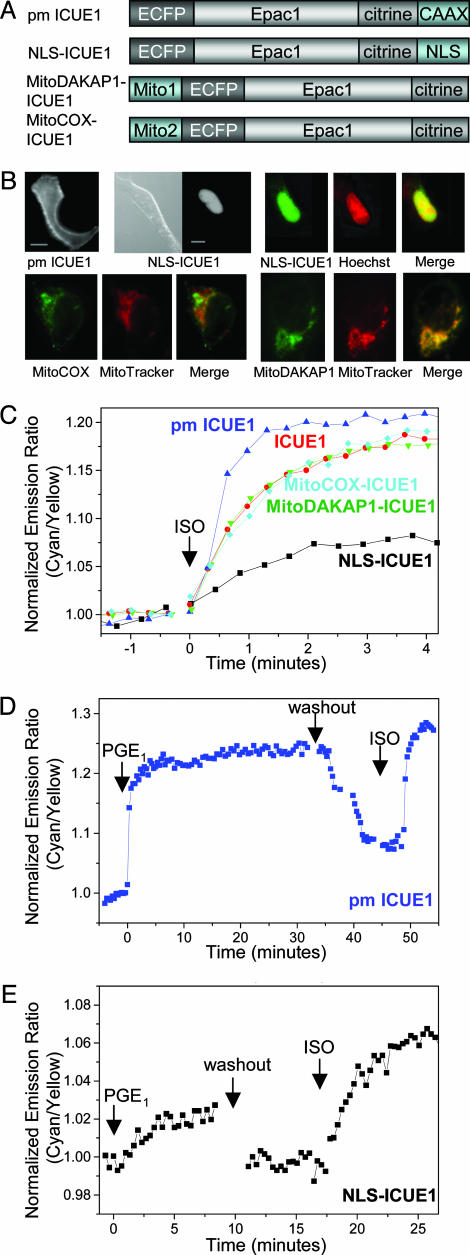
Fig. 3.
Fusions of ICUE1 targeted to various subcellular locations. (A) Domain structures of the fusion constructs. (B) YFP-only images showing plasma membrane and nuclear distributions of various fusions. (Scale bars, 10 μm.) Merged pseudocolor images show colocalization of nuclear localized ICUE1 with Hoechst 33342 cell-permeable dye in nucleus and mitochondria-targeted ICUE1 with MitoTracker at mitochondria. (C) Representative emission ratio time courses for the untagged (ICUE1), plasma membrane-targeted (pm ICUE1), mitochondria-targeted (MitoCOX- and MitoDAKAP1-ICUE1), and nuclear localized reporters (NLS-ICUE1) stimulated with 10 μM ISO. (D) Representative emission ratio time courses for pm ICUE1 stimulated with 10 μM PGE1 followed by the removal of PGE1 and the addition of 10 μM ISO. (E) Representative emission ratio time courses for NLS-ICUE1 in response to 10 μM PGE1 and 10 μM ISO separated by a washing step.
Epac has been shown to localize to mitochondria in a sub-population of cells, but monitoring of cAMP accumulation at mitochondria has not been possible with previous methods. To examine the cAMP dynamics and Epac activation at this sub-cellular location, we fused two different mitochondria-targeting motifs to ICUE1 (Fig. 3A). The first MitoCOX-ICUE1 was generated by fusing the targeting sequence of subunit IV of cytochrome c oxidase (COX) to the N terminus of ICUE1. This COX sequence has been shown to deliver fused proteins to the mitochondrial matrix (32). As shown in Fig. 3B, MitoCOX-ICUE1 was partially targeted to mitochondria (33), showing partial colocalization with a cell-permeable mitochondrial dye, MitoTracker. Activation of β-AR by ISO generated a FRET response (19.0 ± 1.6%; n = 5) in the punctate mitochondria structure within 2–3 min (t1/2 = 40.4 ± 7.3 s; n = 5), indicating that cAMP can enter mitochondria and accumulate in the matrix. In a second mitochondria-targeted ICUE1 (Mito-DAKAP1-ICUE1), a mitochondria-targeting motif taken from the N-terminal sequence of DAKAP1a (34) effectively targeted ICUE1 to mitochondria (Fig. 3B), where the ISO-stimulated FRET response (14.5 ± 1.5%, t1/2 = 42.4 ± 2.5 s; n = 6) is similar to the cytosolic response (Fig. 3C).
When fused to a NLS, ICUE1 was appropriately targeted to the nucleus (Fig. 3B), where its response to ISO stimulation was smaller (5.6 ± 0.5%; n = 12) than the 16.8 ± 1.0% FRET change for cytoplasmically distributed ICUE1 (Fig. 3C). Stimulation with bicarbonate to activate endogenous soluble AC in HEK-293 cells did not generate a cAMP-dependent response in the nucleus (data not shown), presumably because of limited copy numbers of soluble AC in this cell type or the sensitivity of the detection. Interestingly, the ISO-stimulated response in the nucleus is not delayed (t1/2 = 38.5 ± 3.5 s; n = 12) compared with that from untargeted ICUE1. This finding indicates that the available pool of cAMP in the nucleus, although possibly smaller, is not kinetically crippled because of the fast diffusion of cAMP from cytosol to nucleus.
To test whether activation of different receptors leads to production of different pools of cAMP, we compared the cAMP responses induced by PGE1 and ISO at different subcellular sites. At the plasma membrane, addition of PGE1 generated a 12.6 ± 0.9% (n = 8) emission ratio increase within 2–3 min. Surprisingly, sustained stimulation with 10 μM PGE1 did not produce a transient response, as observed previously using cyclic nucleotide-gated ion channels (17). In contrast, the response at the plasma membrane is sustained until removal of PGE1 (Fig. 3D). Variable cellular PDE activities may be responsible for this discrepancy. After removal of PGE1, a second response of similar amplitude was induced by stimulation with ISO. Both untargeted and mitochondria targeted ICUE1 showed similar responses to PGE1 and ISO (data not shown). In the nucleus, PGE1 also stimulated a small response (3.4 ± 0.4%; n = 13), noticeably a few percentages smaller than that induced by ISO in the same cell (Fig. 3E).
Simultaneous Imaging of cAMP Dynamics and PKA Phosphorylation. It has been shown that soluble AC and regulatory subunit (R subunit) and catalytic subunit (C subunit) of PKA coexist in the nucleus of mammalian cells (35). The activation of bicarbonate-responsive soluble AC in the nucleus led to a rapid increase in PKA-dependent phosphorylation, detectable within 2 min. Thus, the immediate presence of a nuclear pool of cAMP after β-AR activation raised the question of whether this pool of cAMP could produce functional PKA responses in the nucleus. Here we take advantage of the targeted cAMP indicators and AKAR (20) to examine the temporal correlation of cAMP dynamics and PKA activation within single living cells.
We coexpressed the plasma membrane-targeted ICUE1 and nuclear-localized AKAR in HEK-293 cells (Fig. 4A). An immediate increase in emission ratios of cyan-to-yellow occurred at the plasma membrane upon stimulation with ISO, indicating an acute rise in cAMP. However, the emission ratio in the nucleus did not increase in the same time frame. After a delay of 5–10 min, in the presence of the sustained cAMP response, a gradual increase in ratio of yellow-to-cyan emissions occurred and reached a plateau in 20–30 min. On the other hand, untargeted AKAR generated acute responses in 2–3 min in the cytosol of HEK-293 cells upon stimulation with ISO (data not shown). This finding is consistent with acute cytosolic AKAR responses upon cAMP elevations we previously reported (20). Therefore, the delay in response indicates that PKA phosphorylation in the nucleus does not occur immediately after cAMP production (Fig. 4B). This delayed nuclear response of PKA phosphorylation is consistent with a slow diffusional translocation of the C subunit of PKA into the nucleus after the dissociation of C and R subunits in the cytoplasm upon cAMP elevation (36, 37).
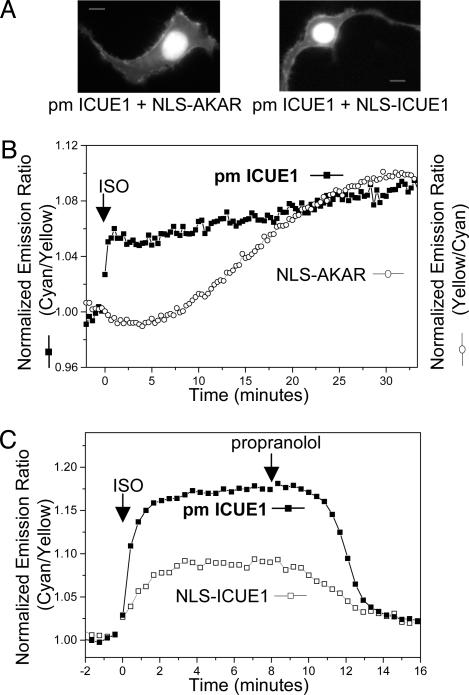
Fig. 4.
Simultaneous imaging of FRET reporters targeted to different sub-cellular locations. (A) Cellular distribution of different fusions. (B) Representative emission ratio time courses for the pm ICUE1 and nuclear localized PKA activity reporter (NLS-AKAR) in the same cell stimulated with 10 μM ISO. Identical results were found in four different cells. The AKAR response was plotted by using a normalized ratio of yellow-to-cyan emissions. (C) Representative emission ratio time courses for pm ICUE1 and NLS-ICUE1 in the same cell stimulated with 10 μM ISO followed by 10 μM propranolol (n = 4).
As a control experiment, we also recorded cAMP responses from single cells coexpressing both plasma membrane-targeted and nuclear-localized ICUE1 for direct comparison of the cAMP dynamics at the plasma membrane and in the nucleus. As shown in Fig. 4C, ISO stimulated an acute cAMP response at the plasma membrane followed by a response in the nucleus, which reached the plateau in 2–3 min. This finding is consistent with data obtained from separate cells expressing either targeted reporter, indicating that ICUE reporter molecules do not notably perturb cAMP distribution throughout the cell. This acute nuclear cAMP response is in sharp contrast to the delayed response of PKA phosphorylation in the nucleus, which requires 20–30 min to reach the maximum. Thus, the presence of this nuclear pool of cAMP immediately after cAMP production is not sufficient to generate a detectable phosphorylation of AKAR by PKA within the nucleus. This lack of immediate PKA response could be caused by either the absence of the PKA holoenzyme in the nucleus or insufficient activation of soluble AC-coupled PKA by this pool of cAMP. In this case, the slow diffusion of the C subunit rather than the fast diffusion of cAMP as the rate-limiting step may provide the temporal control of β-AR-stimulated PKA-dependent phosphorylation in the nucleus.
Discussion
As a cAMP indicator, the present reporter ICUE1 based on Epac1 has some special advantages over previous methods for assessing cAMP dynamics inside cells. Radioimmunoassay (RIA) or enzyme immunoassay for measuring cAMP require destroying large amounts of cells or tissue, have very poor spatial and temporal resolution, and measure total rather than free cAMP. Use of engineered cyclic nucleotide-gated channels to detect free cAMP provides good temporal resolution and quantification but uses indirect calcium measurement or nontrivial patch-clamp technique and lacks the flexibility of measuring cAMP changes within various subcellular compartments (17, 18). Free cAMP can be imaged in single cells microinjected with fluorophore-labeled C and R subunits (15) and, more recently, in cells expressing two colors of GFP mutants fused to the C and R subunits (16), which dissociate from each other and lose FRET upon elevation of cAMP. However, the expression levels of the two fusions have to be carefully matched to allow reliable measurement. Even so, mixed tetramerization may occur between the fluorophore-attached subunits and endogenous partners, reducing the number of functional reporter molecules. Furthermore, it can be difficult to target such bimolecular reporters to different subcellular locations while maintaining appropriate stoichiometry. The current ICUE is a unimolecular cAMP reporter based on Epac1 and can be readily targeted to different subcellular locations or fused to signaling components. In addition, ICUE also functions as an Epac activation reporter and can be used to examine compartmentalized Epac activities and their physiological functions.
While our work was in preparation, Nikolaev et al. (38) published an independent report of single-chain cAMP sensors based on Epac and the R subunit of PKA. Their best reporter consisted of a truncated Epac1 (E157-E316) sandwiched between enhanced YFP and ECFP. In Chinese hamster ovary cells overexpressing adenosine A2B receptors and HEK-293 cells overexpressing β1-AR or β2-AR, this reporter generated a 25–35% decrease in emission ratios of yellow-to-cyan in 1–2 min. This reporter is different from ICUE1, which consists of full-length Epac1, but analogous to other constructs we generated using truncated forms of Epac, including an identically truncated Epac1 (E157-E316). However, these constructs generated only small FRET decreases upon cAMP elevations in HEK-293 cells with endogenous receptors, in comparison with ICUE1 containing full-length Epac1. This discrepancy could be caused by use of different linkers connecting sequences of Epac and fluorescent proteins in individual constructs and the different cell types for testing these constructs.
ICUE1 targeted to plasma membrane, mitochondria, and nucleus revealed differential dynamics of cAMP signaling in response to the activation of β-AR or prostanoid receptor. The response at the plasma membrane, the site of cAMP production, is faster than detected in the cytoplasm, mitochondria, or nucleus. The difference in the response time is in the range of tens of seconds. With a calculated diffusion coefficient of 500 μm2s–1 (38, 39), cAMP would diffuse across a 20-μm cell in <0.5 s, which would presumably blur our observed spatial differences. The role of specific PDE activities in controlling such differential responses needs to be tested in the future. On the other hand, it has been suggested that cAMP is produced in microdomains near the plasma membrane and that diffusion between these domains and a cytosolic compartment is restricted (18). Challenges remain to identify molecular and structural components that control such microdomains. In the nucleus, the response to ISO and PGE1 is small but not delayed compared with that detected in the cytoplasm or mitochondria, indicating a fast diffusion of cAMP from the cytoplasm to nucleus. It remains to be tested whether any factors limit the access to nucleus by cAMP and lead to a reduced pool of cAMP in the nucleus. In fact, in muscle cells, a perinuclear pool of PDE targeted through muscle-selective A-kinase anchoring protein, mAKAP, has been suggested to play a role of restricting nuclear cAMP signaling (40). It is also possible that the local nuclear environment, the presence of binding partners of Epac, or some aggregation of the reporter in the nucleus could account for the small response of the nuclear reporter.
In this study, we demonstrated simultaneous imaging of cAMP dynamics and PKA phosphorylation in single living cells using locus-specific FRET reporters. This method takes advantage of spatial separation of subcellular events and provides unambiguous temporal correlation of these events. We believe that this methodology complements multicolor imaging (41, 42) and is well suited for simultaneously monitoring multiple signaling events and for evaluating the information flow within signaling cascades or crosstalk between different pathways (43). By using this method, we showed that although cAMP production at the plasma membrane immediately produces a pool of cAMP in the nucleus, this pool is not sufficient to generate functional consequences through PKA on the same timescale. The functional role of this nuclear pool of cAMP is not clear; however, it was sufficient to induce a conformational change of Epac1 targeted to the nucleus. Along this line, Epac1 has been shown to localize to mitotic spindle, centrosomes, and nuclear envelop in a cell cycle-dependent fashion (14). While nuclear activities and functions of Epac remain to be analyzed, the availability of fluorescent reporters for cAMP and its effectors PKA and Epac should greatly facilitate mechanistic investigation of compartmentalized cAMP signaling.
Acknowledgments
We thank Dr. Heng Zhu, Qiang Ni, and Dr. Bharath Ananthanarayanan for suggestions and assistance; Drs. Stephen R. Adams, Philip A. Cole, Jun O. Liu, and Roger Y. Tsien for critical reading of the manuscript; and Drs. Susumu Seino and Nobuaki Ozaki for providing Epac2 cDNA. This work was supported by The Johns Hopkins University School of Medicine and The W. M. Keck Center (to J.Z.) and National Institutes of Health Grant GM066170 (to X.C.). L.M.D. is supported by Pharmacology Training Grant GM08763 from the National Institutes of Health.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: β-AR, β-adrenergic receptor; AC, adenylyl cyclase; AKAR, A-kinase activity reporter; C subunit, catalytic subunit; ECFP, enhanced cyan fluorescent protein; ICUE, indicator of cAMP using Epac; ISO, isoproterenol; NLS, nuclear localization signal; PDE, phosphodiesterase; PGE1, prostaglandin E1; PKA, cAMP-dependent protein kinase; R subunit, regulatory subunit; YFP, yellow fluorescent protein.
References
- 1.Kopperud, R., Krakstad, C., Selheim, F. & Doskeland, S. O. (2003) FEBS Lett. 546, 121–126. [DOI] [PubMed] [Google Scholar]
- 2.Chin, K. V., Yang, W. L., Ravatn, R., Kita, T., Reitman, E., Vettori, D., Cvijic, M. E., Shin, M. & Iacono, L. (2002) Ann. N.Y. Acad. Sci. 968, 49–64. [DOI] [PubMed] [Google Scholar]
- 3.Taylor, S. S., Yang, J., Wu, J., Haste, N. M., Radzio-Andzelm, E. & Anand, G. (2004) Biochim. Biophys. Acta 1697, 259–269. [DOI] [PubMed] [Google Scholar]
- 4.de Rooij, J., Zwartkruis, F. J. T., Verheijen, M. H. G., Cool, R. H., Nijman, S. M. B., Wittinghofer, A. & Bos, J. L. (1998) Nature 396, 474–477. [DOI] [PubMed] [Google Scholar]
- 5.Kawasaki, H., Springett, G. M., Mochizuki, N., Toki, S., Nakaya, M., Matsuda, M., Housman, D. E. & Graybiel, A. M. (1998) Science 282, 2275–2279. [DOI] [PubMed] [Google Scholar]
- 6.Bos, J. L. (2003) Nat. Rev. Mol. Cell Biol. 4, 733–738. [DOI] [PubMed] [Google Scholar]
- 7.Steinberg, S. F. & Brunton, L. L. (2001) Annu. Rev. Pharmacol. Toxicol. 41, 751–773. [DOI] [PubMed] [Google Scholar]
- 8.Zippin, J. H., Chen, Y., Nahirney, P., Kamenetsky, M., Wuttke, M. S., Fischman, D. A., Levin, L. R. & Buck, J. (2003) FASEB J. 17, 82–84. [DOI] [PubMed] [Google Scholar]
- 9.Bundey, R. A. & Insel, P. A. (2004) Sci. STKE 2004, e19. [DOI] [PubMed] [Google Scholar]
- 10.Houslay, M. D. & Milligan, G. (1997) Trends Biochem. Sci. 22, 217–224. [DOI] [PubMed] [Google Scholar]
- 11.Hayes, J. S., Brunton, L. L. & Mayer, S. E. (1980) J. Biol. Chem. 255, 5113–5119. [PubMed] [Google Scholar]
- 12.Zaccolo, M. & Pozzan, T. (2002) Science 295, 1711–1715. [DOI] [PubMed] [Google Scholar]
- 13.Michel, J. J. & Scott, J. D. (2002) Annu. Rev. Pharmacol. Toxicol. 42, 235–257. [DOI] [PubMed] [Google Scholar]
- 14.Qiao, J., Mei, F. C., Popov, V. L., Vergara, L. A. & Cheng, X. (2002) J. Biol. Chem. 277, 26581–26586. [DOI] [PubMed] [Google Scholar]
- 15.Adams, S. R., Harootunian, A. T., Buechler, Y. J., Taylor, S. S. & Tsien, R. Y. (1991) Nature 349, 694–697. [DOI] [PubMed] [Google Scholar]
- 16.Zaccolo, M., De Giorgi, F., Cho, C. Y., Feng, L., Knapp, T., Negulescu, P. A., Taylor, S. S., Tsien, R. Y. & Pozzan, T. (2000) Nat. Cell Biol. 2, 25–29. [DOI] [PubMed] [Google Scholar]
- 17.Rich, T. C., Fagan, K. A., Tse, T. E., Schaack, J., Cooper, D. M. & Karpen, J. W. (2001) Proc. Natl. Acad. Sci. USA 98, 13049–13054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rich, T. C., Fagan, K. A., Nakata, H., Schaack, J., Cooper, D. M. & Karpen, J. W. (2000) J. Gen. Physiol. 116, 147–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagai, Y., Miyazaki, M., Aoki, R., Zama, T., Inouye, S., Hirose, K., Iino, M. & Hagiwara, M. (2000) Nat. Biotechnol. 18, 313–316. [DOI] [PubMed] [Google Scholar]
- 20.Zhang, J., Ma, Y., Taylor, S. S. & Tsien, R. Y. (2001) Proc. Natl. Acad. Sci. USA 98, 14997–15002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyawaki, A. (2003) Dev. Cell 4, 295–305. [DOI] [PubMed] [Google Scholar]
- 22.Zhang, J., Campbell, R. E., Ting, A. Y. & Tsien, R. Y. (2002) Nat. Rev. Mol. Cell Biol. 3, 906–918. [DOI] [PubMed] [Google Scholar]
- 23.Ozaki, N., Shibasaki, T., Kashima, Y., Miki, T., Takahashi, K., Ueno, H., Sunaga, Y., Yano, H., Matsuura, Y., Iwanaga, T., et al. (2000) Nat. Cell Biol. 2, 805–811. [DOI] [PubMed] [Google Scholar]
- 24.Miyawaki, A. & Tsien, R. Y. (2000) Methods Enzymol. 327, 472–500. [DOI] [PubMed] [Google Scholar]
- 25.Rehmann, H., Rueppel, A., Bos, J. L. & Wittinghofer, A. (2003) J. Biol. Chem. 278, 23508–23514. [DOI] [PubMed] [Google Scholar]
- 26.Rehmann, H., Prakash, B., Wolf, E., Rueppel, A., de Rooij, J., Bos, J. L. & Wittinghofer, A. (2003) Nat. Struct. Biol. 10, 26–32. [DOI] [PubMed] [Google Scholar]
- 27.Zacharias, D. A., Violin, J. D., Newton, A. C. & Tsien, R. Y. (2002) Science 296, 913–916. [DOI] [PubMed] [Google Scholar]
- 28.Mei, F. C., Qiao, J. B., Tsygankova, O. M., Meinkoth, J. L., Quilliam, L. A. & Cheng, X. D. (2002) J. Biol. Chem. 277, 11497–11504. [DOI] [PubMed] [Google Scholar]
- 29.Enserink, J. M., Christensen, A. E., de Rooij, J., van Triest, M., Schwede, F., Genieser, H. G., Doskeland, S. O., Blank, J. L. & Bos, J. L. (2002) Nat. Cell Biol. 4, 901–906. [DOI] [PubMed] [Google Scholar]
- 30.Nerbonne, J. M., Richard, S., Nargeot, J. & Lester, H. A. (1984) Nature 310, 74–76. [DOI] [PubMed] [Google Scholar]
- 31.Roy, M. O., Leventis, R. & Silvius, J. R. (2000) Biochemistry 39, 8298–8307. [DOI] [PubMed] [Google Scholar]
- 32.Hurt, E. C., Pesold-Hurt, B., Suda, K., Oppliger, W. & Schatz, G. (1985) EMBO J. 4, 2061–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Filippin, L., Magalhaes, P. J., Di Benedetto, G., Colella, M. & Pozzan, T. (2003) J. Biol. Chem. 278, 39224–39234. [DOI] [PubMed] [Google Scholar]
- 34.Ma, Y. & Taylor, S. (2002) J. Biol. Chem. 277, 27328–27336. [DOI] [PubMed] [Google Scholar]
- 35.Zippin, J. H., Farrell, J., Huron, D., Kamenetsky, M., Hess, K. C., Fischman, D. A., Levin, L. R. & Buck, J. (2004) J. Cell Biol. 164, 527–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harootunian, A. T., Adams, S. R., Wen, W., Meinkoth, J. L., Taylor, S. S. & Tsien, R. Y. (1993) Mol. Biol. Cell 4, 993–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meinkoth, J. L., Ji, Y., Taylor, S. S. & Feramisco, J. R. (1990) Proc. Natl. Acad. Sci. USA 87, 9595–9599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nikolaev, V. O., Bunemann, M., Hein, L., Hannawacker, A. & Lohse, M. J. (2004) J. Biol. Chem 279, 37215–37218. [DOI] [PubMed] [Google Scholar]
- 39.Bacskai, B. J., Hochner, B., Mahaut-Smith, M., Adams, S. R., Kaang, B.-K., Kandel, E. R. & Tsien, R. Y. (1993) Science 260, 222–226. [DOI] [PubMed] [Google Scholar]
- 40.Dodge, K. L., Khouangsathiene, S., Kapiloff, M. S., Mouton, R., Hill, E. V., Houslay, M. D., Langeberg, L. K. & Scott, J. D. (2001) EMBO J. 20, 1921–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Violin, J. D., Zhang, J., Tsien, R. Y. & Newton, A. C. (2003) J. Cell Biol. 161, 899–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DeBernardi, M. A. & Brooker, G. (1996) Proc. Natl. Acad. Sci. USA 93, 4577–4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zaccolo, M. (2004) Circ. Res. 94, 866–873. [DOI] [PubMed] [Google Scholar]